The Years 2015–2025 as a Prospective Decade for the Identification of Specific Methylation Biomarkers of Prostate Cancer
Abstract
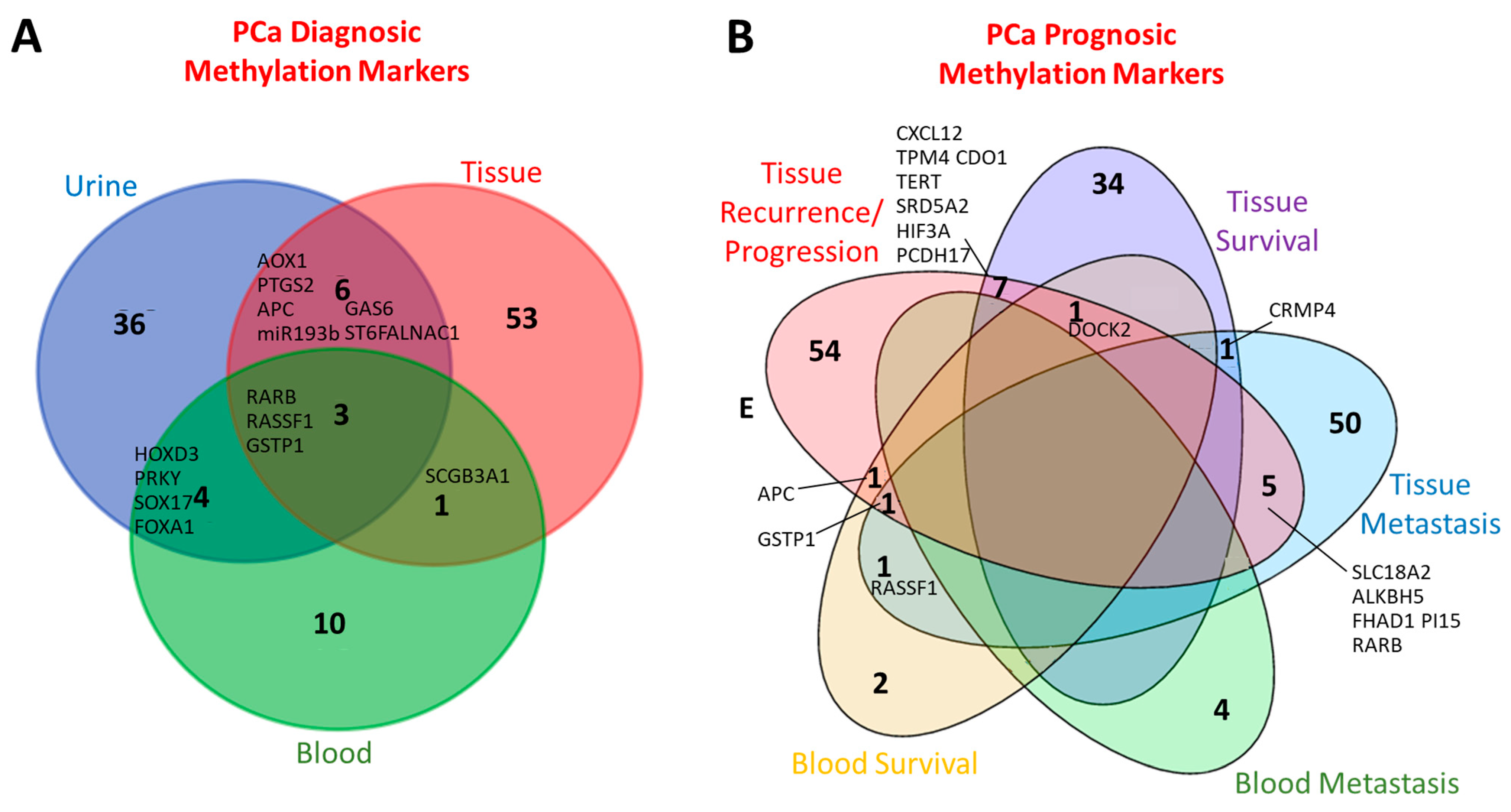
1. State of the Art
2. Diagnosis
2.1. Diagnosis: Leveraging Public Whole-Genome DNA Methylation Profiles for Prostate Cancer Biomarker Discovery
2.2. Diagnosis: Mining Public Whole-Genome Methylomes for Pan-Cancer Biomarkers Applicable to Prostate Cancer Diagnosis
2.3. Diagnosis: Whole-Genome Methylomes from Independent Cohorts for the Identification of New Diagnostic Biomarkers Useful for PCa
2.4. Diagnosis: Performance Tests of Biomarker Sets for PCa Diagnosis in Tissue Biopsies/Tumors
2.5. Diagnosis: Performance Tests of Biomarker Sets for PCa Diagnosis in Liquid Biopsies
2.5.1. Diagnosis: Performance Evaluation of Biomarker Panels for PCa Diagnosis in Blood
2.5.2. Diagnosis: Performance Tests of Biomarker Sets for PCa Diagnosis in Seminal Plasma
Diagnosis: Performance Tests of Biomarker Sets for PCa Diagnosis in Urine
Diagnosis: Whole-Genome Methylomes from Independent Urine Cohorts for the Identification of New Diagnostic Biomarkers Useful for PCa
3. Prognosis
3.1. Prognosis: Whole-Genome Methylomes from Public Databases for the Identification of New Prognostic PCa Biomarkers
3.2. Prognosis: Independent Methylomes Reveal Prognostic Signatures in PCa
3.3. Prognosis: Whole-Genome Methylomes from Public Databases for the Identification of New Prognostic PCa Biomarkers and Independent Validation
3.4. Prognosis: Whole-Genome Methylomes from Independent Cohorts for the Identification of New Prognostic PCa Biomarkers and Independent Validation
3.5. Prognosis: Validation and Prognostic Value of DMG Methylation Panels in PCa
3.6. Prognosis: Performance Tests of PCa Biomarkers in Liquid Biopsies
3.6.1. Prognosis: Value of Liquid Biopsy-Derived Methylation Biomarkers in PCa
3.6.2. Prognosis: Whole-Genome Methylomes from Plasma Samples for the Identification of New Prognostic PCa Biomarkers
3.6.3. Prognosis: Performance Tests of PCa Biomarker Sets in Urine
3.7. Prognosis: Biomarkers for the NE Subtype
3.8. Prognosis: Ethnicity and DNA Methylation in PCa
4. Prediction of the Treatment Response
4.1. Prediction of the Treatment Response: Identification of New Markers
4.2. Prediction of the Treatment Response: Performance of New Markers in Liquid Biopsies
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Hanahan, D. Hallmarks of Cancer: New Dimensions. Cancer Discov. 2022, 12, 31–46. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.-Y.; Koga, H.; Kawaguchi, Y.; Tang, W.; Wong, E.; Gao, Y.-S.; Pandey, U.B.; Kaushik, S.; Tresse, E.; Lu, J.; et al. HDAC6 controls autophagosome maturation essential for ubiquitin-selective quality-control autophagy. EMBO J. 2010, 29, 969–980. [Google Scholar] [CrossRef]
- Peng, W.; Feng, H.; Pang, L.; Zhang, J.; Hao, Y.; Wei, X.; Xia, Q.; Wei, Z.; Song, W.; Wang, S.; et al. Downregulation of CAMK2N1 due to DNA Hypermethylation Mediated by DNMT1 that Promotes the Progression of Prostate Cancer. J. Oncol. 2023, 2023, 1–15. [Google Scholar] [CrossRef]
- Dairo, O.; Oliveira, L.D.; Schaffer, E.; Vidotto, T.; Mendes, A.A.; Lu, J.; Huynh, S.V.; Hicks, J.L.; Sowalsky, A.G.; De Marzo, A.M.; et al. FASN Gene Methylation is Associated with Fatty Acid Synthase Expression and Clinical-genomic Features of Prostate Cancer. Cancer Res. Commun. 2024, 4, 152–163. [Google Scholar] [CrossRef]
- Fiano, V.; Zugna, D.; Grasso, C.; Trevisan, M.; Delsedime, L.; Molinaro, L.; Cassoni, P.; Papotti, M.; Merletti, F.; Akre, O.; et al. DNA methylation in repeat negative prostate biopsies as a marker of missed prostate cancer. Clin. Epigenet. 2019, 11, 152. [Google Scholar] [CrossRef]
- Zhang, W.; Zeng, S.; Gong, L.; Zhang, D.; Hu, X. Gene methylation status in focus of advanced prostate cancer diagnostics and improved individual outcomes. Transl. Androl. Urol. 2023, 12, 1813–1826. [Google Scholar] [CrossRef]
- Liu, W.; Xie, A.; Tu, C.; Liu, W. REX-1 Represses RASSF1a and Activates the MEK/ERK Pathway to Promote Tumorigenesis in Prostate Cancer. Mol. Cancer Res. 2021, 19, 1666–1675. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Meng, X.; Pan, C.; Qu, F.; Gan, W.; Xiang, Z.; Han, X.; Li, D. piR-31470 epigenetically suppresses the expression of glutathione S-transferase pi 1 in prostate cancer via DNA methylation. Cell Signal. 2020, 67, 109501. [Google Scholar] [CrossRef]
- Wang, L.; Wang, B.; Quan, Z. Identification of aberrantly methylated-differentially expressed genes and gene ontology in prostate cancer. Mol. Med. Rep. 2019, 21, 744–758. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Jiang, S.; Gu, Y.; Li, W.; Mo, Z.; Huang, Y.; Li, T.; Hu, Y. Promoter DNA methylation analysis reveals a combined diagnosis of CpG-based biomarker for prostate cancer. Oncotarget 2017, 8, 58199–58209. [Google Scholar] [PubMed]
- Luo, C.; He, S.; Zhang, H.; He, S.; Qi, H.; Wei, A. Clinical and Biological Significance of DNA Methylation-Driven Differentially Expressed Genes in Biochemical Recurrence After Radical Prostatectomy. Front. Genet. 2022, 13, 727307. [Google Scholar] [CrossRef]
- Xu, N.; Wu, Y.-P.; Ke, Z.-B.; Liang, Y.-C.; Cai, H.; Su, W.-T.; Tao, X.; Chen, S.-H.; Zheng, Q.-S.; Wei, Y.; et al. Identification of key DNA methylation-driven genes in prostate adenocarcinoma: An integrative analysis of TCGA methylation data. J. Transl. Med. 2019, 17, 311. [Google Scholar] [CrossRef] [PubMed]
- Nikas, J.B.; Nikas, E.G. Genome-Wide DNA Methylation Model for the Diagnosis of Prostate Cancer. ACS Omega 2019, 4, 14895–14901. [Google Scholar] [CrossRef]
- Tong, Y.; Song, Y.; Deng, S. Combined analysis and validation for DNA methylation and gene expression profiles associated with prostate cancer. Cancer Cell Int. 2019, 19, 50. [Google Scholar] [CrossRef]
- Reynolds, S.R.; Zhang, Z.; Salas, L.A.; Christensen, B.C. Tumor microenvironment deconvolution identifies cell-type-independent aberrant DNA methylation and gene expression in prostate cancer. Clin. Epigenet. 2024, 16, 5. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Liu, Y.; Pan, X.; Li, M.; Yang, S.; Li, S.C. DNA methylation markers for Pan-Cancer prediction by deep learning. Genes 2019, 10, 778. [Google Scholar] [CrossRef]
