A Comprehensive Oncological Biomarker Framework Guiding Precision Medicine
Abstract
1. Introduction
2. Biomarkers Detection
2.1. Immunohistochemistry (IHC) and In Situ Hybridization (ISH)
2.2. Biosensors
2.3. Enzyme-Linked Immunosorbent Assay
2.4. Surface-Enhanced Raman Spectroscopy (SERS)
2.5. ATLAS-seq Technology
3. Types of Biomarkers in Cancer Immunotherapy
3.1. Diagnostic Biomarkers
3.2. Screening/Susceptibility Risk Markers
3.3. Predictive Markers
3.4. Pharmacodynamic Biomarkers
3.5. Preventive Biomarkers
4. Advancing Cancer Immunotherapy Through Companion Diagnostics
5. Gut Microbiota: A Paradigm Shift in Oncology
5.1. The Microbiome: Beyond Bacteria
5.2. Microbiota-Driven Immune Modulation
5.3. Microbiome Modulation as a Therapeutic Strategy
5.4. Biomarker Bank: Unveiling the Interplay of Cancer Immunotherapy and Gut Microbiome
5.5. Integrating Gut Microbiota into Comprehensive Oncological Frameworks
6. Liquid Biopsy: Promises and Hurdles
Implementation in Precision Medicine
7. Biomarker Integration in Combination Therapy Strategies
7.1. Pan-Cancer Approach: Overcoming Challenges of Cancer Heterogeneity and the Tumor Microenvironment
- Microsatellite instability-high (MSI-H): This is an FDA-approved biomarker associated with a high tumor mutational burden and increased neoantigen production. Pembrolizumab has shown efficacy in MSI-H tumors across various cancers [255].
- NTRK gene fusions: There are genetic alterations found across diverse cancers. Larotrectinib targets NTRK fusion-positive tumors with demonstrated clinical success in both adult and pediatric patients [256].
- RET Mutations and Fusions: RET mutations occur in cancers like NSCLC and colorectal cancer. Targeted therapies for RET fusion-positive tumors represent emerging pan-cancer treatment options [269].
- Tumor Mutational Burden (TMB): It predicts enhanced responses to ICIs like pembrolizumab. Its FDA approval for TMB-high solid tumors underscores its importance as a predictive biomarker [256].
7.2. Challenges and Opportunities in Combination Therapies
8. Advancing Biomarker Integration in Precision Oncology: Pathways, Challenges, and Innovations
8.1. Technological Innovations in Biomarker Research
8.2. Addressing Regulatory and Technical Challenges
8.3. Forward Paths and Upcoming Innovations
8.4. The Potential of Comprehensive Biomarker Panels
9. Conclusions
10. Outstanding Questions
- Mechanistic specificity: What molecular pathways enable Akkermansia and microbial metabolites (e.g., inosine, SCFAs) to selectively enhance antitumor CD8+ T cell activity while suppressing Treg-mediated immunosuppression?
- Biomarker validation: Can standardized microbial signatures (e.g., Akkermansia abundance, metabolite profiles) reliably predict ICI response across diverse cancer types and patient demographics?
- Therapeutic optimization: How can microbiome-modulating interventions (e.g., probiotics, fecal transplants) be temporally synchronized with ICI administration to maximize efficacy without exacerbating immune-related adverse events?
- Ecological dynamics: Does sustained ICI efficacy require persistent colonization of therapeutic microbial strains, or can transient microbiome remodeling induce durable immune reprogramming?
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AEs | Adverse Events |
| ASCO | American Society of Clinical Oncology |
| AgNPs | Silver Nanoparticles |
| AuNPs | Gold Nanoparticles |
| BRCA1 | Breast Cancer Gene 1 |
| BRCA2 | Breast Cancer Gene 2 |
| CAR-T | Chimeric Antigen Receptor T-cell |
| CAFs | Cancer-Associated Fibroblasts |
| cfDNA | Cell-Free DNA |
| CIMAC-CIDC | Cancer Immune Monitoring and Analysis Centers and Cancer Immunologic Data Commons |
| CPS | Combined Positive Score |
| CRC | Colorectal Cancer |
| CTCs | Circulating Tumor Cells |
| CTLA-4 | Cytotoxic T Lymphocyte Associated Antigen 4 |
| ctDNA | Circulating Tumor DNA |
| dMMR | Defective DNA Mismatch Repair |
| ELISA | Enzyme-Linked Immunosorbent Assay |
| FITs | Fecal Immunochemical Tests |
| FMT | Fecal Microbiota Transplantation |
| FDA | Food and Drug Administration |
| gFOBTs | Guaiac-Based Fecal Occult Blood Tests |
| GPCRs | G-Protein-Coupled Receptors |
| HLA | Human Leukocyte Antigen |
| HNSCC | Neck Squamous Cell Carcinoma |
| ICB | Immune Checkpoint Blockade |
| ICIs | Immune Checkpoint Inhibitors |
| IFN-γ | Interferon Gamma |
| IHC | Immunohistochemistry |
| IL-8 | Interleukin-8 |
| IrAEs | Immune-Related Events |
| LAG-3 | Lymphocyte-Activation Gene 3 |
| MDSCs | Myeloid-Derived Suppressor Cells |
| ML | Machine Learning |
| MMRd | Mismatch Repair Deficiency |
| MSI | Microsatellite Instability |
| MSI-H | High Microsatellite Instability |
| NCI | National Cancer Institute |
| NK | Natural killer |
| NSCLC | Non-Small Cell Lung Cancer |
| NTRK | Neurotrophic Tyrosine Receptor Kinase |
| ORR | Overall Response Rates |
| PD-1 | Programmed Death-1 |
| PD-L1 | Programmed Death-Ligand 1 |
| PEG | Polyethylene Glycol |
| PET | Positron Emission Tomography |
| PFS | Progression-Free Survival |
| PI3k | Phosphatidylinositol 3-kinase |
| PKB | Phosphorylated Protein Kinase B |
| SCFAs | Short-Chain Fatty Acids |
| SERS | Surface-Enhanced Raman Spectroscopy |
| TCGA | The Cancer Genome Atlas |
| TILs | Tumor-Infiltrating Lymphocytes |
| TMB | Tumor Mutational Burden |
| TME | Tumor Microenvironment |
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Emens, L.A.; Romero, P.J.; Anderson, A.C.; Bruno, T.C.; Capitini, C.M.; Collyar, D.; Gulley, J.L.; Hwu, P.; Posey, A.D., Jr.; Silk, A.W.; et al. Challenges and opportunities in cancer immunotherapy: A Society for Immunotherapy of Cancer (SITC) strategic vision. J. Immunother. Cancer 2024, 12, e009063. [Google Scholar] [CrossRef]
- Oiseth, S.J.; Aziz, M.S. Cancer immunotherapy: A brief review of the history, possibilities, and challenges ahead. J. Cancer Metastasis Treat. 2017, 3, 250–261. [Google Scholar] [CrossRef]
- Tan, S.; Li, D.; Zhu, X. Cancer immunotherapy: Pros, cons and beyond. Biomed. Pharmacother. 2020, 124, 109821. [Google Scholar] [CrossRef]
- Gopalakrishnan, V.; Spencer, C.N.; Nezi, L.; Reuben, A.; Andrews, M.; Karpinets, T.; Prieto, P.; Vicente, D.; Hoffman, K.; Wei, S.C. Gut microbiome modulates response to anti–PD-1 immunotherapy in melanoma patients. Science 2018, 359, 97–103. [Google Scholar] [CrossRef] [PubMed]
