Characterization of a Thermophilic and Acidophilic GH78 α-L-Rhamnosidase from Thermotoga sp. 2812B Capable of Efficiently Hydrolyzing a Variety of Natural Flavonoid Diglycosides
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Reagents
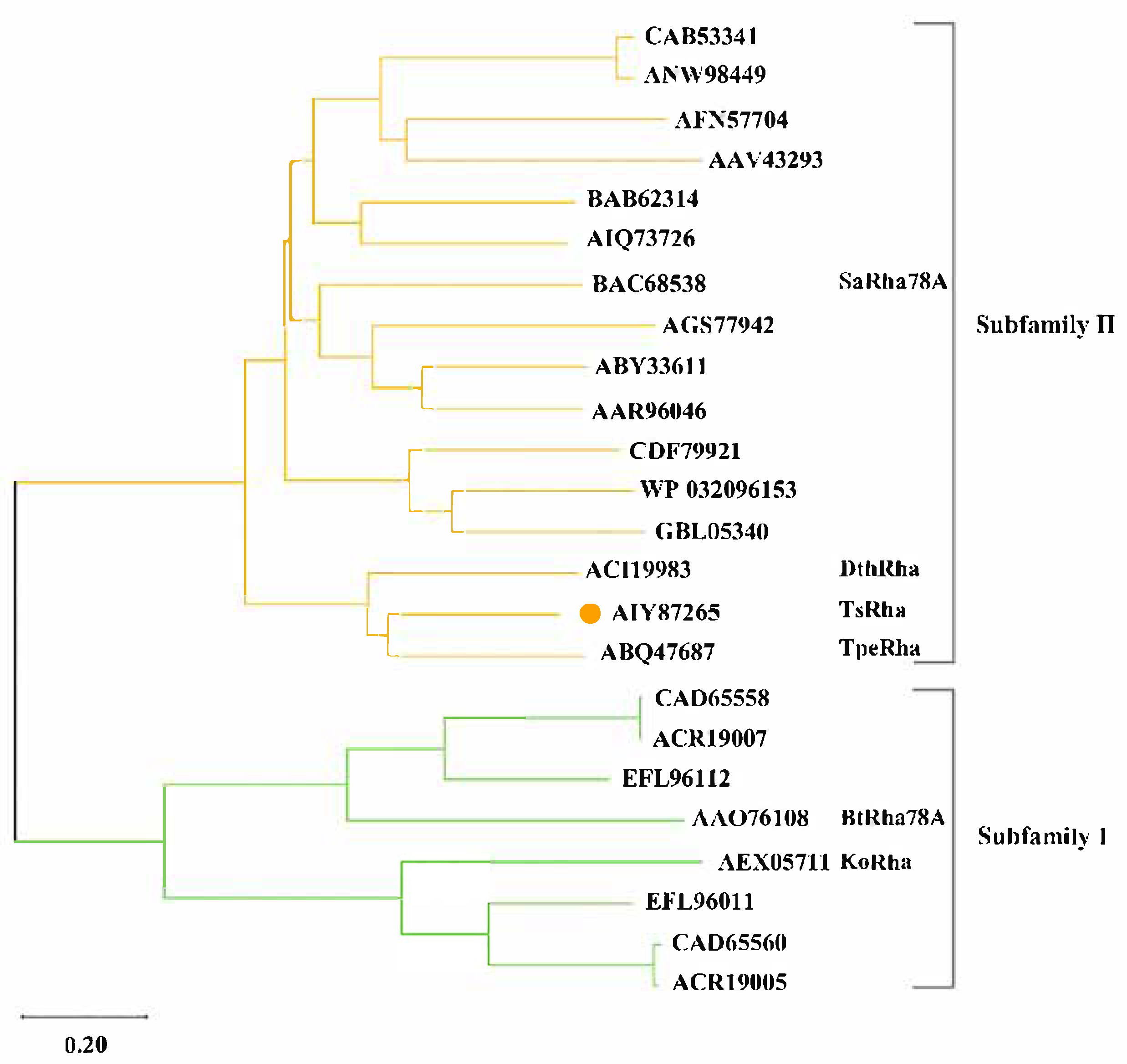
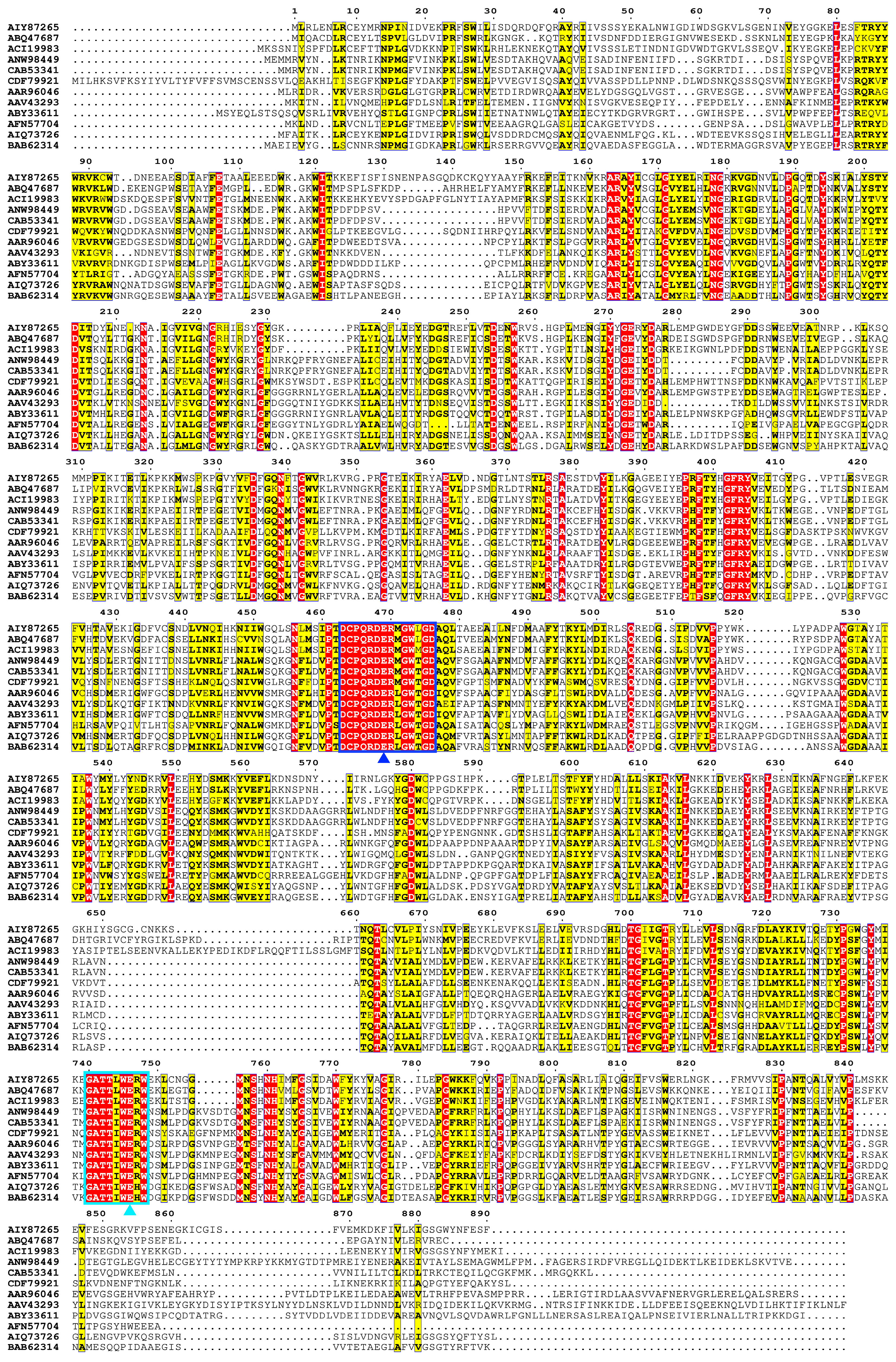
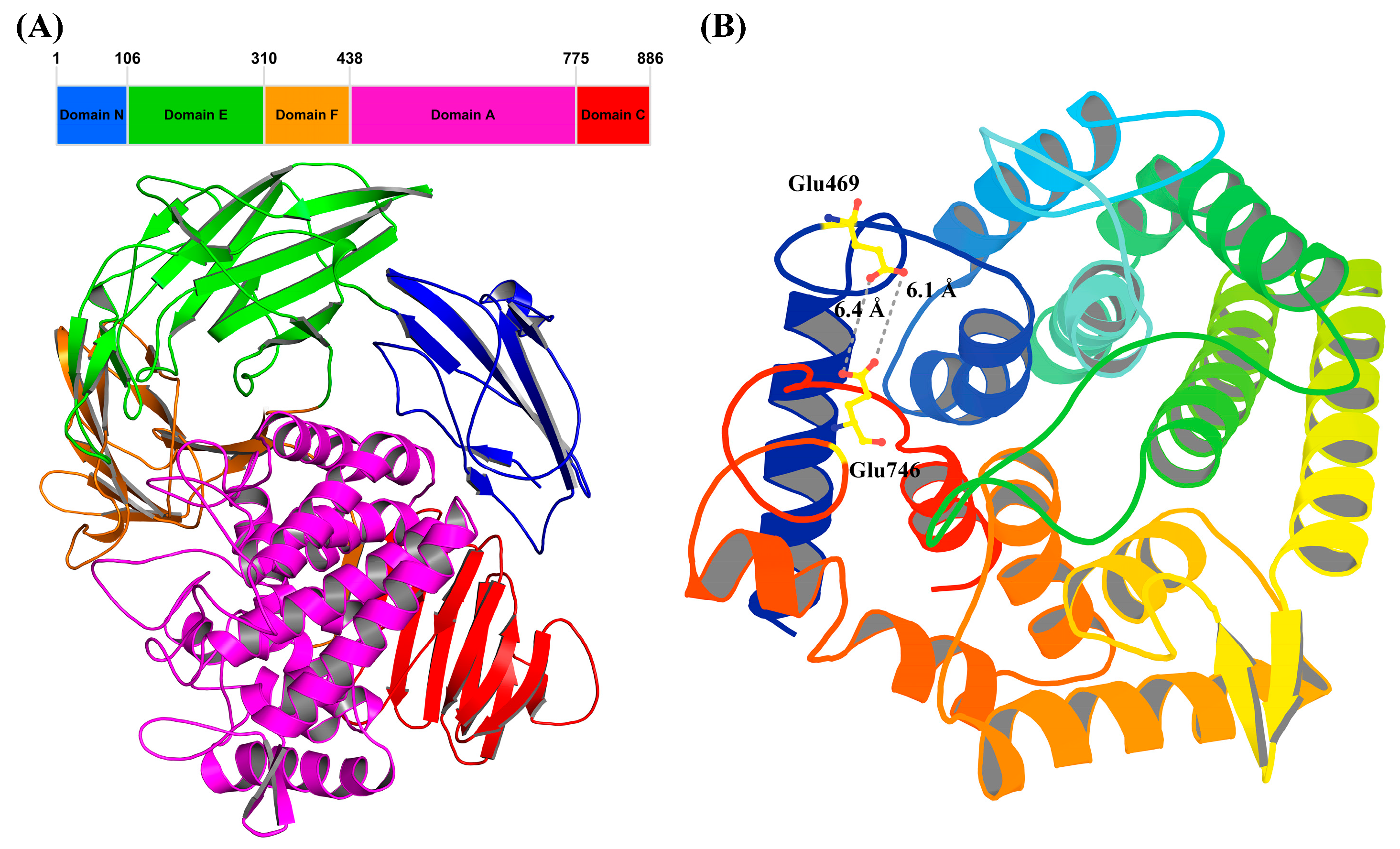
2.2. Bioinformatic and Structural Analysis
2.3. Gene Cloning, Protein Over-Expression and Purification
2.4. Enzymatic Characterization of TsRha
2.4.1. Enzyme Activity Assay
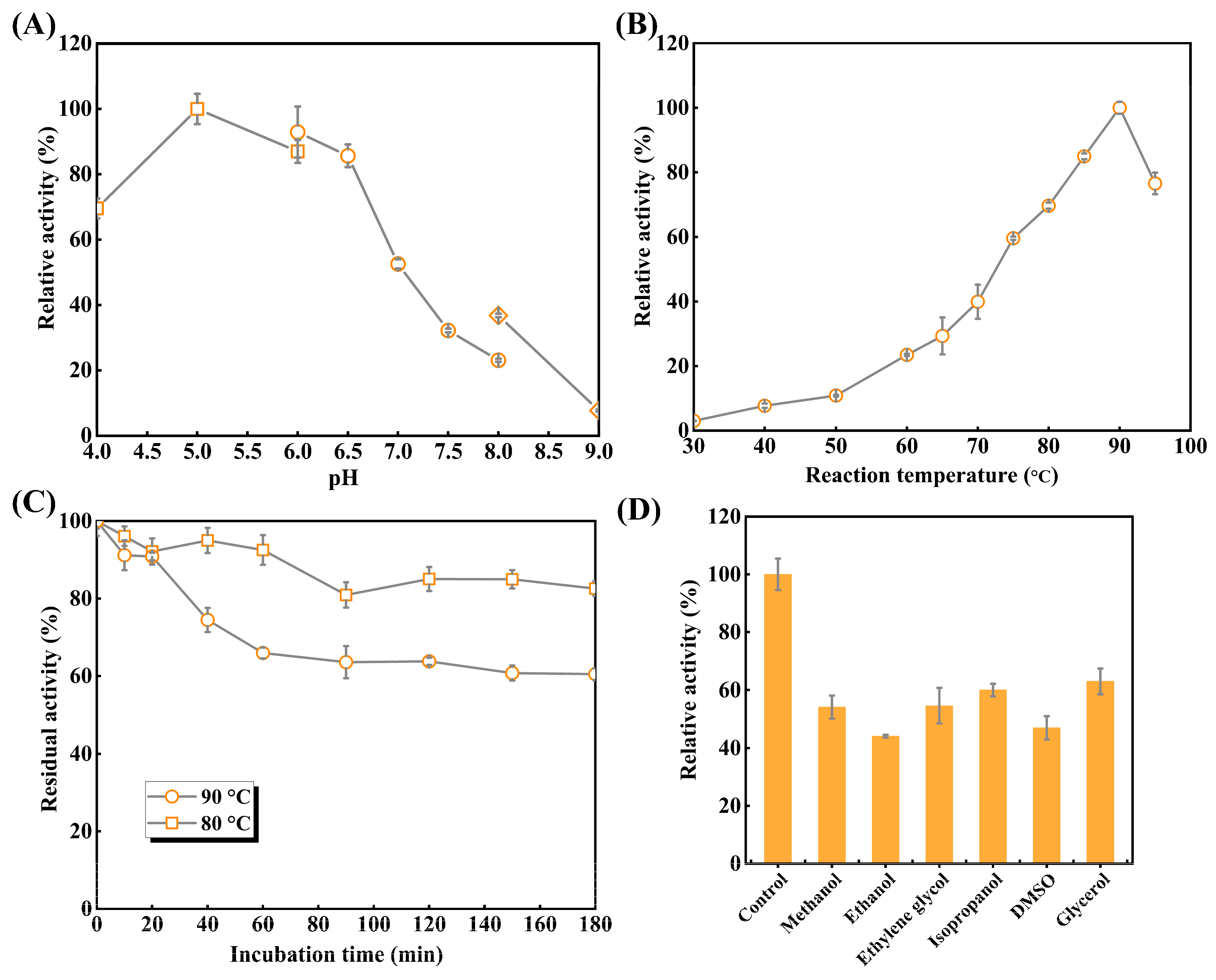
2.4.2. Optimum pH and Temperature
2.4.3. Thermostability and Organic Solvent Tolerance
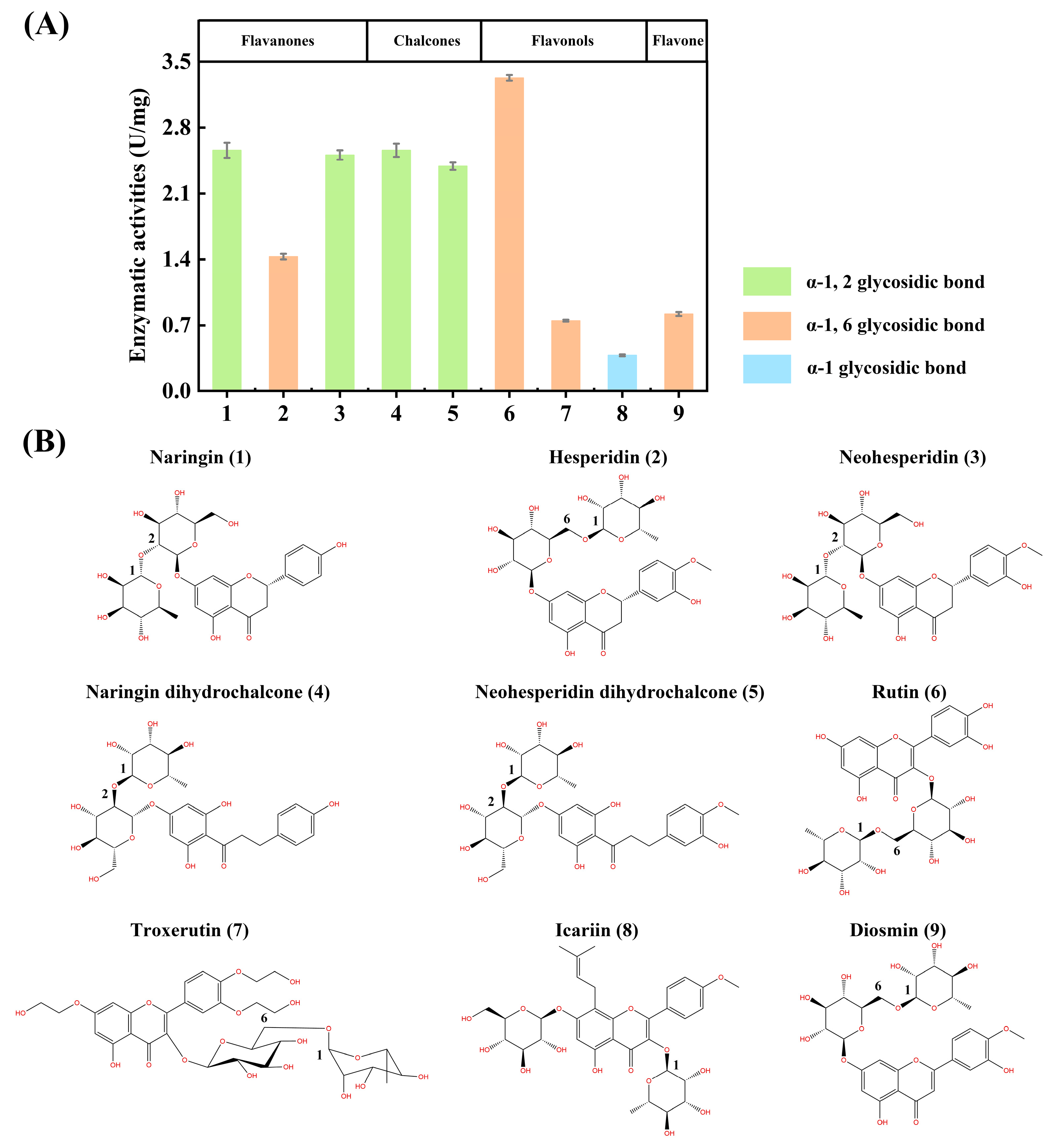
2.4.4. Substrate Selectivity
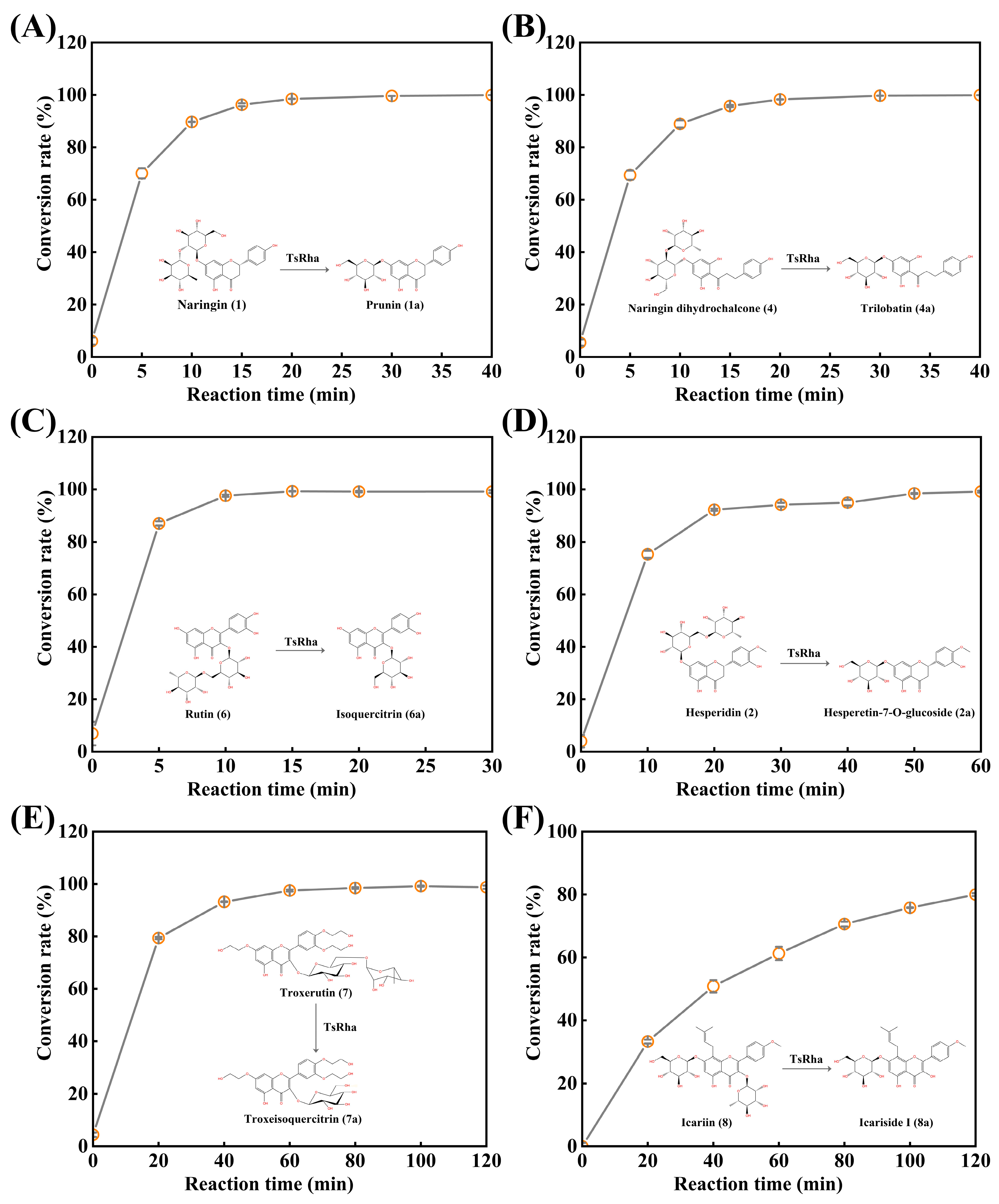
2.5. Biotransformation of Natural Flavonoid Glycosides by TsRha
2.6. High-Performance Liquid Chromatography (HPLC)
3. Results
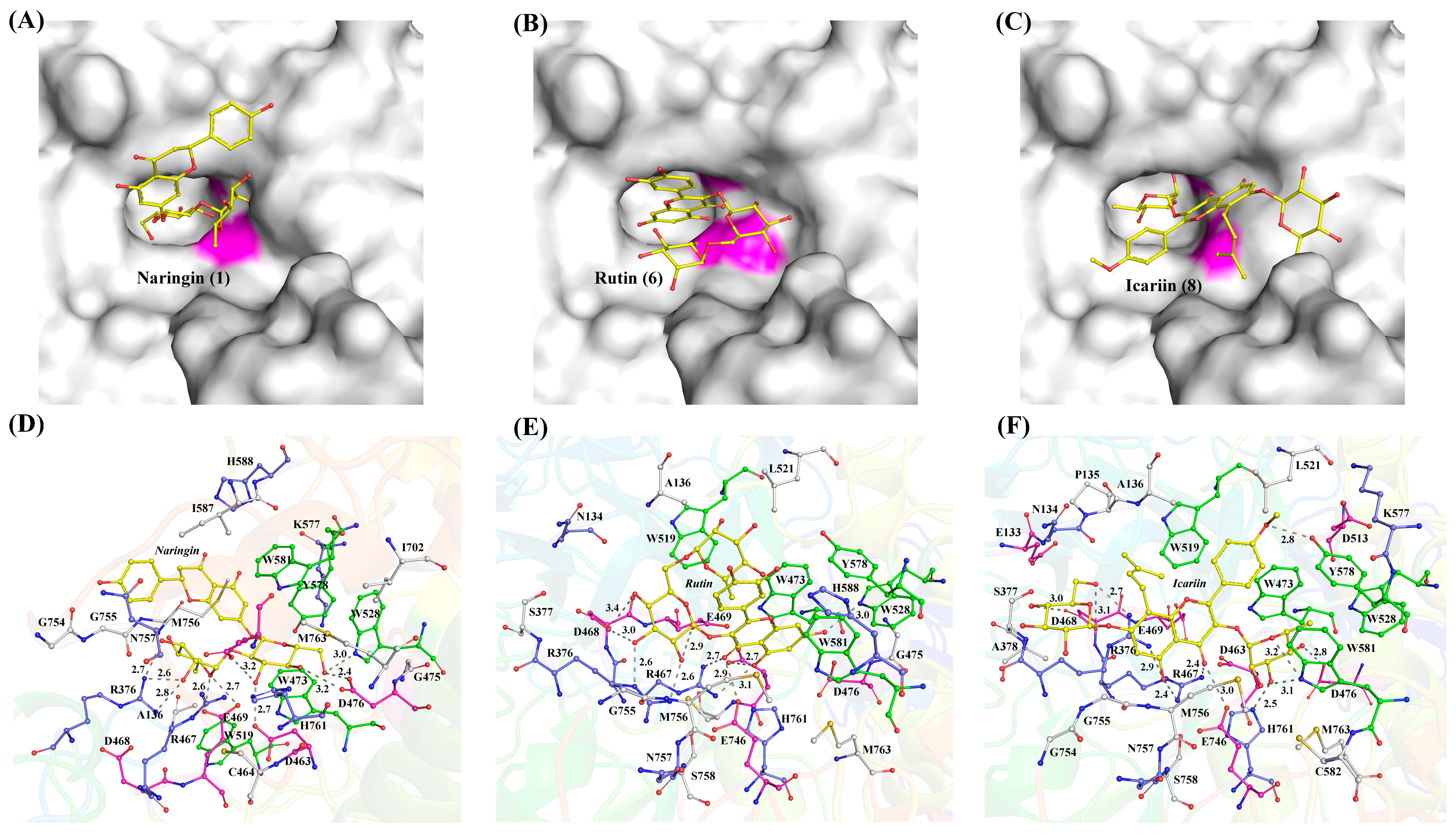
3.1. Bioinformatics and Structural Analysis for TsRha
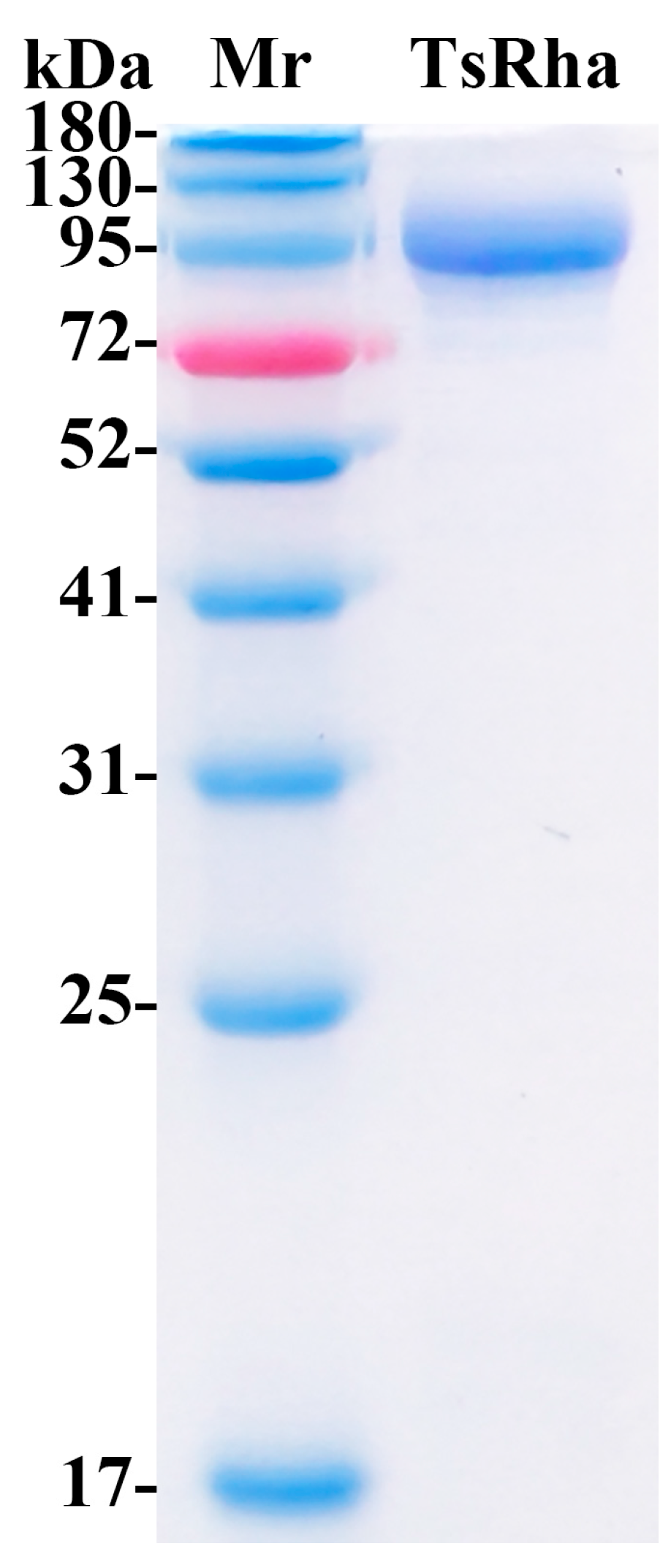
3.2. Heterologous Expression and Purification of TsRha
3.3. Enzymatic Properties of Recombinant TsRha
3.3.1. Optimal pH and Temperature
3.3.2. Thermal Stability and Organic Solvent Tolerance
3.3.3. Substrate Selectivity
3.4. Biotransformation of Natural Flavonoid Glycosides by TsRha
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| CA | Citric acid-sodium citrate |
| PB | NaH2PO4-Na2HPO4 |
| DMSO | Dimethyl sulfoxide |
| pNP | p-Nitrophenol |
| GH | Glycoside hydrolase |
| Rha78 | GH78 α-L-rhamnosidase |
| IPTG | Isopropyl-β-D-thiogalactoside |
| HPLC | High-performance liquid chromatography |
References
- Pan, L.; Zhang, Y.; Zhang, F.; Wang, Z.; Zheng, J. α-L-Rhamnosidase: Production, properties, and applications. World J. Microbiol. Biotechnol. 2023, 39, 191. [Google Scholar] [CrossRef]