- Neefs, I.; De Meulenaere, N.; Vanpoucke, T.; Vandenhoeck, J.; Peeters, D.; Peeters, M.; Van Camp, G.; de Beeck, K.O. Simultaneous detection of eight cancer types using a multiplex droplet digital PCR assay. Mol. Oncol. 2024, 19, 188–203. [Google Scholar] [CrossRef]
- Geybels, M.S.; Zhao, S.; Wong, C.-J.; Bibikova, M.; Klotzle, B.; Wu, M.; Ostrander, E.A.; Fan, J.-B.; Feng, Z.; Stanford, J.L. Epigenomic profiling of DNA methylation in paired prostate cancer versus adjacent benign tissue. Prostate 2015, 75, 1941–1950. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.S.; Lee, S.C.; Lim, B.; Shin, S.H.; Kim, M.Y.; Kim, S.Y.; Lim, H.; Charton, C.; Shin, D.; Moon, H.W.; et al. DNA methylation biomarkers distinguishing early-stage prostate cancer from benign prostatic hyperplasia. Prostate Int. 2023, 11, 113–121. [Google Scholar] [CrossRef]
- Martignano, F.; Gurioli, G.; Salvi, S.; Calistri, D.; Costantini, M.; Gunelli, R.; De Giorgi, U.; Foca, F.; Casadio, V. GSTP1Methylation and Protein Expression in Prostate Cancer: Diagnostic Implications. Dis. Markers 2016, 2016, 1–6. [Google Scholar] [CrossRef]
- Vo, T.T.L.; Ta, B.T.; Ta, V.T.; Vuong, D.L.; Nguyen, Q.U. Promoter methylation profile of GSTP1 and RASSF1A in prostate cancerand benign hyperplasia in Vietnamese men. Turk. J. Med. Sci. 2016, 46, 228–235. [Google Scholar] [CrossRef]
- Patel, P.G.; Wessel, T.; Kawashima, A.; Okello, J.B.A.; Jamaspishvili, T.; Guérard, K.; Lee, L.; Lee, A.Y.; How, N.E.; Dion, D.; et al. A three-gene DNA methylation biomarker accurately classifies early stage prostate cancer. Prostate 2019, 79, 1705–1714. [Google Scholar] [CrossRef]
- Gurioli, G.; Salvi, S.; Martignano, F.; Foca, F.; Gunelli, R.; Costantini, M.; Cicchetti, G.; De Giorgi, U.; Sbarba, P.D.; Calistri, D.; et al. Methylation pattern analysis in prostate cancer tissue: Identification of biomarkers using an MS-MLPA approach. J. Transl. Med. 2016, 14, 249. [Google Scholar] [CrossRef] [PubMed]
- Yang, B.; Etheridge, T.; McCormick, J.; Schultz, A.; Khemees, T.A.; Damaschke, N.; Leverson, G.; Woo, K.; Sonn, G.A.; Klein, E.A.; et al. Validation of an epigenetic field of susceptibility to detect significant prostate cancer from non-tumor biopsies. Clin. Epigenet. 2019, 11, 168. [Google Scholar] [CrossRef]
- Kwabi-Addo, B.; Wang, S.; Chung, W.; Jelinek, J.; Patierno, S.R.; Wang, B.-D.; Andrawis, R.; Lee, N.H.; Apprey, V.; Issa, J.-P.; et al. Identification of Differentially Methylated Genes in Normal Prostate Tissues from African American and Caucasian Men. Clin. Cancer Res. 2010, 16, 3539–3547. [Google Scholar] [CrossRef]
- Moses-Fynn, E.; Tang, W.; Beyene, D.; Apprey, V.; Copeland, R.; Kanaan, Y.; Kwabi-Addo, B.; Chuu, C.-P. Correlating blood-based DNA methylation markers and prostate cancer risk in African-American men. PLoS ONE 2018, 13, e0203322. [Google Scholar] [CrossRef]
- Barry, K.H.; Moore, L.E.; Liao, L.M.; Huang, W.; Andreotti, G.; Poulin, M.; Berndt, S.I. Prospective study of DNA methylation at LINE-1 and Alu in peripheral blood and the risk of prostate cancer. Prostate 2015, 75, 1718–1725. [Google Scholar] [CrossRef] [PubMed]
- Aykanli, E.; Arisan, S.; Arisan, E.D.; Yavuzsan, A.H. Diagnostic Value of GSTP1, RASSF1, and RASSF2 Methylation in Serum of Prostate Cancer Patients. Urol. J. 2024, 21, 182–188. [Google Scholar] [CrossRef]
- Bryzgunova, O.; Bondar, A.; Ruzankin, P.; Laktionov, P.; Tarasenko, A.; Kurilshikov, A.; Epifanov, R.; Zaripov, M.; Kabilov, M.; Laktionov, P. Locus-Specific Methylation of GSTP1, RNF219, and KIAA1539 Genes with Single Molecule Resolution in Cell-Free DNA from Healthy Donors and Prostate Tumor Patients: Application in Diagnostics. Cancers 2021, 13, 6234. [Google Scholar] [CrossRef] [PubMed]
- Reis, I.M.; Ramachandran, K.; Speer, C.; Gordian, E.; Singal, R. Serum GADD45a methylation is a useful biomarker to distinguish benign vs malignant prostate disease. Br. J. Cancer 2015, 113, 460–468. [Google Scholar] [CrossRef]
- Friedemann, M.; Jandeck, C.; Tautz, L.; Gutewort, K.; von Rein, L.; Sukocheva, O.; Fuessel, S.; Menschikowski, M. Blood-Based DNA Methylation Analysis by Multiplexed OBBPA-ddPCR to Verify Indications for Prostate Biopsies in Suspected Prostate Cancer Patients. Cancers 2024, 16, 1324. [Google Scholar] [CrossRef]
- Dai, Z.; Chen, H.; Feng, K.; Li, T.; Liu, W.; Zhou, Y.; Yang, D.; Xue, B.; Zhu, J. Promoter hypermethylation of Y-chromosome gene PRKY as a potential biomarker for the early diagnosis of prostate cancer. Epigenomics 2024, 16, 835–850. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Liu, W.; Chen, Z.; Zang, Y.; Xu, L.; Dai, Z.; Zhou, Y.; Zhu, J. A noninvasive method for predicting clinically significant prostate cancer using magnetic resonance imaging combined with PRKY promoter methylation level: A machine learning study. BMC Med. Imaging 2024, 24, 60. [Google Scholar] [CrossRef]
- Škara, L.; Vodopić, T.; Pezelj, I.; Abramovic, I.; Vrhovec, B.; Vrtarić, A.; Sincic, N.; Tomas, D.; Bulimbašić, S.; Kuliš, T.; et al. Methylation pattern of caveolin-1 in prostate cancer as potential cfDNA biomarker. Biomol. Biomed. 2023, 23, 176–186. [Google Scholar] [CrossRef] [PubMed]
- Lleshi, E.; Milne-Clark, T.; Yu, H.L.; Martin, H.W.; Hanson, R.; Lach, R.; Rossi, S.H.; Riediger, A.L.; Görtz, M.; Sültmann, H.; et al. Prostate cancer detection through unbiased capture of methylated cell-free DNA. iScience 2024, 27, 110330. [Google Scholar] [CrossRef] [PubMed]
- Minciu, R.; Dumache, R.; Gheorghe, P.; Daminescu, L.; Rogobete, A.; Ionescu, D. Molecular Diagnostic of Prostate Cancer from Body Fluids Using Methylation-Specific PCR (MS-PCR) Method. Clin. Lab. 2016, 62, 1183–1186. [Google Scholar] [CrossRef]
- The Movember Urine Biomarker Consortium; Zhao, F.; Olkhov-Mitsel, E.; Kamdar, S.; Jeyapala, R.; Garcia, J.; Hurst, R.; Hanna, M.Y.; Mills, R.; Tuzova, A.V.; et al. A urine-based DNA methylation assay, ProCUrE, to identify clinically significant prostate cancer. Clin. Epigenet. 2018, 10, 147. [Google Scholar] [CrossRef]
- Moreira-Barbosa, C.; Barros-Silva, D.; Costa-Pinheiro, P.; Torres-Ferreira, J.; Constâncio, V.; Freitas, R.; Oliveira, J.; Antunes, L.; Henrique, R.; Jerónimo, C. Comparing diagnostic and prognostic performance of two-gene promoter methylation panels in tissue biopsies and urines of prostate cancer patients. Clin. Epigenet. 2018, 10, 132. [Google Scholar] [CrossRef]