- Mokhtari, R.B.; Sambi, M.; Qorri, B.; Baluch, N.; Ashayeri, N.; Kumar, S.; Cheng, H.M.; Yeger, H.; Das, B.; Szewczuk, M.R. The Next-Generation of Combination Cancer Immunotherapy: Epigenetic Immunomodulators Transmogrify Immune Training to Enhance Immunotherapy. Cancers 2021, 13, 3596. [Google Scholar] [CrossRef] [PubMed]
- Schwabe, R.F.; Greten, T.F. Gut microbiome in HCC—Mechanisms, diagnosis and therapy. J. Hepatol. 2020, 72, 230–238. [Google Scholar] [CrossRef]
- Routy, B.; Le Chatelier, E.; Derosa, L.; Duong, C.P.M.; Alou, M.T.; Daillère, R.; Fluckiger, A.; Messaoudene, M.; Rauber, C.; Roberti, M.P.; et al. Gut microbiome influences efficacy of PD-1–based immunotherapy against epithelial tumors. Science 2018, 359, 91–97. [Google Scholar] [CrossRef]
- Haslam, A.; Gill, J.; Prasad, V. Estimation of the Percentage of US Patients With Cancer Who Are Eligible for Immune Checkpoint Inhibitor Drugs. JAMA Netw. Open 2020, 3, e200423. [Google Scholar] [CrossRef]
- Subbiah, V.; Solit, D.B.; Chan, T.A.; Kurzrock, R. The FDA approval of pembrolizumab for adult and pediatric patients with tumor mutational burden (TMB) ≥10: A decision centered on empowering patients and their physicians. Ann. Oncol. 2020, 31, 1115–1118. [Google Scholar] [CrossRef]
- Emens, L.A.; Ascierto, P.A.; Darcy, P.K.; Demaria, S.; Eggermont, A.M.M.; Redmond, W.L.; Seliger, B.; Marincola, F.M. Cancer immunotherapy: Opportunities and challenges in the rapidly evolving clinical landscape. Eur. J. Cancer 2017, 81, 116–129. [Google Scholar] [CrossRef]
- Jardim, D.L.; Goodman, A.; de Melo Gagliato, D.; Kurzrock, R. The Challenges of Tumor Mutational Burden as an Immunotherapy Biomarker. Cancer Cell 2021, 39, 154–173. [Google Scholar] [CrossRef]
- Herbst, R.S.; Baas, P.; Kim, D.-W.; Felip, E.; Pérez-Gracia, J.L.; Han, J.-Y.; Molina, J.; Kim, J.-H.; Arvis, C.D.; Ahn, M.-J.; et al. Pembrolizumab versus docetaxel for previously treated, PD-L1-positive, advanced non-small-cell lung cancer (KEYNOTE-010): A randomised controlled trial. Lancet 2016, 387, 1540–1550. [Google Scholar] [CrossRef]
- Tsimberidou, A.M.; Levit, L.A.; Schilsky, R.L.; Averbuch, S.D.; Chen, D.; Kirkwood, J.M.; McShane, L.M.; Sharon, E.; Mileham, K.F.; Postow, M.A. Trial Reporting in Immuno-Oncology (TRIO): An American Society of Clinical Oncology-Society for Immunotherapy of Cancer Statement. J. Clin. Oncol. 2019, 37, 72–80. [Google Scholar] [CrossRef]
- Kelly, R.J.; Ajani, J.A.; Kuzdzal, J.; Zander, T.; Van Cutsem, E.; Piessen, G.; Mendez, G.; Feliciano, J.; Motoyama, S.; Lièvre, A.; et al. Adjuvant Nivolumab in Resected Esophageal or Gastroesophageal Junction Cancer. N. Engl. J. Med. 2021, 384, 1191–1203. [Google Scholar] [CrossRef]
- Marabelle, A.; Le, D.T.; Ascierto, P.A.; Di Giacomo, A.M.; De Jesus-Acosta, A.; Delord, J.P.; Geva, R.; Gottfried, M.; Penel, N.; Hansen, A.R.; et al. Efficacy of Pembrolizumab in Patients With Noncolorectal High Microsatellite Instability/Mismatch Repair-Deficient Cancer: Results From the Phase II KEYNOTE-158 Study. J. Clin. Oncol. 2020, 38, 1–10. [Google Scholar] [CrossRef]
- Colombo, N.; Dubot, C.; Lorusso, D.; Caceres, M.V.; Hasegawa, K.; Shapira-Frommer, R.; Tewari, K.S.; Salman, P.; Hoyos Usta, E.; Yañez, E.; et al. Pembrolizumab for Persistent, Recurrent, or Metastatic Cervical Cancer. N. Engl. J. Med. 2021, 385, 1856–1867. [Google Scholar] [CrossRef]
- Powles, T.; Park, S.H.; Voog, E.; Caserta, C.; Valderrama, B.P.; Gurney, H.; Kalofonos, H.; Radulović, S.; Demey, W.; Ullén, A.; et al. Avelumab Maintenance Therapy for Advanced or Metastatic Urothelial Carcinoma. N. Engl. J. Med. 2020, 383, 1218–1230. [Google Scholar] [CrossRef]
- André, T.; Shiu, K.K.; Kim, T.W.; Jensen, B.V.; Jensen, L.H.; Punt, C.; Smith, D.; Garcia-Carbonero, R.; Benavides, M.; Gibbs, P.; et al. Pembrolizumab in Microsatellite-Instability-High Advanced Colorectal Cancer. N. Engl. J. Med. 2020, 383, 2207–2218. [Google Scholar] [CrossRef]
- Lefler, D.S.; Manobianco, S.A.; Bashir, B. Immunotherapy resistance in solid tumors: Mechanisms and potential solutions. Cancer Biol. Ther. 2024, 25, 2315655. [Google Scholar] [CrossRef]
- Yuan, Z.; Li, Y.; Zhang, S.; Wang, X.; Dou, H.; Yu, X.; Zhang, Z.; Yang, S.; Xiao, M. Extracellular matrix remodeling in tumor progression and immune escape: From mechanisms to treatments. Mol. Cancer 2023, 22, 48. [Google Scholar] [CrossRef]
- Koustas, E.; Sarantis, P.; Papavassiliou, A.G.; Karamouzis, M.V. The resistance mechanisms of checkpoint inhibitors in solid tumors. Biomolecules 2020, 10, 666. [Google Scholar] [CrossRef]
- Wang, Y.; Tong, Z.; Zhang, W.; Zhang, W.; Buzdin, A.; Mu, X.; Yan, Q.; Zhao, X.; Chang, H.H.; Duhon, M.; et al. FDA-Approved and Emerging Next Generation Predictive Biomarkers for Immune Checkpoint Inhibitors in Cancer Patients. Front. Oncol. 2021, 11, 683419. [Google Scholar] [CrossRef]
- Ling, S.P.; Ming, L.C.; Dhaliwal, J.S.; Gupta, M.; Ardianto, C.; Goh, K.W.; Hussain, Z.; Shafqat, N. Role of Immunotherapy in the Treatment of Cancer: A Systematic Review. Cancers 2022, 14, 5205. [Google Scholar] [CrossRef]
- Barber, F.D. Adverse events of oncologic immunotherapy and their management. Asia-Pac. J. Oncol. Nurs. 2019, 6, 212–226. [Google Scholar] [CrossRef] [PubMed]
- Brown, T.J.; Mamtani, R.; Bange, E.M. Immunotherapy adverse effects. JAMA Oncol. 2021, 7, 1908. [Google Scholar] [CrossRef] [PubMed]
- Olsen, T.A.; Zhuang, T.Z.; Caulfield, S.; Martini, D.J.; Brown, J.T.; Carthon, B.C.; Kucuk, O.; Harris, W.; Bilen, M.A.; Nazha, B. Advances in Knowledge and Management of Immune-Related Adverse Events in Cancer Immunotherapy. Front. Endocrinol. 2022, 13, 779915. [Google Scholar] [CrossRef]
- Zhou, C.; Peng, S.; Lin, A.; Jiang, A.; Peng, Y.; Gu, T.; Liu, Z.; Cheng, Q.; Zhang, J.; Luo, P. Psychiatric disorders associated with immune checkpoint inhibitors: A pharmacovigilance analysis of the FDA Adverse Event Reporting System (FAERS) database. EClinicalMedicine 2023, 59, 101967. [Google Scholar] [CrossRef]
- Cappelli, L.C.; Shah, A.A.; Bingham, C.O., 3rd. Cancer immunotherapy-induced rheumatic diseases emerge as new clinical entities. RMD Open 2016, 2, e000321. [Google Scholar] [CrossRef]
- Kumar, V.; Chaudhary, N.; Garg, M.; Floudas, C.S.; Soni, P.; Chandra, A.B. Current Diagnosis and Management of Immune Related Adverse Events (irAEs) Induced by Immune Checkpoint Inhibitor Therapy. Front. Pharmacol. 2017, 8, 49. [Google Scholar] [CrossRef]
- Yan, T.; Yu, L.; Zhang, J.; Chen, Y.; Fu, Y.; Tang, J.; Liao, D. Achilles’ Heel of currently approved immune checkpoint inhibitors: Immune related adverse events. Front. Immunol. 2024, 15, 1292122. [Google Scholar] [CrossRef]
