Progress of Research on the Metabolic Regulation of Lactylation in Muscle Tissues and Its Disease Associations
Abstract
1. Introduction
2. Discovery and Characterization of Lactylation
3. Chemical Basis and Mechanisms of Lactylation
3.1. Chemical Basis of Lactylation
3.2. Lactylation Mechanisms
4. Characteristics of Lactylation in Muscle Tissues
4.1. Dynamic Regulation of Lactylation in Muscle Tissue
4.2. Specificity of Lactylation in Muscle Tissue
4.3. Functional Diversity of Lactylation in Muscle Tissue
4.3.1. Lactylation Regulates Glucose and Lipid Energy Metabolism in Muscle Cells
4.3.2. Lactylation Regulates Muscle Cell Proliferation and Differentiation
4.3.3. Lactylation Contributes the Adaptation of Skeletal Muscle to Exercise
4.4. Central Mechanisms of Lactylation in Muscle Tissue Under Pathological Conditions
4.4.1. Lactylation in Muscle Aging
4.4.2. Lactylation in Metabolic Myopathies
4.4.3. Lactylation in Cardiovascular Diseases
- (1)
- Atherosclerosis: The progression of atherosclerosis is closely linked to metabolic dysregulation and phenotypic changes in VSMCs and macrophages [93]. In VSMCs, cellular senescence is a key driver of atherosclerosis [94]. Senescent VSMCs undergo a metabolic shift from oxidative phosphorylation to aerobic glycolysis. The mitochondrial protein tumor necrosis factor receptor-associated protein 1 (TRAP1) is overexpressed in senescent VSMCs, which promotes glycolysis and suppresses the tricarboxylic acid cycle. Conversely, TRAP1 knockout reduces lactate levels and histone H4 lactylation. Further investigations have revealed that TRAP1 promotes H4K12 lactylation by downregulating HDAC3 via lactate, thereby activating senescence-associated secretory phenotype (SASP) transcription and accelerating VSMC senescence and atherosclerosis progression [59]. In advanced atherosclerotic plaques, chronic inflammation induces the transdifferentiation of some VSMCs into macrophage-like cells, a process associated with sex-determining region Y (SRY)-related HMG-box gene 10 (Sox10) lactylation, which exacerbates intimal inflammation and promotes vulnerable plaque formation [60].
- (2)
- Myocardial Infarction: Myocardial infarction (MI), as a severe clinical manifestation of atherosclerosis, is characterized by coronary artery occlusion that drastically reduces myocardial blood supply and impairs peripheral organ perfusion. This condition frequently induces hyperlactatemia, a metabolic disorder closely associated with progressive myocardial damage [95]. During the early phase of MI (day 1), lactate promotes H3K18 lactylation in monocytes and macrophages, thereby enhancing the transcription of repair-related genes (e.g., Lrg1, Vegf-α, and IL-10). Through their anti-inflammatory and pro-angiogenic activities, these genes foster a microenvironment conducive to tissue repair. Elevated H3K18 lactylation suppresses detrimental inflammation and improves cardiac function post-MI [71]. However, lactate is not entirely beneficial. In the later stage of MI (day 6), it may also promote Snail family transcriptional repressor 1 (Snail1) lactylation, activating the TGF-β/Smad2 signaling pathway and driving endothelial–mesenchymal transition (EndMT), which ultimately exacerbates myocardial fibrosis [32].
- (3)
- Myocardial Ischemia-Reperfusion Injury: Myocardial ischemia-reperfusion injury (MIRI) is a secondary injury that occurs after blood flow restoration after post-myocardial infarction [96]. During MIRI, the cardiac energy metabolism shifts toward glycolysis, resulting in ATP depletion and lactate accumulation [97]. This subsequently induces protein lactylation, which exerts complex effects on cardiomyocyte survival. On the one hand, lactylation exhibits protective roles; heat shock protein A12A supports cardiomyocyte survival under hypoxia/reoxygenation by maintaining histone H3 lactylation and aerobic glycolytic homeostasis, and its deficiency exacerbates cardiac dysfunction [72]. Fibroblast-derived lactylated Serpina3k (SA3K) inhibits the WNT signaling pathway and activates the reperfusion injury salvage kinase (RISK) and survivor activating factor enhancement (SAFE) signaling pathways, reducing cardiomyocyte apoptosis [73].
- (4)
- Heart Failure: Lactylation exerts dual regulatory effects in heart failure development, with its impact being highly target- and stage-specific. During heart failure progression, metabolic reprogramming increases lactate efflux and reduces intracellular lactate levels, consequently diminishing the lactylation of key proteins [98]. In both murine models and human patients, decreased lactylation at K1897 of α-myosin heavy chain (α-MHC) impairs its interaction with titin, compromising sarcomeric integrity and reducing contractility. Moreover, this modification deficiency upregulates the expression of the fibrotic markers alpha-smooth muscle actin and type I collagen, further deteriorating cardiac function. Experimentally, restoring α-MHC-K1897 lactylation by elevating lactate concentration or inhibiting lactate efflux effectively improves cardiac performance [78].
- (5)
- Pulmonary Hypertension: In pulmonary hypertension, lactylation contributes to pulmonary vascular remodeling by directly promoting smooth muscle proliferation and regulating cellular senescence and inflammatory phenotypes [54,55,56]. Lactylation modulates the senescence of PASMCs in pulmonary hypertension models. The accumulation of the senescence-associated prelamin A and the subsequent increase in Interleukin-6 (IL-6) secretion collectively create a microenvironment that promotes PASMC proliferation, and histone lactylation is implicated in driving this process. Lactylation plays a critical role in regulating inflammation [62]. The long non-coding RNA UNC5B-AS1 remodels cellular metabolism by suppressing glycolysis and enhancing oxidative phosphorylation, thereby reducing lactate levels and H3K18 lactylation enrichment at the pro-inflammatory genes, such as IL-1β, IL-6, and TNF-α. This mechanism attenuates the transition of PASMCs to a pro-inflammatory phenotype and inhibits the formation of a pro-inflammatory vascular microenvironment [63].
