1. Introduction
Wheat, the third largest grain crop of China, is cultivated worldwide as a cereal crop. As one of the most important rations in various countries and regions worldwide [
1], wheat is also the most important ration of Northern China. Therefore, the importance of its stable development is evident. With the global population increase and the natural environment’s deterioration, soil salinisation has become an increasingly serious global problem [
2]. Approximately 7% of the world’s land area (more than 900 million hectares) is threatened by climate change. The area of saline–alkali land in China has reached 100 million hectares, including 3.73 million hectares in the Songnen Plain in Northeast China [
3], which is one of the three typical saline–alkali lands in the global soil distribution area. Therefore, soil salinisation is a widespread source of abiotic stress and has become a major limiting factor in global crop production.
After long-term saline–alkali stress, plants change their morphology to better adapt to the environment [
4]. Leaf anatomical structure analysis revealed that salt-tolerant
Atriplex glauca L. has thicker leaves than other plants, due to increased epidermal and mesophyll thickness and enhanced mesophyll density [
5]. Under saline–alkali stress, the accumulation of sodium ions in the soil solution leads to higher osmotic pressure of the soil solution than that of the plant cell sap, and water flows out of plant cells, leading to osmotic stress and physiological drought [
6]. Plant cells synthesise and accumulate several small-molecule organic compounds, such as proline, soluble proteins, betaine, sugar, polyols, and polyamines, to maintain their water potential in cells and cope with this stress [
7]. Under saline–alkali stress, high concentrations of sodium ions in the soil destroy the dynamic balance of ions in cells, leading to a series of destructive effects on plants, such as the destruction of cell membrane structure, abnormal metabolism in cells, and ion toxicity [