- Lau, S.C.M.; Leighl, N.B. Hyperprogressive disease with immunotherapy: New directions. J. Thorac. Dis. 2019, 11, S1877–S1880. [Google Scholar] [CrossRef]
- Smithy, J.W.; Faleck, D.M.; Postow, M.A. Facts and Hopes in Prediction, Diagnosis, and Treatment of Immune-Related Adverse Events. Clin. Cancer Res. 2022, 28, 1250–1257. [Google Scholar] [CrossRef]
- Yuan, M.; Zhang, C.; Von Feilitzen, K.; Zwahlen, M.; Shi, M.; Li, X.; Yang, H.; Song, X.; Turkez, H.; Uhlén, M. The Human Pathology Atlas for deciphering the prognostic features of human cancers. EBioMedicine 2025, 111, 105495. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, S.; Sharma, G.; Karmakar, S.; Banerjee, S. Multi-OMICS approaches in cancer biology: New era in cancer therapy. Biochim. Biophys. Acta Mol. Basis Dis. 2024, 1870, 167120. [Google Scholar] [CrossRef] [PubMed]
- Liang, A.; Kong, Y.; Chen, Z.; Qiu, Y.; Wu, Y.; Zhu, X.; Li, Z. Advancements and applications of single-cell multi-omics techniques in cancer research: Unveiling heterogeneity and paving the way for precision therapeutics. Biochem. Biophys. Reports 2024, 37, 101589. [Google Scholar] [CrossRef]
- Acharya, D.; Mukhopadhyay, A. A comprehensive review of machine learning techniques for multi-omics data integration: Challenges and applications in precision oncology. Brief. Funct. Genom. 2024, 23, 549–560. [Google Scholar] [CrossRef]
- Shi, H.-h.; Mugaanyi, J.; Lu, C.; Li, Y.; Huang, J.; Dai, L. A paradigm shift in cancer research based on integrative multi-omics approaches: Glutaminase serves as a pioneering cuproptosis-related gene in pan-cancer. BMC Women’s Health 2024, 24, 213. [Google Scholar] [CrossRef] [PubMed]
- Rehman, Z.U.; Ahmad Fauzi, M.F.; Wan Ahmad, W.S.H.M.; Abas, F.S.; Cheah, P.L.; Chiew, S.F.; Looi, L.-M. Review of in situ hybridization (ISH) stain images using computational techniques. Diagnostics 2024, 14, 2089. [Google Scholar] [CrossRef]
- Eskuri, M.; Birkman, E.M.; Kauppila, J.H. Gastric cancer molecular classification based on immunohistochemistry and in-situ hybridisation and mortality. Histopathology 2024, 85, 327–337. [Google Scholar] [CrossRef]
- Khanmohammadi, A.; Aghaie, A.; Vahedi, E.; Qazvini, A.; Ghanei, M.; Afkhami, A.; Hajian, A.; Bagheri, H. Electrochemical biosensors for the detection of lung cancer biomarkers: A review. Talanta 2020, 206, 120251. [Google Scholar] [CrossRef]
- El Aamri, M.; Yammouri, G.; Mohammadi, H.; Amine, A.; Korri-Youssoufi, H. Electrochemical Biosensors for Detection of MicroRNA as a Cancer Biomarker: Pros and Cons. Biosensors 2020, 10, 186. [Google Scholar] [CrossRef]
- Yuan, X.; Lin, B.; Liu, T.; Zhang, W.; Chu, Z.; Gu, X.; Ma, Z.; Jin, W. Recent Progress on Nanomaterial-Facilitated Electrochemical Strategies for Cancer Diagnosis. Adv. Healthc Mater. 2023, 12, e2203029. [Google Scholar] [CrossRef]
- Kivrak, E.; Kara, P. Simultaneous detection of ovarian cancer related miRNA biomarkers with carboxylated graphene oxide modified electrochemical biosensor platform. Bioelectrochemistry 2025, 161, 108806. [Google Scholar] [CrossRef] [PubMed]
- Mahalakshmi, D.; Nandhini, J.; Meenaloshini, G.; Karthikeyan, E.; Karthik, K.; Sujaritha, J.; Vandhana, V.; Ragavendran, C. Graphene Nanomaterial-Based Electrochemical Biosensors for Salivary Biomarker Detection: A Translational Approach to Oral Cancer Diagnostics. Nano TransMed 2025, 4, 100073. [Google Scholar] [CrossRef]
- Zouleh, R.S.; Rahimnejad, M.; Najafpour-Darzi, G.; Sabour, D. Design of a microneedle-based enzyme biosensor using a simple and cost-effective electrochemical strategy to monitor superoxide anion released from cancer cells. Anal. Biochem. 2025, 697, 115710. [Google Scholar] [CrossRef]
- Kanagavalli, P.; Elkaffas, R.A.; Mohideen, M.I.H.; Eissa, S. Electrochemical immunosensor for the predictive cancer biomarker SLFN11 using reduced graphene oxide/MIL-101 (Cr)-NH2 composite. Int. J. Biol. Macromol. 2025, 285, 138174. [Google Scholar] [CrossRef]
- Öndeş, B.; Kilimci, U.; Uygun, M.; Uygun, D.A. Determination of carcinoembryonic antigen (CEA) by label-free electrochemical immunosensor using functionalized boron nitride nanosheets. Bioelectrochemistry 2024, 157, 108676. [Google Scholar] [CrossRef]
- Aydin, S. A short history, principles, and types of ELISA, and our laboratory experience with peptide/protein analyses using ELISA. Peptides 2015, 72, 4–15. [Google Scholar] [CrossRef]
- Shah, K.; Maghsoudlou, P. Enzyme-linked immunosorbent assay (ELISA): The basics. Br. J. Hosp. Med. 2016, 77, C98–C101. [Google Scholar] [CrossRef]
- Kim, D.; Herr, A.E. Protein immobilization techniques for microfluidic assays. Biomicrofluidics 2013, 7, 41501. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.E.; Seo, J.H.; Kim, C.S.; Kwon, Y.; Ha, J.H.; Choi, S.S.; Cha, H.J. A comparative study on antibody immobilization strategies onto solid surface. Korean J. Chem. Eng. 2013, 30, 1934–1938. [Google Scholar] [CrossRef]
- Hirsch, J.D.; Eslamizar, L.; Filanoski, B.J.; Malekzadeh, N.; Haugland, R.P.; Beechem, J.M.; Haugland, R.P. Easily reversible desthiobiotin binding to streptavidin, avidin, and other biotin-binding proteins: Uses for protein labeling, detection, and isolation. Anal. Biochem. 2002, 308, 343–357. [Google Scholar] [CrossRef]
- Dobaño, C.; Santano, R.; Jiménez, A.; Vidal, M.; Chi, J.; Melero, N.R.; Popovic, M.; López-Aladid, R.; Fernández-Barat, L.; Tortajada, M. Immunogenicity and crossreactivity of antibodies to the nucleocapsid protein of SARS-CoV-2: Utility and limitations in seroprevalence and immunity studies. Transl. Res. 2021, 232, 60–74. [Google Scholar] [CrossRef]
- Nadeem, R.; Barakat, A.B.; Bahgat, M.M. Cross-reaction between mouse and rat immunoglobulin G: Does it matter in sandwich ELISA? J. Genet. Eng. Biotechnol. 2021, 19, 1–7. [Google Scholar] [CrossRef]
- Wang, X.; Zhao, M.; Nolte, D.D. Prostate-specific antigen immunoassays on the BioCD. Anal. Bioanal. Chem. 2009, 393, 1151–1156. [Google Scholar] [CrossRef]
- Wang, X.; Zhao, M.; Nolte, D.D.; Ratliff, T.L. Prostate specific antigen detection in patient sera by fluorescence-free BioCD protein array. Biosens. Bioelectron. 2011, 26, 1871–1875. [Google Scholar] [CrossRef][Green Version]
- Simpson, K.L.; Whetton, A.D.; Dive, C. Quantitative mass spectrometry-based techniques for clinical use: Biomarker identification and quantification. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2009, 877, 1240–1249. [Google Scholar] [CrossRef]