5. Novel Strategies Targeting Lactylation for Muscular Disorders
6. Conclusions and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| HK | Hexokinase |
| LDHA | Lactate dehydrogenase A |
| ACSS2 | Acetyl-CoA synthetase 2 |
| GTPSCS | Succinyl-CoA synthetase |
| GLO1 | Glyoxalase 1 |
| GLO2 | Glyoxalase 2 |
| KAT2A | Lysine acetyltransferase 2A |
| P300 | E1A binding protein p300 |
| CBP | CREB-binding protein |
| GCN5 | General control non-depressible 5 |
| TIP60 | Tat-interactive protein 60 |
| AARS1/2 | Aminoacyl-tRNA synthetase 1/2 |
| HDAC1-3 | Histone deacetylase 1-3 |
| SIRT1-3 | Sirtuin1-3 |
| Brg1 | Brahma-related gene 1 |
| TRIM33 | Tripartite motif-containing 33 |
| DPF2 | Double PHD fingers 2 |
| MCT4 | Monocarboxylate transporter 4 |
| PFKM | Phosphofructokinase-M |
| PKM2 | Pyruvate kinase M2 |
| FASN | Fatty acid synthase |
| HIF-1α | Hypoxia-inducible factor-1α |
| ACLY | ATP-citrate lyase |
| ACADS | Short-chain acyl-CoA dehydrogenase |
| ACAA2 | Acetyl-CoA acyltransferase 2 |
| HADHA | Rifunctional enzyme subunit alph |
| PDHA1 | pyruvate dehydrogenase E1 alpha 1 Subunit |
| CPT2 | Carnitine palmitoyltransferase 2 |
| MDH2 | Malate dehydrogenase 2 |
| MT-ATP8 | Mitochondrially encoded ATP synthase membrane subunit 8 |
| ATP5MG | ATP synthase membrane subunit g |
| ATP5PO | ATP synthase peripheral stalk subunit oscp |
| OXPHOS | Oxidative phosphorylation |
| Neu2 | Neuraminidase 2 |
| MyHC | Myosin heavy chain |
| Mex3b | Mex-3bRNA binding family member b |
| E2f2 | E2F transcription factor 2 |
| Rfc3 | Replication factor C subunit 3 |
| Cdk2 | Cyclin-dependent kinase 2 |
| P21 | Cyclin-dependent kinase 1A |
| P53 | Tumor protein p53 |
| Bmp5 | Bone morphogenetic protein 5 |
| Trpc5 | Transient receptor potential cation channel subfamily C member 5 |
| Kit | V-kit Hardy–Zuckerman 4 feline sarcoma viral oncogene homolog |
| Gbe1 | 1,4-α-Glucan branching enzyme |
| Pgf | Placental growth factor |
| Mt2A | Metallothionein 2A |
| ACADS | Acyl-CoA dehydrogenase, short chain |
| TOP1 | Topoisomerase I |
| EMILIN-1 | Elastin microfibril interface protein 1 |
| VPS34 | Vacuolar protein sorting 34 |
| BECN1 | Beclin 1 |
| AGT14 | Autophagy related angiotensinogen14 |
| UVRIG | UV radiation resistance associated gene |
| SKMC | Skeletal muscle cell |
| Mφ | Macrophage |
| VSMC | Vascular smooth muscle cell |
| Pds5b | Pds5 cohesin associated factor b |
| Park7 | Parkinsonism associated deglycase 7 |
| Eya1 | Eyes absent homolog 1 |
| Nedd4 | Neural precursor cell expressed development down-regulated 4 |
| Wwp1 | Ww domain containing E3 ubiquitin protein ligase |
| Ecpas | Erythrocyte coproporphyrinogen oxidase |
| CHI3L1 | Chitinase-3-like protein 1 |
| Phospho1 | Phosphoethanolamine |
| TRAP1 | Tumor necrosis factor receptor-associated protein 1 |
| SASP | Senescence-associated secretory phenotype |
| Sox10 | Sex-determining region Y (SRY)-related HMG-box gene 10 |
| IL-10 | Interleukin-10 |
| PDHA | Pyruvate dehydrogenase alpha |
| Cd74 | Cluster of differentiation 74 |
| C3 | Complement component 3 |
| Lyz2 | lysozyme 2 |
| MI | Myocardial infarction |
| Lrg1 | Leucine-rich alpha-2-glycoprotein 1 |
| EC | Endothelial cell |
| Snail1 | Snail family transcriptional repressor 1 |
| TGF-β | Transforming growth factor beta |
| Smad2 | Smad family member 2 |
| EndMT | Endothelial–mesenchymal transition |
| MIRI | Myocardial ischemia-reperfusion injury |
| FB | Fibroblast |
| SA3K | Serpina3k |
| WNT | Wingless-type MMTV integration site family, Member |
| RISK | Reperfusion injury salvage kinase |
| SAFE | Survivor activating factor enhancement |
| CM | Cardiac muscle cell |
| ACSL4 | Acyl-CoA synthetase long chain family member 4 |
| GPX4 | Glutathione peroxidase 4 |
| NLRP3 | NOD-like receptor protein 3 |
| GSDMD | Gasdermin D |
| YTHDF2 | YTH domain-containing family protein 2 |
| G3BP1 | Ras GTPase-activating protein-binding protein 1 |
| PASMC | Pulmonary arterial smooth muscle cell |
| α-MCH | α-Myosin heavy chain |
| IL-1β | Interleukin-1β |
| IL-6 | Interleukin-6 |
| TNF-α | Tumor necrosis factor alpha |
References
- Rabinowitz, J.D.; Enerbäck, S. Lactate: The ugly duckling of energy metabolism. Nat. Metab. 2020, 2, 566–571. [Google Scholar] [CrossRef]
- Zhang, D.; Tang, Z.; Huang, H.; Zhou, G.; Cui, C.; Weng, Y.; Liu, W.; Kim, S.; Lee, S.; Perez-Neut, M. Metabolic regulation of gene expression by histone lactylation. Nature 2019, 574, 575–580. [Google Scholar] [CrossRef]
- Seheult, J.; Fitzpatrick, G.; Boran, G. Lactic acidosis: An update. Clin. Chem. Lab. Med. 2017, 55, 322–333. [Google Scholar] [CrossRef]
- Dartiguelongue, J.B. Biological significance and clinical utility of lactate in sepsis. Arch. Argent. Pediatr. 2024, 122, e202310149. [Google Scholar]
- Dietrich, D.; Elzinga, G. ATP formation and energy demand in anoxic heart muscle of the rabbit. Am. J. Physiol. Heart Circ. Physiol. 1992, 263, H526–H532. [Google Scholar] [CrossRef] [PubMed]