- Torres-Ferreira, J.; Ramalho-Carvalho, J.; Gomez, A.; Menezes, F.D.; Freitas, R.; Oliveira, J.; Antunes, L.; Bento, M.J.; Esteller, M.; Henrique, R.; et al. MiR-193b promoter methylation accurately detects prostate cancer in urine sediments and miR-34b/c or miR-129-2 promoter methylation define subsets of clinically aggressive tumors. Mol. Cancer 2017, 16, 26. [Google Scholar] [CrossRef]
- Kaukoniemi, K.M.; Rauhala, H.E.; Scaravilli, M.; Latonen, L.; Annala, M.; Vessella, R.L.; Nykter, M.; Tammela, T.L.J.; Visakorpi, T. Epigenetically altered miR-193b targets cyclin D1 in prostate cancer. Cancer Med. 2015, 4, 1417–1425. [Google Scholar] [CrossRef]
- Brikun, I.; Nusskern, D.; Decatus, A.; Harvey, E.; Li, L.; Freije, D. A panel of DNA methylation markers for the detection of prostate cancer from FV and DRE urine DNA. Clin. Epigenet. 2018, 10, 91. [Google Scholar] [CrossRef]
- Shah, P.; Taylor, W.R.; Negaard, B.J.; Gochanour, B.R.; Mahoney, D.W.; Then, S.S.; Devens, M.E.; Foote, P.H.; Doering, K.A.; Burger, K.N.; et al. Methylated DNA Markers in Voided Urine for the Identification of Clinically Significant Prostate Cancer. Life 2024, 14, 1024. [Google Scholar] [CrossRef]
- Khemees, T.A.; Yang, B.; Schultz, A.; Allen, G.O.; Gawdzik, J.; Nihal, A.; Richards, K.A.; Abel, E.J.; Jarrard, D.F. Epigenetic field alterations in non-tumor prostate tissues detect prostate cancer in urine. Am. J. Clin. Exp. Urol. 2021, 9, 479–488. [Google Scholar] [CrossRef]
- Yao, L.; Ren, S.; Zhang, M.; Du, F.; Zhu, Y.; Yu, H.; Zhang, C.; Li, X.; Yang, C.; Liu, H.; et al. Identification of specific DNA methylation sites on the Y-chromosome as biomarker in prostate cancer. Oncotarget 2015, 6, 40611–40621. [Google Scholar] [CrossRef][Green Version]
- Constâncio, V.; Nunes, S.P.; Moreira-Barbosa, C.; Freitas, R.; Oliveira, J.; Pousa, I.; Oliveira, J.; Soares, M.; Dias, C.G.; Dias, T.; et al. Early detection of the major male cancer types in blood-based liquid biopsies using a DNA methylation panel. Clin. Epigenet. 2019, 11, 175. [Google Scholar] [CrossRef] [PubMed]
- Jarrard, W.E.; Schultz, A.; Etheridge, T.; Damodaran, S.; Allen, G.O.; Jarrard, D.; Yang, B.; Suzuki, H. Screening of urine identifies PLA2G16 as a field defect methylation biomarker for prostate cancer detection. PLoS ONE 2019, 14, e0218950. [Google Scholar] [CrossRef]
- Kiełb, P.; Kowalczyk, K.; Gurwin, A.; Nowak, Ł.; Krajewski, W.; Sosnowski, R.; Szydełko, T.; Małkiewicz, B. Novel Histopathological Biomarkers in Prostate Cancer: Implications and Perspectives. Biomedicines 2023, 11, 1552. [Google Scholar] [CrossRef] [PubMed]
- Fiorentino, V.; Martini, M.; Dell’aquila, M.; Musarra, T.; Orticelli, E.; Larocca, L.M.; Rossi, E.; Totaro, A.; Pinto, F.; Lenci, N.; et al. Histopathological Ratios to Predict Gleason Score Agreement between Biopsy and Radical Prostatectomy. Diagnostics 2020, 11, 10. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Yin, L.; Huang, K.; Zhao, Y.; Zhang, H.; Cai, C.; Xu, Y.; Huang, L.; Wang, X.; Lan, T.; et al. Downregulation of DACT-2 by Promoter Methylation and its Clinicopathological Significance in Prostate Cancer. J. Cancer 2019, 10, 1755–1763. [Google Scholar] [CrossRef]
- Su, Y.; Huang, Q.; Lu, L.; Qu, H.; Wang, D.; Qiu, J.; Li, W.; Lin, M.; Liu, H.; Wang, Z.; et al. Promoter Methylation-Mediated NPTX2 Silencing Promotes Tumor Growth in Human Prostate Cancer. J. Cancer 2022, 13, 706–714. [Google Scholar] [CrossRef]
- Chen, L.; Wang, Y.; Zhang, B. Hypermethylation in the promoter region inhibits AJAP1 expression and activates the JAK/STAT pathway to promote prostate cancer cell migration and stem cell sphere formation. Pathol.-Res. Pract. 2022, 241, 154224. [Google Scholar] [CrossRef]
- Yin, W.; He, P.; Zou, Z.; Lin, J.; Liang, Z.; Wu, Z.; Ye, J.; Lu, J.; Zhong, W. SLC15A2 Serves as a Novel Prognostic Biomarker and Target for Prostate Cancer. Anticancer Res. 2024, 45, 153–172. [Google Scholar] [CrossRef]
- Ma, J.; Xue, K.; Jiang, Y.; Wang, X.; He, D.; Guo, P. Down-regulation of SLC14A1 in prostate cancer activates CDK1/CCNB1 and mTOR pathways and promotes tumor progression. Sci. Rep. 2024, 14, 14914. [Google Scholar] [CrossRef] [PubMed]
- Ke, S.; Liu, Z.; Wang, Q.; Zhai, G.; Shao, H.; Yu, X.; Guo, J. FAM107A Inactivation Associated with Promoter Methylation Affects Prostate Cancer Progression through the FAK/PI3K/AKT Pathway. Cancers 2022, 14, 3915. [Google Scholar] [CrossRef] [PubMed]
- Meng, X.; Yuan, H.; Li, W.; Xiong, Z.; Dong, W.; Xiao, W.; Zhang, X. Solute carrier family 16 member 5 downregulation and its methylation might serve as a prognostic indicator of prostate cancer. IUBMB Life 2021, 73, 1363–1377. [Google Scholar] [CrossRef]
- Sun, J.; Wang, F.; Zhou, H.; Zhao, C.; Li, K.; Fan, C.; Wang, J. Downregulation of PGM5 expression correlates with tumor progression and poor prognosis in human prostate cancer. Discov. Oncol. 2022, 13, 63. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Zhu, Y.; Ma, C.; Qiu, Z.; Zhang, X.; Kang, Z.; Wu, Z.; Wang, H.; Xu, X.; Zhang, H.; et al. Downregulation of EphA5 by promoter methylation in human prostate cancer. BMC Cancer 2015, 15, 18. [Google Scholar] [CrossRef]
- Zhang, P.; Wang, H.; Wang, J.; Liu, Q.; Wang, Y.; Feng, F.; Shi, L. Association between protocadherin 8 promoter hypermethylation and the pathological status of prostate cancer. Oncol. Lett. 2017, 14, 1657–1664. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Pugongchai, A.; Bychkov, A.; Sampatanukul, P. Promoter hypermethylation of SOX11 correlates with adverse clinicopathological features of human prostate cancer. Int. J. Exp. Pathol. 2017, 98, 341–346. [Google Scholar] [CrossRef]
- Lounglaithong, K.; Bychkov, A.; Sampatanukul, P. Aberrant promoter methylation of the PAQR3 gene is associated with prostate cancer. Pathol.-Res. Pract. 2018, 214, 126–129. [Google Scholar] [CrossRef]
- Liu, T.; Qiu, X.; Zhao, X.; Yang, R.; Lian, H.; Qu, F.; Li, X.; Guo, H. Hypermethylation of the SPARC promoter and its prognostic value for prostate cancer. Oncol. Rep. 2017, 39, 659–666. [Google Scholar] [CrossRef]
- Qin, X.-P.; Lu, Q.-J.; Yang, C.-H.; Wang, J.; Chen, J.-F.; Liu, K.; Chen, X.; Zhou, J.; Pan, Y.-H.; Li, Y.-H.; et al. CRMP4 CpG Hypermethylation Predicts Upgrading to Gleason Score ≥ 8 in Prostate Cancer. Front. Oncol. 2022, 12, 840950. [Google Scholar] [CrossRef]
- Gao, X.; Huang, Q.-X.; Xiao, C.-T.; Chen, Z.; Lu, M.-H.; Pang, J.; Di, J.-M.; Luo, Z.-H. Combined analysis of CRMP4 methylation levels and CAPRA-S score predicts metastasis and outcomes in prostate cancer patients. Asian J. Androl. 2018, 20, 56–61. [Google Scholar] [CrossRef]