- Hawkridge, A.M.; Muddiman, D.C. Mass spectrometry-based biomarker discovery: Toward a global proteome index of individuality. Annu. Rev. Anal. Chem. 2009, 2, 265–277. [Google Scholar] [CrossRef] [PubMed]
- Plou, J.; Valera, P.S.; García, I.; de Albuquerque, C.D.L.; Carracedo, A.; Liz-Marzán, L.M. Prospects of Surface-Enhanced Raman Spectroscopy for Biomarker Monitoring toward Precision Medicine. ACS Photonics 2022, 9, 333–350. [Google Scholar] [CrossRef]
- Zong, C.; Xu, M.; Xu, L.-J.; Wei, T.; Ma, X.; Zheng, X.-S.; Hu, R.; Ren, B. Surface-enhanced Raman spectroscopy for bioanalysis: Reliability and challenges. Chem. Rev. 2018, 118, 4946–4980. [Google Scholar] [CrossRef]
- Rodal-Cedeira, S.; Vazquez-Arias, A.; Bodelon, G.; Skorikov, A.; Nunez-Sanchez, S.; Laporta, A.; Polavarapu, L.; Bals, S.; Liz-Marzan, L.M.; Perez-Juste, J. An expanded surface-enhanced Raman scattering tags library by combinatorial encapsulation of reporter molecules in metal nanoshells. ACS Nano 2020, 14, 14655–14664. [Google Scholar] [CrossRef]
- Xi, W.; Shrestha, B.K.; Haes, A.J. Promoting intra-and intermolecular interactions in surface-enhanced Raman scattering. Anal. Chem. 2017, 90, 128–143. [Google Scholar] [CrossRef]
- Shen, W.; Lin, X.; Jiang, C.; Li, C.; Lin, H.; Huang, J.; Wang, S.; Liu, G.; Yan, X.; Zhong, Q. Reliable quantitative SERS analysis facilitated by core—Shell nanoparticles with embedded internal standards. Angew. Chem. Int. Ed. 2015, 54, 7308–7312. [Google Scholar] [CrossRef]
- Zheng, Y.; Thai, T.; Reineck, P.; Qiu, L.; Guo, Y.; Bach, U. DNA-directed self-assembly of core-satellite plasmonic nanostructures: A highly sensitive and reproducible near-IR SERS sensor. Adv. Funct. Mater. 2013, 23, 1519–1526. [Google Scholar] [CrossRef]
- Choi, N.; Dang, H.; Das, A.; Sim, M.S.; Chung, I.Y.; Choo, J. SERS biosensors for ultrasensitive detection of multiple biomarkers expressed in cancer cells. Biosens. Bioelectron. 2020, 164, 112326. [Google Scholar] [CrossRef]
- Liu, X.; Jia, Y.; Zheng, C. Recent progress in Surface-Enhanced Raman Spectroscopy detection of biomarkers in liquid biopsy for breast cancer. Front. Oncol. 2024, 14, 1400498. [Google Scholar] [CrossRef]
- Wu, Y.; Wang, Y.; Mo, T.; Liu, Q. Surface-Enhanced Raman Scattering-based strategies for tumor markers detection: A review. Talanta 2024, 280, 126717. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Yang, G.; Zhang, S.; Ren, H. Advancing clinical cancer care: Unveiling the power of surface-enhanced Raman spectroscopy. J. Raman Spectrosc. 2024, 55, 429–444. [Google Scholar] [CrossRef]
- Huang, Y.; Wang, X.; Lai, K.; Fan, Y.; Rasco, B.A. Trace analysis of organic compounds in foods with surface-enhanced Raman spectroscopy: Methodology, progress, and challenges. Compr. Rev. Food Sci. Food Saf. 2020, 19, 622–642. [Google Scholar] [CrossRef] [PubMed]
- Luo, S.; Notaro, A.; Lin, L. ATLAS-seq: A microfluidic single-cell TCR screen for antigen-reactive TCRs. Nat. Commun. 2025, 16, 216. [Google Scholar] [CrossRef]
- Group, F.-N.B.W. BEST (Biomarkers, EndpointS, and other Tools) Resource. Available online: https://www.ncbi.nlm.nih.gov/books/NBK326791/ (accessed on 25 August 2025).
- Califf, R.M. Biomarker definitions and their applications. Exp. Biol. Med. 2018, 243, 213–221. [Google Scholar] [CrossRef]
- Gu, X.; Wei, S.; Lv, X. Circulating tumor cells: From new biological insights to clinical practice. Signal Transduct. Target. Ther. 2024, 9, 226. [Google Scholar] [CrossRef]
- García-Silva, S.; Peinado, H. Mechanisms of lymph node metastasis: An extracellular vesicle perspective. Eur. J. Cell Biol. 2024, 103, 151447. [Google Scholar] [CrossRef]
- Jerabkova-Roda, K.; Dupas, A.; Osmani, N.; Hyenne, V.; Goetz, J.G. Circulating extracellular vesicles and tumor cells: Sticky partners in metastasis. Trends Cancer 2022, 8, 799–805. [Google Scholar] [CrossRef]
- Muinao, T.; Boruah, H.P.D.; Pal, M. Multi-biomarker panel signature as the key to diagnosis of ovarian cancer. Heliyon 2019, 5, e02826. [Google Scholar] [CrossRef]
- Rossi, G.; Russo, A.; Tagliamento, M.; Tuzi, A.; Nigro, O.; Vallome, G.; Sini, C.; Grassi, M.; Dal Bello, M.G.; Coco, S.; et al. Precision Medicine for NSCLC in the Era of Immunotherapy: New Biomarkers to Select the Most Suitable Treatment or the Most Suitable Patient. Cancers 2020, 12, 1125. [Google Scholar] [CrossRef]
- Garon, E.B.; Hellmann, M.D.; Rizvi, N.A.; Carcereny, E.; Leighl, N.B.; Ahn, M.J.; Eder, J.P.; Balmanoukian, A.S.; Aggarwal, C.; Horn, L.; et al. Five-Year Overall Survival for Patients With Advanced Non-Small-Cell Lung Cancer Treated With Pembrolizumab: Results From the Phase I KEYNOTE-001 Study. J. Clin. Oncol. 2019, 37, 2518–2527. [Google Scholar] [CrossRef]
- Shitara, K.; Van Cutsem, E.; Bang, Y.J.; Fuchs, C.; Wyrwicz, L.; Lee, K.W.; Kudaba, I.; Garrido, M.; Chung, H.C.; Lee, J.; et al. Efficacy and Safety of Pembrolizumab or Pembrolizumab Plus Chemotherapy vs Chemotherapy Alone for Patients With First-line, Advanced Gastric Cancer: The KEYNOTE-062 Phase 3 Randomized Clinical Trial. JAMA Oncol. 2020, 6, 1571–1580. [Google Scholar] [CrossRef]
- Emens, L.A.; Molinero, L.; Loi, S.; Rugo, H.S.; Schneeweiss, A.; Dieras, V.; Iwata, H.; Barrios, C.H.; Nechaeva, M.; Nguyen-Duc, A.; et al. Atezolizumab and nab-Paclitaxel in Advanced Triple-Negative Breast Cancer: Biomarker Evaluation of the IMpassion130 Study. J. Natl. Cancer Inst. 2021, 113, 1005–1016. [Google Scholar] [CrossRef]
- Song, R.; Liu, F.; Ping, Y.; Zhang, Y.; Wang, L. Potential non-invasive biomarkers in tumor immune checkpoint inhibitor therapy: Response and prognosis prediction. Biomark. Res. 2023, 11, 57. [Google Scholar] [CrossRef]
- Kong, J.; Ha, D.; Lee, J.; Kim, I.; Park, M.; Im, S.H.; Shin, K.; Kim, S. Network-based machine learning approach to predict immunotherapy response in cancer patients. Nat. Commun. 2022, 13, 3703. [Google Scholar] [CrossRef] [PubMed]
- Boicean, A.; Boeras, I.; Birsan, S.; Ichim, C.; Todor, S.B.; Onisor, D.M.; Brusnic, O.; Bacila, C.; Dura, H.; Roman-Filip, C. In Pursuit of Novel Markers: Unraveling the Potential of miR-106, CEA and CA 19-9 in Gastric Adenocarcinoma Diagnosis and Staging. Int. J. Mol. Sci. 2024, 25, 7898. [Google Scholar] [CrossRef]
- Kildusiene, I.; Dulskas, A.; Smailyte, G. Value of combined serum CEA, CA72-4, and CA19-9 marker detection in diagnosis of colorectal cancer. Tech. Coloproctol 2024, 28, 33. [Google Scholar] [CrossRef]
- Kamada, T.; Ohdaira, H.; Takahashi, J.; Aida, T.; Nakashima, K.; Ito, E.; Hata, T.; Yoshida, M.; Eto, K.; Suzuki, Y. Novel tumor marker index using carcinoembryonic antigen and carbohydrate antigen 19-9 is a significant prognostic factor for resectable colorectal cancer. Sci. Rep. 2024, 14, 4192. [Google Scholar] [CrossRef]
