Gene Duplication, Translocation, and Molecular Evolution of Dmrt1 and Related Sex-Determining Genes in Anurans
Abstract
1. Introduction
2. Materials and Methods
2.1. Anuran Genome Retrieval and Quality Assessment
2.2. Phylogeny of Species and DM Domain Sequences Across Anurans
2.3. Chromosome-Level Synteny Analysis Across Anuran Genomes
2.4. Analyses on Gene Duplications of Dmrt1, Foxl2, Sox3 and Bod1l
2.5. Analyses of Gene Translocation of Dmrt1, Foxl2, Sox3, and Bod1l
2.6. Molecular Evolution and Selection Analysis for Dmrt1, Foxl2, and Sox3
2.7. Evolution of Conserved DM Domain in Dmrt1
3. Results
3.1. Strong Preservation of Chromosome-Level Synteny Across Anuran Genomes
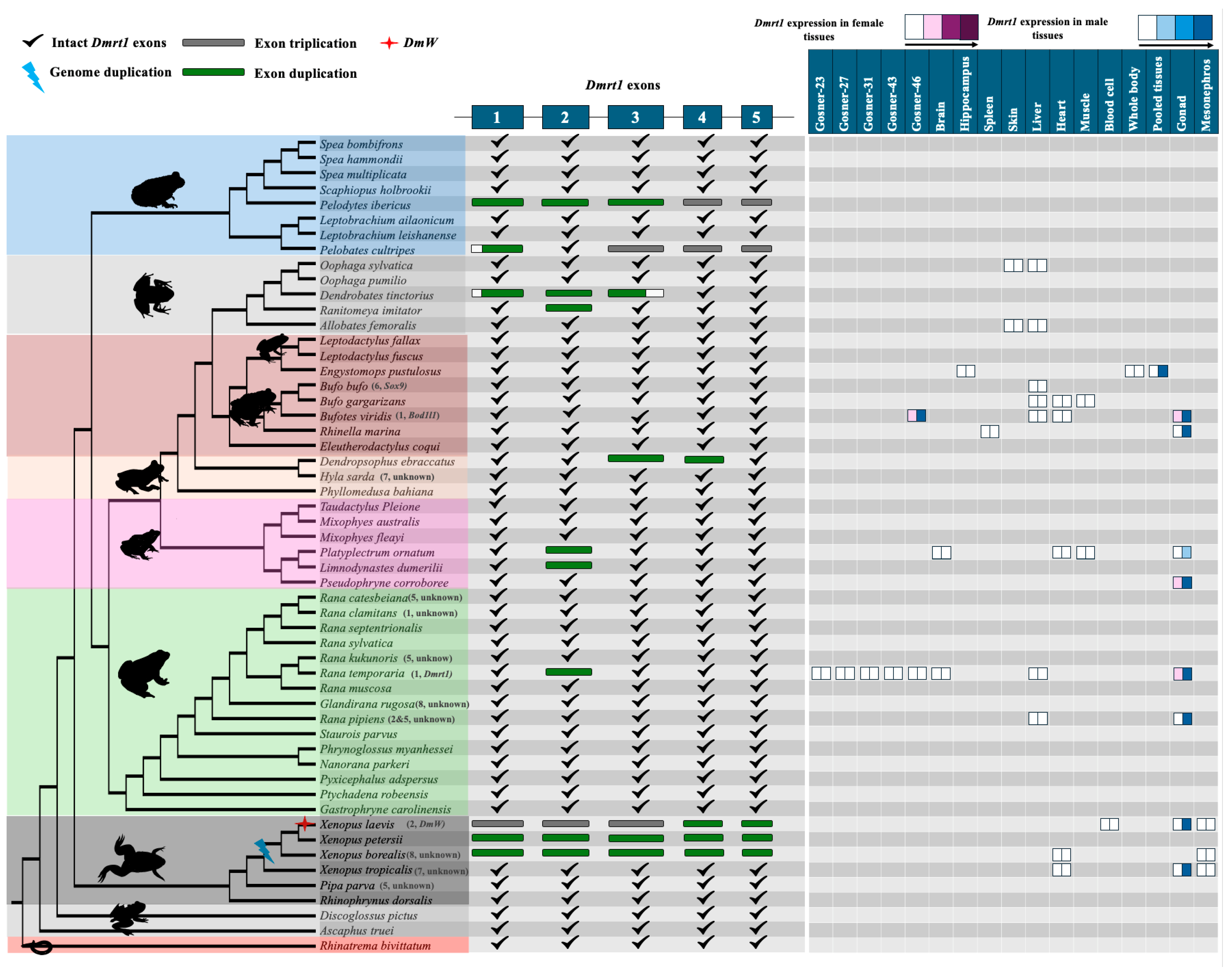
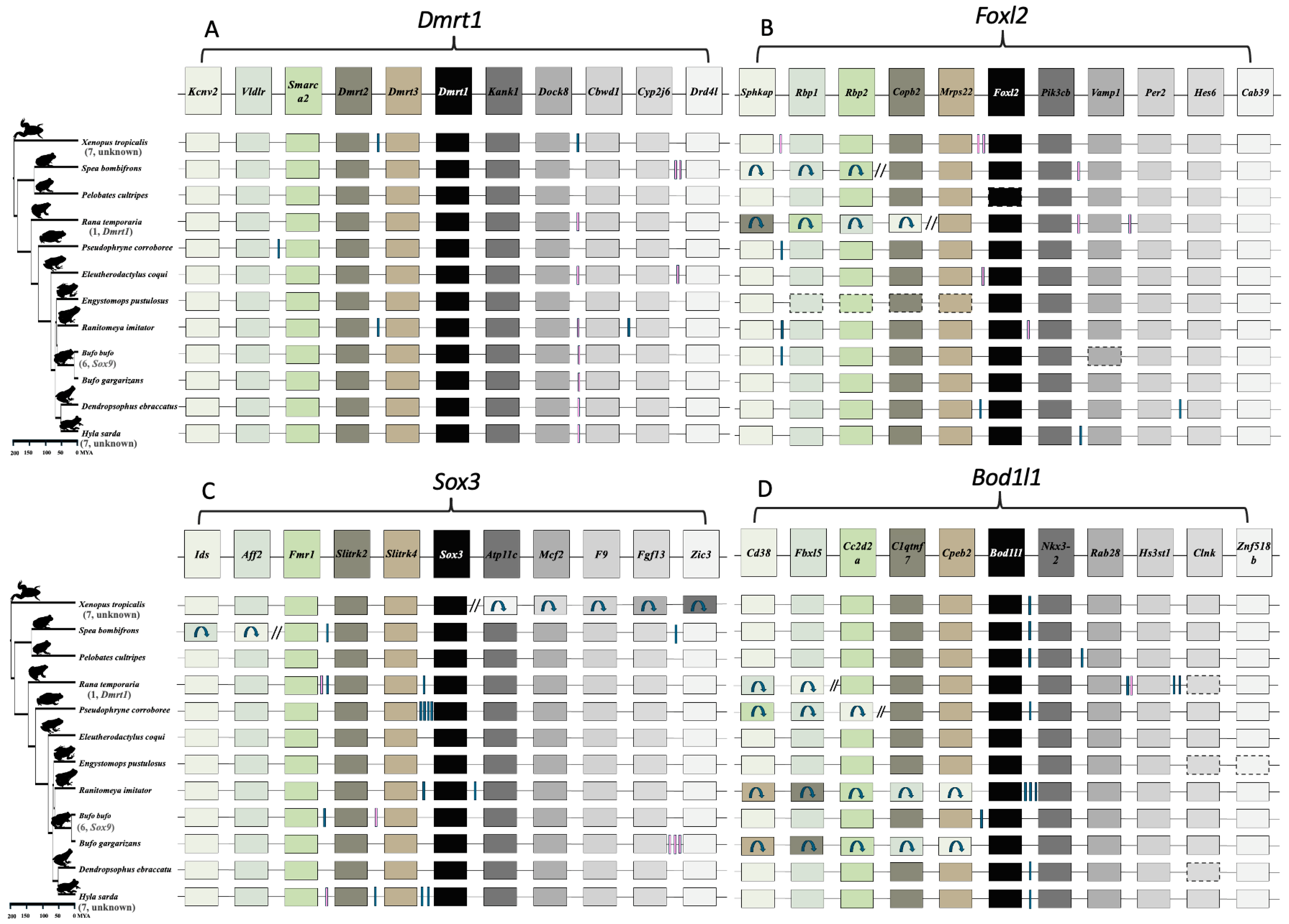
3.2. Dmrt1 Rarely Duplicated in Various Anuran Lineages with Fully Sequenced Genomes
3.3. No Evidence for Gene Translocation of Key Frog Sex-Determining Genes Driving Sex Chromosome Turnover in Frogs
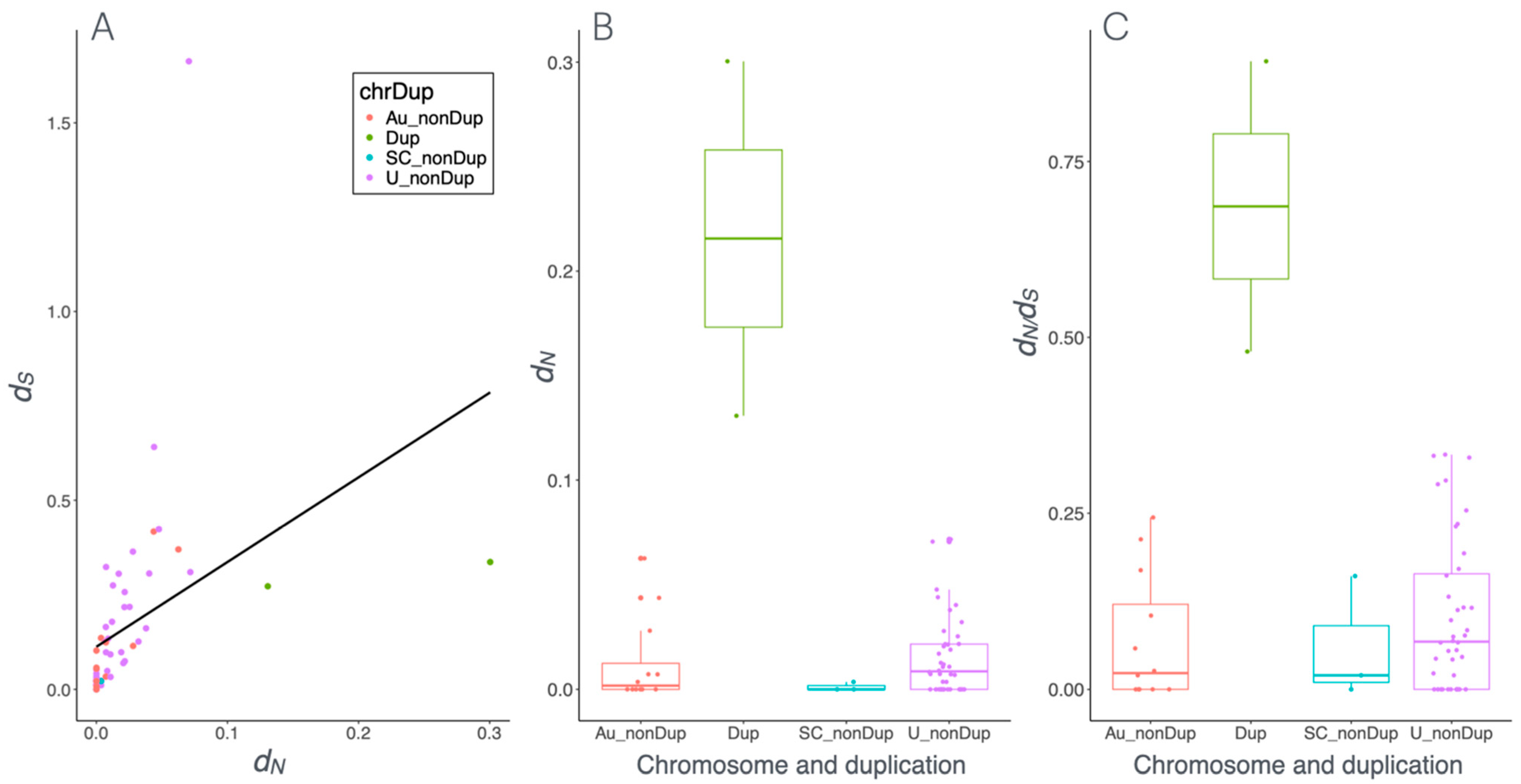
3.4. Strong Purifying Selection Acting on Dmrt1 and Other Sex-Determining Genes
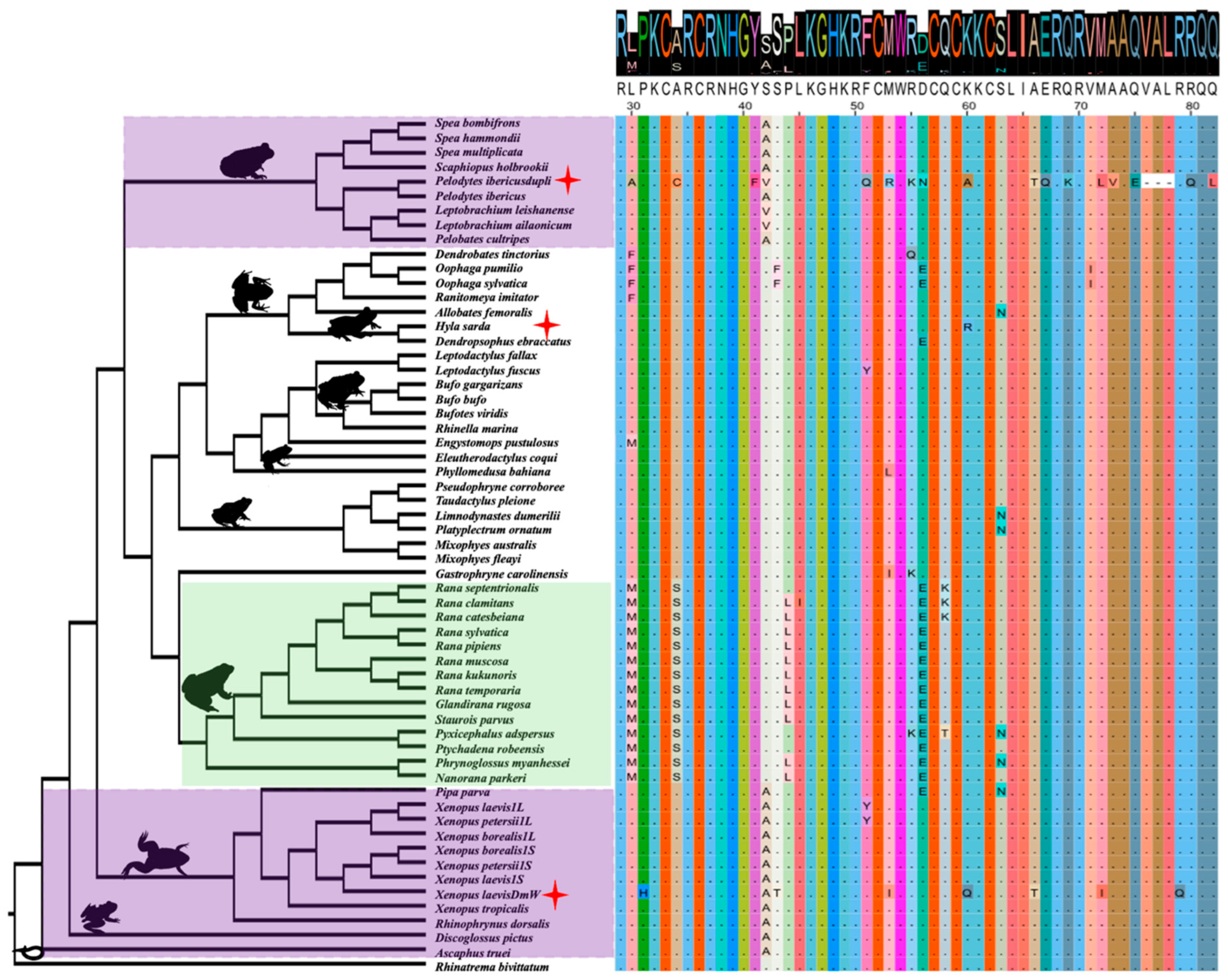
3.5. Conservation and Lineage-Specific Mutations on the DM Domain
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bachtrog, D.; Mank, J.E.; Peichel, C.L.; Kirkpatrick, M.; Otto, S.P.; Ashman, T.; Hahn, M.W.; Kitano, J.; Mayrose, I.; Ming, R.; et al. Sex determination: Why so many ways of doing it? PLoS Biol. 2014, 12, e1001899. [Google Scholar] [CrossRef]
- Beukeboom, L.W.; Perrin, N. The Evolution of Sex Determination; Oxford University Press: Oxford, UK; New York, NY, USA, 2014; ISBN 9780226107547. [Google Scholar]
- Ezaz, T.; Graves, J.A.M. Foreword: Sex and sex chromosomes-new clues from nonmodel species. Chromosome Res. 2012, 20, 1–5. [Google Scholar] [CrossRef]
- Ohno, S. Sex Chromosomes and Sex-Linked Genes; Springer: Berlin/Heidelberg, Germany, 1966. [Google Scholar]
- Peichel, C.L. Convergence and divergence in sex-chromosome evolution. Nat. Genet. 2017, 49, 321–322. [Google Scholar] [CrossRef]
- Furman, B.L.S.; Metzger, D.C.H.; Darolti, I.; Wright, A.E.; Sandkam, B.A.; Almeida, P.; Shu, J.J.; Mank, J.E. Sex chromosome evolution: So many exceptions to the rules. Genome Biol. Evol. 2020, 12, 750–763. [Google Scholar] [CrossRef]
- Mank, J.E. Small but mighty: The evolutionary dynamics of W and Y sex chromosomes. Chromosome Res. 2012, 20, 21–33. [Google Scholar] [CrossRef]
- Veltsos, P.; Shinde, S.; Ma, W.-J. Sex Chromosome Evolution: The Classical Paradigm and So Much Beyond. In Reference Module in Life Sciences; Elsevier: Amsterdam, The Netherlands, 2024; ISBN 9780128096338. [Google Scholar] [CrossRef]
- McDaniel, S.F.; Neubig, K.M.; Payton, A.C.; Quatrano, R.S.; Cove, D.J. Recent gene-capture on the UV sex chromosomes of the moss ceratodon purpureus. Evolution 2013, 67, 2811–2822. [Google Scholar] [CrossRef] [PubMed]