- Gao, X.; Li, L.-Y.; Rassler, J.; Pang, J.; Chen, M.-K.; Liu, W.-P.; Chen, Z.; Ren, S.-C.; Zhou, F.-J.; Xie, K.-J.; et al. Prospective Study ofCRMP4Promoter Methylation in Prostate Biopsies as a Predictor for Lymph Node Metastases. JNCI J. Natl. Cancer Inst. 2017, 109, djw282. [Google Scholar] [CrossRef]
- Li, K.; Pang, J.; Cheng, H.; Liu, W.-P.; Di, J.-M.; Xiao, H.-J.; Luo, Y.; Zhang, H.; Huang, W.-T.; Chen, M.-K.; et al. Manipulation of prostate cancer metastasis by locus-specific modification of the CRMP4 promoter region using chimeric TALE DNA methyltransferase and demethylase. Oncotarget 2015, 6, 10030–10044. [Google Scholar] [CrossRef]
- Goltz, D.; Gevensleben, H.; Dietrich, J.; Ellinger, J.; Landsberg, J.; Kristiansen, G.; Dietrich, D. Promoter methylation of the immune checkpoint receptor PD-1 (PDCD1) is an independent prognostic biomarker for biochemical recurrence-free survival in prostate cancer patients following radical prostatectomy. OncoImmunology 2016, 5, e1221555. [Google Scholar] [CrossRef]
- Castelo-Branco, P.; Leão, R.; Lipman, T.; Campbell, B.; Lee, D.; Price, A.; Zhang, C.; Heidari, A.; Stephens, D.; Boerno, S.; et al. A cancer specific hypermethylation signature of the TERT promoter predicts biochemical relapse in prostate cancer: A retrospective cohort study. Oncotarget 2016, 7, 57726–57736. [Google Scholar] [CrossRef] [PubMed]
- Ashour, N.; Angulo, J.C.; González-Corpas, A.; Orea, M.J.; Lobo, M.V.T.; Colomer, R.; Colás, B.; Esteller, M.; Ropero, S. Epigenetic Regulation of Gfi1 in Endocrine-Related Cancers: A Role Regulating Tumor Growth. Int. J. Mol. Sci. 2020, 21, 4687. [Google Scholar] [CrossRef] [PubMed]
- Haldrup, C.; Lynnerup, A.-S.; Storebjerg, T.M.; Vang, S.; Wild, P.; Visakorpi, T.; Arsov, C.; Schulz, W.A.; Lindberg, J.; Grönberg, H.; et al. Large-scale evaluation of SLC18A2 in prostate cancer reveals diagnostic and prognostic biomarker potential at three molecular levels. Mol. Oncol. 2016, 10, 825–837. [Google Scholar] [CrossRef]
- Meller, S.; Zipfel, L.; Gevensleben, H.; Dietrich, J.; Ellinger, J.; Majores, M.; Stein, J.; Sailer, V.; Jung, M.; Kristiansen, G.; et al. CDO1 promoter methylation is associated with gene silencing and is a prognostic biomarker for biochemical recurrence-free survival in prostate cancer patients. Epigenetics 2016, 11, 871–880. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Kim, C.; Liu, W.; Zhu, J.; Chin, K.; Rodriguez-Canales, J.; Rodgers, G.P. Olfactomedin 4 downregulation is associated with tumor initiation, growth and progression in human prostate cancer. Int. J. Cancer 2019, 146, 1346–1358. [Google Scholar] [CrossRef]
- Wang, Z.; Deng, T.; Long, X.; Lin, X.; Wu, S.; Wang, H.; Ge, R.; Zhang, Z.; Wu, C.-L.; Taplin, M.-E.; et al. Methylation of SRD5A2 promoter predicts a better outcome for castration-resistant prostate cancer patients undergoing androgen deprivation therapy. PLoS ONE 2020, 15, e0229754. [Google Scholar] [CrossRef]
- Gevensleben, H.; Holmes, E.E.; Goltz, D.; Dietrich, J.; Sailer, V.; Ellinger, J.; Dietrich, D.; Kristiansen, G. PD-L1promoter methylation is a prognostic biomarker for biochemical recurrence-free survival in prostate cancer patients following radical prostatectomy. Oncotarget 2016, 7, 79943–79955. [Google Scholar] [CrossRef] [PubMed]
- Saleem, M.A.; Mustafa, M.S. Promoter Hypermethylation of the BRCA1 Gene as a Novel Biomarker for Prostate Cancer. Cureus 2024, 16, e66467. [Google Scholar] [CrossRef]
- Uhl, B.; Gevensleben, H.; Tolkach, Y.; Sailer, V.; Majores, M.; Jung, M.; Meller, S.; Stein, J.; Ellinger, J.; Dietrich, D.; et al. PITX2 DNA Methylation as Biomarker for Individualized Risk Assessment of Prostate Cancer in Core Biopsies. J. Mol. Diagn. 2017, 19, 107–114. [Google Scholar] [CrossRef]
- Jiang, Q.; Xie, M.; He, M.; Yan, F.; Chen, M.; Xu, S.; Zhang, X.; Shen, P. PITX2 methylation: A novel and effective biomarker for monitoring biochemical recurrence risk of prostate cancer. Medicine 2019, 98, e13820. [Google Scholar] [CrossRef] [PubMed]
- Luan, Z.; Zhang, H.; Qu, X. Prediction efficiency of PITX2 DNA methylation for prostate cancer survival. Genet. Mol. Res. 2016, 15, 10-4238. [Google Scholar] [CrossRef] [PubMed]
- Holmes, E.E.; Goltz, D.; Sailer, V.; Jung, M.; Meller, S.; Uhl, B.; Dietrich, J.; Röhler, M.; Ellinger, J.; Kristiansen, G.; et al. PITX3 promoter methylation is a prognostic biomarker for biochemical recurrence-free survival in prostate cancer patients after radical prostatectomy. Clin. Epigenet. 2016, 8, 104. [Google Scholar] [CrossRef]
- Rocha, S.M.; Sousa, I.; Gomes, I.M.; Arinto, P.; Costa-Pinheiro, P.; Coutinho, E.; Santos, C.R.; Jerónimo, C.; Lemos, M.C.; Passarinha, L.A.; et al. Promoter Demethylation Upregulates STEAP1 Gene Expression in Human Prostate Cancer: In Vitro and In Silico Analysis. Life 2021, 11, 1251. [Google Scholar] [CrossRef]
- Wang, C.; Leavenworth, J.; Zhang, C.; Liu, Z.; Yuan, K.Y.; Wang, Y.; Zhang, G.; Wang, S.; Cui, X.; Zhang, Y.; et al. Epigenetic regulation of EIF4A1 through DNA methylation and an oncogenic role of eIF4A1 through BRD2 signaling in prostate cancer. Oncogene 2022, 41, 2778–2785. [Google Scholar] [CrossRef]
- Fiano, V.; Zugna, D.; Grasso, C.; Trevisan, M.; Delsedime, L.; Molinaro, L.; Gillio-Tos, A.; Merletti, F.; Richiardi, L. LINE-1 methylation status in prostate cancer and non-neoplastic tissue adjacent to tumor in association with mortality. Epigenetics 2016, 12, 11–18. [Google Scholar] [CrossRef][Green Version]
- Zhang, W.; Shu, P.; Wang, S.; Song, J.; Liu, K.; Wang, C.; Ran, L. ZNF154 is a promising diagnosis biomarker and predicts biochemical recurrence in prostate cancer. Gene 2018, 675, 136–143. [Google Scholar] [CrossRef]
- Tonmoy, M.I.Q.; Fariha, A.; Hami, I.; Kar, K.; Al Reza, H.; Bahadur, N.M.; Hossain, S. Computational epigenetic landscape analysis reveals association of CACNA1G-AS1, F11-AS1, NNT-AS1, and MSC-AS1 lncRNAs in prostate cancer progression through aberrant methylation. Sci. Rep. 2022, 12, 10260. [Google Scholar] [CrossRef]
- Li, X.; Zhang, W.; Song, J.; Zhang, X.; Ran, L.; He, Y. SLCO4C1 promoter methylation is a potential biomarker for prognosis associated with biochemical recurrence-free survival after radical prostatectomy. Clin. Epigenet. 2019, 11, 99. [Google Scholar] [CrossRef] [PubMed]