8]. Saline–alkali stress promotes the formation and accumulation of reactive oxygen species (ROS), which affects the physiological function of cells and leads to metabolic disorders. Eliminating the effects of ROS metabolism is of great significance in alleviating damage to wheat seedlings under saline–alkali stress.
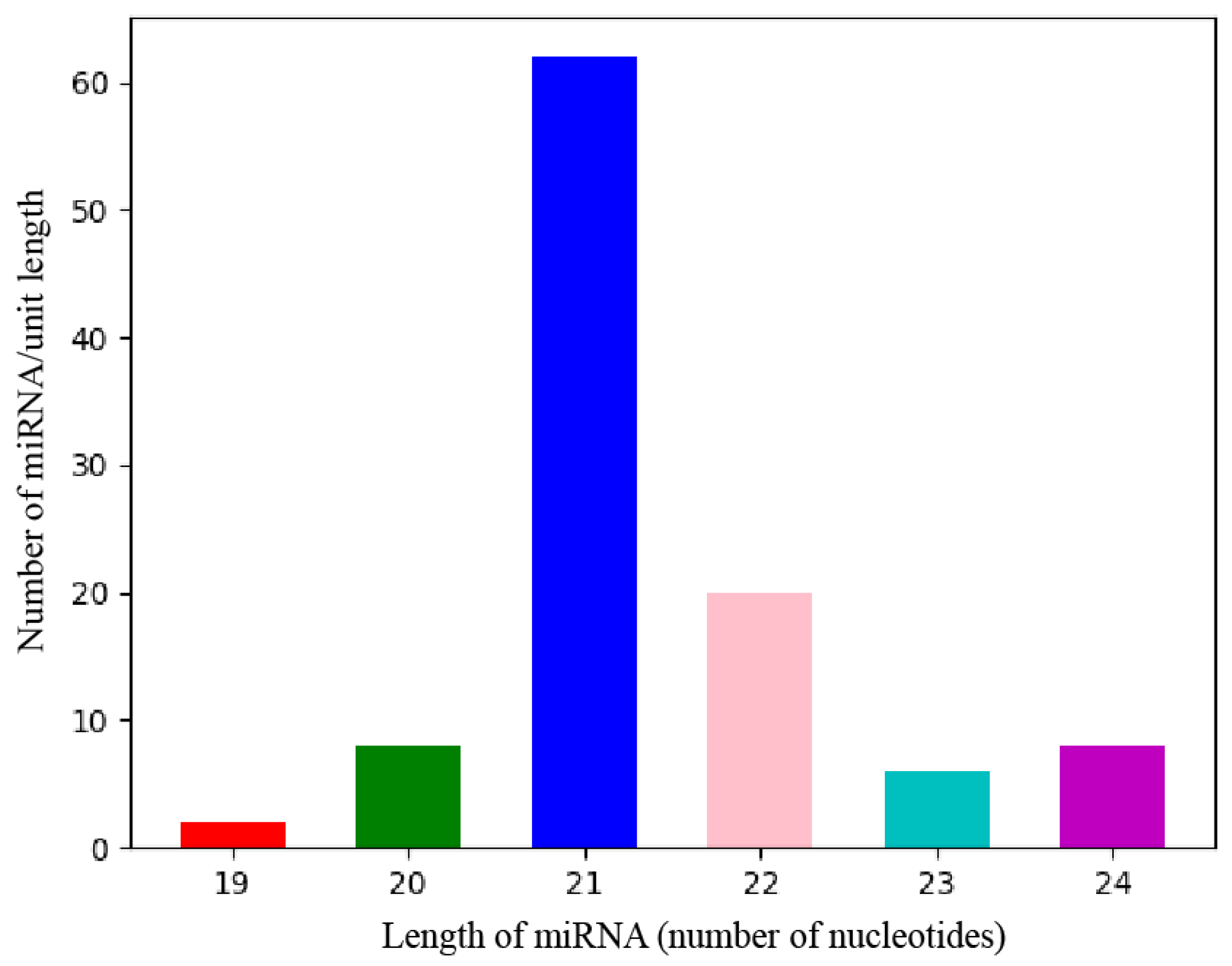
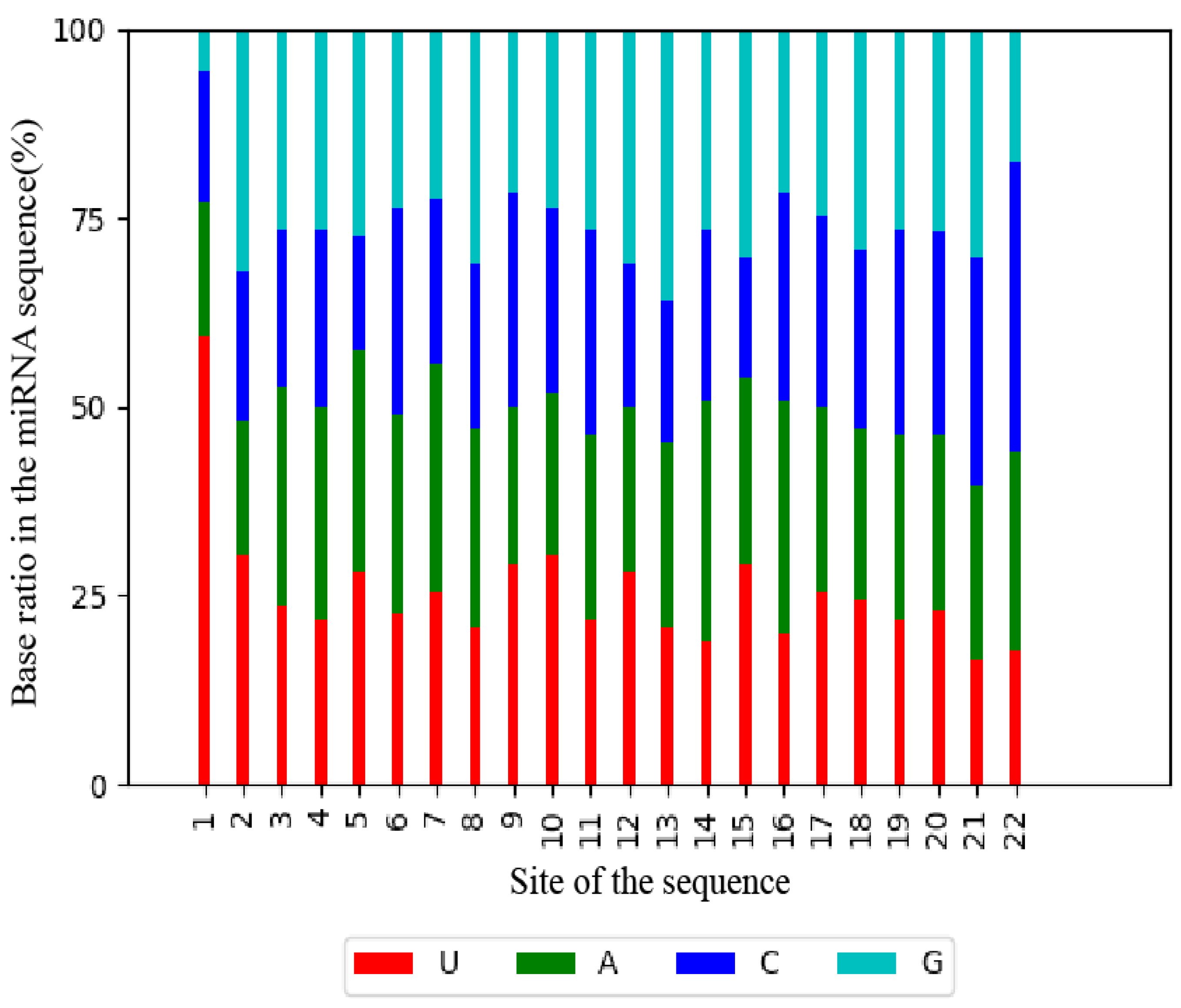
MiRNAs are a class of non-coding RNAs approximately 20–24 nucleotides (nt) in length. In plants, the abundance and diversity of miRNAs enable them to readily regulate most biological processes in the organism via one or more specific miRNAs [
9]. MiRNAs play regulatory roles in key plant physiological processes, such as plant growth and development, as well as responses to biotic and abiotic environmental stresses [
10]. It has been reported that miRNAs are differently regulated in different plant species, such as wheat [
11], corn [
3], soybean [
12], and
Arabidopsis [
13], under saline–alkali stress. Specific post-transcriptional regulation mediated by miRNAs is critical for improving crop resistance to abiotic stresses [
14]. Previous studies have confirmed that, under high salt stress, specific wheat miRNAs enhance salt tolerance by modulating the expression levels of genes related to antioxidation, nutrient absorption, and lipid metabolic balance [
15]. The molecular mechanism of salt tolerance in the roots of salt-tolerant wheat may involve the regulation of expression of miRNAs that target auxin response factors, which are involved in cell growth, ion homeostasis, and hormone signal transduction, thereby contributing to the enhancement of salt tolerance [
16]. However, there are a few studies on the dynamic expression profiles of miRNAs in wheat varieties with contrasting saline–alkali tolerance under different concentrations of saline–alkali stress.
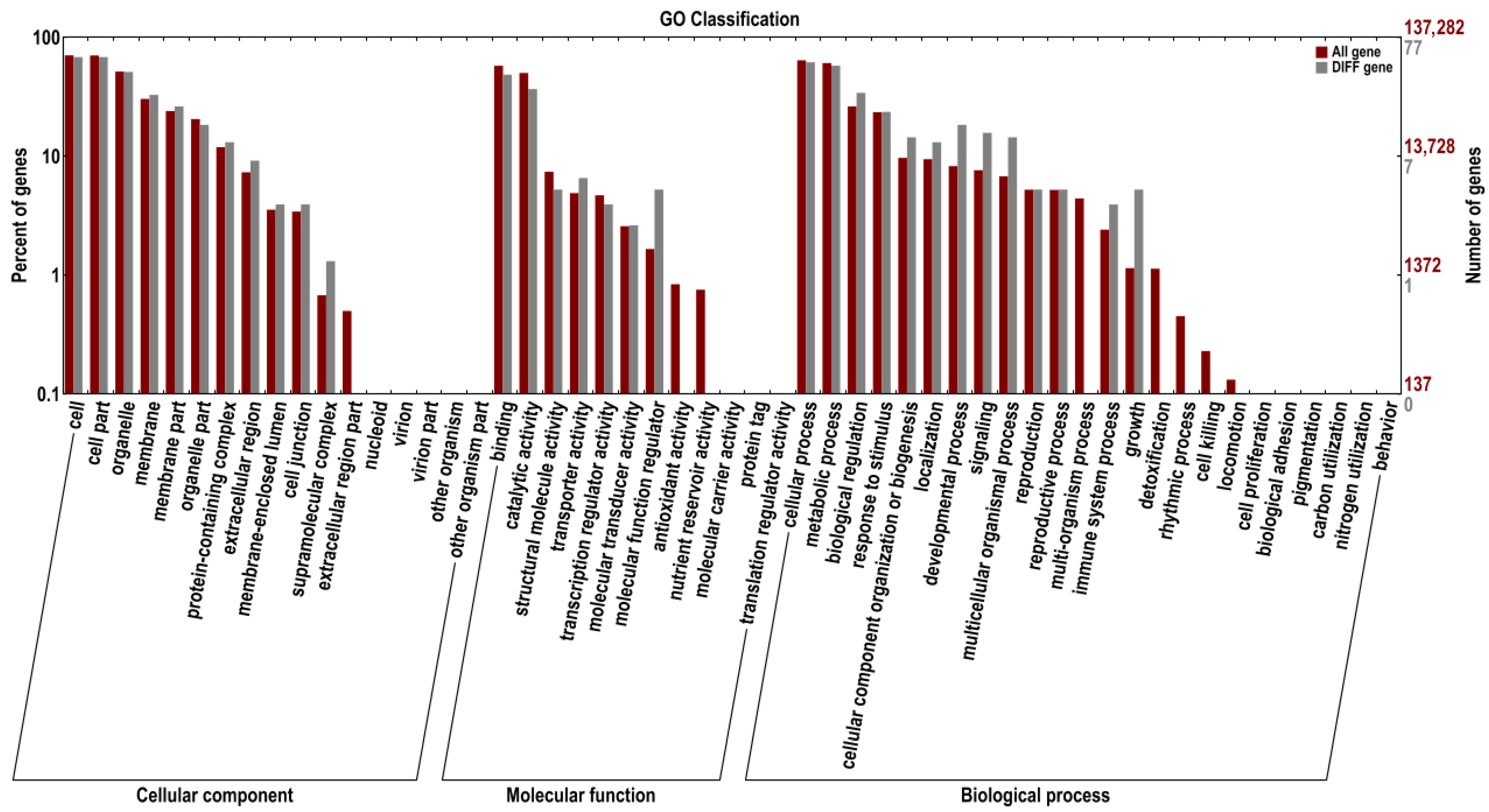