- Xu, C.; Jun, E.; Okugawa, Y.; Toiyama, Y.; Borazanci, E.; Bolton, J.; Taketomi, A.; Kim, S.C.; Shang, D.; Von Hoff, D. A circulating panel of circRNA biomarkers for the noninvasive and early detection of pancreatic ductal adenocarcinoma. Gastroenterology 2024, 166, 178–190.e116. [Google Scholar] [CrossRef]
- Heydari, R.; Abdollahpour-Alitappeh, M.; Shekari, F.; Meyfour, A. Emerging Role of Extracellular Vesicles in Biomarking the Gastrointestinal Diseases. Expert Rev. Mol. Diagn. 2021, 21, 939–962. [Google Scholar] [CrossRef]
- Zhou, Y.; Tao, L.; Qiu, J.; Xu, J.; Yang, X.; Zhang, Y.; Tian, X.; Guan, X.; Cen, X.; Zhao, Y. Tumor biomarkers for diagnosis, prognosis and targeted therapy. Signal Transduct. Target. Ther. 2024, 9, 132. [Google Scholar] [CrossRef] [PubMed]
- Ye, Q.; Ling, S.; Zheng, S.; Xu, X. Liquid biopsy in hepatocellular carcinoma: Circulating tumor cells and circulating tumor DNA. Mol. Cancer 2019, 18, 114. [Google Scholar] [CrossRef] [PubMed]
- Kapoor, A.; Bayat Mokhtari, R.; Sonti, S.S.; Patel, R.; George, A.; Attwood, K.; Iyer, R.; Chakraborty, S. Circulatory Agrin Serves as a Prognostic Indicator for Hepatocellular Carcinoma. Cancers 2024, 16, 2719. [Google Scholar] [CrossRef]
- Jia, L.; Li, G.; Ma, N.; Zhang, A.; Zhou, Y.; Ren, L.; Dong, D. Soluble POSTN is a novel biomarker complementing CA153 and CEA for breast cancer diagnosis and metastasis prediction. BMC Cancer 2022, 22, 760. [Google Scholar] [CrossRef]
- Burton, H.; Chowdhury, S.; Dent, T.; Hall, A.; Pashayan, N.; Pharoah, P. Public health implications from COGS and potential for risk stratification and screening. Nat. Genet. 2013, 45, 349–351. [Google Scholar] [CrossRef]
- Przybytkowski, E.; Davis, T.; Hosny, A.; Eismann, J.; Matulonis, U.A.; Wulf, G.M.; Nabavi, S. An immune-centric exploration of BRCA1 and BRCA2 germline mutation related breast and ovarian cancers. BMC Cancer 2020, 20, 197. [Google Scholar] [CrossRef]
- Chowell, D.; Morris, L.G.T.; Grigg, C.M.; Weber, J.K.; Samstein, R.M.; Makarov, V.; Kuo, F.; Kendall, S.M.; Requena, D.; Riaz, N.; et al. Patient HLA class I genotype influences cancer response to checkpoint blockade immunotherapy. Science 2018, 359, 582–587. [Google Scholar] [CrossRef] [PubMed]
- Zwezerijnen-Jiwa, F.H.; Sivov, H.; Paizs, P.; Zafeiropoulou, K.; Kinross, J. A systematic review of microbiome-derived biomarkers for early colorectal cancer detection. Neoplasia 2023, 36, 100868. [Google Scholar] [CrossRef]
- Wölffer, M.; Battke, F.; Schulze, M.; Feldhahn, M.; Flatz, L.; Martus, P.; Forschner, A. Biomarkers associated with immune-related adverse events under checkpoint inhibitors in metastatic melanoma. Cancers 2022, 14, 302. [Google Scholar] [CrossRef]
- Duffy, M.J.; Diamandis, E.P.; Crown, J. Circulating tumor DNA (ctDNA) as a pan-cancer screening test: Is it finally on the horizon? Clin. Chem. Lab. Med. 2021, 59, 1353–1361. [Google Scholar] [CrossRef] [PubMed]
- Kohaar, I.; Hodges, N.A.; Srivastava, S. Biomarkers in Cancer Screening: Promises and Challenges in Cancer Early Detection. Hematol. Oncol. Clin. North. Am. 2024, 38, 869–888. [Google Scholar] [CrossRef]
- Tarighati, E.; Keivan, H.; Mahani, H. A review of prognostic and predictive biomarkers in breast cancer. Clin. Exp. Med. 2023, 23, 1–16. [Google Scholar] [CrossRef]
- Satgunaseelan, L.; Sy, J.; Shivalingam, B.; Sim, H.-W.; Alexander, K.L.; Buckland, M.E. Prognostic and predictive biomarkers in central nervous system tumours: The molecular state of play. Pathology 2024, 56, 158–169. [Google Scholar] [CrossRef] [PubMed]
- Glembocki, A.I.; Somers, G.R. Prognostic and predictive biomarkers in paediatric solid tumours. Pathology 2024, 56, 283–296. [Google Scholar] [CrossRef]
- Larkin, J.; Chiarion-Sileni, V.; Gonzalez, R.; Grob, J.J.; Rutkowski, P.; Lao, C.D.; Cowey, C.L.; Schadendorf, D.; Wagstaff, J.; Dummer, R.; et al. Five-Year Survival with Combined Nivolumab and Ipilimumab in Advanced Melanoma. N. Engl. J. Med. 2019, 381, 1535–1546. [Google Scholar] [CrossRef]
- Hellmann, M.D.; Paz-Ares, L.; Bernabe Caro, R.; Zurawski, B.; Kim, S.-W.; Carcereny Costa, E.; Park, K.; Alexandru, A.; Lupinacci, L.; de la Mora Jimenez, E.; et al. Nivolumab plus Ipilimumab in Advanced Non–Small-Cell Lung Cancer. N. Engl. J. Med. 2019, 381, 2020–2031. [Google Scholar] [CrossRef] [PubMed]
- Motzer, R.J.; Penkov, K.; Haanen, J.; Rini, B.; Albiges, L.; Campbell, M.T.; Venugopal, B.; Kollmannsberger, C.; Negrier, S.; Uemura, M.; et al. Avelumab plus Axitinib versus Sunitinib for Advanced Renal-Cell Carcinoma. N. Engl. J. Med. 2019, 380, 1103–1115. [Google Scholar] [CrossRef]
- Reck, M.; Rodríguez-Abreu, D.; Robinson, A.G.; Hui, R.; Csőszi, T.; Fülöp, A.; Gottfried, M.; Peled, N.; Tafreshi, A.; Cuffe, S.; et al. Pembrolizumab versus Chemotherapy for PD-L1-Positive Non-Small-Cell Lung Cancer. N. Engl. J. Med. 2016, 375, 1823–1833. [Google Scholar] [CrossRef] [PubMed]
- Maiorano, B.A.; Di Maio, M.; Cerbone, L.; Maiello, E.; Procopio, G.; Roviello, G.; Group, M. Significance of PD-L1 in Metastatic Urothelial Carcinoma Treated With Immune Checkpoint Inhibitors: A Systematic Review and Meta-Analysis. JAMA Netw. Open 2024, 7, e241215. [Google Scholar] [CrossRef]
- Yamaguchi, H.; Hsu, J.M.; Sun, L.; Wang, S.C.; Hung, M.C. Advances and prospects of biomarkers for immune checkpoint inhibitors. Cell Rep. Med. 2024, 5, 101621. [Google Scholar] [CrossRef]
- Herbst, R.S.; Giaccone, G.; de Marinis, F.; Reinmuth, N.; Vergnenegre, A.; Barrios, C.H.; Morise, M.; Felip, E.; Andric, Z.; Geater, S.; et al. Atezolizumab for First-Line Treatment of PD-L1-Selected Patients with NSCLC. N. Engl. J. Med. 2020, 383, 1328–1339. [Google Scholar] [CrossRef]
- Anagnostou, V.; Landon, B.V.; Medina, J.E.; Forde, P.; Velculescu, V.E. Translating the evolving molecular landscape of tumors to biomarkers of response for cancer immunotherapy. Sci. Transl. Med. 2022, 14, eabo3958. [Google Scholar] [CrossRef] [PubMed]
- Batis, N.; Brooks, J.M.; Payne, K.; Sharma, N.; Nankivell, P.; Mehanna, H. Lack of predictive tools for conventional and targeted cancer therapy: Barriers to biomarker development and clinical translation. Adv. Drug Deliv. Rev. 2021, 176, 113854. [Google Scholar] [CrossRef]
- Wang, T.; Denman, D.; Bacot, S.M.; Feldman, G.M. Challenges and the evolving landscape of assessing blood-based PD-L1 expression as a biomarker for anti-PD-(L) 1 immunotherapy. Biomedicines 2022, 10, 1181. [Google Scholar] [CrossRef]