- Geybels, M.S.; Wright, J.L.; Bibikova, M.; Klotzle, B.; Fan, J.-B.; Zhao, S.; Feng, Z.; Ostrander, E.A.; Lin, D.W.; Nelson, P.S.; et al. Epigenetic signature of Gleason score and prostate cancer recurrence after radical prostatectomy. Clin. Epigenet. 2016, 8, 97. [Google Scholar] [CrossRef]
- Aldakheel, F.M.; Alnajran, H.; Alduraywish, S.A.; Mateen, A.; Alqahtani, M.S.; Syed, R. Analysing DNA methylation and transcriptomic signatures to predict prostate cancer recurrence risk. Discov. Oncol. 2025, 16, 110. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Wei, Y.; Li, T.; Lin, L.; Yang, Z.; Ye, L. DNA Methylation-Mediated Lowly Expressed AOX1 Promotes Cell Migration and Invasion of Prostate Cancer. Urol. Int. 2022, 107, 517–525. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Zhang, L. Construction of DNA methylation-based nomogram for predicting biochemical-recurrence-free survival in prostate cancer. Medicine 2022, 101, e32205. [Google Scholar] [CrossRef]
- Liao, Y.; Wu, M.; Jia, Y.; Mou, R.; Li, X. EpCAM as a Novel Biomarker for Survivals in Prostate Cancer Patients. Front. Cell Dev. Biol. 2022, 10, 843604. [Google Scholar] [CrossRef]
- Creighton, C.J.; Zhang, F.; Zhang, Y.; Castro, P.; Hu, R.; Islam; Ghosh, S.; Ittmann, M.; Kwabi-Addo, B. Comparative and integrative analysis of transcriptomic and epigenomic-wide DNA methylation changes in African American prostate cancer. Epigenetics 2023, 18, 2180585. [Google Scholar] [CrossRef]
- Liu, Q.; Reed, M.; Zhu, H.; Cheng, Y.; Almeida, J.; Fruhbeck, G.; Ribeiro, R.; Hu, P. Epigenome-wide DNA methylation and transcriptome profiling of localized and locally advanced prostate cancer: Uncovering new molecular markers. Genomics 2022, 114, 110474. [Google Scholar] [CrossRef]
- Ylitalo, E.B.; Thysell, E.; Landfors, M.; Brattsand, M.; Jernberg, E.; Crnalic, S.; Widmark, A.; Hultdin, M.; Bergh, A.; Degerman, S.; et al. A novel DNA methylation signature is associated with androgen receptor activity and patient prognosis in bone metastatic prostate cancer. Clin. Epigenet. 2021, 13, 133. [Google Scholar] [CrossRef]
- FitzGerald, L.M.; Jung, C.-H.; Wong, E.M.; Joo, J.E.; Bassett, J.K.; Dowty, J.G.; Wang, X.; Dai, J.Y.; Stanford, J.L.; O’callaghan, N.; et al. Detection of differentially methylated CpGs between tumour and adjacent benign cells in diagnostic prostate cancer samples. Sci. Rep. 2024, 14, 17877. [Google Scholar] [CrossRef]
- Nørgaard, M.; Haldrup, C.; Bjerre, M.T.; Høyer, S.; Ulhøi, B.; Borre, M.; Sørensen, K.D. Epigenetic silencing of MEIS2 in prostate cancer recurrence. Clin. Epigenet. 2019, 11, 147. [Google Scholar] [CrossRef]
- Mundbjerg, K.; Chopra, S.; Alemozaffar, M.; Duymich, C.; Lakshminarasimhan, R.; Nichols, P.W.; Aron, M.; Siegmund, K.D.; Ukimura, O.; Aron, M.; et al. Identifying aggressive prostate cancer foci using a DNA methylation classifier. Genome Biol. 2017, 18, 3. [Google Scholar] [CrossRef]
- Habeshian, T.S.; Cannavale, K.; Slezak, J.M.; Shu, Y.H.; Chien, G.W.; Chen, X.; Shi, F.; Siegmund, K.D.; Van Den Eeden, S.K.; Huang, J.; et al. DNA methylation markers for risk of metastasis in a cohort of men with localized prostate cancer. Epigenetics 2024, 19, 2308920. [Google Scholar] [CrossRef] [PubMed]
- Chao, C.R.; Slezak, J.; Siegmund, K.; Cannavale, K.; Shu, Y.; Chien, G.W.; Chen, X.; Shi, F.; Song, N.; Eeden, S.K.V.D.; et al. Genome-wide methylation profiling of diagnostic tumor specimens identified DNA methylation markers associated with metastasis among men with untreated localized prostate cancer. Cancer Med. 2023, 12, 18837–18849. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Geybels, M.S.; Leonardson, A.; Rubicz, R.; Kolb, S.; Yan, Q.; Klotzle, B.; Bibikova, M.; Hurtado-Coll, A.; Troyer, D.; et al. Epigenome-Wide Tumor DNA Methylation Profiling Identifies Novel Prognostic Biomarkers of Metastatic-Lethal Progression in Men Diagnosed with Clinically Localized Prostate Cancer. Clin. Cancer Res. 2017, 23, 311–319. [Google Scholar] [CrossRef]
- Wilkinson, E.J.; Raspin, K.; Malley, R.C.; Donovan, S.; Nott, L.M.; Holloway, A.F.; Dickinson, J.L. WNT5A is a putative epi-driver of prostate cancer metastasis to the bone. Cancer Med. 2024, 13, e70122. [Google Scholar] [CrossRef]
- Angulo, J.C.; Lopez, J.I.; Dorado, J.F.; Sanchez-Chapado, M.; Colas, B.; Ropero, S. A DNA Hypermethylation Profile Independently Predicts Biochemical Recurrence Following Radical Prostatectomy. Urol. Int. 2016, 97, 16–25. [Google Scholar] [CrossRef] [PubMed]
- Rubicz, R.; Zhao, S.; Geybels, M.; Wright, J.L.; Kolb, S.; Klotzle, B.; Bibikova, M.; Troyer, D.; Lance, R.; Ostrander, E.A.; et al. DNA methylation profiles in African American prostate cancer patients in relation to disease progression. Genomics 2019, 111, 10–16. [Google Scholar] [CrossRef]
- Dowty, J.G.; Yu, C.; Hosseinpour, M.; Joo, J.E.; Wong, E.M.; Nguyen-Dumont, T.; Rosenbluh, J.; Giles, G.G.; Milne, R.L.; MacInnis, R.J.; et al. Heritable methylation marks associated with prostate cancer risk. Fam. Cancer 2023, 22, 313–317. [Google Scholar] [CrossRef] [PubMed]
- Bjerre, M.T.; Strand, S.H.; Nørgaard, M.; Kristensen, H.; Rasmussen, A.K.; Mortensen, M.M.; Fredsøe, J.; Mouritzen, P.; Ulhøi, B.; Ørntoft, T.; et al. Aberrant DOCK2, GRASP, HIF3A and PKFP Hypermethylation has Potential as a Prognostic Biomarker for Prostate Cancer. Int. J. Mol. Sci. 2019, 20, 1173. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Davison, J.; Qu, X.; Morrissey, C.; Storer, B.; Brown, L.; Vessella, R.; Nelson, P.; Fang, M. Methylation profiling identified novel differentially methylated markers includingOPCMLandFLRT2in prostate cancer. Epigenetics 2016, 11, 247–258. [Google Scholar] [CrossRef]
- Strand, S.H.; Switnicki, M.; Moller, M.; Haldrup, C.; Storebjerg, T.M.; Hedegaard, J.; Nordentoft, I.; Hoyer, S.; Borre, M.; Pedersen, J.S.; et al. RHCG and TCAF1 promoter hypermethylation predicts biochemical recurrence in prostate cancer patients treated by radical prostatectomy. Oncotarget 2016, 8, 5774–5788. [Google Scholar] [CrossRef]
- Wang, X.; Jordahl, K.M.; Zhu, C.; Livingstone, J.; Rhie, S.K.; Wright, J.L.; Grady, W.M.; Boutros, P.C.; Stanford, J.L.; Dai, J.Y. Methylation Subtypes of Primary Prostate Cancer Predict Poor Prognosis. Cancer Epidemiol. Biomark. Prev. 2022, 31, 1473–1482. [Google Scholar] [CrossRef]