- Müller, D.G.; Gaschet, E.; Godfroy, O.; Gueno, J.; Cossard, G.; Peters, A.F.; Westermeier, R.; Boland, W.; Cock, J.M.; Agnieszka, P.; et al. A partially sex-reversed giant kelp sheds light into the mechanisms of sexual differentiation in a UV sexual system. New Phytol. 2021, 232, 252–263. [Google Scholar] [CrossRef]
- Lipinska, A.P.; Cormier, A.; Peters, A.F.; Kogame, K.; Cock, J.M.; Coelho, S.M. Rapid turnover of life-cycle-related genes in the brown algae. Genome Biol. 2019, 20, 35. [Google Scholar] [CrossRef]
- Hood, M.E.; Petit, E.; Giraud, T. Extensive divergence between mating-type chromosomes of the anther-smut fungus. Genetics 2013, 193, 309–315. [Google Scholar] [CrossRef] [PubMed]
- Idnurm, A.; Hood, M.E.; Johannesson, H.; Giraud, T. Contrasted patterns in mating-type chromosomes in fungi: Hotspots versus coldspots of recombination. Fungal Biol. Rev. 2015, 29, 220–229. [Google Scholar] [CrossRef] [PubMed]
- Muralidhar, P.; Veller, C. Sexual antagonism and the instability of environmental sex determination. Nat. Ecol. Evol. 2018, 2, 343–351. [Google Scholar] [CrossRef]
- Baroiller, J.F.; D’Cotta, H.; Saillant, E. Environmental effects on fish sex determination and differentiation. Sex. Dev. 2009, 3, 118–135. [Google Scholar] [CrossRef]
- Akashi, H.; Hasui, D.; Ueda, K.; Ishikawa, M.; Takeda, M.; Miyagawa, S. Understanding the role of environmental temperature on sex determination through comparative studies in reptiles and amphibians. J. Exp. Zool. A Ecol. Integr. Physiol. 2023, 341, 48–59. [Google Scholar] [CrossRef]
- Warner, R.R. The adaptive significance of sequential hermaphroditism in animals. Am. Nat. 1975, 109, 61–82. [Google Scholar] [CrossRef]
- Georges, A.; Holleley, C.E. How does temperature determine sex? Science 2018, 360, 601–602. [Google Scholar] [CrossRef]
- Valenzuela, N.; Lance, V.A. Temperature-Dependent Sex Determination in Vertebrates; Valenzuela, N., Lance, V.A., Eds.; Smithsonian Books: Washington, DC, USA, 2004; ISBN 1588342034. [Google Scholar]
- Cutting, A.; Chue, J.; Smith, C.A. Just how conserved is vertebrate sex determination? Dev. Dyn. 2013, 242, 380–387. [Google Scholar] [CrossRef]
- Smith, C.A.; Roeszler, K.N.; Hudson, Q.J.; Sinclair, A.H. Avian sex determination: What, when and where? Cytogenet. Genome Res. 2007, 117, 165–173. [Google Scholar] [CrossRef] [PubMed]
- Augstenová, B.; Ma, W. Decoding Dmrt1: Insights into vertebrate sex determination and gonadal sex differentiation. J. Evol. Biol. 2025, 38, 811–831. [Google Scholar] [CrossRef] [PubMed]
- Ellegren, H. Sex-chromosome evolution: Recent progress and the influence of male and female heterogamety. Nat. Rev. Genet. 2011, 12, 157–166. [Google Scholar] [CrossRef] [PubMed]
- Ma, W.-J.; Rovatsos, M. Sex chromosome evolution: The remarkable diversity in the evolutionary rates and mechanisms. J. Evol. Biol. 2022, 35, 1581–1588. [Google Scholar] [CrossRef] [PubMed]