- Liu, Z.; Li, Z.; Qu, J.; Zhang, R.; Zhou, X.; Li, L.; Sun, K.; Tang, Z.; Jiang, H.; Li, H.; et al. Radiomics of Multiparametric MRI for Pretreatment Prediction of Pathologic Complete Response to Neoadjuvant Chemotherapy in Breast Cancer: A Multicenter Study. Clin. Cancer Res. 2019, 25, 3538–3547. [Google Scholar] [CrossRef]
- Addeo, A.; Friedlaender, A.; Banna, G.L.; Weiss, G.J. TMB or not TMB as a biomarker: That is the question. Crit. Rev. Oncol. Hematol. 2021, 163, 103374. [Google Scholar] [CrossRef]
- Merino, D.M.; McShane, L.M.; Fabrizio, D.; Funari, V.; Chen, S.J.; White, J.R.; Wenz, P.; Baden, J.; Barrett, J.C.; Chaudhary, R.; et al. Establishing guidelines to harmonize tumor mutational burden (TMB): In silico assessment of variation in TMB quantification across diagnostic platforms: Phase I of the Friends of Cancer Research TMB Harmonization Project. J. Immunother. Cancer 2020, 8, e000147. [Google Scholar] [CrossRef]
- Shi, S.; Wang, Y.; Liu, Y.; Yang, Y.; Kong, J.; Gao, S.; Cui, H.; Huangfu, L.; Sun, X.; Li, Z. Predictive value of PD-L1 and TMB for short-term efficacy prognosis in non-small cell lung cancer and construction of prediction models. Front. Oncol. 2024, 14, 1342262. [Google Scholar] [CrossRef]
- Hellmann, M.D.; Ciuleanu, T.E.; Pluzanski, A.; Lee, J.S.; Otterson, G.A.; Audigier-Valette, C.; Minenza, E.; Linardou, H.; Burgers, S.; Salman, P.; et al. Nivolumab plus Ipilimumab in Lung Cancer with a High Tumor Mutational Burden. N. Engl. J. Med. 2018, 378, 2093–2104. [Google Scholar] [CrossRef]
- Dora, D.; Bokhari, S.M.Z.; Aloss, K.; Takacs, P.; Desnoix, J.Z.; Szklenárik, G.; Hurley, P.D.; Lohinai, Z. Implication of the Gut Microbiome and Microbial-Derived Metabolites in Immune-Related Adverse Events: Emergence of Novel Biomarkers for Cancer Immunotherapy. Int. J. Mol. Sci. 2023, 24, 2769. [Google Scholar] [CrossRef]
- Chung, M.W.; Kim, M.J.; Won, E.J.; Lee, Y.J.; Yun, Y.W.; Cho, S.B.; Joo, Y.E.; Hwang, J.E.; Bae, W.K.; Chung, I.J.; et al. Gut microbiome composition can predict the response to nivolumab in advanced hepatocellular carcinoma patients. World J. Gastroenterol. 2021, 27, 7340–7349. [Google Scholar] [CrossRef]
- Nomura, M.; Nagatomo, R.; Doi, K.; Shimizu, J.; Baba, K.; Saito, T.; Matsumoto, S.; Inoue, K.; Muto, M. Association of short-chain fatty acids in the gut microbiome with clinical response to treatment with nivolumab or pembrolizumab in patients with solid cancer tumors. JAMA Netw. Open 2020, 3, e202895. [Google Scholar] [CrossRef]
- Sun, J.-Y.; Zhang, D.; Wu, S.; Xu, M.; Zhou, X.; Lu, X.-J.; Ji, J. Resistance to PD-1/PD-L1 blockade cancer immunotherapy: Mechanisms, predictive factors, and future perspectives. Biomark. Res. 2020, 8, 35. [Google Scholar] [CrossRef]
- Ren, D.; Hua, Y.; Yu, B.; Ye, X.; He, Z.; Li, C.; Wang, J.; Mo, Y.; Wei, X.; Chen, Y. Predictive biomarkers and mechanisms underlying resistance to PD1/PD-L1 blockade cancer immunotherapy. Mol. Cancer 2020, 19, 19. [Google Scholar] [CrossRef]
- Diskin, B.; Adam, S.; Cassini, M.F.; Sanchez, G.; Liria, M.; Aykut, B.; Buttar, C.; Li, E.; Sundberg, B.; Salas, R.D. PD-L1 engagement on T cells promotes self-tolerance and suppression of neighboring macrophages and effector T cells in cancer. Nat. Immunol. 2020, 21, 442–454. [Google Scholar] [CrossRef]
- Wang, Z.; Wu, X. Study and analysis of antitumor resistance mechanism of PD1/PD-L1 immune checkpoint blocker. Cancer Med. 2020, 9, 8086–8121. [Google Scholar] [CrossRef]
- John, P.; Pulanco, M.C.; Galbo Jr, P.M.; Wei, Y.; Ohaegbulam, K.C.; Zheng, D.; Zang, X. The immune checkpoint B7x expands tumor-infiltrating Tregs and promotes resistance to anti-CTLA-4 therapy. Nat. Commun. 2022, 13, 2506. [Google Scholar] [CrossRef]
- Zhang, H.; Dai, Z.; Wu, W.; Wang, Z.; Zhang, N.; Zhang, L.; Zeng, W.-J.; Liu, Z.; Cheng, Q. Regulatory mechanisms of immune checkpoints PD-L1 and CTLA-4 in cancer. J. Exp. Clin. Cancer Res. 2021, 40, 184. [Google Scholar] [CrossRef]
- He, Y.; Sun, M.M.; Zhang, G.G.; Yang, J.; Chen, K.S.; Xu, W.W.; Li, B. Targeting PI3K/Akt signal transduction for cancer therapy. Signal Transduct. Target. Ther. 2021, 6, 425. [Google Scholar] [CrossRef]
- Conciatori, F.; Bazzichetto, C.; Falcone, I.; Ciuffreda, L.; Ferretti, G.; Vari, S.; Ferraresi, V.; Cognetti, F.; Milella, M. PTEN function at the interface between cancer and tumor microenvironment: Implications for response to immunotherapy. Int. J. Mol. Sci. 2020, 21, 5337. [Google Scholar] [CrossRef]
- O’Donnell, J.S.; Massi, D.; Teng, M.W.; Mandala, M. PI3K-AKT-mTOR inhibition in cancer immunotherapy, redux. Semin. Cancer Biol. 2018, 48, 91–103. [Google Scholar] [CrossRef] [PubMed]
- van den Heuvel, M.; Holdenrieder, S.; Schuurbiers, M.; Cigoianu, D.; Trulson, I.; van Rossum, H.; Lang, D. Serum tumor markers for response prediction and monitoring of advanced lung cancer: A review focusing on immunotherapy and targeted therapies. Tumor Biol. 2024, 46, S233–S268. [Google Scholar] [CrossRef]
- Liao, G.J.; Clark, A.S.; Schubert, E.K.; Mankoff, D.A. 18F-Fluoroestradiol PET: Current Status and Potential Future Clinical Applications. J. Nucl. Med. 2016, 57, 1269–1275. [Google Scholar] [CrossRef]
- Morschhauser, F.; Machiels, J.P.; Salles, G.; Rottey, S.; Rule, S.A.J.; Cunningham, D.; Peyrade, F.; Fruchart, C.; Arkenau, H.T.; Genvresse, I.; et al. On-Target Pharmacodynamic Activity of the PI3K Inhibitor Copanlisib in Paired Biopsies from Patients with Malignant Lymphoma and Advanced Solid Tumors. Mol. Cancer Ther. 2020, 19, 468–478. [Google Scholar] [CrossRef]
- Lambert, S.L.; Zhang, C.; Guo, C.; Turan, T.; Masica, D.L.; Englert, S.; Fang, Y.; Sheridan, J.; McLaughlin, R.T.; Tribouley, C.; et al. Association of Baseline and Pharmacodynamic Biomarkers With Outcomes in Patients Treated With the PD-1 Inhibitor Budigalimab. J. Immunother. 2022, 45, 167–179. [Google Scholar] [CrossRef]
- Salawu, A.; Hernando-Calvo, A.; Chen, R.Y.; Araujo, D.V.; Oliva, M.; Liu, Z.A.; Siu, L.L. Impact of pharmacodynamic biomarkers in immuno-oncology phase 1 clinical trials. Eur. J. Cancer 2022, 173, 167–177. [Google Scholar] [CrossRef]
- Hou, M.; Yu, Q.Q.; Yang, L.; Zhao, H.; Jiang, P.; Qin, L.; Zhang, Q. The role of short-chain fatty acid metabolism in the pathogenesis, diagnosis and treatment of cancer. Front. Oncol. 2024, 14, 1451045. [Google Scholar] [CrossRef]
- Li, S.; Duan, Y.; Luo, S.; Zhou, F.; Wu, Q.; Lu, Z. Short-chain fatty acids and cancer. Trends Cancer 2025, 11, 154–168. [Google Scholar] [CrossRef]