- Daniunaite, K.; Bakavicius, A.; Zukauskaite, K.; Rauluseviciute, I.; Lazutka, J.R.; Ulys, A.; Jankevicius, F.; Jarmalaite, S. Promoter Methylation of PRKCB, ADAMTS12, and NAALAD2 Is Specific to Prostate Cancer and Predicts Biochemical Disease Recurrence. Int. J. Mol. Sci. 2021, 22, 6091. [Google Scholar] [CrossRef]
- Zhao, C.; Zhang, W.; Zhu, X.; Xu, Y.; Yang, K.; Wei, D.; Liang, S.; Zhao, F.; Zhang, Y.; Chen, X.; et al. TWIST2: A new candidate tumor suppressor in prostate cancer. Prostate 2019, 79, 1647–1657. [Google Scholar] [CrossRef]
- Ruiz-Deya, G.; Matta, J.; Encarnación-Medina, J.; Ortiz-Sanchéz, C.; Dutil, J.; Putney, R.; Berglund, A.; Dhillon, J.; Kim, Y.; Park, J.Y. Differential DNA Methylation in Prostate Tumors from Puerto Rican Men. Int. J. Mol. Sci. 2021, 22, 733. [Google Scholar] [CrossRef] [PubMed]
- Pidsley, R.; Lam, D.; Qu, W.; Peters, T.J.; Luu, P.; Korbie, D.; Stirzaker, C.; Daly, R.J.; Stricker, P.; Kench, J.G.; et al. Comprehensive methylome sequencing reveals prognostic epigenetic biomarkers for prostate cancer mortality. Clin. Transl. Med. 2022, 12, e1030. [Google Scholar] [CrossRef]
- Haldrup, C.; Mundbjerg, K.; Vestergaard, E.M.; Lamy, P.; Wild, P.; Schulz, W.A.; Arsov, C.; Visakorpi, T.; Borre, M.; Høyer, S.; et al. DNA Methylation Signatures for Prediction of Biochemical Recurrence After Radical Prostatectomy of Clinically Localized Prostate Cancer. J. Clin. Oncol. 2013, 31, 3250–3258. [Google Scholar] [CrossRef]
- Haldrup, C.; Pedersen, A.L.; Øgaard, N.; Strand, S.H.; Høyer, S.; Borre, M.; Ørntoft, T.F.; Sørensen, K.D. Biomarker potential of ST6GALNAC3 and ZNF660 promoter hypermethylation in prostate cancer tissue and liquid biopsies. Mol. Oncol. 2018, 12, 545–560. [Google Scholar] [CrossRef] [PubMed]
- Van Neste, L.; Partin, A.W.; Stewart, G.D.; Epstein, J.I.; Harrison, D.J.; Van Criekinge, W. Risk score predicts high-grade prostate cancer in DNA-methylation positive, histopathologically negative biopsies. Prostate 2016, 76, 1078–1087. [Google Scholar] [CrossRef] [PubMed]
- Savio, A.J.; Kamdar, S.; Jeyapala, R.; Olkhov-Mitsel, E.; Cuizon, C.; Finelli, A.; Zlotta, A.R.; Toi, A.; Fleshner, N.E.; van der Kwast, T.; et al. Methylation Markers in Prostate Biopsies Are Prognosticators for Late Biochemical Recurrence and Therapy after Surgery in Prostate Cancer Patients. J. Mol. Diagn. 2020, 22, 30–39. [Google Scholar] [CrossRef]
- Witt, J.H.; Friedrich, M.; Jandrig, B.; Porsch, M.; Baumunk, D.; Liehr, U.; Wendler, J.J.; Schostak, M. Molecular margin status after radical prostatectomy using glutathione S-transferase P1 (GSTP1) promoter hypermethylation. BJU Int. 2021, 130, 454–462. [Google Scholar] [CrossRef] [PubMed]
- Litovkin, K.; Van Eynde, A.; Joniau, S.; Lerut, E.; Laenen, A.; Gevaert, T.; Gevaert, O.; Spahn, M.; Kneitz, B.; Gramme, P.; et al. DNA Methylation-Guided Prediction of Clinical Failure in High-Risk Prostate Cancer. PLoS ONE 2015, 10, e0130651. [Google Scholar] [CrossRef]
- Eismann, L.; von Walter, P.; Jung, A.; Chaloupka, M.; Rodler, S.; Westhofen, T.; Buchner, A.; Stief, C.G.; Stadler, T.; Schlenker, B. Methylation status of various gene loci in localized prostate cancer: Novel biomarkers for diagnostics and biochemical recurrence. Urol. Oncol. Semin. Orig. Investig. 2023, 41, 325.e1–325.e8. [Google Scholar] [CrossRef]
- Jeyapala, R.; Kamdar, S.; Olkhov-Mitsel, E.; Savio, A.J.; Zhao, F.; Cuizon, C.; Liu, R.S.; Zlotta, A.; Fleshner, N.; van der Kwast, T.; et al. An integrative DNA methylation model for improved prognostication of postsurgery recurrence and therapy in prostate cancer patients. Urol. Oncol. Semin. Orig. Investig. 2020, 38, 39.e1–39.e9. [Google Scholar] [CrossRef]
- Ahmad, A.S.; Vasiljević, N.; Carter, P.; Berney, D.M.; Møller, H.; Foster, C.S.; Cuzick, J.; Lorincz, A.T. A novel DNA methylation score accurately predicts death from prostate cancer in men with low to intermediate clinical risk factors. Oncotarget 2016, 7, 71833–71840. [Google Scholar] [CrossRef]
- Rybicki, B.A.; Rundle, A.; Kryvenko, O.N.; Mitrache, N.; Do, K.C.; Jankowski, M.; Chitale, D.A.; Trudeau, S.; Belinsky, S.A.; Tang, D. Methylation in benign prostate and risk of disease progression in men subsequently diagnosed with prostate cancer. Int. J. Cancer 2016, 138, 2884–2893. [Google Scholar] [CrossRef]
- Møller, M.; Strand, S.H.; Mundbjerg, K.; Liang, G.; Gill, I.; Haldrup, C.; Borre, M.; Høyer, S.; Ørntoft, T.F.; Sørensen, K.D. Heterogeneous patterns of DNA methylation-based field effects in histologically normal prostate tissue from cancer patients. Sci. Rep. 2017, 7, 40636. [Google Scholar] [CrossRef]
- Lin, Y.-L.; Deng, Q.-K.; Wang, Y.-H.; Fu, X.-L.; Ma, J.-G.; Li, W.-P. Aberrant Protocadherin17 (PCDH17) Methylation in Serum is a Potential Predictor for Recurrence of Early-Stage Prostate Cancer Patients After Radical Prostatectomy. Med. Sci. Monit. 2015, 21, 3955. [Google Scholar] [CrossRef]
- Lin, Y.-L.; Li, Y.-L.; Ma, J.-G. Aberrant Promoter Methylation of Protocadherin8 (PCDH8) in Serum is a Potential Prognostic Marker for Low Gleason Score Prostate Cancer. Med. Sci. Monit. 2017, 23, 4895–4900. [Google Scholar] [CrossRef] [PubMed]
- Deng, Q.-K.; Lei, Y.-G.; Lin, Y.-L.; Ma, J.-G.; Li, W.-P. Prognostic Value of Protocadherin10 (PCDH10) Methylation in Serum of Prostate Cancer Patients. Med. Sci. Monit. 2016, 22, 516–521. [Google Scholar] [CrossRef] [PubMed]
- Londra, D.; Mastoraki, S.; Bournakis, E.; Zavridou, M.; Thanos, A.; Rampias, T.; Lianidou, E.S. USP44 Promoter Methylation in Plasma Cell-Free DNA in Prostate Cancer. Cancers 2021, 13, 4607. [Google Scholar] [CrossRef] [PubMed]
- Lan, T.; Yin, L.; Zhang, H.; Gu, B.; Ma, P.; Li, S.; Li, H. Diagnostic value of DACT-2 methylation in serum of prostate cancer patients. Ann. Palliat. Med. 2021, 10, 2421–2428. [Google Scholar] [CrossRef]
- Horning, A.M.; Awe, J.A.; Wang, C.; Liu, J.; Lai, Z.; Wang, V.Y.; Jadhav, R.R.; Louie, A.D.; Lin, C.; Kroczak, T.; et al. DNA methylation screening of primary prostate tumors identifies SRD5A2 and CYP11A1 as candidate markers for assessing risk of biochemical recurrence. Prostate 2015, 75, 1790–1801. [Google Scholar] [CrossRef]
- Dillinger, T.; Sheibani-Tezerji, R.; Pulverer, W.; Stelzer, I.; Hassler, M.R.; Scheibelreiter, J.; Malla, C.U.P.; Kuroll, M.; Domazet, S.; Redl, E.; et al. Identification of tumor tissue-derived DNA methylation biomarkers for the detection and therapy response evaluation of metastatic castration resistant prostate cancer in liquid biopsies. Mol. Cancer 2022, 21, 7. [Google Scholar] [CrossRef]