- Pieczywek, P.M.; Cybulska, J.; Zdunek, A. An Atomic Force Microscopy Study on the Effect of β-Galactosidase, α-L-Rhamnosidase and α-L-Arabinofuranosidase on the Structure of Pectin Extracted from Apple Fruit Using Sodium Carbonate. Int. J. Mol. Sci. 2020, 21, 4064. [Google Scholar] [CrossRef]
- Cheng, L.; Zhang, H.; Cui, H.; Cheng, J.; Wang, W.; Wei, B.; Liu, F.; Liang, H.; Shen, X.; Yuan, Q. A novel α-L-Rhamnosidase renders efficient and clean production of icaritin. J. Clean. Prod. 2022, 341, 130903. [Google Scholar] [CrossRef]
- Cheng, L.; Zhang, H.; Cui, H.; Davari, M.D.; Wei, B.; Wang, W.; Yuan, Q. Efficient enzyme-catalyzed production of diosgenin: Inspired by the biotransformation mechanisms of steroid saponins in Talaromyces stollii CLY-6. Green Chem. 2021, 23, 5896–5910. [Google Scholar] [CrossRef]
- Cui, C.H.; Shin, D.; Hurh, B.S.; Im, W.T. A Novel Ginsenoside-Transforming α-L-Rhamnosidase from Bifidobacterium: Screening, Characterization and Application. Biomolecules 2024, 14, 1611. [Google Scholar] [CrossRef] [PubMed]
- Li, L.J.; Wu, Z.Y.; Yu, Y.; Zhang, L.J.; Zhu, Y.B.; Ni, H.; Chen, F. Development and characterization of an α-L-rhamnosidase mutant with improved thermostability and a higher efficiency for debittering orange juice. Food Chem. 2018, 245, 1070–1078. [Google Scholar] [CrossRef]
- Slámová, K.; Kapešová, J.; Valentová, K. “Sweet Flavonoids”: Glycosidase-Catalyzed Modifications. Int. J. Mol. Sci. 2018, 19, 2126. [Google Scholar] [CrossRef] [PubMed]
- Park, H.Y.; Choi, H.D.; Eom, H.; Choi, I. Enzymatic modification enhances the protective activity of citrus flavonoids against alcohol-induced liver disease. Food Chem. 2013, 139, 231–240. [Google Scholar] [CrossRef] [PubMed]