- Perrin, N. Sex reversal: A fountain of youth for sex chromosomes? Evolution 2009, 63, 3043–3049. [Google Scholar] [CrossRef]
- Vicoso, B. Molecular and evolutionary dynamics of animal sex-chromosome turnover. Nat. Ecol. Evol. 2019, 3, 1632–1641. [Google Scholar] [CrossRef] [PubMed]
- Van Doorn, G.S.; Kirkpatrick, M. Turnover of sex chromosomes induced by sexual conflict. Nature 2007, 449, 909–912. [Google Scholar] [CrossRef]
- Saunders, P.A. Sex chromosome turnovers in evolution. In eLS; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2019. [Google Scholar] [CrossRef]
- Jeffries, D.L.; Lavanchy, G.; Sermier, R.; Sredl, M.J.; Miura, I.; Borzée, A.; Barrow, L.N.; Canestrelli, D.; Crochet, P.; Dufresnes, C.; et al. A rapid rate of sex-chromosome turnover and non-random transitions in true frogs. Nat. Commun. 2018, 9, 4088. [Google Scholar] [CrossRef]
- Dufresnes, C.; Borzee, A.; Horn, A.; Stock, M.; Ostini, M.; Sermier, R.; Wassef, J.; Litvinchuck, S.N.; Kosch, T.A.; Waldman, B.; et al. Sex-chromosome homomorphy in palearctic tree frogs results from both turnovers and X – Y recombination. Mol. Biol. Evol. 2015, 32, 2328–2337. [Google Scholar] [CrossRef] [PubMed]
- Kitano, J.; Peichel, C.L. Turnover of sex chromosomes and speciation in fishes. Environ. Biol. Fishes 2012, 94, 549–558. [Google Scholar] [CrossRef]
- Sessions, S.K.; Bizjak Mali, L.; Green, D.M.; Trifonov, V.; Ferguson-Smith, M. Evidence for sex chromosome turnover in proteid salamanders. Cytogenet. Genome Res. 2016, 148, 305–313. [Google Scholar] [CrossRef]
- Myosho, T.; Takehana, Y.; Hamaguchi, S.; Sakaizumi, M. Turnover of sex chromosomes in Celebensis group medaka fishes. G3 2015, 5, 2685–2691. [Google Scholar] [CrossRef]
- Tennessen, J.A.; Wei, N.; Straub, S.; Govindarajulu, R.; Liston, A.; Ashman, T.; Colleges, W.S. Repeated translocation of a gene cassette drives sex chromosome turnover in strawberries. PLoS Genet. 2017, 16, e2006062. [Google Scholar] [CrossRef]
- Behrens, K.A.; Zimmermann, H.; Blažek, R.; Reichard, M.; Koblmüller, S.; Kocher, T.D. Turnover of sex chromosomes in the lake tanganyika cichlid tribe Tropheini (Teleostei: Cichlidae). Sci. Rep. 2024, 14, 2471. [Google Scholar] [CrossRef] [PubMed]
- Kabir, A.; Ieda, R.; Hosoya, S.; Fujikawa, D.; Atsumi, K.; Tajima, S.; Nozawa, A.; Koyama, T.; Hirase, S.; Nakamura, O.; et al. Repeated translocation of a supergene underlying rapid sex chromosome turnover in takifugu pufferfish. Proc. Natl. Acad. Sci. USA 2022, 119, e2121469119. [Google Scholar] [CrossRef]
- Lubieniecki, K.P.; Lin, S.; Cabana, E.I.; Li, J.; Lai, Y.Y.Y.; Davidson, W.S. Genomic instability of the sex-determining locus in Atlantic salmon (Salmo salar). G3 2015, 5, 2513–2522. [Google Scholar] [CrossRef]
- Ma, W.-J.; Veltsos, P. The diversity and evolution of sex chromosomes in frogs. Genes 2021, 12, 483. [Google Scholar] [CrossRef]