- Stephens, N.L.; Kroeger, E.A.; Loh, W. Intracellular pH in hypoxic smooth muscle. Am. J. Physiol.-Endocrinol. Metab. 1977, 232, E330. [Google Scholar] [CrossRef]
- Huang, W.; Su, J.; Chen, X.; Li, Y.; Xing, Z.; Guo, L.; Li, S.; Zhang, J. High-intensity interval training induces protein lactylation in different tissues of mice with specificity and time dependence. Metabolites 2023, 13, 647. [Google Scholar] [CrossRef] [PubMed]
- Mao, Y.; Zhang, J.; Zhou, Q.; He, X.; Zheng, Z.; Wei, Y.; Zhou, K.; Lin, Y.; Yu, H.; Zhang, H. Hypoxia induces mitochondrial protein lactylation to limit oxidative phosphorylation. Cell Res. 2024, 34, 13–30. [Google Scholar] [CrossRef]
- Wu, Z.; Chai, Z.; Cai, X.; Wang, J.; Wang, H.; Yue, B.; Zhang, M.; Wang, J.; Wang, H.; Zhong, J. Protein Lactylation Profiles Provide Insights into Molecular Mechanisms Underlying Metabolism in Yak. J. Agric. Food Chem. 2024, 72, 14057–14066. [Google Scholar] [CrossRef] [PubMed]
- Galle, E.; Wong, C.-W.; Ghosh, A.; Desgeorges, T.; Melrose, K.; Hinte, L.C.; Castellano-Castillo, D.; Engl, M.; de Sousa, J.A.; Ruiz-Ojeda, F.J. H3K18 lactylation marks tissue-specific active enhancers. Genome Biol. 2022, 23, 207. [Google Scholar] [CrossRef]
- Meng, F.; He, J.; Zhang, X.; Lyu, W.; Wei, R.; Wang, S.; Du, Z.; Wang, H.; Bi, J.; Hua, X. Histone Lactylation Antagonizes Senescence and Skeletal Muscle Aging by Modulating Aging-Related Pathways. Adv. Sci. 2025, 12, e2412747. [Google Scholar] [CrossRef]
- Maschari, D.; Saxena, G.; Law, T.D.; Walsh, E.; Campbell, M.C.; Consitt, L.A. Lactate-induced lactylation in skeletal muscle is associated with insulin resistance in humans. Front. Physiol. 2022, 13, 951390. [Google Scholar] [CrossRef]
- Xie, M.; Kong, Y.; Tan, W.; May, H.; Battiprolu, P.K.; Pedrozo, Z.; Wang, Z.V.; Morales, C.; Luo, X.; Cho, G. Histone deacetylase inhibition blunts ischemia/reperfusion injury by inducing cardiomyocyte autophagy. Circulation 2014, 129, 1139–1151. [Google Scholar] [CrossRef]
- Angata, T.; Varki, A. Chemical diversity in the sialic acids and related α-keto acids: An evolutionary perspective. Chem. Rev. 2002, 102, 439–470. [Google Scholar] [CrossRef]
- Muthana, S.M.; Campbell, C.T.; Gildersleeve, J.C. Modifications of glycans: Biological significance and therapeutic opportunities. ACS Chem. Biol. 2012, 7, 31–43. [Google Scholar] [CrossRef] [PubMed]
- Fu, Q.; Cat, A.; Zheng, Y.G. New histone lysine acylation biomarkers and their roles in epigenetic regulation. Curr. Protoc. 2023, 3, e746. [Google Scholar] [CrossRef]
- Wang, Z.; Zhu, L. New insights into lactate in exercise adaptations: Does protein lactylation play a role? Am. J. Physiol.-Endocrinol. Metab. 2025, 329, E405–E419. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Gao, J.; Zhu, Z.; Mao, Q.; Xu, Z.; Singh, P.K.; Rimayi, C.C.; Moreno-Yruela, C.; Xu, S.; Li, G. Lysine L-lactylation is the dominant lactylation isomer induced by glycolysis. Nat. Chem. Biol. 2025, 21, 91–99. [Google Scholar] [CrossRef]
- Zhao, L.; Qi, H.; Lv, H.; Liu, W.; Zhang, R.; Yang, A. Lactylation in health and disease: Physiological or pathological? Theranostics 2025, 15, 1787. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.M.; Yu, Q.W.; Wang, C.; Wang, S.H.; Wang, P.; Zhang, L.R.; Han, S.N. Lactylation in Cardiovascular Diseases: Current Progress and Perspectives. J. Am. Heart Assoc. 2025, 14, e043801. [Google Scholar] [CrossRef]
- Zhao, Q.; Wang, Q.; Yao, Q.; Yang, Z.; Li, W.; Cheng, X.; Wen, Y.; Chen, R.; Xu, J.; Wang, X. Nonenzymatic lysine D-lactylation induced by glyoxalase II substrate SLG dampens inflammatory immune responses. Cell Res. 2025, 35, 97–116. [Google Scholar] [CrossRef]
- Zang, Y.; Zhang, J.; Xia, M.; Wang, A.; Fan, Z.; Han, Y.; Zhang, H.; Wang, S.; Niu, Z.; Wu, J. D-lactate derived from intestinal bacteria drives lysine D-lactylation to modulate transcription in liver cells. bioRxiv 2024. [Google Scholar] [CrossRef]
- Zhu, R.; Ye, X.; Lu, X.; Xiao, L.; Yuan, M.; Zhao, H.; Guo, D.; Meng, Y.; Han, H.; Luo, S.; et al. ACSS2 acts as a lactyl-CoA synthetase and couples KAT2A to function as a lactyltransferase for histone lactylation and tumor immune evasion. Cell Metab. 2025, 37, 361–376.e367. [Google Scholar] [CrossRef]
- Liu, R.; Ren, X.; Park, Y.E.; Feng, H.; Sheng, X.; Song, X.; AminiTabrizi, R.; Shah, H.; Li, L.; Zhang, Y. Nuclear GTPSCS functions as a lactyl-CoA synthetase to promote histone lactylation and gliomagenesis. Cell Metab. 2025, 37, 377–394.e379. [Google Scholar] [CrossRef]
- Zong, Z.; Xie, F.; Wang, S.; Wu, X.; Zhang, Z.; Yang, B.; Zhou, F. Alanyl-tRNA synthetase, AARS1, is a lactate sensor and lactyltransferase that lactylates p53 and contributes to tumorigenesis. Cell 2024, 187, 2375–2392.e2333. [Google Scholar] [CrossRef] [PubMed]
- Moreno-Yruela, C.; Zhang, D.; Wei, W.; Bæk, M.; Liu, W.; Gao, J.; Danková, D.; Nielsen, A.L.; Bolding, J.E.; Yang, L. Class I histone deacetylases (HDAC1–3) are histone lysine delactylases. Sci. Adv. 2022, 8, eabi6696. [Google Scholar] [CrossRef]