- de Araújo, M.E.; Moreira Franco, Y.E.; Alberto, T.G.; Sobreiro, M.A.; Conrado, M.A.; Priolli, D.G.; Frankland Sawaya, A.C.; Ruiz, A.L.; de Carvalho, J.E.; de Oliveira Carvalho, P. Enzymatic de-glycosylation of rutin improves its antioxidant and antiproliferative activities. Food Chem. 2013, 141, 266–273. [Google Scholar] [CrossRef]
- Lee, Y.S.; Huh, J.Y.; Nam, S.H.; Moon, S.K.; Lee, S.B. Enzymatic bioconversion of citrus hesperidin by Aspergillus sojae naringinase: Enhanced solubility of hesperetin-7-O-glucoside with in vitro inhibition of human intestinal maltase, HMG-CoA reductase, and growth of Helicobacter pylori. Food Chem. 2012, 135, 2253–2259. [Google Scholar] [CrossRef]
- Chang, H.Y.; Lee, Y.B.; Bae, H.A.; Huh, J.Y.; Nam, S.H.; Sohn, H.S.; Lee, H.J.; Lee, S.B. Purification and characterisation of Aspergillus sojae naringinase: The production of prunin exhibiting markedly enhanced solubility with in vitro inhibition of HMG-CoA reductase. Food Chem. 2011, 124, 234–241. [Google Scholar] [CrossRef]
- Makino, T.; Shimizu, R.; Kanemaru, M.; Suzuki, Y.; Moriwaki, M.; Mizukami, H. Enzymatically modified isoquercitrin, alpha-oligoglucosyl quercetin 3-O-glucoside, is absorbed more easily than other quercetin glycosides or aglycone after oral administration in rats. Biol. Pharm. Bull. 2009, 32, 2034–2040. [Google Scholar] [CrossRef]
- Luo, C.M.; Ke, L.F.; Huang, X.Y.; Zhuang, X.Y.; Guo, Z.W.; Xiao, Q.; Chen, J.; Chen, F.Q.; Yang, Q.M.; Ru, Y.; et al. Efficient biosynthesis of prunin in methanol cosolvent system by an organic solvent-tolerant α-L-rhamnosidase from Spirochaeta thermophila. Enzyme Microb. Technol. 2024, 175, 110410. [Google Scholar] [PubMed]