- Ma, W.-J.; Rodrigues, N.; Sermier, R.; Brelsford, A.; Perrin, N. Dmrt1 polymorphism covaries with sex-determination patterns in Rana temporaria. Ecol. Evol. 2016, 6, 5017–5117. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, N.; Studer, T.; Dufresnes, C.; Ma, W.-J.; Veltsos, P.; Perrin, N. Dmrt1 polymorphism and sex-chromosome differentiation in Rana temporaria. Mol. Ecol. 2017, 26, 4897–4905. [Google Scholar] [CrossRef] [PubMed]
- Raymond, C.S.; Murphy, M.W.; O’Sullivan, M.G.; Bardwell, V.J.; Zarkower, D. Dmrt1, a gene related to worm and fly sexual regulators, is required for mammalian testis differentiation. Genes Dev. 2000, 14, 2587–2595. [Google Scholar] [CrossRef]
- Zarkower, D.; Murphy, M.W. DMRT1: An ancient sexual regulator required for human gonadogenesis. Sex. Dev. 2022, 16, 112–125. [Google Scholar] [CrossRef]
- Kopp, A. Dmrt genes in the development and evolution of sexual dimorphism. Trends Genet. 2012, 28, 175–184. [Google Scholar] [CrossRef] [PubMed]
- Mawaribuchi, S.; Ito, Y.; Ito, M. Independent evolution for sex determination and differentiation in the Dmrt family in animals. Biol. Open 2019, 8, bio041962. [Google Scholar] [CrossRef]
- Yoshimoto, S.; Okada, E.; Umemoto, H.; Tamura, K.; Uno, Y.; Nishida-umehara, C.; Matsuda, Y.; Takamatsu, N.; Shiba, T.; Ito, M. A W-linked DM-domain gene, DM-W, participatees in primary ovary development in Xenopus laevis. Proc. Natl. Acad. Sci. USA 2008, 105, 2469–2474. [Google Scholar] [CrossRef]
- Bewick, A.J.; Anderson, D.W.; Evans, B.J. Evolution of the closely related, sex-related genes dm-w and dmrt1 in african clawed frogs (Xenopus). Evolution 2011, 65, 698–712. [Google Scholar] [CrossRef]
- Mawaribuchi, S.; Musashijima, M.; Wada, M.; Izutsu, Y.; Kurakata, E.; Park, M.K.; Takamatsu, N.; Ito, M. Molecular evolution of two distinct Dmrt1 promoters for germ and somatic cells in vertebrate gonads. Mol. Biol. Evol. 2017, 34, 724–733. [Google Scholar] [CrossRef]
- Yoshimoto, S.; Ikeda, N.; Izutsu, Y.; Shiba, T.; Takamatsu, N.; Ito, M. Opposite roles of dmrt1 and its W-linked paralogue, DM-W, in sexual dimorphism of Xenopus laevis: Implications of a ZZ/ZW-type sex-determining system. Development 2010, 137, 2519–2526. [Google Scholar] [CrossRef]
- Cauret, C.M.S.; Gansauge, M.T.; Tupper, A.S.; Furman, B.L.S.; Knytl, M.; Song, X.Y.; Greenbaum, E.; Meyer, M.; Evans, B.J.; Wilson, M. Developmental systems drift and the drivers of sex chromosome evolution. Mol. Biol. Evol. 2020, 37, 799–810. [Google Scholar] [CrossRef]
- Cauret, C.M.S.; Jordan, D.C.; Kukoly, L.M.; Burton, S.R.; Anele, E.U.; Kwiecien, J.M.; Gansauge, M.T.; Senthillmohan, S.; Greenbaum, E.; Meyer, M.; et al. Functional dissection and assembly of a small, newly evolved, W chromosome-specific genomic region of the African clawed frog Xenopus laevis. PLoS Genet. 2023, 19, e1010990. [Google Scholar] [CrossRef]