In this study, we aimed to characterise the expression patterns of saline-alkali stress-responsive wheat miRNAs via high-throughput sequencing and further determined the differentially expressed miRNAs (DEMs) in response to saline–alkali stress in wheat roots. Subsequently, the predicted target genes of these DEMs were functionally annotated using Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses to clarify their functional roles in the plant’s saline–alkali stress response. Additionally, we investigated the regulatory mechanisms underlying reactive oxygen species (ROS) metabolism in saline–alkali-tolerant and -sensitive wheat cultivars under saline–alkali stress. Collectively, this study provides a systematic and comprehensive analysis of DEMs in wheat roots in response to saline–alkali stress and offers valuable insights into the differences in saline–alkali tolerance among wheat varieties, thereby aiding in elucidating the molecular mechanisms governing wheat seedling roots’ response to saline–alkali stress.
2. Materials and Methods
2.1. Biological Material
The experimental materials were two wheat (
Triticum aestivum L.) varieties: Qingmai 6 (abbreviated as QM, saline–alkali-tolerant genotype, donated by Shandong Academy of Agricultural Sciences, Jinan, China) [
17] and Meisheng 0308 (abbreviated as MS, saline–alkali-sensitive genotype, provided by professor Fu Zhaolin from our laboratory).
2.2. Cultivation of Wheat Under Laboratory Conditions
After the surface of the seeds were disinfected for 5 min (0.1% mercuric chloride), the residual mercuric chloride was washed with distilled water, and 50 plump, uniform caryopses were used per experimental variant. The caryopses were first sown in sterile Petri dishes with moist filter paper for 2 days of germination, then transplanted into plastic pots (10 cm × 10 cm × 12 cm) containing 1/5 Hoagland nutrient solution for 7 days. Cultivation was conducted in an intelligent artificial climate chamber (Model: RXZ-500D, Ningbo Jiangnan Instrument Factory, Ningbo, China) with the specified 12 h light/12 h dark cycle and 23 ± 2 °C temperature. For stress treatment, seedlings were exposed to 1/5 strength Hoagland nutrient solution supplemented with a saline–alkali mixture (Na2SO4:NaCl:Na2CO3:NaHCO3 = 9:3:3:1, molar ratio) at two total salt concentrations: 150 mmol·L−1 and 300 mmol·L−1. Meanwhile, the control group was treated with 1/5 strength Hoagland nutrient solution (without additional saline–alkali).