- Xie, J.; Zhao, J.; Zhang, N.; Xu, H.; Yang, J.; Ye, J.; Jiang, J. Efficient production of isoquercitin, icariin and icariside II by a novel thermostable α-L-rhamnosidase PodoRha from Paenibacillus odorifer with high α-1, 6-/α-1, 2- glycoside specificity. Enzyme Microb. Technol. 2022, 158, 110039. [Google Scholar] [CrossRef]
- Lou, H.; Liu, X.; Liu, S.; Chen, Q. Purification and Characterization of a Novel α-L-Rhamnosidase from Papiliotrema laurentii ZJU-L07 and Its Application in Production of Icariin from Epimedin C. J. Fungi. 2022, 8, 644. [Google Scholar] [CrossRef]
- Zhang, J.; Huang, J.; Yang, Y.; Zhao, D.; Wang, C.; Zhong, P.; Jia, J.; Dang, W.; Lu, Q.; Zhang, C.; et al. Icaritin production from Epimedium folium extract by a one-pot enzymatic cascade of a multifunctional glycosidase and rhamnosidase. Int. J. Biol. Macromol. 2024, 283, 137784. [Google Scholar] [CrossRef]
- Drula, E.; Garron, M.L.; Dogan, S.; Lombard, V.; Henrissat, B.; Terrapon, N. The carbohydrate-active enzyme database: Functions and literature. Nucleic Acids Res. 2022, 50, D571–D577. [Google Scholar] [PubMed]
- Guillotin, L.; Kim, H.; Traore, Y.; Moreau, P.; Lafite, P.; Coquoin, V.; Nuccio, S.; de Vaumas, R.; Daniellou, R. Biochemical Characterization of the α-L-Rhamnosidase DtRha from Dictyoglomus thermophilum: Application to the Selective Derhamnosylation of Natural Flavonoids. ACS Omega 2019, 4, 1916–1922. [Google Scholar] [CrossRef]
- Zverlov, V.V.; Hertel, C.; Bronnenmeier, K.; Hroch, A.; Kellermann, J.; Schwarz, W.H. The thermostable alpha-L-rhamnosidase RamA of Clostridium stercorarium: Biochemical characterization and primary structure of a bacterial alpha-L-rhamnoside hydrolase, a new type of inverting glycoside hydrolase. Mol. Microbiol. 2000, 35, 173–179. [Google Scholar] [CrossRef] [PubMed]