- Brelsford, A.; Dufresnes, C.; Perrin, N. Trans-species variation in Dmrt1 is associated with sex determination in four european tree-frog species. Evolution 2016, 70, 840–847. [Google Scholar] [CrossRef] [PubMed]
- Kuhl, H.; Tan, W.H.; Klopp, C.; Kleiner, W.; Koyun, B.; Ciorpac, M.; Feron, R.; Knytl, M.; Kloas, W.; Schartl, M.; et al. A candidate sex determination locus in amphibians which evolved by structural variation between X- and Y-chromosomes. Nat. Commun. 2024, 15, 4781. [Google Scholar] [CrossRef] [PubMed]
- Oshima, Y.; Naruse, K.; Nakamura, Y.; Nakamura, M. Sox3: A transcription factor for Cyp19 expression in the frog Rana rugosa. Gene 2009, 445, 38–48. [Google Scholar] [CrossRef]
- Li, M.H.; Yang, H.H.; Li, M.R.; Sun, Y.L.; Jiang, X.L.; Xie, Q.P.; Wang, T.R.; Shi, H.J.; Sun, L.N.; Zhou, L.Y.; et al. Antagonistic roles of Dmrt1 and Foxl2 in sex differentiation via estrogen production in tilapia as demonstrated by TALENs. Endocrinology 2013, 154, 4814–4825. [Google Scholar] [CrossRef] [PubMed]
- Kossack, M.E.; Draper, B.W. Genetic regulation of sex determination and maintenance in zebrafish (Danio rerio). Curr. Top. Dev. Biol. 2019, 134, 119–149. [Google Scholar] [CrossRef]
- Zhang, J. Evolution of DMY, a newly emergent male sex-determination gene of medaka fish. Genetics 2004, 166, 1887–1895. [Google Scholar] [CrossRef] [PubMed]
- Matsuda, M.; Nagahama, Y.; Shinomiya, A.; Sato, T.; Matsuda, C.; Kobayashi, T.; Morrey, C.E.; Shibata, N.; Asakawa, S.; Shimizu, N.; et al. DMY is a Y-specific DM-domain gene required for male development in the medaka fish. Nature 2002, 417, 559–563. [Google Scholar] [CrossRef]
- Ogita, Y.; Mawaribuchi, S.; Nakasako, K.; Tamura, K.; Matsuda, M.; Katsumura, T.; Oota, H.; Watanabe, G.; Yoneda, S.; Takamatsu, N.; et al. Parallel evolution of two Dmrt1-derived genes, Dmy and Dm-W, for vertebrate sex determination. iScience 2020, 23, 100757. [Google Scholar] [CrossRef] [PubMed]
- Vicoso, B.; Charlesworth, B. Effective population size and the Faster-X effect: An extended model. Evolution 2009, 63, 2413–2426. [Google Scholar] [CrossRef] [PubMed]
- Mank, J.E.; Vicoso, B.; Berlin, S.; Charlesworth, B. Effective population size and the Faster-X effect: Empirical results and their interpretation. Evolution 2009, 64, 663–674. [Google Scholar] [CrossRef]
- Ellegren, H.; Parsch, J. The evolution of sex-biased genes and sex-biased gene expression. Nat. Rev. Genet. 2007, 8, 689–698. [Google Scholar] [CrossRef]
- Mullon, C.; Pomiankowski, A.; Reuter, M. Molecular evolution of Drosophila sex-lethal and related sex determining genes. BMC Evol. Biol. 2012, 12, 5. [Google Scholar] [CrossRef]
- Huang, S.; Ye, L.; Chen, H. Sex determination and maintenance: The role of DMRT1 and FOXL2. Asian J. Androl. 2017, 19, 619–624. [Google Scholar] [CrossRef]
- Alam, M.A.; Kobayashi, Y.; Horiguchi, R.; Hirai, T.; Nakamura, M. molecular cloning and quantitative expression of sexually dimorphic markers Dmrt1 and Foxl2 during female-to-male sex change in Epinephelus merra. Gen. Comp. Endocrinol. 2008, 157, 75–85. [Google Scholar] [CrossRef]