- Hu, X.; Huang, X.; Yang, Y.; Sun, Y.; Zhao, Y.; Zhang, Z.; Qiu, D.; Wu, Y.; Wu, G.; Lei, L. Dux activates metabolism-lactylation-MET network during early iPSC reprogramming with Brg1 as the histone lactylation reader. Nucleic Acids Res. 2024, 52, 5529–5548. [Google Scholar] [CrossRef] [PubMed]
- Nuñez, R.; Sidlowski, P.F.; Steen, E.A.; Wynia-Smith, S.L.; Sprague, D.J.; Keyes, R.F.; Smith, B.C. The TRIM33 bromodomain recognizes histone lysine lactylation. ACS Chem. Biol. 2024, 19, 2418–2428. [Google Scholar] [CrossRef]
- Zhai, G.; Niu, Z.; Jiang, Z.; Zhao, F.; Wang, S.; Chen, C.; Zheng, W.; Wang, A.; Zang, Y.; Han, Y. DPF2 reads histone lactylation to drive transcription and tumorigenesis. Proc. Natl. Acad. Sci. USA 2024, 121, e2421496121. [Google Scholar] [CrossRef]
- Li, S.; Xu, C.; Fu, Y.; Lei, P.-J.; Yao, Y.; Yang, W.; Zhang, Y.; Washburn, M.P.; Florens, L.; Jaiswal, M.; et al. DYRK1A interacts with histone acetyl transferase p300 and CBP and localizes to enhancers. Nucleic Acids Res. 2018, 46, 11202–11213. [Google Scholar] [CrossRef] [PubMed]
- Kikuchi, M.; Morita, S.; Wakamori, M.; Sato, S.; Uchikubo-Kamo, T.; Suzuki, T.; Dohmae, N.; Shirouzu, M.; Umehara, T. Epigenetic mechanisms to propagate histone acetylation by p300/CBP. Nat. Commun. 2023, 14, 4103. [Google Scholar] [CrossRef]
- Fan, M.; Yang, K.; Wang, X.; Chen, L.; Gill, P.S.; Ha, T.; Liu, L.; Lewis, N.H.; Williams, D.L.; Li, C. Lactate promotes endothelial-to-mesenchymal transition via Snail1 lactylation after myocardial infarction. Sci. Adv. 2023, 9, eadc9465. [Google Scholar] [CrossRef]
- Porter, N.J.; Christianson, D.W. Structure, mechanism, and inhibition of the zinc-dependent histone deacetylases. Curr. Opin. Struct. Biol. 2019, 59, 9–18. [Google Scholar] [CrossRef]
- Latham, T.; Mackay, L.; Sproul, D.; Karim, M.; Culley, J.; Harrison, D.J.; Hayward, L.; Langridge-Smith, P.; Gilbert, N.; Ramsahoye, B.H. Lactate, a product of glycolytic metabolism, inhibits histone deacetylase activity and promotes changes in gene expression. Nucleic Acids Res. 2012, 40, 4794–4803. [Google Scholar] [CrossRef]
- Miranda-Gonçalves, V.; Lameirinhas, A.; Macedo-Silva, C.; Lobo, J.C.; Dias, P.; Ferreira, V.; Henrique, R.; Jerónimo, C. Lactate increases renal cell carcinoma aggressiveness through sirtuin 1-dependent epithelial mesenchymal transition axis regulation. Cells 2020, 9, 1053. [Google Scholar] [CrossRef] [PubMed]
- Levine, D.C.; Kuo, H.-Y.; Hong, H.-K.; Cedernaes, J.; Hepler, C.; Wright, A.G.; Sommars, M.A.; Kobayashi, Y.; Marcheva, B.; Gao, P. NADH inhibition of SIRT1 links energy state to transcription during time-restricted feeding. Nat. Metab. 2021, 3, 1621–1632. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Wang, X.; Zhu, Y.; Li, Z.; Zhu, Y.T.; Wu, J.C.; Qin, Z.H.; Xiang, M.; Lin, F. Exercise activates lysosomal function in the brain through AMPK-SIRT1-TFEB pathway. CNS Neurosci. Ther. 2019, 25, 796–807. [Google Scholar] [CrossRef]
- Zhang, C.; Zhou, T.; Li, C.; Wang, D.; Tao, J.; Zhu, X.; Lu, J.; Ni, J.; Yao, Y.-F. Deciphering novel enzymatic and non-enzymatic lysine lactylation in Salmonella. Emerg. Microbes Infect. 2025, 14, 2475838. [Google Scholar] [CrossRef] [PubMed]
- Kong, L.R.; Gupta, K.; Wu, A.J.; Perera, D.; Ivanyi-Nagy, R.; Ahmed, S.M.; Tan, T.Z.; Tan, S.L.-W.; Fuddin, A.; Sundaramoorthy, E. A glycolytic metabolite bypasses “two-hit” tumor suppression by BRCA2. Cell 2024, 187, 2269–2287.e2216. [Google Scholar] [CrossRef]
- Gaffney, D.O.; Jennings, E.Q.; Anderson, C.C.; Marentette, J.O.; Shi, T.; Oxvig, A.-M.S.; Streeter, M.D.; Johannsen, M.; Spiegel, D.A.; Chapman, E. Non-enzymatic lysine lactoylation of glycolytic enzymes. Cell Chem. Biol. 2020, 27, 206–213.e206. [Google Scholar] [CrossRef]
- Henderson, G.C.; Horning, M.A.; Wallis, G.A.; Brooks, G.A. Pyruvate metabolism in working human skeletal muscle. Am. J. Physiol.-Endocrinol. Metab. 2007, 292, E366. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Li, S.; Cui, Q.; Guo, B.; Ding, W.; Liu, J.; Quan, L.; Li, X.; Xie, P.; Jin, L. Activation of GPR81 by lactate drives tumour-induced cachexia. Nat. Metab. 2024, 6, 708–723. [Google Scholar] [CrossRef]
- Chen, G.; Liu, J.; Guo, Y.; Sun, P. Mechanisms for Regulatory Effects of Exercise on Metabolic Diseases from the Lactate–Lactylation Perspective. Int. J. Mol. Sci. 2025, 26, 3469. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Xing, T.; Zhang, L.; Zhao, L.; Gao, F. Dynamic changes of protein lactylation and their correlations with the glycolytic process during the postmortem acidification of broiler breast. Poult. Sci. 2024, 103, 104354. [Google Scholar] [CrossRef]
- Liu, X.; Xu, Y.; Zhao, X.; Bai, Y.; Ren, C.; Li, X.; Hou, C.; Zhang, D. The role of lactate in meat beyond pH regulation: A study on lactylation and its effects on meat metabolism. Food Chem. 2025, 489, 144975. [Google Scholar] [CrossRef]
- Essén-Gustavsson, B.; Henriksson, J. Enzyme levels in pools of microdissected human muscle fibres of identified type: Adaptive response to exercise. Acta Physiol. Scand. 1984, 120, 505–515. [Google Scholar] [CrossRef]