- Ge, L.; Liu, Y.; Zhou, F.; Zhan, L.; Zhao, L. Heterologous Expression and Characterization of a Thermostable α-L-Rhamnosidase from Thermoclostridium stercorarium subsp. thermolacticum DSM 2910 and Its Application in the Biotransformation of Rutin. J. Microbiol. Biotechnol. 2023, 33, 1521–1530. [Google Scholar] [CrossRef]
- Birgisson, H.; Hreggvidsson, G.O.; Fridjónsson, O.H.; Mort, A.; Kristjánsson, J.K.; Mattiasson, B. Two new thermostable α-L-rhamnosidases from a novel thermophilic bacterium. Enzyme Microb. Technol. 2004, 34, 561–571. [Google Scholar] [CrossRef]
- Xie, J.; Zhang, S.; Tong, X.; Wu, T.; Pei, J.; Zhao, L. Biochemical characterization of a novel hyperthermophilic α-L-rhamnosidase from Thermotoga petrophila and its application in production of icaritin from epimedin C with a thermostable β-glucosidase. Process Biochem. 2020, 93, 115–124. [Google Scholar] [CrossRef]
- Xu, J.; Shi, X.; Zhang, X.; Wang, Z.; Xiao, W.; Zhao, L. Immobilization of GH78 α-L-Rhamnosidase from Thermotoga petrophilea with High-Temperature-Resistant Magnetic Particles Fe3O4-SiO2-NH2-Cellu-ZIF8 and Its Application in the Production of Prunin Form Naringin. J. Microbiol. Biotechnol. 2021, 31, 419–428. [Google Scholar] [CrossRef] [PubMed]
- Beekwilder, J.; Marcozzi, D.; Vecchi, S.; de Vos, R.; Janssen, P.; Francke, C.; van Hylckama Vlieg, J.; Hall, R.D. Characterization of Rhamnosidases from Lactobacillus plantarum and Lactobacillus acidophilus. Appl. Environ. Microbiol. 2009, 75, 3447–3454. [Google Scholar] [CrossRef] [PubMed]
- Michlmayr, H.; Brandes, W.; Eder, R.; Schümann, C.; del Hierro, A.M.; Kulbe, K.D. Characterization of two distinct glycosyl hydrolase family 78 alpha-L-rhamnosidases from Pediococcus acidilactici. Appl. Environ. Microbiol. 2011, 77, 6524–6530. [Google Scholar] [CrossRef]
- Baudrexl, M.; Schwarz, W.H.; Zverlov, V.V.; Liebl, W. Biochemical characterisation of four rhamnosidases from thermophilic bacteria of the genera Thermotoga, Caldicellulosiruptor and Thermoclostridium. Sci. Rep. 2019, 9, 15924. [Google Scholar] [CrossRef]