- Simão, F.A.; Waterhouse, R.M.; Ioannidis, P.; Kriventseva, E.V.; Zdobnov, E.M. BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 2015, 31, 3210–3212. [Google Scholar] [CrossRef]
- Portik, D.M.; Streicher, J.W.; Wiens, J.J. Frog Phylogeny: A time-calibrated, species-level tree based on hundreds of loci and 5242 Species. Mol. Phylogenet. Evol. 2023, 188, 107907. [Google Scholar] [CrossRef]
- Darriba, D.; Posada, D.; Kozlov, A.M.; Stamatakis, A.; Morel, B.; Flouri, T. ModelTest-NG: A new and scalable tool for the selection of dna and protein evolutionary models. Mol. Biol. Evol. 2020, 37, 291–294. [Google Scholar] [CrossRef] [PubMed]
- Xia, X.; Xie, Z.; Salemi, M.; Chen, L.; Wang, Y. An index of substitution saturation and its application. Mol. Phylogenet. Evol. 2003, 26, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Xia, X.; Lemey, P. Assessing substitution saturation with DAMBE. In The Phylogenetic Handbook: A Practical Approach to Phylogenetic Analysis and Hypothesis Testing; Cambridge University Press: Cambridge, UK, 2012; pp. 615–630. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, H.; Debarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Wu, Y.; Li, J.; Wang, X.; Zeng, Z.; Xu, J.; Liu, Y.; Feng, J.; Chen, H.; He, Y.; et al. TBtools-II: A “one for all, all for one” bioinformatics platform for biological big-data mining. Mol. Plant 2023, 16, 1733–1742. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef] [PubMed]
- Ma, W.-J.; Veltsos, P.; Sermier, R.; Parker, D.J.; Perrin, N. Evolutionary and developmental dynamics of sex-biased gene expression in common frogs with proto-Y chromosomes. Genome Biol. 2018, 19, 156. [Google Scholar] [CrossRef]
- Ma, W.-J.; Veltsos, P.; Toups, M.A.; Rodrigues, N.; Sermier, R.; Jeffries, D.L.; Perrin, N. Tissue specificity and dynamics of sex-biased gene expression in a common frog population with differentiated, yet homomorphic, sex chromosomes. Genes 2018, 9, 294. [Google Scholar] [CrossRef]
- Toups, M.A.; Rodrigues, N.; Perrin, N.; Kirkpatrick, M. A reciprocal translocation radically reshapes sex-linked inheritance in the common frog. Mol. Ecol. 2019, 28, 1877–1889. [Google Scholar] [CrossRef]
- Brůna, T.; Hoff, K.J.; Lomsadze, A.; Stanke, M.; Borodovsky, M. BRAKER2: Automatic eukaryotic genome annotation with GeneMark-EP+ and AUGUSTUS supported by a protein database. NAR Genom. Bioinform. 2021, 3, lqaa108. [Google Scholar] [CrossRef] [PubMed]
- Cantarel, B.L.; Korf, I.; Robb, S.M.C.; Parra, G.; Ross, E.; Moore, B.; Holt, C.; Alvarado, A.S.; Yandell, M. MAKER: An easy-to-use annotation pipeline designed for emerging model organism genomes. Genome Res. 2008, 18, 188–196. [Google Scholar] [CrossRef]