- Ediriweera, M.K. Fatty acids as histone deacetylase inhibitors: Old biochemistry tales in a new life sciences town. Drug Discov. Today 2023, 28, 103569. [Google Scholar] [CrossRef]
- Thulasinathan, B.; Suvilesh, K.N.; Maram, S.; Grossmann, E.; Ghouri, Y.; Teixeiro, E.P.; Chan, J.; Kaifi, J.T.; Rachagani, S. The impact of gut microbial short-chain fatty acids on colorectal cancer development and prevention. Gut Microbes 2025, 17, 2483780. [Google Scholar] [CrossRef]
- Moniri, N.H.; Farah, Q. Short-chain free-fatty acid G protein-coupled receptors in colon cancer. Biochem. Pharmacol. 2021, 186, 114483. [Google Scholar] [CrossRef]
- Li, Y.; Huang, Y.; Liang, H.; Wang, W.; Li, B.; Liu, T.; Huang, Y.; Zhang, Z.; Qin, Y.; Zhou, X.; et al. The roles and applications of short-chain fatty acids derived from microbial fermentation of dietary fibers in human cancer. Front. Nutr. 2023, 10, 1243390. [Google Scholar] [CrossRef]
- Kalyanaraman, B.; Cheng, G.; Hardy, M. The role of short-chain fatty acids in cancer prevention and cancer treatment. Arch. Biochem. Biophys. 2024, 761, 110172. [Google Scholar] [CrossRef]
- Son, M.Y.; Cho, H.S. Anticancer Effects of Gut Microbiota-Derived Short-Chain Fatty Acids in Cancers. J. Microbiol. Biotechnol. 2023, 33, 849–856. [Google Scholar] [CrossRef]
- Donohoe, D.R.; Collins, L.B.; Wali, A.; Bigler, R.; Sun, W.; Bultman, S.J. The Warburg effect dictates the mechanism of butyrate-mediated histone acetylation and cell proliferation. Mol. Cell 2012, 48, 612–626. [Google Scholar] [CrossRef]
- Oncel, S.; Safratowich, B.D.; Zeng, H. The Protective Potential of Butyrate against Colon Cancer Cell Migration and Invasion Is Critically Dependent on Cell Type. Mol. Nutr. Food Res. 2024, 68, 2400421. [Google Scholar] [CrossRef]
- Kaufman, H.L.; Russell, J.; Hamid, O.; Bhatia, S.; Terheyden, P.; D’Angelo, S.P.; Shih, K.C.; Lebbé, C.; Linette, G.P.; Milella, M.; et al. Avelumab in patients with chemotherapy-refractory metastatic Merkel cell carcinoma: A multicentre, single-group, open-label, phase 2 trial. Lancet Oncol. 2016, 17, 1374–1385. [Google Scholar] [CrossRef]
- Su, J.; Fu, Y.; Cui, Z.; Abidin, Z.; Yuan, J.; Zhang, X.; Li, R.; Zhao, C. Relatlimab: A novel drug targeting immune checkpoint LAG-3 in melanoma therapy. Front. Pharmacol. 2023, 14, 1349081. [Google Scholar] [CrossRef]
- Patel, T.H.; Brewer, J.R.; Fan, J.; Cheng, J.; Shen, Y.L.; Xiang, Y.; Zhao, H.; Lemery, S.J.; Pazdur, R.; Kluetz, P.G.; et al. FDA Approval Summary: Tremelimumab in Combination with Durvalumab for the Treatment of Patients with Unresectable Hepatocellular Carcinoma. Clin. Cancer Res. 2024, 30, 269–273. [Google Scholar] [CrossRef] [PubMed]
- Shaukat, A.; Khan, A.; Riaz, R.; Batool, U.E.A.; Malikzai, A. Retifanlimab: A breakthrough for the management of Merkel cell carcinoma. IJS Oncol. 2023, 8, 18–20. [Google Scholar] [CrossRef]
- Incorvaia, L.; Fanale, D.; Badalamenti, G.; Barraco, N.; Bono, M.; Corsini, L.R.; Galvano, A.; Gristina, V.; Listì, A.; Vieni, S. Programmed death ligand 1 (PD-L1) as a predictive biomarker for pembrolizumab therapy in patients with advanced non-small-cell lung cancer (NSCLC). Adv. Ther. 2019, 36, 2600–2617. [Google Scholar] [CrossRef]
- Ding, J.; Yeong, C. Advances in DLL3-targeted therapies for small cell lung cancer: Challenges, opportunities, and future directions. Front. Oncol. 2024, 14, 1504139. [Google Scholar] [CrossRef]
- Paz-Ares, L.; Champiat, S.; Lai, W.V.; Izumi, H.; Govindan, R.; Boyer, M.; Hummel, H.-D.; Borghaei, H.; Johnson, M.L.; Steeghs, N.; et al. Tarlatamab, a First-in-Class DLL3-Targeted Bispecific T-Cell Engager, in Recurrent Small-Cell Lung Cancer: An Open-Label, Phase I Study. J. Clin. Oncol. 2023, 41, 2893–2903. [Google Scholar] [CrossRef]
- Shanehbandi, D.; Majidi, J.; Kazemi, T.; Baradaran, B.; Aghebati-Maleki, L. CD20-based immunotherapy of B-cell derived hematologic malignancies. Curr. Cancer Drug Targets 2017, 17, 423–444. [Google Scholar] [CrossRef]
- Lica, J.J.; Pradhan, B.; Safi, K.; Jakóbkiewicz-Banecka, J.; Hellmann, A. Promising therapeutic strategies for hematologic malignancies: Innovations and potential. Molecules 2024, 29, 4280. [Google Scholar] [CrossRef] [PubMed]
- Freitas, M.B.; Gullo, I.; Leitão, D.; Águas, L.; Oliveira, C.; Polónia, A.; Gomes, J.; Carneiro, F.; Reis, C.A.; Duarte, H.O. HER2 and PD-L1 Expression in Gastric and Gastroesophageal Junction Cancer: Insights for Combinatorial Targeting Approaches. Cancers 2024, 16, 1227. [Google Scholar] [CrossRef] [PubMed]
- Skórzewska, M.; Gęca, K.; Polkowski, W.P. A Clinical Viewpoint on the Use of Targeted Therapy in Advanced Gastric Cancer. Cancers 2023, 15, 5490. [Google Scholar] [CrossRef]
- Carlino, F.; Diana, A.; Piccolo, A.; Ventriglia, A.; Bruno, V.; De Santo, I.; Letizia, O.; De Vita, F.; Daniele, B.; Ciardiello, F.; et al. Immune-Based Therapy in Triple-Negative Breast Cancer: From Molecular Biology to Clinical Practice. Cancers 2022, 14, 2102. [Google Scholar] [CrossRef]
- Hou, J.; Yang, X.; Xie, S.; Zhu, B.; Zha, H. Circulating T cells: A promising biomarker of anti-PD-(L)1 therapy. Front. Immunol. 2024, 15, 1371559. [Google Scholar] [CrossRef]
- Wang, J.; Hou, Y.; Mu, L.; Yang, M.; Ai, X. Gut microbiota contributes to the intestinal and extraintestinal immune homeostasis by balancing Th17/Treg cells. Int. Immunopharmacol. 2024, 143, 113570. [Google Scholar] [CrossRef]
- Wu, X.Q.; Ying, F.; Chung, K.P.S.; Leung, C.O.N.; Leung, R.W.H.; So, K.K.H.; Lei, M.M.L.; Chau, W.K.; Tong, M.; Yu, J.; et al. Intestinal Akkermansia muciniphila complements the efficacy of PD1 therapy in MAFLD-related hepatocellular carcinoma. Cell Rep. Med. 2025, 6, 101900. [Google Scholar] [CrossRef]
- Cao, M.; Deng, Y.; Hao, Q.; Yan, H.; Wang, Q.L.; Dong, C.; Wu, J.; He, Y.; Huang, L.B.; Xia, X.; et al. Single-cell transcriptomic analysis reveals gut microbiota-immunotherapy synergy through modulating tumor microenvironment. Signal Transduct. Target. Ther. 2025, 10, 140. [Google Scholar] [CrossRef]
- Sun, J.; Song, S.; Liu, J.; Chen, F.; Li, X.; Wu, G. Gut microbiota as a new target for anticancer therapy: From mechanism to means of regulation. NPJ Biofilms Microbiomes 2025, 11, 43. [Google Scholar] [CrossRef]
- Koulouris, A.; Tsagkaris, C.; Messaritakis, I.; Gouvas, N.; Sfakianaki, M.; Trypaki, M.; Spyrou, V.; Christodoulakis, M.; Athanasakis, E.; Xynos, E. Resectable colorectal cancer: Current perceptions on the correlation of recurrence risk, microbiota and detection of genetic mutations in liquid biopsies. Cancers 2021, 13, 3522. [Google Scholar] [CrossRef]