- Cui, Z.; Maruyama, Y.; Mikami, B.; Hashimoto, W.; Murata, K. Crystal structure of glycoside hydrolase family 78 α-L-Rhamnosidase from Bacillus sp. GL1. J. Mol. Biol. 2007, 374, 384–398. [Google Scholar] [CrossRef]
- Li, B.; Ji, Y.; Li, Y.; Ding, G. Characterization of a glycoside hydrolase family 78 α-L-rhamnosidase from Bacteroides thetaiotaomicron VPI-5482 and identification of functional residues. 3 Biotech 2018, 8, 120. [Google Scholar] [CrossRef]
- Velankar, S.; Burley, S.K.; Kurisu, G.; Hoch, J.C.; Markley, J.L. The Protein Data Bank Archive. Methods Mol. Biol. 2021, 2305, 3–21. [Google Scholar]
- O’Neill, E.C.; Stevenson, C.E.; Paterson, M.J.; Rejzek, M.; Chauvin, A.L.; Lawson, D.M.; Field, R.A. Crystal structure of a novel two domain GH78 family α-rhamnosidase from Klebsiella oxytoca with rhamnose bound. Proteins 2015, 83, 1742–1749. [Google Scholar] [CrossRef]
- Fujimoto, Z.; Jackson, A.; Michikawa, M.; Maehara, T.; Momma, M.; Henrissat, B.; Gilbert, H.J.; Kaneko, S. The structure of a Streptomyces avermitilis α-L-rhamnosidase reveals a novel carbohydrate-binding module CBM67 within the six-domain arrangement. J. Biol. Chem. 2013, 288, 12376–12385. [Google Scholar] [CrossRef]
- Makabe, K.; Ishida, N.; Kanezaki, N.; Shiono, Y.; Koseki, T. Aspergillus oryzae α-l-rhamnosidase: Crystal structure and insight into the substrate specificity. Proteins 2024, 92, 236–245. [Google Scholar] [CrossRef]
- Pachl, P.; Škerlová, J.; Šimčíková, D.; Kotik, M.; Křenková, A.; Mader, P.; Brynda, J.; Kapešová, J.; Křen, V.; Otwinowski, Z.; et al. Crystal structure of native α-L-rhamnosidase from Aspergillus terreus. Acta Crystallogr. D Struct. Biol. 2018, 74, 1078–1084. [Google Scholar] [CrossRef]