- Kobayashi, M. Fiber type-specific localization of monocarboxylate transporters MCT1 and MCT4 in rat skeletal muscle. Kurume Med. J. 2004, 51, 253–261. [Google Scholar] [CrossRef]
- Vøillestad, N.; Tabata, I.; Medbø, J. Glycogen breakdown in different human muscle fibre types during exhaustive exercise of short duration. Acta Physiol. Scand. 1992, 144, 135–141. [Google Scholar] [CrossRef] [PubMed]
- Mattingly, M.L.; Anglin, D.A.; Ruple, B.A.; Scarpelli, M.C.; Bergamasco, J.G.; Godwin, J.S.; Mobley, C.B.; Frugé, A.D.; Libardi, C.A.; Roberts, M.D. Acute and Chronic Resistance Training, Acute Endurance Exercise, nor Physiologically Plausible Lactate In Vitro Affect Skeletal Muscle Lactylation. Int. J. Mol. Sci. 2024, 25, 12216. [Google Scholar] [CrossRef]
- Chang, J.; Wu, W.; Qian, P.; Lu, Z.; He, X.; Wang, F.; Zhang, T. Multi-omics study on the effect of moderate-intensity exercise on protein lactylation in mouse muscle tissue. Front. Cell Dev. Biol. 2025, 12, 1472338. [Google Scholar] [CrossRef]
- Dai, W.; Wu, G.; Liu, K.; Chen, Q.; Tao, J.; Liu, H.; Shen, M. Lactate promotes myogenesis via activating H3K9 lactylation-dependent up-regulation of Neu2 expression. J. Cachexia Sarcopenia Muscle 2023, 14, 2851–2865. [Google Scholar] [CrossRef]
- Desgeorges, T.; Galle, E.; Zhang, J.; von Meyenn, F.; De Bock, K. Histone lactylation in macrophages is predictive for gene expression changes during ischemia induced-muscle regeneration. Mol. Metab. 2024, 83, 101923. [Google Scholar] [CrossRef]
- Sun, W.; Jia, M.; Feng, Y.; Cheng, X. Lactate is a bridge linking glycolysis and autophagy through lactylation. Autophagy 2023, 19, 3240–3241. [Google Scholar] [CrossRef]
- Chen, J.; Zhang, M.; Liu, Y.; Zhao, S.; Wang, Y.; Wang, M.; Niu, W.; Jin, F.; Li, Z. Histone lactylation driven by mROS-mediated glycolytic shift promotes hypoxic pulmonary hypertension. J. Mol. Cell Biol. 2022, 14, mjac073. [Google Scholar] [CrossRef]
- Chen, A.; Chen, Z.; Huang, B.; Lian, G.; Luo, L.; Xie, L. Hypoxia-induced histone lactylation promotes pulmonary arterial smooth muscle cells proliferation in pulmonary hypertension. Mol. Cell Biol. 2025, 480, 5685–5697. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; Zhyvylo, I.; Goncharov, D.; Teos, L.; Lin, D.; Franzi, L.; Saiyed, A.; Neeli, S.; Kenyon, N.; Wolters, P. LDHA-Lactate Promotes Smooth Muscle Remodeling and Pulmonary Hypertension Through Lactylation of TOP1 and EMILIN1. Circulation 2023, 148, A14485. [Google Scholar] [CrossRef]
- Zhu, Y.; Chen, J.-C.; Zhang, J.-L.; Wang, F.-F.; Liu, R.-P. A new mechanism of arterial calcification in diabetes: Interaction between H3K18 lactylation and CHI3L1. Clin. Sci. 2025, 139, 115–130. [Google Scholar] [CrossRef]
- Ma, W.; Jia, K.; Cheng, H.; Xu, H.; Li, Z.; Zhang, H.; Xie, H.; Sun, H.; Yi, L.; Chen, Z. Orphan nuclear receptor NR4A3 promotes vascular calcification via histone lactylation. Circ. Res. 2024, 134, 1427–1447. [Google Scholar] [CrossRef]
- Li, X.; Chen, M.; Chen, X.; He, X.; Li, X.; Wei, H.; Tan, Y.; Min, J.; Azam, T.; Xue, M. TRAP1 drives smooth muscle cell senescence and promotes atherosclerosis via HDAC3-primed histone H4 lysine 12 lactylation. Eur. Heart J. 2024, 45, 4219–4235. [Google Scholar] [CrossRef]
- Xu, X.; Zhang, D.-D.; Kong, P.; Gao, Y.-K.; Huang, X.-F.; Song, Y.; Zhang, W.-D.; Guo, R.-J.; Li, C.-L.; Chen, B.-W. Sox10 escalates vascular inflammation by mediating vascular smooth muscle cell transdifferentiation and pyroptosis in neointimal hyperplasia. Cell Rep. 2023, 42, 112869. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Jiang, H.; Dong, M.; Min, J.; He, X.; Tan, Y.; Liu, F.; Chen, M.; Chen, X.; Yin, Q. Macrophage MCT4 inhibition activates reparative genes and protects from atherosclerosis by histone H3 lysine 18 lactylation. Cell Rep. 2024, 43, 114180. [Google Scholar] [CrossRef]
- Zhang, J.; Luo, M.-Y.; Li, N.-P.; Liang, N.; Yang, Y.-H.; Zhang, Y.-R.; Tan, G.-K.; Xie, T.; Gong, S.-X.; Wang, A.-P. Histone lactylation-derived prelamin A accelerates pulmonary arterial smooth muscle cells senescence in hypoxia-induced pulmonary hypertension rats. Int. Immunopharmacol. 2025, 159, 114871. [Google Scholar] [CrossRef]
- Zhu, X.; Pang, X.; Wang, X.; Guan, X.; Tang, Y.; Wang, Z.; Zhang, L.; Zheng, X.; Li, F.; Mei, J. Super-Enhancer–Driven LncRNA UNC5B-AS1 Inhibits Inflammatory Phenotypic Transition in Pulmonary Artery Smooth Muscle Cells via Lactylation. Arterioscler. Thromb. Vasc. Biol. 2025, 45, e307–e323. [Google Scholar] [CrossRef]
- Zhang, T.; Zhu, Y.; Wang, X.; Chong, D.; Wang, H.; Bu, D.; Zhao, M.; Fang, L.; Li, C. The characterization of protein lactylation in relation to cardiac metabolic reprogramming in neonatal mouse hearts. J. Genet. Genom. 2024, 51, 735–748. [Google Scholar] [CrossRef]
- Li, R.; Wang, J.; Zhao, J.; Liu, J.; Qin, Y.; Wang, Y.; Yuan, Y.; Kang, N.; Yao, L.; Yang, F. Altered Lactylation Myocardial Tissue May Contribute to a More Severe Energy-Deprived State of the Tissue and Left Ventricular Outflow Tract Obstruction in HOCM. Bioengineering 2025, 12, 379. [Google Scholar] [CrossRef]