- Id, E.S.D.; Id, J.V.; Id, T.J.P.V.D.; Snel, B. Measuring the impact of gene prediction on gene loss estimates in eukaryotes by quantifying falsely inferred absences. PLoS Comput. Biol. 2019, 15, e1007301. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-Seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq-A python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef]
- Trapnell, C.; Roberts, A.; Goff, L.; Pertea, G.; Kim, D.; Kelley, D.R.; Pimentel, H.; Salzberg, S.L.; Rinn, J.L.; Pachter, L. Differential gene and transcript expression analysis of rna-seq experiments with tophat and cufflinks. Nat. Protoc. 2012, 7, 562–578. [Google Scholar] [CrossRef]
- Yang, Z. PAML 4: Phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 2007, 24, 1586–1591. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z. User Guide PAML: Phylogenetic Analysis by Maximum Likelihood, version 4.6; University College London: London, UK, 2014. [Google Scholar]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Arvestad, L. Alv: A console-based viewer for molecular sequence alignments. J. Open Source Softw. 2018, 3, 955. [Google Scholar] [CrossRef]
- Roco, Á.S.; Olmstead, A.W.; Degitz, S.J.; Amano, T.; Zimmerman, L.B.; Bullejos, M. Coexistence of Y, W, and Z sex chromosomes in Xenopus tropicalis. Proc. Natl. Acad. Sci. USA 2015, 112, E4752–E4761. [Google Scholar] [CrossRef]
- Dufresnes, C.; Brelsford, A.; Baier, F.; Perrin, N. When sex chromosomes recombine only in the heterogametic sex: Heterochiasmy and heterogamety in hyla tree frogs. Mol. Biol. Evol. 2020, 38, 192–200. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, N.; Studer, T.; Dufresnes, C.; Perrin, N. Sex-chromosome recombination in common frogs brings water to the fountain-of-youth. Mol. Biol. Evol. 2018, 35, 942–948. [Google Scholar] [CrossRef] [PubMed]
- Matson, C.K.; Murphy, M.W.; Griswold, M.D.; Yoshida, S.; Bardwell, V.J.; Zarkower, D. The mammalian Doublesex homolog DMRT1 is a transcriptional gatekeeper that controls the mitosis versus meiosis decision in male germ cells. Dev. Cell 2010, 19, 612–624. [Google Scholar] [CrossRef]
- Lindeman, R.E.; Gearhart, M.D.; Minkina, A.; Krentz, A.D.; Bardwell, V.J.; Zarkower, D. sexual cell fate reprogramming in the ovary by DMRT1. Curr. Biol. 2015, 25, 764–771. [Google Scholar] [CrossRef] [PubMed]
- Krentz, A.D.; Murphy, M.W.; Sarver, A.L.; Griswold, M.D.; Bardwell, V.J.; Zarkower, D. DMRT1 promotes oogenesis by transcriptional activation of stra8 in the mammalian fetal ovary. Dev. Biol. 2011, 356, 63–70. [Google Scholar] [CrossRef]
- Kosch, T.A.; Crawford, A.J.; Mueller, R.L.; Valero, K.C.W.; Power, M.L.; Rodríguez, A.; O’Connell, L.A.; Young, N.D.; Skerratt, L.F. Comparative analysis of amphibian genomes: An emerging resource for basic and applied research. Mol. Ecol. Resour. 2024, 25, e14025. [Google Scholar] [CrossRef]
- Bredeson, J.V.; Mudd, A.B.; Medina-Ruiz, S.; Mitros, T.; Smith, O.K.; Miller, K.E.; Lyons, J.B.; Batra, S.S.; Park, J.; Berkoff, K.C.; et al. Conserved chromatin and repetitive patterns reveal slow genome evolution in frogs. Nat. Commun. 2024, 15, 579. [Google Scholar] [CrossRef]