- Ban, Y.; Yang, H.; Jiang, J.; Wang, C.; Lv, B.; Feng, Y. A α-L-rhamnosidase from Echinacea purpurea endophyte Simplicillium sinense EFF1 and its application in production of Calceorioside B. Int. J. Biol. Macromol. 2024, 270, 132090. [Google Scholar] [CrossRef] [PubMed]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef]
- Robert, X.; Gouet, P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014, 42, W320–W324. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- WeMol, Wecomput Technology Co., Ltd. Room 2104, No. 9, North 4th Ring West Road, Haidian District, Beijing. Available online: https://wemol.wecomput.com (accessed on 18 July 2025).
- The PyMOL Molecular Graphics System, Version 3.0 Schrödinger, LLC. Available online: https://pymol.org/ (accessed on 24 October 2024).
- Ding, J.; Gao, T.; Liu, S.; Li, Z.; Hu, B.; Zheng, J.; Yao, X.; Liu, H.; Hu, H. Rhamnosidase from Parabacteroides distasonis exhibit the catabolism of epimedin C in the human gut microbiota. Int. J. Biol. Macromol. 2025, 309, 142481. [Google Scholar] [CrossRef]
- Vieille, C.; Zeikus, G.J. Hyperthermophilic enzymes: Sources, uses, and molecular mechanisms for thermostability. Microbiol. Mol. Biol. Rev. 2001, 65, 1–43. [Google Scholar] [CrossRef] [PubMed]
- Vila-Real, H.; Alfaia, A.J.; Calado, A.R.; Ribeiro, M.H.L. Improvement of activity and stability of soluble and sol–gel immobilized naringinase in co-solvent systems. J. Mol. Catal. B-Enzym. 2010, 65, 91–101. [Google Scholar] [CrossRef]
- Shen, Y.; Wang, H.; Lu, Y.; Xu, L.; Yin, H.; Tam, J.P.; Yang, H.; Jia, X. Construction of a novel catalysis system for clean and efficient preparation of baohuoside I from icariin based on biphase enzymatic hydrolysis. J. Clean. Prod. 2018, 170, 727–734. [Google Scholar] [CrossRef]