Biological replicates were defined as independent experimental units; each replicate consisted of 50 seedlings grown in separate pots under identical environmental conditions (same climate chamber, nutrient solution, and stress treatment). Six experimental variants were established in this study, following the order: QMCK (control group for saline–alkali-tolerant wheat variety Qingmai 6), QM150 (Qingmai 6 treated with 150 mmol·L−1 saline–alkali mixture), QM300 (Qingmai 6 treated with 300 mmol·L−1 saline–alkali mixture), MSCK (control group for saline–alkali-sensitive wheat variety Meisheng 0308), MS150 (Meisheng 0308 treated with 150 mmol·L−1 saline–alkali mixture), and MS300 (Meisheng 0308 treated with 300 mmol·L−1 saline–alkali mixture).
Three biological replicates were established for each experimental variant (QMCK, QM150, QM300, MSCK, MS150, and MS300) and used for all analyses, including miRNA library sequencing, physiological index determination, and phenotypic trait measurement. After 7 days of growth, seedlings were collected, and roots were isolated from these seedlings for the measurement of physiological indicators, phenotypic analysis, and miRNA sequencing, except for the morphological indicators of leaves. The length of the leaves was measured as the straight-line distance from the ligule (the membranous structure at the junction of the leaf base and the leaf sheath) to the leaf tip. Average root length was determined by randomly selecting 10 plants per replicate and measuring the longest root of each selected plant with a vernier caliper (accuracy: 0.1 mm). Average root dry weight was determined by randomly selecting 10 plants per replicate and measuring all roots of each selected plant. All roots were blotted dry with absorbent paper, then oven-dried at 80 °C to a constant weight, and the dry weight was measured using an electronic balance (accuracy: 0.001 g).