- Zavridou, M.; Strati, A.; Bournakis, E.; Smilkou, S.; Tserpeli, V.; Lianidou, E. Prognostic Significance of Gene Expression and DNA Methylation Markers in Circulating Tumor Cells and Paired Plasma Derived Exosomes in Metastatic Castration Resistant Prostate Cancer. Cancers 2021, 13, 780. [Google Scholar] [CrossRef]
- Friedemann, M.; Horn, F.; Gutewort, K.; Tautz, L.; Jandeck, C.; Bechmann, N.; Sukocheva, O.; Wirth, M.P.; Fuessel, S.; Menschikowski, M. Increased Sensitivity of Detection of RASSF1A and GSTP1 DNA Fragments in Serum of Prostate Cancer Patients: Optimisation of Diagnostics Using OBBPA-ddPCR. Cancers 2021, 13, 4459. [Google Scholar] [CrossRef]
- Bjerre, M.T.; Nørgaard, M.; Larsen, O.H.; Jensen, S.Ø.; Strand, S.H.; Østergren, P.; Fode, M.; Fredsøe, J.; Ulhøi, B.P.; Mortensen, M.M.; et al. Epigenetic Analysis of Circulating Tumor DNA in Localized and Metastatic Prostate Cancer: Evaluation of Clinical Biomarker Potential. Cells 2020, 9, 1362. [Google Scholar] [CrossRef]
- Silva, R.; Moran, B.; Russell, N.M.; Fahey, C.; Vlajnic, T.; Manecksha, R.P.; Finn, S.P.; Brennan, D.J.; Gallagher, W.M.; Perry, A.S. Evaluating liquid biopsies for methylomic profiling of prostate cancer. Epigenetics 2020, 15, 715–727. [Google Scholar] [CrossRef] [PubMed]
- Wu, A.; Cremaschi, P.; Wetterskog, D.; Conteduca, V.; Franceschini, G.M.; Kleftogiannis, D.; Jayaram, A.; Sandhu, S.; Wong, S.Q.; Benelli, M.; et al. Genome-wide plasma DNA methylation features of metastatic prostate cancer. J. Clin. Investig. 2020, 130, 1991–2000. [Google Scholar] [CrossRef]
- Chen, S.; Petricca, J.; Ye, W.; Guan, J.; Zeng, Y.; Cheng, N.; Gong, L.; Shen, S.Y.; Hua, J.T.; Crumbaker, M.; et al. The cell-free DNA methylome captures distinctions between localized and metastatic prostate tumors. Nat. Commun. 2022, 13, 6467. [Google Scholar] [CrossRef]
- A Jatkoe, T.; Karnes, R.J.; Freedland, S.J.; Wang, Y.; Le, A.; Baden, J. A urine-based methylation signature for risk stratification within low-risk prostate cancer. Br. J. Cancer 2015, 112, 802–808. [Google Scholar] [CrossRef] [PubMed]
- Bakavicius, A.; Daniunaite, K.; Zukauskaite, K.; Barisiene, M.; Jarmalaite, S.; Jankevicius, F. Urinary DNA methylation biomarkers for prediction of prostate cancer upgrading and upstaging. Clin. Epigenet. 2019, 11, 115. [Google Scholar] [CrossRef] [PubMed]
- O’reilly, E.; Tuzova, A.V.; Walsh, A.L.; Russell, N.M.; O’brien, O.; Kelly, S.; Ni Dhomhnallain, O.; DeBarra, L.; Dale, C.M.; Brugman, R.; et al. epiCaPture: A Urine DNA Methylation Test for Early Detection of Aggressive Prostate Cancer. JCO Precis. Oncol. 2019, 3, 1–18. [Google Scholar] [CrossRef]
- Brikun, I.; Nusskern, D.; Freije, D. An expanded biomarker panel for the detection of prostate cancer from urine DNA. Exp. Hematol. Oncol. 2019, 8, 13. [Google Scholar] [CrossRef]
- Nekrasov, K.A.; Vikarchuk, M.V.; Rudenko, E.E.; Ivanitskiy, I.V.; Grygorenko, V.M.; Danylets, R.O.; Kondratov, A.G.; Stoliar, L.A.; Sharopov, B.R.; Kashuba, V.I. 6-gene promoter methylation assay is potentially applicable for prostate cancer clinical staging based on urine collection following prostatic massage. Oncol. Lett. 2019, 18, 6917–6925. [Google Scholar] [CrossRef]
- Berchuck, J.E.; Baca, S.C.; McClure, H.M.; Korthauer, K.; Tsai, H.K.; Nuzzo, P.V.; Kelleher, K.M.; He, M.; Steinharter, J.A.; Zacharia, S.; et al. Detecting Neuroendocrine Prostate Cancer Through Tissue-Informed Cell-Free DNA Methylation Analysis. Clin. Cancer Res. 2022, 28, 928–938. [Google Scholar] [CrossRef]
- Franceschini, G.M.; Quaini, O.; Mizuno, K.; Orlando, F.; Ciani, Y.; Ku, S.-Y.; Sigouros, M.; Rothmann, E.; Alonso, A.; Benelli, M.; et al. Noninvasive Detection of Neuroendocrine Prostate Cancer through Targeted Cell-free DNA Methylation. Cancer Discov. 2023, 14, 424–445. [Google Scholar] [CrossRef]
- Westaby, D.; Jiménez-Vacas, J.M.; Figueiredo, I.; Rekowski, J.; Pettinger, C.; Gurel, B.; Lundberg, A.; Bogdan, D.; Buroni, L.; Neeb, A.; et al. BCL2 expression is enriched in advanced prostate cancer with features of lineage plasticity. J. Clin. Investig. 2024, 134. [Google Scholar] [CrossRef] [PubMed]
- Stevens, C.; Hightower, A.; Buxbaum, S.G.; Falzarano, S.M.; Rhie, S.K. Genomic, epigenomic, and transcriptomic signatures of prostate cancer between African American and European American patients. Front. Oncol. 2023, 13, 1079037. [Google Scholar] [CrossRef] [PubMed]
- Chernoff, M.; Demanelis, K.; Gillard, M.; Delgado, D.; Gleason, K.J.; Oliva, M.; Chen, L.; Williams, A.; Szmulewitz, R.Z.; Vander Griend, D.J.; et al. Differential DNA Methylation in the Benign and Cancerous Prostate Tissue of African American and European American Men. Cancer Epidemiol. Biomark. Prev. 2025, 34, 428–438. [Google Scholar] [CrossRef]
- Ferrari, M.G.; Ganaie, A.A.; Shabenah, A.; Mansini, A.P.; Wang, L.; Murugan, P.; Davicioni, E.; Wang, J.; Deng, Y.; Hoeppner, L.H.; et al. Identifying and treating ROBO1-ve /DOCK1+ve prostate cancer: An aggressive cancer subtype prevalent in African American patients. Prostate 2020, 80, 1045–1057. [Google Scholar] [CrossRef]
- Shiina, M.; Hashimoto, Y.; Kato, T.; Yamamura, S.; Tanaka, Y.; Majid, S.; Saini, S.; Varahram, S.; Kulkarni, P.; Dasgupta, P.; et al. Differential expression of miR-34b and androgen receptor pathway regulate prostate cancer aggressiveness between African-Americans and Caucasians. Oncotarget 2016, 8, 8356–8368. [Google Scholar] [CrossRef]
- Apprey, V.; Wang, S.; Tang, W.; Kittles, R.A.; Southerland, W.M.; Ittmann, M.; Kwabi-Addo, B. Association of Genetic Ancestry with DNA Methylation Changes in Prostate Cancer Disparity. Anticancer Res. 2019, 39, 5861–5866. [Google Scholar] [CrossRef]
- Farah, E.; Zhang, Z.; Utturkar, S.M.; Liu, J.; Ratliff, T.L.; Liu, X. Targeting DNMTs to Overcome Enzalutamide Resistance in Prostate Cancer. Mol. Cancer Ther. 2021, 21, 193–205. [Google Scholar] [CrossRef]
- Peter, M.R.; Bilenky, M.; Davies, A.; Isserlin, R.; Bader, G.D.; Fleshner, N.E.; Hirst, M.; Zoubeidi, A.; Bapat, B. Distinct DNA methylation patterns associated with treatment resistance in metastatic castration resistant prostate cancer. Sci. Rep. 2021, 11, 6630. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Huang, C.-C.; Huang, S.; Tian, Y.; Huang, J.; Bitaraf, A.; Dong, X.; Nevalainen, M.T.; Patel, M.; Wong, J.; et al. 5-hydroxymethylcytosine sequencing of plasma cell-free DNA identifies epigenomic features in prostate cancer patients receiving androgen deprivation therapies. Commun. Med. 2025, 5, 61. [Google Scholar] [CrossRef]