- Zhang, T.-N.; Huang, X.-M.; Li, L.; Li, Y.; Liu, Y.-P.; Shi, X.-L.; Wu, Q.-J.; Wen, R.; Yang, Y.-H.; Zhang, T. Lactylation of HADHA promotes sepsis-induced myocardial depression. Circ. Res. 2025, 137, e65–e87. [Google Scholar] [CrossRef]
- She, H.; Hu, Y.; Zhao, G.; Du, Y.; Wu, Y.; Chen, W.; Li, Y.; Wang, Y.; Tan, L.; Zhou, Y. Dexmedetomidine Ameliorates Myocardial Ischemia-Reperfusion Injury by Inhibiting MDH2 Lactylation via Regulating Metabolic Reprogramming. Adv. Sci. 2024, 11, 2409499. [Google Scholar] [CrossRef] [PubMed]
- Luan, X.; Du, R.; Su, G.; Yan, C.; Ren, X.; Ju, K.; Jin, Y.; An, Y.; Guo, D.; Tian, Z. Epigenetic regulation of cardiac tissue development by lysine lactylation. hLife 2025, 3, 82–97. [Google Scholar] [CrossRef]
- Zhang, H.; Feng, Z.; Tang, K.; Zhang, L.; Qiu, Z.; Qian, L. 12-HEPE promotes cardiomyocyte proliferation by activating glycolysis and histone lactylation via Hippo signaling. Eur. J. Pharmacol. 2025, 1003, 177880. [Google Scholar] [CrossRef]
- Ma, X.M.; Geng, K.; Wang, P.; Jiang, Z.; Law, B.Y.-K.; Xu, Y. MCT4-dependent lactate transport: A novel mechanism for cardiac energy metabolism injury and inflammation in type 2 diabetes mellitus. Cardiovasc. Diabetol. 2024, 23, 96. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; Wang, W.; Wang, X.; Mang, G.; Chen, J.; Yan, X.; Tong, Z.; Yang, Q.; Wang, M.; Chen, L. Histone lactylation boosts reparative gene activation post–myocardial infarction. Circ. Res. 2022, 131, 893–908. [Google Scholar] [CrossRef] [PubMed]
- Yu, W.; Kong, Q.; Jiang, S.; Li, Y.; Wang, Z.; Mao, Q.; Zhang, X.; Liu, Q.; Zhang, P.; Li, Y.; et al. HSPA12A maintains aerobic glycolytic homeostasis and Histone3 lactylation in cardiomyocytes to attenuate myocardial ischemia/reperfusion injury. JCI Insight 2024, 9, e169125. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Li, D.; Yao, F.; Feng, S.; Tong, C.; Rao, R.; Zhong, M.; Wang, X.; Feng, W.; Hu, Z. Serpina3k lactylation protects from cardiac ischemia reperfusion injury. Nat. Commun. 2025, 16, 1012. [Google Scholar] [CrossRef]
- Lv, J.; Yin, M.; Jin, H. Hypoxia Aggravates Myocardial Ischemia/Reperfusion Injury Through the Promotion of Ferroptosis via ACSL4 Lactylation. J. Cardiovasc. Transl. Res. 2025, 18, 1132–1145. [Google Scholar] [CrossRef]
- Wang, Y.; Yue, Q.; Song, X.; Du, W.; Liu, R. Hypoxia/reoxygenation-induced Glycolysis Mediates Myocardial Ischemia–reperfusion Injury Through Promoting the Lactylation of GPX4. J. Cardiovasc. Transl. Res. 2025, 18, 762–774. [Google Scholar] [CrossRef]
- Fang, L.; Yu, Z.; Qian, X.; Fang, H.; Wang, Y. LDHA exacerbates myocardial ischemia-reperfusion injury through inducing NLRP3 lactylation. BMC Cardiovasc. Disord. 2024, 24, 651. [Google Scholar] [CrossRef]
- Xu, G.-E.; Yu, P.; Hu, Y.; Wan, W.; Shen, K.; Cui, X.; Wang, J.; Wang, T.; Cui, C.; Chatterjee, E. Exercise training decreases lactylation and prevents myocardial ischemia–reperfusion injury by inhibiting YTHDF2. Basic Res. Cardiol. 2024, 119, 651–671. [Google Scholar] [CrossRef]
- Zhang, N.; Zhang, Y.; Xu, J.; Wang, P.; Wu, B.; Lu, S.; Lu, X.; You, S.; Huang, X.; Li, M. α-myosin heavy chain lactylation maintains sarcomeric structure and function and alleviates the development of heart failure. Cell Res. 2023, 33, 679–698. [Google Scholar] [CrossRef]
- Zhao, S.S.; Liu, J.; Wu, Q.C.; Zhou, X.L. Lactate regulates pathological cardiac hypertrophy via histone lactylation modification. J. Cell. Mol. Med. 2024, 28, e70022. [Google Scholar] [CrossRef]
- Wang, B.; Ma, J.; Yang, D. Role of PFKM lactylation in glycolysis regulation in endometrial cancer cells. Genes Dis. 2025, 12, 101400. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Yang, P.; Yu, T.; Gao, M.; Liu, D.; Zhang, J.; Lu, C.; Chen, X.; Zhang, X.; Liu, Y. Lactylation of PKM2 suppresses inflammatory metabolic adaptation in pro-inflammatory macrophages. Int. J. Biol. Sci. 2022, 18, 6210. [Google Scholar] [CrossRef]
- Pan, R.-Y.; He, L.; Zhang, J.; Liu, X.; Liao, Y.; Gao, J.; Liao, Y.; Yan, Y.; Li, Q.; Zhou, X. Positive feedback regulation of microglial glucose metabolism by histone H4 lysine 12 lactylation in Alzheimer’s disease. Cell Metab. 2022, 34, 634–648.e636. [Google Scholar] [CrossRef]
- Chen, X.; Huang, W.; Zhang, J.; Li, Y.; Xing, Z.; Guo, L.; Jiang, H.; Zhang, J. High-intensity interval training induces lactylation of fatty acid synthase to inhibit lipid synthesis. BMC Biol. 2023, 21, 196. [Google Scholar] [CrossRef]
- Wang, Y.; Sun, J.; Xue, L.; Sun, Y.; Zhang, K.; Fan, M.; Qian, H.; Li, Y.; Wang, L. Chlorogenic Acid Improves High-Fat Diet-Induced Skeletal Muscle Metabolic Disorders by Regulating Mitochondrial Function and Lactate Metabolism. J. Agric. Food Chem. 2025, 73, 10347–10357. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wang, F.; Sun, J.; Xue, L.; Sun, Y.; Zhang, K.; Fan, M.; Qian, H.; Yang, B.; Du, J. Gallic Acid Ameliorates Skeletal Muscle Metabolic Inflexibility by Regulating Lactate Metabolism and Promoting Mitochondrial Function. Mol. Nutr. Food Res. 2025, 69, e70106. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Q. On the indirect relationship between protein dynamics and enzyme activity. Prog. Biophys. Mol. Biol. 2017, 125, 52–60. [Google Scholar] [CrossRef] [PubMed]