2.3. Construction and Sequencing of miRNA Library
First, total RNA was extracted from the roots using the TRIzol (Invitrogen, CA, USA) method. The total RNA extracted was detected using Nanodrop, Qubit 2.0 (Thermo Fisher Scientific, CA, USA), and Agilent 2100 Bioanalyzer (Agilent Technologies, USA) methods to detect the purity, concentration, and integrity of RNA samples to ensure that qualified samples were used for sequencing. The samples were submitted to Nanjing Jisi Huiyuan Biotechnology Co., Ltd. (Nanjing, China). for testing. After the sample passed the quality control test, total RNA was used as the starting sample, and a small RNA sample prep kit (TIANGEN Biotech, Beijing, China)was used to construct the library. Since the small RNA has a phosphate group at the 5’ end and a hydroxyl group at the 3’ end, T4 RNA Ligase 1 and T4 RNA Ligase 2 (truncated) were used to add connectors at the 5’ and 3’ ends of the small RNA, respectively. Reverse transcription was used to synthesise cDNA and PCR amplification was performed. Target fragments were screened using PAGE analysis, and the fragments recovered by gel cutting were used as small RNA libraries. After the construction of the library, a Qubit 2.0 was used to determine the concentration of the library, an Agilent 2100 Bioanalyzer, and qRT-PCR were used to determine the insert size and effective concentration of the library, respectively, to ensure its quality. After library inspection, the NovaSeq 6000 platform (Illumina, Inc., San Diego, CA, USA) was used for high-throughput sequencing with a single-end (SE) reading length of 50 bp (base pairs).
2.4. Prediction and Analysis of miRNA
The unclassified ncRNA sequences were aligned to the reference sequence (depth ≥ 2) using Bowtie (v1.2.2) [
18], generating mapped sequences with positional annotations. The reads corresponding to the reference genome (IWGSC RefSeq v2.1) were compared with miRBase to obtain annotation information for known miRNAs. The Rfam database was used to annotate the reads that did not conform to known miRNAs, filter ribosomal RNA (rRNA), transfer RNA (tRNA), intranuclear small RNA (snRNA), nucleolar small RNA (snoRNA), and other ncRNAs (non-coding RNAs) and repetitive sequences and obtained uncommented reads containing potential miRNAs. The sRNAs of Rfam and miRBase was then compared to the reference genome, the surrounding sequences intercepted, and miRDeep2 software (Version 2.0.1.2) was used to predict the secondary structure [
19]. Based on the predicted results, they were filtered using Dicer digestion site information, energy values, and other characteristics to identify new miRNAs. The miRNA expression was standardised to tags per million (TPM), and differentially expressed miRNAs were detected using DESeq2 software (v1.38.0, Bioconductor,
https://bioconductor.org/packages/DESeq2/, accessed on 25 January 2026) [
20] with the following criteria: log
2FC (log
2 Fold Change) ≥ 1 (significantly upregulated) or log
2FC ≤ −1 (significantly downregulated), and FDR (False Discovery Rate) < 0.05. FDR is a statistical measure to control the proportion of false positives among significant results, while log
2FC represents the log
2-transformed ratio of miRNA expression levels between stress and control groups.
2.5. Determination of Plant Physiological Indexes
The superoxide radical (O
2−) content was determined by the hydroxylamine oxidation method [
21]. The hydrogen peroxide (H
2O
2) content was measured using the titanium chloride method [
22]. Malondialdehyde (MDA) content was assessed using the method described and improved [
23]. Ascorbic acid (AsA) content was assayed according to the described method [
24]. Superoxide dismutase (SOD) activity was measured based on nitrate blue tetrazolium (NBT) photoreduction [
25]. The peroxidase (POD) activity was determined using guaiacol as the substrate [
26]. Catalase (CAT) activity was mainly measured using the spectrophotometric assays [
27] and ascorbate peroxidase (APX) activity was mainly assayed using the UV absorption method [
28]. Glutathione reductase (GR) activity was determined according to a modified version of the method described [
29]. Dehydroascorbate (DHA) content was determined by the method [
30]. Oxidized glutathione (GSSG) and reduced glutathione (GSH) were estimated following the method [
31]. Root activity was determined using a modified triphenyltetrazolium chloride (TTC) reduction method [
32]. Briefly, fresh root samples (0.5 g) were incubated in TTC solution at 37 °C for 2 h, and the formazan produced was extracted with ethanol. The absorbance was measured at 485 nm, and root activity was expressed as μg formazan g
−1 fresh weight h
−1.