- Schagdarsurengin, U.; Breiding, V.; Loose, M.; Wagenlehner, F.; Dansranjav, T. Interleukin-1 receptor associated kinase 1 (IRAK1) is epigenetically activated in luminal epithelial cells in prostate cancer. Front. Oncol. 2022, 12, 991368. [Google Scholar] [CrossRef]
- Labbé, M.; Chang, M.; Saintpierre, B.; Letourneur, F.; de Beaurepaire, L.; Véziers, J.; Deshayes, S.; Cotinat, M.; Fonteneau, J.-F.; Blanquart, C.; et al. Loss of miR-200c-3p promotes resistance to radiation therapy via the DNA repair pathway in prostate cancer. Cell Death Dis. 2024, 15, 751. [Google Scholar] [CrossRef]
- He, H.; Zhou, Q.; Zhang, Y.; Li, Y.; Ding, L.; Shen, T.; Liu, S.; Peng, S.; Huang, M.; Zhou, H.; et al. PTBP1 Regulates DNMT3B Alternative Splicing by Interacting with RALY to Enhance the Radioresistance of Prostate Cancer. Adv. Sci. 2024, 11, e2405997. [Google Scholar] [CrossRef]
- Zhang, C.; Ma, S.; Hao, X.; Wang, Z.; Sun, Z. Methylation status of TK1 correlated with immune infiltrates in prostate cancer. Front. Genet. 2022, 13, 899384. [Google Scholar] [CrossRef] [PubMed]
- Büttner, T.; Dietrich, D.; Zarbl, R.; Klümper, N.; Ellinger, J.; Krausewitz, P.; Ritter, M. Feasibility of Monitoring Response to Metastatic Prostate Cancer Treatment with a Methylation-Based Circulating Tumor DNA Approach. Cancers 2024, 16, 482. [Google Scholar] [CrossRef] [PubMed]
- Hendriks, R.J.; Dijkstra, S.; Smit, F.P.; Vandersmissen, J.; Van de Voorde, H.; Mulders, P.F.A.; van Oort, I.M.; Van Criekinge, W.; Schalken, J.A. Epigenetic markers in circulating cell-free DNA as prognostic markers for survival of castration-resistant prostate cancer patients. Prostate 2018, 78, 336–342. [Google Scholar] [CrossRef]
- Peter, M.R.; Bilenky, M.; Isserlin, R.; Bader, G.D.; Shen, S.Y.; De Carvalho, D.D.; Hansen, A.R.; Hu, P.; E Fleshner, N.; Joshua, A.M.; et al. Dynamics of the Cell-Free DNA Methylome of Metastatic Prostate Cancer During Androgen-Targeting Treatment. Epigenomics 2020, 12, 1317–1332. [Google Scholar] [CrossRef]
- Peter, M.R.; Bilenky, M.; Shi, Y.; Pu, J.; Kamdar, S.; Hansen, A.R.; E Fleshner, N.; Sridhar, S.S.; Joshua, A.M.; Hirst, M.; et al. A novel methylated cell-free DNA marker panel to monitor treatment response in metastatic prostate cancer. Epigenomics 2022, 14, 811–822. [Google Scholar] [CrossRef] [PubMed]
- Gordevičius, J.; Kriščiūnas, A.; Groot, D.E.; Yip, S.M.; Susic, M.; Kwan, A.; Kustra, R.; Joshua, A.M.; Chi, K.N.; Petronis, A.; et al. Cell-Free DNA Modification Dynamics in Abiraterone Acetate-Treated Prostate Cancer Patients. Clin. Cancer Res. 2018, 24, 3317–3324. [Google Scholar] [CrossRef]
- Silva, R.; Moran, B.; Baird, A.-M.; O’rOurke, C.J.; Finn, S.P.; McDermott, R.; Watson, W.; Gallagher, W.M.; Brennan, D.J.; Perry, A.S. Longitudinal analysis of individual cfDNA methylome patterns in metastatic prostate cancer. Clin. Epigenet. 2021, 13, 168. [Google Scholar] [CrossRef]
- Overs, A.; Peixoto, P.; Hervouet, E.; Molimard, C.; Monnien, F.; Durand, J.; Guittaut, M.; Vienot, A.; Viot, J.; Herfs, M.; et al. COL25A1 and METAP1D DNA methylation are promising liquid biopsy epigenetic biomarkers of colorectal cancer using digital PCR. Clin. Epigenet. 2024, 16, 146. [Google Scholar] [CrossRef] [PubMed]

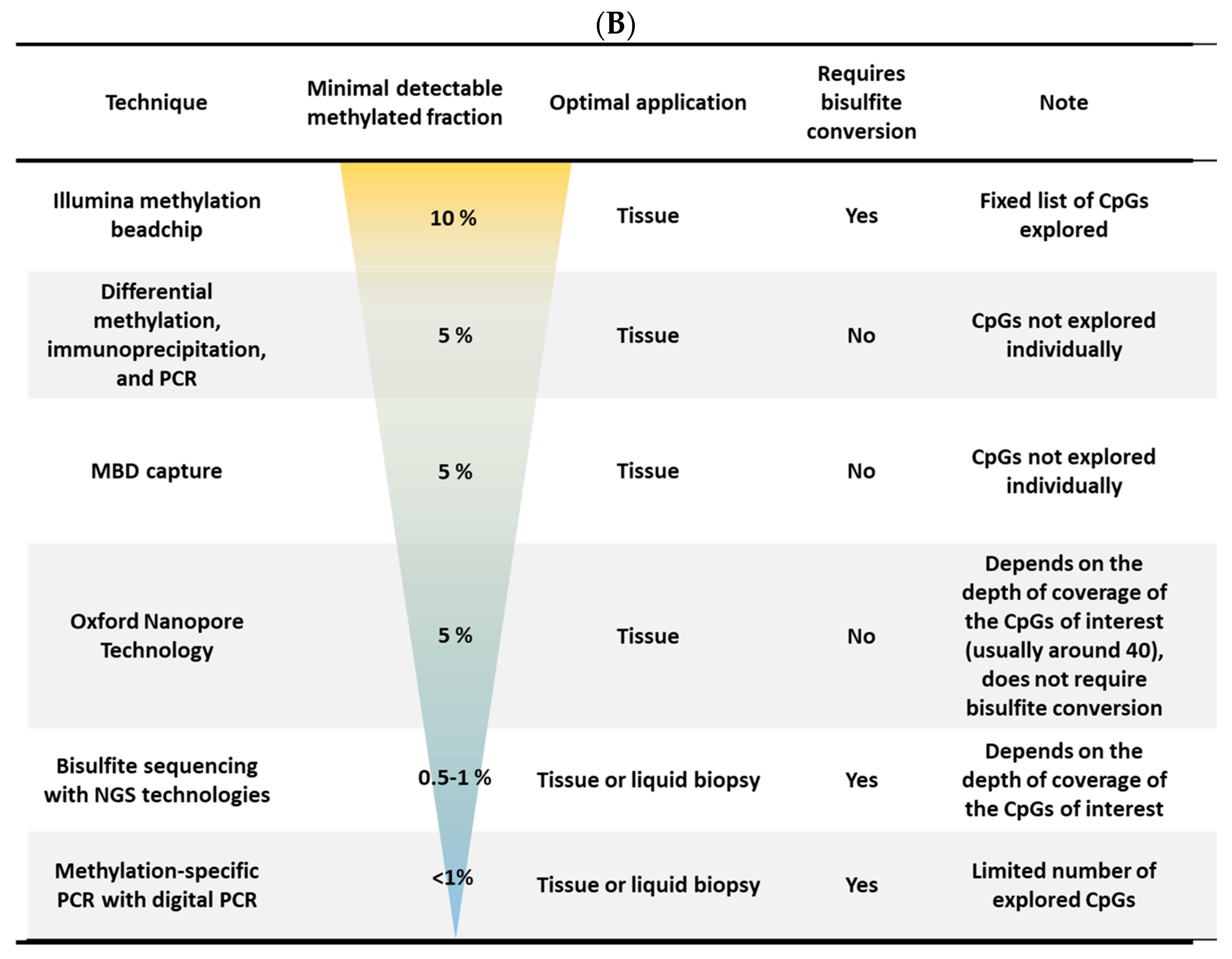
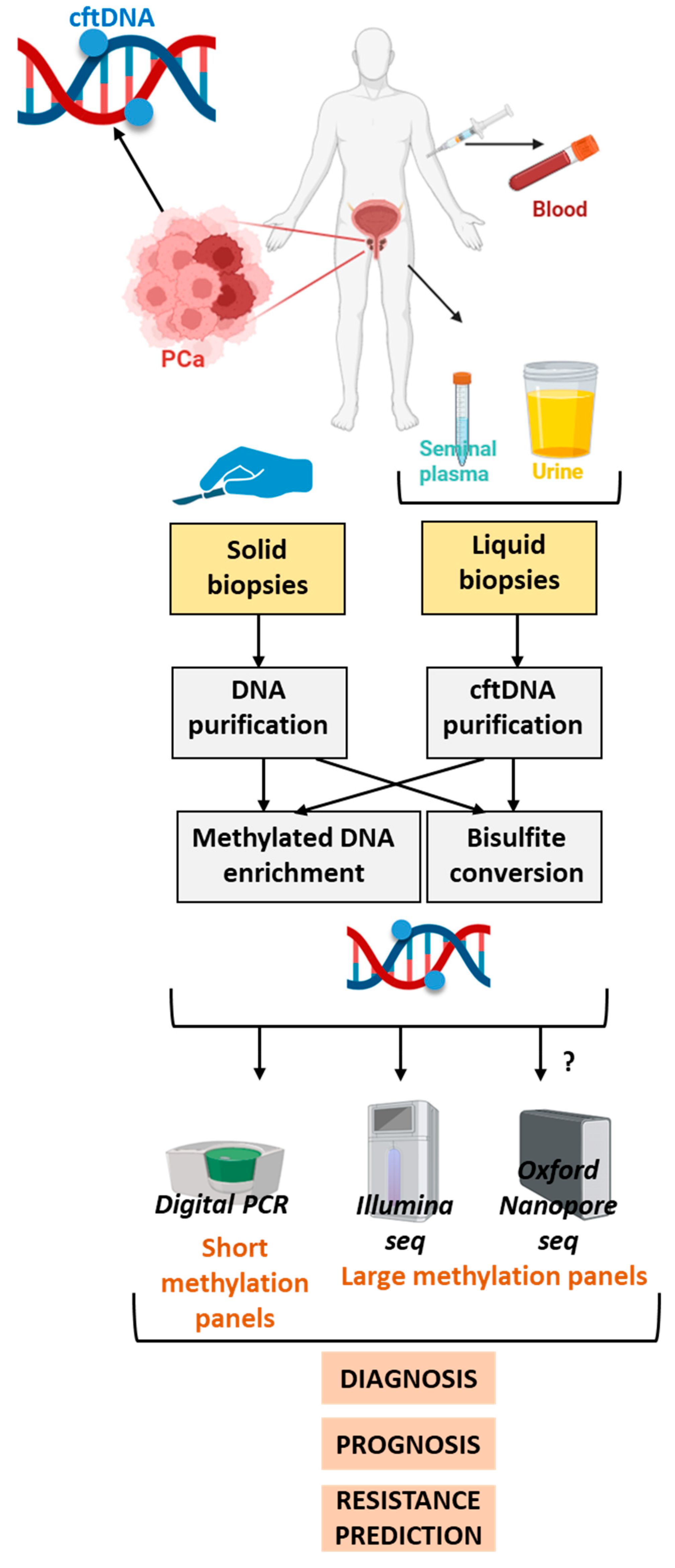
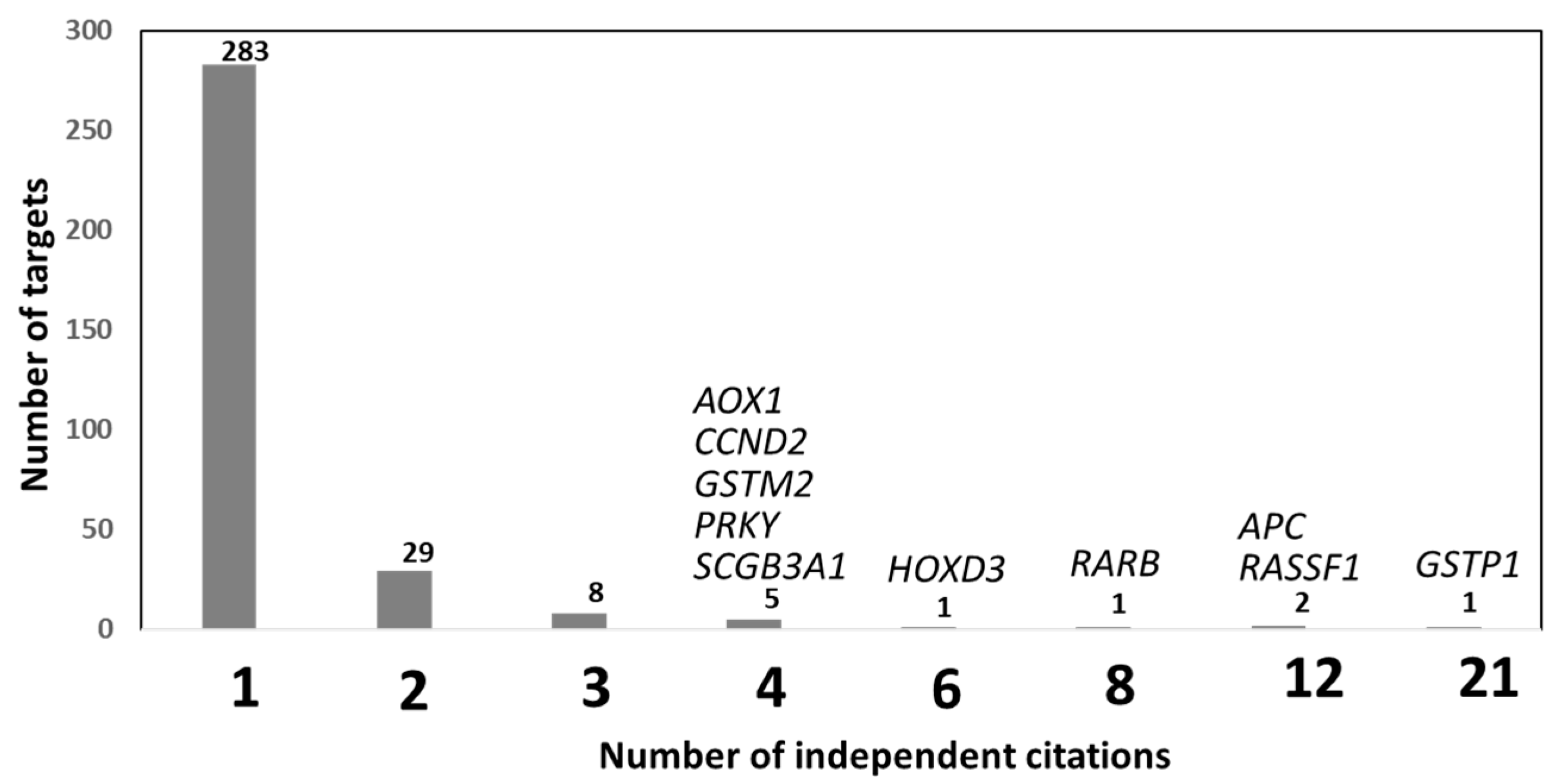
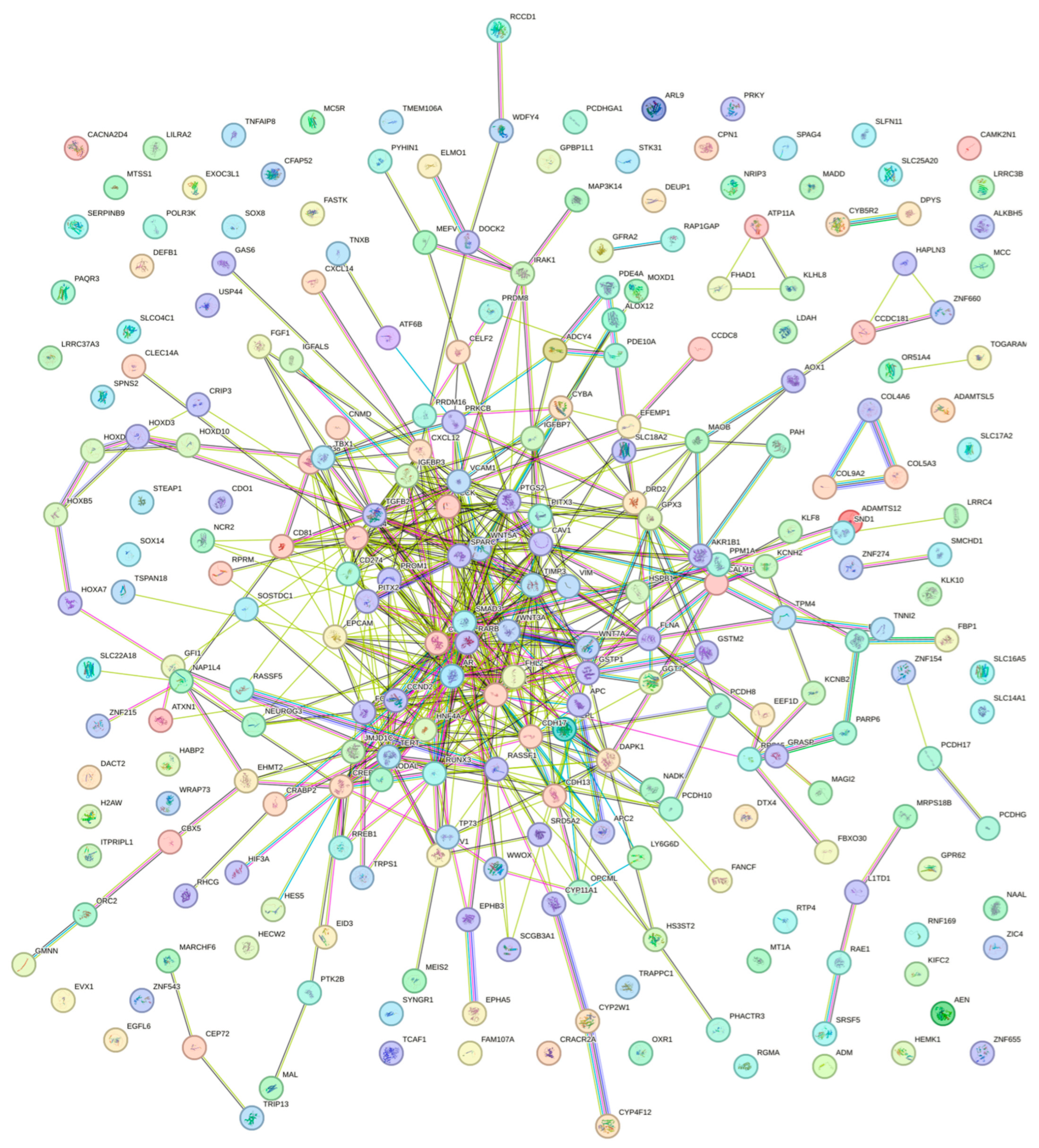
| Function | Fdr |
|---|---|
| Molecular function | |
| Transcription factor binding | 0.0004 |
| RNA polymerase II cis-regulatory region sequence-specific DNA binding | 0.0007 |
| DNA-binding transcriptional factor | 0.002 |
| RNA polymerase II transcription regulatory region sequence-specific DNA binding | 0.002 |
| Biological process | |
| Regulation of epithelial cell proliferation | 2 × 10−7 |
| Response to endogenous stimulus | 8.8 × 10−10 |
| Epithelial cell differentiation | 2.8 × 10−6 |
| Biomarker | Methylation Panel | Liquid Biopsy | Cohort | Technique | Specificity | Sensitivity | Reference |
|---|---|---|---|---|---|---|---|
| Diagnosis | GSTP1, RASSF1, RASSF2 | Plasma | PCa n = 13; HG neoplasia n = 3; BPH n = 20; ASAP n = 3; HD n = 15 | MSP | 83% | 8% | [28] |
| Diagnosis | GSTP1, HOXD3 | Urine | PCa n = 408 and BPH n = 182 | MSP | 97% | 57% | [37] |
| Diagnosis | miR34c, miR193b | Urine | PCa n = 87; HD n = 32 | MSP | 92% | 95% | [38] |
| Diagnosis | miR193b | Urine | PCa n = 95; non-urological cancer n = 29; HD n = 17 | MBD capture-PCR | 96% | 92% | [40] |
| Diagnosis | AOX1rc, APC2, CXCL14, EPHX3, KIFC2, GFRA2, GSTP1, NEUROG3, NODAL, RASSF5, HEMK1, HOXA7, HOXB5, HOXD3a, HOXD3b, HOXD10, MOXD1 | Urine | PCa n = 42; controls n = 50 | MSP | 70% | 90% | [41] |
| Diagnosis | AKR1B1HES5, CHST11, GAS6, GRASP, ITPRIPL1, KCNB2, MAX.chr3.6187, AX.chr3.8.28, SCOL3A1, SERPIN B9, ST6GALNAC2, WNT3A, ZNF655 | Urine | PCa n = 24; HD n = 24 | bisulfite sequencing | 100% | 59% | [42] |
| Diagnosis | APC, FOXA1, GSTP1, HOXD3, RARB2, RASSF1A, SEPT9, SOX17 | Plasma | PCa n = 121 | MSP | 72% | 72% | [46] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Selmani, Z.; Peixoto, P.; Overs, A.; Hervouet, E. The Years 2015–2025 as a Prospective Decade for the Identification of Specific Methylation Biomarkers of Prostate Cancer. Biomolecules 2025, 15, 1334. https://doi.org/10.3390/biom15091334
Selmani Z, Peixoto P, Overs A, Hervouet E. The Years 2015–2025 as a Prospective Decade for the Identification of Specific Methylation Biomarkers of Prostate Cancer. Biomolecules. 2025; 15(9):1334. https://doi.org/10.3390/biom15091334
Chicago/Turabian StyleSelmani, Zohair, Paul Peixoto, Alexis Overs, and Eric Hervouet. 2025. "The Years 2015–2025 as a Prospective Decade for the Identification of Specific Methylation Biomarkers of Prostate Cancer" Biomolecules 15, no. 9: 1334. https://doi.org/10.3390/biom15091334
APA StyleSelmani, Z., Peixoto, P., Overs, A., & Hervouet, E. (2025). The Years 2015–2025 as a Prospective Decade for the Identification of Specific Methylation Biomarkers of Prostate Cancer. Biomolecules, 15(9), 1334. https://doi.org/10.3390/biom15091334