- Mattingly, M.L.; Ruple, B.A.; Sexton, C.L.; Godwin, J.S.; McIntosh, M.C.; Smith, M.A.; Plotkin, D.L.; Michel, J.M.; Anglin, D.A.; Kontos, N.J. Resistance training in humans and mechanical overload in rodents do not elevate muscle protein lactylation. Front. Physiol. 2023, 14, 1281702. [Google Scholar] [CrossRef]
- Nowacka, A.; Śniegocki, M.; Śniegocka, M.; Ziółkowska, E.A. Nicotinamide and Pyridoxine in Muscle Aging: Nutritional Regulation of Redox, Inflammation, and Regeneration. Antioxidants 2025, 14, 911. [Google Scholar] [CrossRef]
- Torres-Méndez, J.K.; Niño-Narvión, J.; Martinez-Santos, P.; Diarte-Añazco, E.M.G.; Méndez-Lara, K.A.; Del Olmo, T.V.; Rotllan, N.; Julián, M.T.; Alonso, N.; Mauricio, D. Nicotinamide Prevents Diabetic Brain Inflammation via NAD+-Dependent Deacetylation Mechanisms. Nutrients 2023, 15, 3083. [Google Scholar] [CrossRef]
- Zhang, X.; Meng, F.; Lyu, W.; He, J.; Wei, R.; Du, Z.; Zhang, C.; Guan, Y.; Huang, X.; Lyu, G. Histone lactylation antagonizes senescence and skeletal muscle aging via facilitating gene expression reprogramming. bioRxiv 2023. [Google Scholar] [CrossRef]
- Zou, K.; Hinkley, J.M.; Park, S.; Zheng, D.; Jones, T.E.; Pories, W.J.; Hornby, P.J.; Lenhard, J.; Dohm, G.L.; Houmard, J.A. Altered tricarboxylic acid cycle flux in primary myotubes from severely obese humans. Int. J. Obes. 2019, 43, 895–905. [Google Scholar]
- Zhou, R.; Ding, R.-C.; Yu, Q.; Qiu, C.-Z.; Zhang, H.-Y.; Yin, Z.-J.; Ren, D.-L. Metformin attenuates neutrophil recruitment through the H3K18 lactylation/reactive oxygen species pathway in zebrafish. Antioxidants 2024, 13, 176. [Google Scholar] [CrossRef]
- Xu, R.; Yuan, W.; Wang, Z. Advances in glycolysis metabolism of atherosclerosis. J. Cardiovasc. Transl. Res. 2023, 16, 476–490. [Google Scholar] [CrossRef]
- Grootaert, M.O.; Moulis, M.; Roth, L.; Martinet, W.; Vindis, C.; Bennett, M.R.; De Meyer, G.R. Vascular smooth muscle cell death, autophagy and senescence in atherosclerosis. Cardiovasc. Res. 2018, 114, 622–634. [Google Scholar] [CrossRef] [PubMed]
- Ouyang, J.; Wang, H.; Huang, J. The role of lactate in cardiovascular diseases. Cell Commun. Signal. 2023, 21, 317. [Google Scholar] [CrossRef]
- Tsao, C.W.; Aday, A.W.; Almarzooq, Z.I.; Alonso, A.; Beaton, A.Z.; Bittencourt, M.S.; Boehme, A.K.; Buxton, A.E.; Carson, A.P.; Commodore-Mensah, Y. Heart disease and stroke statistics—2022 update: A report from the American Heart Association. Circulation 2022, 145, e153–e639. [Google Scholar] [CrossRef]
- Bei, Y.; Zhu, Y.; Zhou, J.; Ai, S.; Yao, J.; Yin, M.; Hu, M.; Qi, W.; Spanos, M.; Li, L. Inhibition of Hmbox1 promotes cardiomyocyte survival and glucose metabolism through Gck activation in ischemia/reperfusion injury. Circulation 2024, 150, 848–866. [Google Scholar] [CrossRef]
- Bosso, G.; Mercurio, V.; Diab, N.; Pagano, A.; Porta, G.; Allegorico, E.; Serra, C.; Guiotto, G.; Numis, F.G.; Tocchetti, C.G. Time-weighted lactate as a predictor of adverse outcome in acute heart failure. ESC Heart Fail. 2021, 8, 539–545. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Fan, W.; Li, N.; Ma, Y.; Yao, M.; Wang, G.; He, S.; Li, W.; Tan, J.; Lu, Q. YY1 lactylation in microglia promotes angiogenesis through transcription activation-mediated upregulation of FGF2. Genome Biol. 2023, 24, 87. [Google Scholar] [CrossRef] [PubMed]
- Ferguson, B.S.; Rogatzki, M.J.; Goodwin, M.L.; Kane, D.A.; Rightmire, Z.; Gladden, L.B. Lactate metabolism: Historical context, prior misinterpretations, and current understanding. Eur. J. Appl. Physiol. 2018, 118, 691–728. [Google Scholar] [CrossRef]
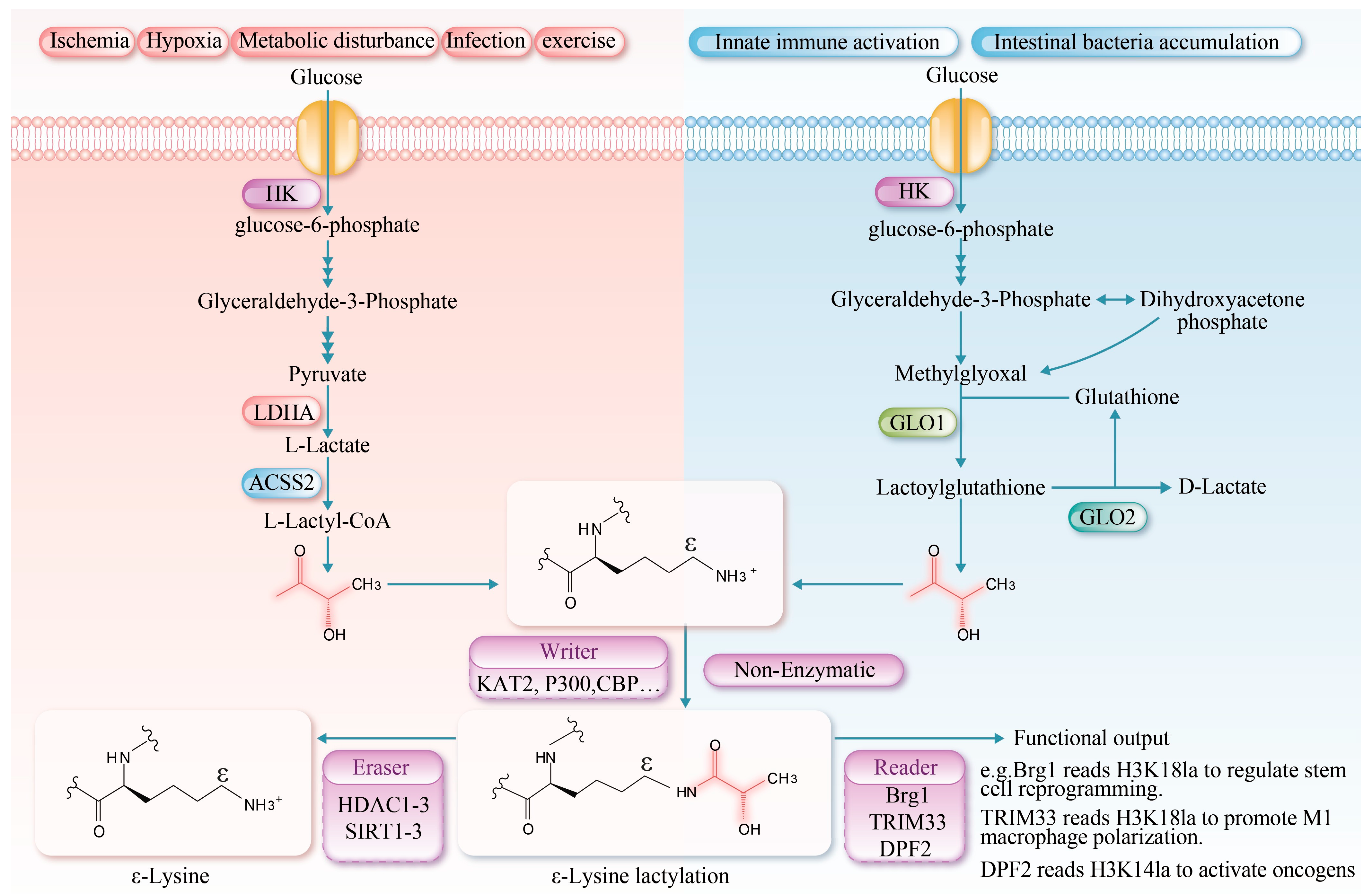
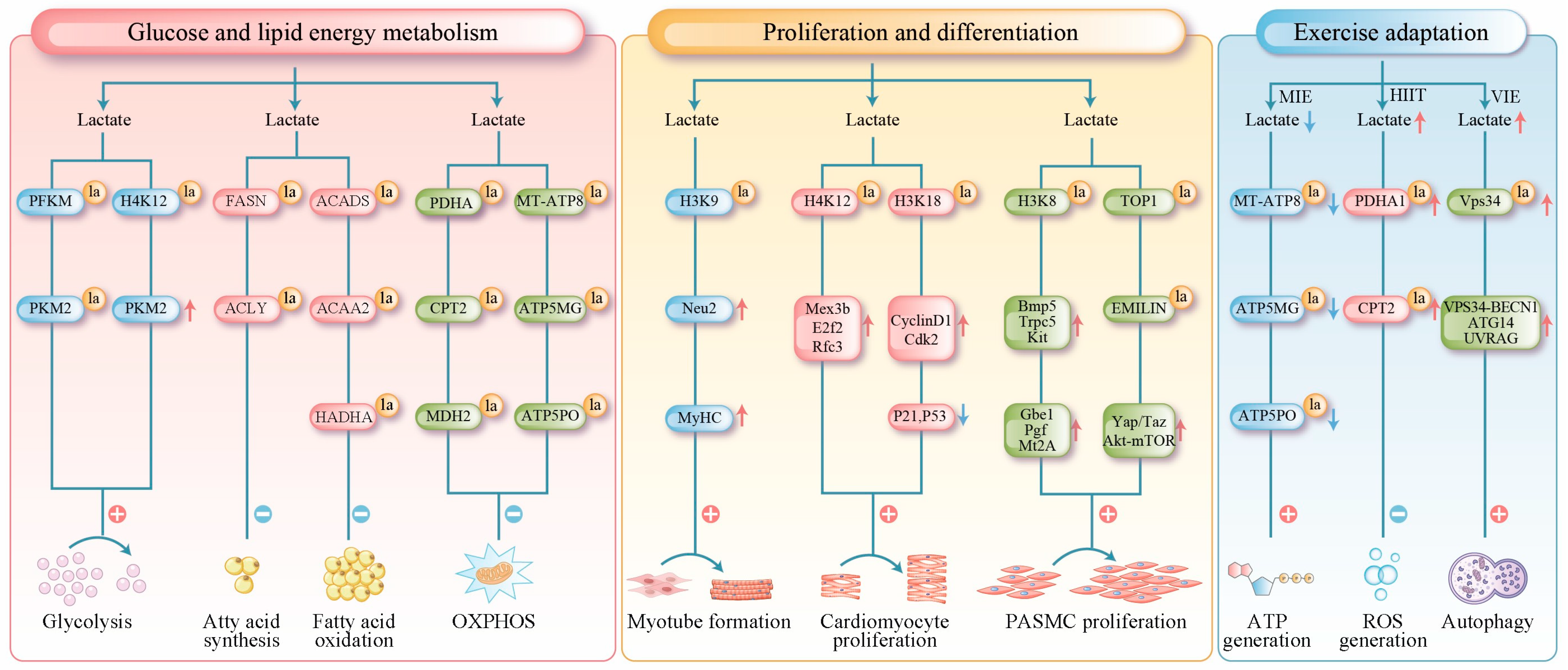
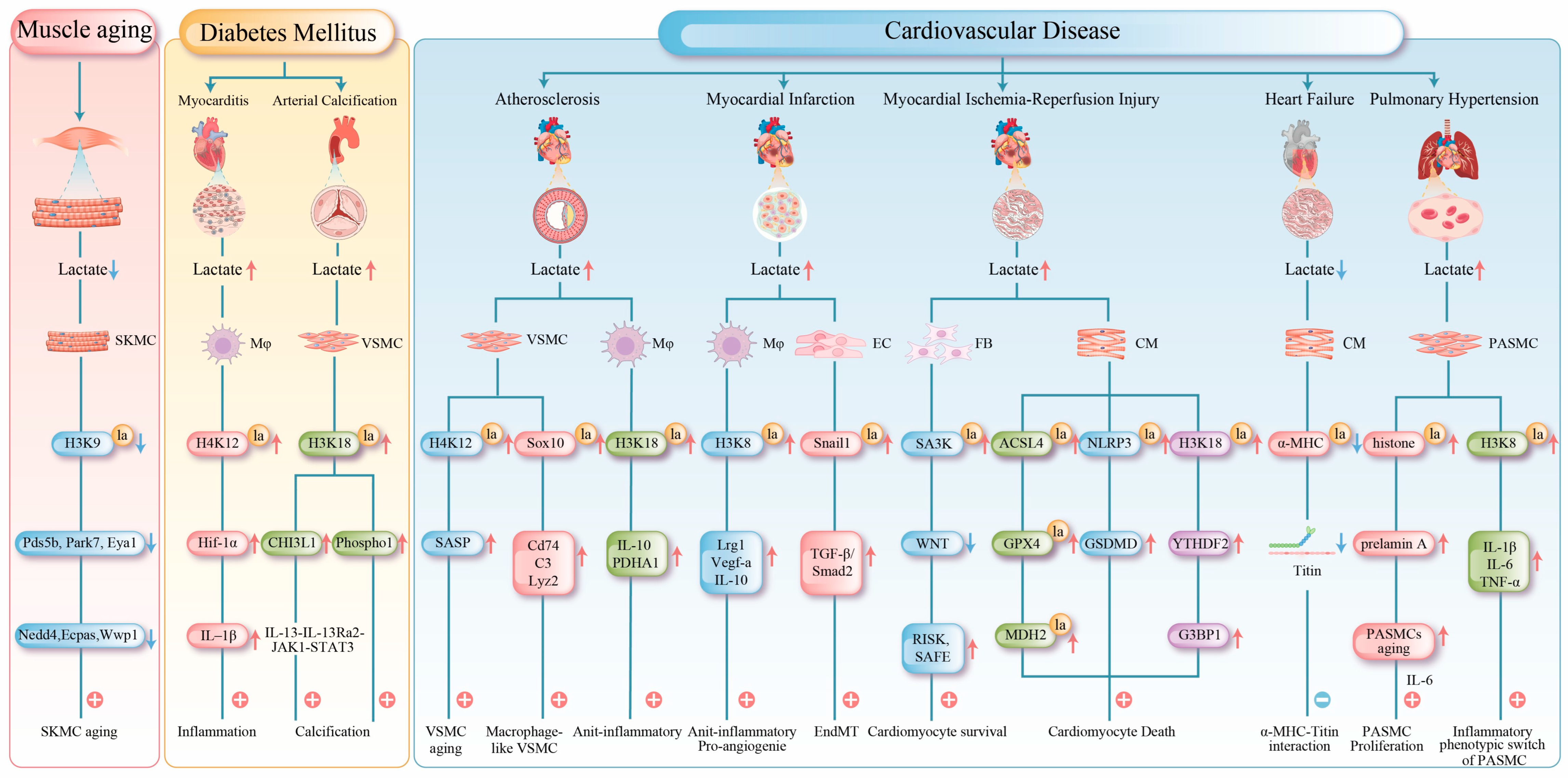
| Enzymes | Regulatory Factors | Response to Physiological /Pathological States | References |
|---|---|---|---|
| Lactyl-CoA synthetase | |||
| ACSS2 |
| High metabolic demand: In glycolytically active cells (tumor cells), they can respond to tumor microenvironmental changes such as growth factor signaling, hypoxia, and lactate accumulation. | [23] |
| GTPSCS |
| High metabolic demand: In glycolysis-active tumor cells, lactate accumulation and increased levels of GTP/CoA lead to substantially enhanced activity. | [24] |
| Lysine lactyltransferase | |||
| P300/CBP |
| Metabolic reprogramming: When cellular metabolism shifts from oxidative phosphorylation to glycolysis, the enzyme may show a preferential tendency to catalyze lactylation. | [30,31,32] |
| AARS1/2 |
|
| [8,25] |
| Lysine delactylase | |||
| HDAC1-3 |
|
| [26,33,34] |
| SIRT1-3 |
|
| [35,36,37] |
| Cell Type | Lactylation Site | Target Gene/ Pathway | Function | Reference |
|---|---|---|---|---|