- Li, B.C.; Wu, B.; Hou, X.; Ding, G.B. Substrate Selectivities of GH78 α-L-Rhamnosidases from Human Gut Bacteria on Dietary Flavonoid Glycosides. Molecules 2025, 30, 980. [Google Scholar] [CrossRef] [PubMed]
| pNP Glycosides | Enzymatic Activities (U/mg) |
|---|---|
| pNPαRha | 31.7 ± 1.0 |
| pNPαGlc | NA |
| pNPαGal | NA |
| pNPαMan | NA |
| pNPβGlc | 3.5 × 10−2 ± 0.1 × 10−2 |
| pNPβGal | 2.1 × 10−2 ± 0.3 × 10−2 |
| pNPβXyl | NA |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license.
Share and Cite
Li, B.-C.; Dong, W.; Wu, B.; Liu, Y.; Han, N.; Ding, G.-B. Characterization of a Thermophilic and Acidophilic GH78 α-L-Rhamnosidase from Thermotoga sp. 2812B Capable of Efficiently Hydrolyzing a Variety of Natural Flavonoid Diglycosides. Biomolecules 2026, 16, 68. https://doi.org/10.3390/biom16010068
Li B-C, Dong W, Wu B, Liu Y, Han N, Ding G-B. Characterization of a Thermophilic and Acidophilic GH78 α-L-Rhamnosidase from Thermotoga sp. 2812B Capable of Efficiently Hydrolyzing a Variety of Natural Flavonoid Diglycosides. Biomolecules. 2026; 16(1):68. https://doi.org/10.3390/biom16010068
Chicago/Turabian StyleLi, Bin-Chun, Weijuan Dong, Bingbing Wu, Yanlong Liu, Na Han, and Guo-Bin Ding. 2026. "Characterization of a Thermophilic and Acidophilic GH78 α-L-Rhamnosidase from Thermotoga sp. 2812B Capable of Efficiently Hydrolyzing a Variety of Natural Flavonoid Diglycosides" Biomolecules 16, no. 1: 68. https://doi.org/10.3390/biom16010068
APA StyleLi, B.-C., Dong, W., Wu, B., Liu, Y., Han, N., & Ding, G.-B. (2026). Characterization of a Thermophilic and Acidophilic GH78 α-L-Rhamnosidase from Thermotoga sp. 2812B Capable of Efficiently Hydrolyzing a Variety of Natural Flavonoid Diglycosides. Biomolecules, 16(1), 68. https://doi.org/10.3390/biom16010068


_Kwok.png)