2.6. Statistical Analysis
All experimental data were analyzed using SPSS 26.0 software (IBM Corporation, Chicago, IL, USA). Significant differences between groups were determined by one-way analysis of variance (ANOVA) followed by Duncan’s multiple range test (p < 0.05). Error bars in figures and tables represent the standard deviation (SD) of three biological replicates.
4. Discussion
MiRNAs can regulate the normal growth and differentiation of plant organisms, embryonic development and growth, and other biochemical processes, as well as the regulatory response to external stress or external stress signals [
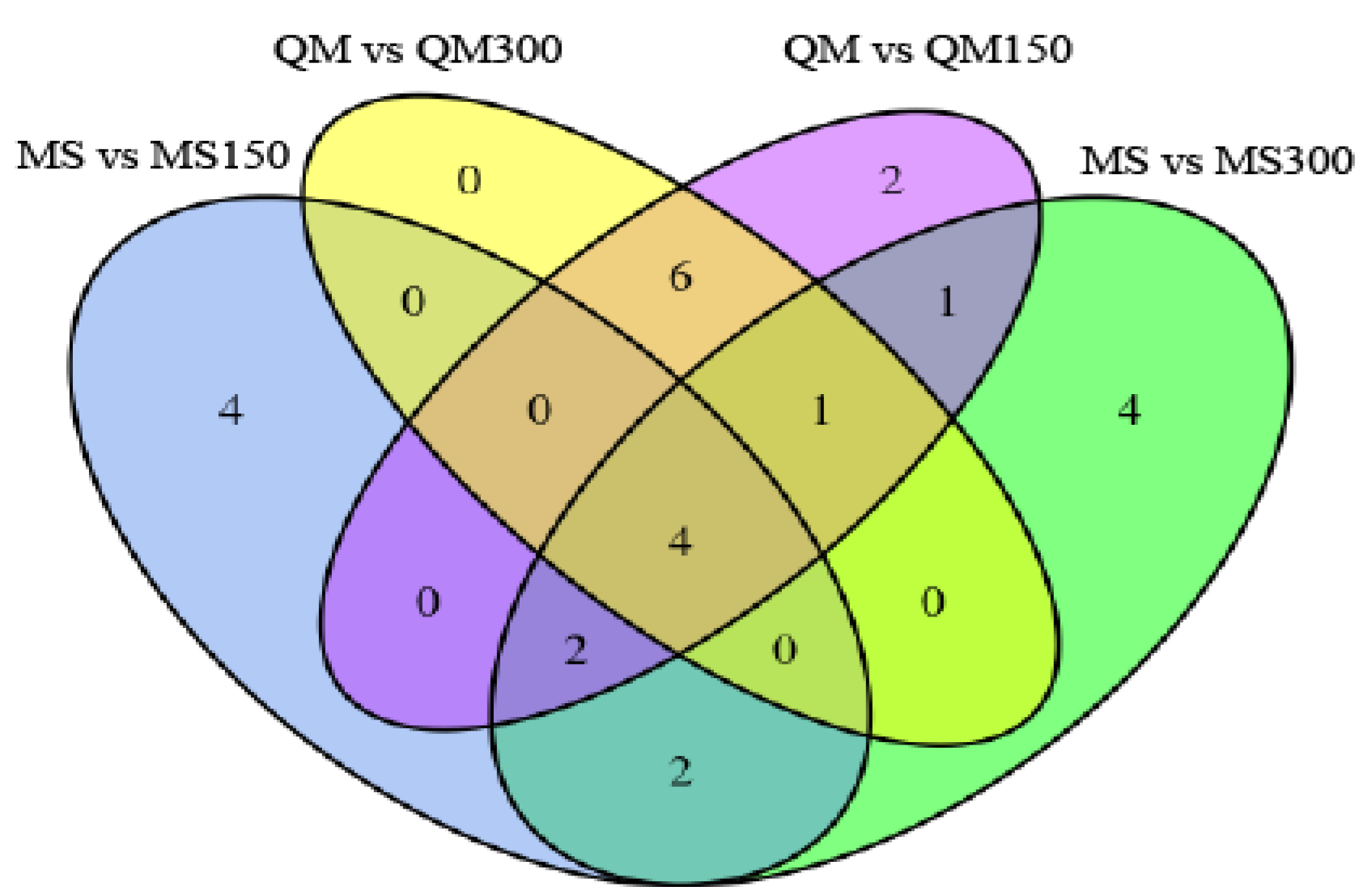
16]. In our study, 56 miRNAs were identified in the two wheat varieties under different alkali stresses, and 4 miRNAs (miR9653b, miR5384-3p, miR9777, and miR531) showed a common trend.
MiR159 targets the MYB transcription factor [
33]. The transcription factor
R2R3 MYB is involved in plant growth and stress response [
34].