| SKMC | PKM2, PKFM | / | Linked to glycolytic metabolism. | [9] |
| SKMC | PDHA1 K336, CPT2 K457/458 | / | Inhibiting acetyl-CoA generation from both glucose and fatty acids. | [8] |
| SKMC | MT-ATP8, ATP5MG, ATP5PO | / | Suppressing the expression of related proteins to restrict oxidative phosphorylation. | [50] |
| SKMC | H3K9 | Neu2 | Driving myoblast differentiation into myotubes. | [51] |
| Macrophage | H3K18 | / | Forecasting muscle regeneration potential based on injury biomarkers. | [52] |
| SKMC | VPS34 K356/K781 | VPS34-BECNI, ATG14, UVRAG | Promoting autophagy and endo-lysosomal degradation. | [53] |
| SKMC | H3K9 | Pds5b, Park7b, Eya1, Nedd4, Ubc, Wwp1 | Activating DNA repair and proteostasis pathways to counteract muscle aging. | [11] |
| HSKMC | / | / | Linked to the development of insulin resistance. | [12] |
| PASMC | H3K18 | Bmp5, Trpc5, Gbe1 | Driving the proliferation of PASMCs and vascular remodeling. | [54] |
| PASMC | H3K18 | Gbe1, Pgf, Mt2A, Ythdf2, Gys1 | Driving the proliferation of PASMCs. | [55] |
| PASMC | TOP1, EMILIN-1 | Yap/Taz, Akt-mTOR | Promoting PASMCs proliferation and survival. | [56] |
| VSMC | H3K18 | CHI3L1 | Inducing osteogenic transdifferentiation of VSMC. | [57] |
| VSMC | H3K18 | Phospho1 | Driving vascular calcification and regulating related gene expression. | [58] |
| VSMC | H4K12 | SASP | Driving smooth muscle cell senescence and promoting disease development. | [59] |
| VSMC | Sox10 | Cd74, C3, Lyz2 | Inducing macrophage-like phenotypic switching in vascular smooth muscle cells | [60] |
| Macrophage | H3K18 | PDHA, IL-10 | Inducing M1 macrophage polarization. | [61] |
| PASMC | Histone | prelamin A | Elevating IL-6 secretion which drives proliferation of PASMC. | [62], |
| PASMC | H3K18 | IL-1β, IL-6, TNF-α | Attenuating pro-inflammatory phenotypic switching in pulmonary artery smooth muscle cells. | [63] |
| Cardiomyocyte | PKM2, PKFM | / | Linked to glycolytic metabolism. | [64] |
| Cardiomyocyte | SCAD, ACAA2 | / | Suppressing mitochondrial β-oxidation of fatty acids. | [65] |
| Cardiomyocyte | HADHA | / | Suppressing mitochondrial β-oxidation of fatty acids. | [66] |
| Cardiomyocyte | MDH2 K241 | / | Participating in the regulation of mitochondrial function and cell death. | [67] |
| Cardiomyocyte | H4K12 | Mex3b, E2f2, Rfc3 | Promoting proliferative capacity of cardiomyocytes. | [68] |
| Cardiomyocyte | H3K18 | CyclinD1, Cdk2 P21, P53 | Activating the proliferation program in cardiomyocytes. | [69] |
| Macrophage | H4K12 | HIF-1α | Reduced infiltration of HIF-1α-positive and IL-1β-positive inflammatory macrophages in the heart. | [70] |
| Cardiomyocyte | H3K18 | Lrg1, Vegf-a, IL-10 | Regulating monocyte–macrophage function; participating in the early and remote activation of genes. | [71] |
| Endothelial cell | Snail1 | TGF-β/Smad2 | Activating TGF-β/Smad2pathway. | [32] |
| Cardiomyocyte | H3K56 | / | Supports cardiomyocyte survival under hypoxia/reoxygenation | [72] |
| Fibroblast | SA3K K351 | RISK/SAFE, WNT | Enhancing the protein stability of SA3K and reducing cardiomyocyte apoptosis. | [73] |
| Cardiomyocyte | ACSL4 K83 | / | Inducing lipid peroxidation-driven ferroptosis in cardiomyocytes. | [74] |
| Cardiomyocyte | GPX4 K21/K228 | / | Sensitizing cardiomyocytes to ferroptosis through GPX4 inhibition. | [75] |
| Cardiomyocyte | NLRP3 K245 | GSDMD | Promoting cardiomyocyte pyroptosis and aggravating injury. Mediating injury through the LDHA-NLRP3 pathway. | [76] |
| Cardiomyocyte | H3K18 | YTHDF2 | Participating in exercise-induced physiological cardiac hypertrophy and I/R injury repair. | [77] |
| Cardiomyocyte | α-MHC K1897 | Titin | Maintaining sarcomere structure and function. | [78] |
| Cardiomyocyte | H3K18 | / | Driving pathological cardiac hypertrophy. | [79] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2026 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license.
Share and Cite
Wang, Z.; Zhang, J.; Wu, J.; Liu, G.; He, Y.; Zhao, H.; Jiang, X.; Yang, S. Progress of Research on the Metabolic Regulation of Lactylation in Muscle Tissues and Its Disease Associations. Biomolecules 2026, 16, 212. https://doi.org/10.3390/biom16020212
Wang Z, Zhang J, Wu J, Liu G, He Y, Zhao H, Jiang X, Yang S. Progress of Research on the Metabolic Regulation of Lactylation in Muscle Tissues and Its Disease Associations. Biomolecules. 2026; 16(2):212. https://doi.org/10.3390/biom16020212
Chicago/Turabian StyleWang, Zhihang, Ji Zhang, Junxi Wu, Guangrun Liu, Yun He, Hongbo Zhao, Xiaolin Jiang, and Shengbo Yang. 2026. "Progress of Research on the Metabolic Regulation of Lactylation in Muscle Tissues and Its Disease Associations" Biomolecules 16, no. 2: 212. https://doi.org/10.3390/biom16020212
APA StyleWang, Z., Zhang, J., Wu, J., Liu, G., He, Y., Zhao, H., Jiang, X., & Yang, S. (2026). Progress of Research on the Metabolic Regulation of Lactylation in Muscle Tissues and Its Disease Associations. Biomolecules, 16(2), 212. https://doi.org/10.3390/biom16020212