- Adolfi, M.C.; Herpin, A.; Schartl, M. The replaceable master of sex determination: Bottom-up hypothesis revisited. Philos. Trans. R. Soc. B 2021, 376, 20200090. [Google Scholar] [CrossRef]
- Herpin, A.; Schartl, M. Dmrt1 genes at the crossroads: A widespread and central class of sexual development factors in fish. FEBS J. 2011, 278, 1010–1019. [Google Scholar] [CrossRef]
- Herpin, A.; Braasch, I.; Kraeussling, M.; Schmidt, C.; Thoma, E.C.; Nakamura, S.; Tanaka, M.; Schartl, M. Transcriptional rewiring of the sex determining Dmrt1 gene duplicate by transposable elements. PLoS Genet. 2010, 6, e1000844. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Sun, F.; Wan, Z.Y.; Yang, Z.; Tay, Y.X.; Lee, M.; Ye, B.; Wen, Y.; Meng, Z.; Fan, B.; et al. Transposon-induced epigenetic silencing in the x chromosome as a novel form of Dmrt1 expression regulation during sex determination in the fighting fish. BMC Biol. 2022, 20, 5. [Google Scholar] [CrossRef] [PubMed]
- Fisher, R.A. The evolution of dominance. Biol. Rev. 1931, 6, 345–368. [Google Scholar] [CrossRef]
- Nanda, I.; Kondo, M.; Hornung, U.; Asakawa, S.; Winkler, C.; Shimizu, A.; Shan, Z.; Haaf, T.; Shimizu, N.; Shima, A.; et al. A duplicated copy of DMRT1 in the sex-determining region of the y chromosome of the medaka, Oryzias latipes. Proc. Nat. Acad. Sci. USA 2002, 99, 11778–11783. [Google Scholar] [CrossRef]
- Geuverink, E.; Beukeboom, L.W. Phylogenetic distribution and evolutionary dynamics of the sex determination genes Doublesex and Transformer in Insects. Sex. Dev. 2014, 8, 38–49. [Google Scholar] [CrossRef]
- Suzuki, M.G.; Ohbayashi, F.; Mita, K.; Shimada, T. The mechanism of sex-specific splicing at the Doublesex gene is different between Drosophila melanogaster and Bombyx mori. Insect Biochem. Mol. Biol. 2001, 31, 1201–1211. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shinde, S.S.; Veltsos, P.; Ma, W.-J. Gene Duplication, Translocation, and Molecular Evolution of Dmrt1 and Related Sex-Determining Genes in Anurans. Biomolecules 2025, 15, 1306. https://doi.org/10.3390/biom15091306
Shinde SS, Veltsos P, Ma W-J. Gene Duplication, Translocation, and Molecular Evolution of Dmrt1 and Related Sex-Determining Genes in Anurans. Biomolecules. 2025; 15(9):1306. https://doi.org/10.3390/biom15091306
Chicago/Turabian StyleShinde, Sagar S., Paris Veltsos, and Wen-Juan Ma. 2025. "Gene Duplication, Translocation, and Molecular Evolution of Dmrt1 and Related Sex-Determining Genes in Anurans" Biomolecules 15, no. 9: 1306. https://doi.org/10.3390/biom15091306
APA StyleShinde, S. S., Veltsos, P., & Ma, W.-J. (2025). Gene Duplication, Translocation, and Molecular Evolution of Dmrt1 and Related Sex-Determining Genes in Anurans. Biomolecules, 15(9), 1306. https://doi.org/10.3390/biom15091306