MsMYB2L is also rapidly induced by NaCl, indicating that MYB is involved in the resilience response [
35]. Similarly,
BnaMYB21,
BnaMYB141 and
BnaMYB148 are involved in the regulation of salt tolerance [
36]. Our data also showed that miR159 was not expressed in QM but was promoted in MS (
Table 2 and
Table 3), thus reducing the expression of target genes by encoding MYB-related transcription factors in MS, leading to different resistances between varieties.
MiR164 is a conserved miRNA unique to plants, and its target gene is primarily the NAC transcription factor [
37]. Induction by heavy metal ions, radiation, and other factors can significantly upregulate the expression of miR164a, miR164c, and miR164d in hybrid rice [
38]. Compared to other saline-alkali-sensitive varieties, miR164a, miR164c, and miR164d gene expression in saline–alkali-tolerant cotton was upregulated under saline–alkali stress [
39]. Our experiments also showed that QM had a higher level of miR164 (
Table 4 and
Table 5). Thus, miR164 and its target gene, NAC, may be involved in regulating the normal growth and development of plants and in stress resistance.
MiR171 is a highly conserved miRNA gene family and one of the oldest miRNA families in the plant kingdom [
40]. The target genes of different members of the same miRNA family are the same [
41]. Plants overexpressing miR171 show phenotypic changes, such as reduction of lateral branches and shortening of the main roots [
42]. In addition, overexpression of miR171 reduces salt tolerance in rice [
43]. These findings suggest that differences in miR171 expression may be linked to oxidases, which play important roles in promoting biosynthesis, metabolism, detoxification, and stress-resistant growth in plant cells [
44]. In addition, our data showed that under high saline–alkali stress, miR171a was downregulated in QM seedlings (
Table 5), and miR171b was upregulated in MS seedlings (
Table 2); thus, QM wheat seedlings had relatively longer roots (
Table 2). Therefore, the target genes of miR171a and miR171b in the miR171 family include the GRAS family transcription factors and different wheat varieties may be an important factor in the formation of different saline–alkali tolerances.
MiR5384 is a drought-responsive miRNA in wheat [
45]. Under drought stress, miR5384-3p affects transport and cellular redox homeostasis [
46]. We predicted that the target of miR5384-3p is
CYP450 (
Table 6).
CYP450 inhibitors are multifunctional [
47]. The GRAS protein family is a plant-specific transcription factor that regulates the normal development of plant root organs, meristem formation, plant signal pathway transduction, stress resistance responses, and other plant growth and development processes [
48]. Downregulation of
OsCYP707A7 induces an increase in the ABA content and antioxidant enzyme activity in rice [
49]. The upregulated expression of
CYP85A1 in spinach improves the drought resistance of transgenic tobacco [
50]. Therefore, we speculated that miR5384-3p may respond to antioxidant stress in wheat by regulating the target gene
CYP450 and improving the salt–alkali resistance of wheat.
MiR408 is a highly conserved miRNA in plants that targets genes encoding copper-containing proteins [
51]. An increase in miR408 gene expression further enhances the tolerance in plant cells to salt, low temperature, and oxidative stress [
52]. Overexpression of miR408 enhances drought tolerance in chickpea [
53]. Compared to the wild type, overexpression of miR408-3p (OX-amiR408) in transgenic cowpeas increased chlorophyll content, decreased cell H
2O
2 levels, and showed stronger drought resistance and salt tolerance [
45]. Our results also showed that miR408 was not expressed in MS but was downregulated in QM (
Table 4 and
Table 5). In summary, miR408 may play a role in saline–alkali tolerance by regulating chlorophyll and ROS metabolism, which is an important reason for the differences in saline–alkali tolerance in wheat varieties.
MiR1135 can be considered a putative regulator of gene expression at the protein level. miR1135 is upregulated under saline–alkali stress, and its target is the MAPK signalling pathway. MAPK is a multifunctional signalling molecule that interacts with ROS and hormones to form an adaptive response [
54]. ROS are important and common messengers that are generated under various environmental pressures. It also activates several MAPKs [
55]. ABA-activated MAPK components are also activated by ROS, indicating that ABA and active oxygen may polymerise at the MAPK level to regulate stomatal closure [
56]. miR1135 was highly expressed in QM under high saline–alkali stress and was not expressed in the other three treatment groups (
Table 5). Thus, the expression of target genes encoding MAPK pathway-related transcription factors in QM was reduced, leading to differences in saline–alkali resistance among the varieties. In conclusion, miR1135 may make QM more tolerant to saline–alkali stress than MS by activating the expression of MAPK-related transcription factors in ROS in wheat.
In summary, the differential expression of many miRNAs (miR5384-3p, miR408, and miR1135) is related to ROS production, which may be involved in the differences in salt and alkaline tolerance between the two cultivars. ROS between plant cells is maintained at a low level under natural conditions [
57], but various environmental pressures disrupt the balance between ROS production and elimination, leading to the continuous accumulation of ROS [
58]. High ROS levels are cytotoxic [
59]. ROS can seriously damage normal metabolism through oxidative damage to carbohydrates, lipids, proteins, and nucleic acids, causing damage to the membrane system and increasing MDA content [
60]. Therefore, MDA is considered as a reliable indicator of oxidative stress [
61]. In this study, the rate of formation of O
2− and H
2O
2 content gradually increased with the increase of treatment concentration, which was consistent with the increase in MDA content under saline–alkali stress (
Supplementary Figure S2). In a highly stressful environment, green plants can increase their antioxidant levels and antioxidant enzyme activities to eliminate ROS [
62]. SOD is the first line of defence against oxygen free radicals. SOD catalyses O
2− into O
2 and H
2O
2. Subsequently, H
2O
2 is decomposed into H
2O and O
2 by APX to protect organelles and cell membranes from damage by active oxygen. Under saline–alkali stress, the activities of SOD, POD, and CAT in the two wheat varieties with different saline–alkali tolerances showed a significant decreasing trend (
Supplementary Table S2) owing to the increase in saline–alkali content. Under saline–alkali stress, the reaction of the enzymatic defence systems of different salt-tolerant varieties in the same plant is also quite different, which further indicates that the higher salt tolerance in plants is due to the improvement in protective enzyme activity.
In recent years, research has mainly focused on changes in AsA–GSH cycle activity under environmental stress as a good indicator of plant stress levels [
63]. Various components of the AsA–GSH cycle have been detected in the cytoplasm, chloroplasts, mitochondria, and peroxidase [
64]. The AsA–GSH cycle is an important and effective pathway for removing H
2O
2 from plants using AsA and GSH [
65]. Transgenic plants over-expressing key AsA–GSH cycle enzyme genes can improve stress resistance by increasing AsA and GSH levels [
66]. In the AsA–GSH cycle, APX uses AsA as the reductant to catalyse the reduction of H
2O
2 to H
2O. The root system of the salt-tolerant variety QM showed higher APX activity (
Supplementary Figure S1), indicating that under saline–alkali stress, QM can effectively control ROS levels by increasing APX activity, thereby limiting the oxidative tissue damage caused by saline–alkali stress. APX uses AsA to process H
2O
2 and generate DHA and MDHA. As an electron donor, glutathione (GSH) participates in the transformation of DHA into AsA. GR catalyses the oxidation of GSSG to regenerate GSH. The results showed that under saline–alkali stress, the root GR activity of the salt-tolerant variety QM significantly increased, and the GR activity was higher (
Supplementary Figure S1). Therefore, increased GR activity plays a key role in the antioxidant stress response in wheat. The increase in GR activity explains the increase in AsA/DHA and GSH/GSSG ratios. These results indicate that the increase in APX and GR enzyme activities may be one of the reasons why the roots maintained high levels of AsA and GSH under saline–alkali stress.
5. Conclusions
This study systematically analysed miRNA expression profiles and ROS metabolism in two wheat varieties under saline–alkaline stress. Saline–alkali stress (150/300 mmol·L−1) induces differential expression of miRNAs in the roots of tolerant (QM) and sensitive (MS) wheat varieties. Four core miRNAs (miR9653b, miR5384-3p, miR9777, and miR531) show consistent expression trends across both the varieties and stress concentrations. Among these, miR5384-3p targets the cytochrome P450 pathway, while miR408 (upregulated in QM only) and miR1135 (highly expressed in QM under high stress) regulate the plant hormone signal transduction and the MAPK pathway, respectively. These miRNA-mediated pathways collectively modulate the antioxidant system: QM maintains higher activities of APX and GR (key enzymes in the AsA-GSH cycle), elevated levels of non-enzymatic antioxidants (AsA, GSH), and balanced AsA/DHA and GSH/GSSG ratios, which reduce ROS (O2−, H2O2) accumulation and MDA-induced oxidative damage. In contrast, MS exhibits weak activation of these miRNA pathways, leading to reduced antioxidant capacity, excessive ROS accumulation, and severe growth inhibition (e.g., reduced root length, root activity). This coordinated interplay between miRNA regulation, redox homeostasis, and stress tolerance is the core finding of our study.
Future research should focus on: (1) Functional verification of core miRNAs (miR5384-3p, miR408, miR1135) using genetic engineering techniques (e.g., overexpression or CRISPR/Cas9-mediated knockout) to confirm their roles in saline–alkaline stress response; (2) exploration of the interaction between miRNA target genes (e.g., CYP450, MAPK) and ROS scavenging systems at the protein level; (3) development of molecular markers based on core miRNAs for breeding saline-alkali-tolerant wheat varieties.