- Herrera-Quintana, L.; Vázquez-Lorente, H.; Lopez-Garzon, M.; Cortés-Martín, A.; Plaza-Diaz, J. Cancer and the Microbiome of the Human Body. Nutrients 2024, 16, 2790. [Google Scholar] [CrossRef]
- Haque, H.; Zehra, S.W.; Shahzaib, M.; Abbas, S.; Jaffar, N. Beyond bacteria: Role of non-bacterial gut microbiota species in inflammatory bowel disease and colorectal cancer progression. World J. Gastroenterol. 2024, 30, 4078–4082. [Google Scholar] [CrossRef]
- Jaswal, K.; Todd, O.A.; Behnsen, J. Neglected gut microbiome: Interactions of the non-bacterial gut microbiota with enteric pathogens. Gut Microbes 2023, 15, 2226916. [Google Scholar] [CrossRef]
- Zong, Z.; Zhou, F.; Zhang, L. The fungal mycobiome: A new hallmark of cancer revealed by pan-cancer analyses. Signal Transduct. Target. Ther. 2023, 8, 50. [Google Scholar] [CrossRef]
- Gamal, A.; Elshaer, M.; Alabdely, M.; Kadry, A.; McCormick, T.S.; Ghannoum, M. The Mycobiome: Cancer Pathogenesis, Diagnosis, and Therapy. Cancers 2022, 14, 2875. [Google Scholar] [CrossRef] [PubMed]
- Tamayo-Trujillo, R.; Guevara-Ramírez, P.; Cadena-Ullauri, S.; Paz-Cruz, E.; Ruiz-Pozo, V.A.; Zambrano, A.K. Human virome: Implications in cancer. Heliyon 2023, 9, e14086. [Google Scholar] [CrossRef]
- Stern, J.; Miller, G.; Li, X.; Saxena, D. Virome and bacteriome: Two sides of the same coin. Curr. Opin. Virol. 2019, 37, 37–43. [Google Scholar] [CrossRef]
- Konatala, A.; Parackel, F.; Sudhakar, P. Recent Advancements in Microbiome–Immune Homeostasis and their Involvement in Cancer Immunotherapy. In Microbiome in Human Health and Disease; Springer: Singapore, 2021; pp. 239–258. [Google Scholar] [CrossRef]
- Matson, V.; Chervin, C.S.; Gajewski, T.F. Cancer and the microbiome—Influence of the commensal microbiota on cancer, immune responses, and immunotherapy. Gastroenterology 2021, 160, 600–613. [Google Scholar] [CrossRef] [PubMed]
- Zitvogel, L.; Ma, Y.; Raoult, D.; Kroemer, G.; Gajewski, T.F. The microbiome in cancer immunotherapy: Diagnostic tools and therapeutic strategies. Science 2018, 359, 1366–1370. [Google Scholar] [CrossRef]
- Chung, I.-Y.; Kim, J.; Koh, A. The microbiome matters: Its impact on cancer development and therapeutic responses. J. Microbiol. 2024, 62, 137–152. [Google Scholar] [CrossRef]
- Yoo, J.Y.; Groer, M.; Dutra, S.V.O.; Sarkar, A.; McSkimming, D.I. Gut microbiota and immune system interactions. Microorganisms 2020, 8, 1587. [Google Scholar] [CrossRef] [PubMed]
- Ge, Y.; Wang, X.; Guo, Y.; Yan, J.; Abuduwaili, A.; Aximujiang, K.; Yan, J.; Wu, M. Gut microbiota influence tumor development and Alter interactions with the human immune system. J. Exp. Clin. Cancer Res. 2021, 40, 42. [Google Scholar] [CrossRef]
- Zhou, C.-B.; Zhou, Y.-L.; Fang, J.-Y. Gut microbiota in cancer immune response and immunotherapy. Trends Cancer 2021, 7, 647–660. [Google Scholar] [CrossRef] [PubMed]
- Abidin, Z.Z.; Kamaruddin, N.S.; Hein, Z.M.; Nassir, C.M.N.C.M.; Ramli, M.D.C. The Gut-Brain Axis and Depression: Understanding Microbiota-Driven Mechanisms and Novel Therapeutic Strategies. Preprints 2025. [Google Scholar] [CrossRef]
- Aron-Wisnewsky, J.; Vigliotti, C.; Witjes, J.; Le, P.; Holleboom, A.G.; Verheij, J.; Nieuwdorp, M.; Clément, K. Gut microbiota and human NAFLD: Disentangling microbial signatures from metabolic disorders. Nat. Rev. Gastroenterol. Hepatol. 2020, 17, 279–297. [Google Scholar] [CrossRef]
- Tao, J.; Li, S.; Gan, R.-Y.; Zhao, C.-N.; Meng, X.; Li, H.-B. Targeting gut microbiota with dietary components on cancer: Effects and potential mechanisms of action. Crit. Rev. Food Sci. Nutr. 2020, 60, 1025–1037. [Google Scholar] [CrossRef]
- Oey, O.; Liu, Y.Y.; Sunjaya, A.F.; Simadibrata, D.M.; Khattak, M.A.; Gray, E. Gut microbiota diversity and composition in predicting immunotherapy response and immunotherapy-related colitis in melanoma patients: A systematic review. World J. Clin. Oncol. 2022, 13, 929–942. [Google Scholar] [CrossRef]
- Lalani, A.-K.A.; Xie, W.; Braun, D.A.; Kaymakcalan, M.; Bossé, D.; Steinharter, J.A.; Martini, D.J.; Simantov, R.; Lin, X.; Wei, X.X. Effect of antibiotic use on outcomes with systemic therapies in metastatic renal cell carcinoma. Eur. Urol. Oncol. 2020, 3, 372–381. [Google Scholar] [CrossRef] [PubMed]
- Pinato, D.J.; Howlett, S.; Ottaviani, D.; Urus, H.; Patel, A.; Mineo, T.; Brock, C.; Power, D.; Hatcher, O.; Falconer, A. Association of prior antibiotic treatment with survival and response to immune checkpoint inhibitor therapy in patients with cancer. JAMA Oncol. 2019, 5, 1774–1778. [Google Scholar] [CrossRef]
- Tinsley, N.; Zhou, C.; Tan, G.; Rack, S.; Lorigan, P.; Blackhall, F.; Krebs, M.; Carter, L.; Thistlethwaite, F.; Graham, D. Cumulative antibiotic use significantly decreases efficacy of checkpoint inhibitors in patients with advanced cancer. Oncologist 2020, 25, 55–63. [Google Scholar] [CrossRef]
- Wilson, B.E.; Routy, B.; Nagrial, A.; Chin, V.T. The effect of antibiotics on clinical outcomes in immune-checkpoint blockade: A systematic review and meta-analysis of observational studies. Cancer Immunol. Immunother. 2020, 69, 343–354. [Google Scholar] [CrossRef]
- Said, S.S.; Ibrahim, W.N. Gut Microbiota–Tumor Microenvironment Interactions: Mechanisms and Clinical Implications for Immune Checkpoint Inhibitor Efficacy in Cancer. Cancer Manag. Res. 2025, 17, 171–192. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Xu, B.; Zhang, Y.; Chen, G.; Zhao, P.; Gao, Q.; Yuan, L. The role of intestinal flora on tumorigenesis, progression, and the efficacy of PD-1/PD-L1 antibodies in colorectal cancer. Cancer Biol. Med. 2024, 21, 65–82. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.-H.; Cho, S.-Y.; Yoon, Y.; Park, C.; Sohn, J.; Jeong, J.-J.; Jeon, B.-N.; Jang, M.; An, C.; Lee, S. Bifidobacterium bifidum strains synergize with immune checkpoint inhibitors to reduce tumour burden in mice. Nat. Microbiol. 2021, 6, 277–288. [Google Scholar] [CrossRef]
- Kaźmierczak-Siedlecka, K.; Roviello, G.; Catalano, M.; Polom, K. Gut microbiota modulation in the context of immune-related aspects of Lactobacillus spp. and Bifidobacterium spp. in gastrointestinal cancers. Nutrients 2021, 13, 2674. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.A.; Shaw, H.M.; Bataille, V.; Nathan, P.; Spector, T.D. Role of the gut microbiome for cancer patients receiving immunotherapy: Dietary and treatment implications. Eur. J. Cancer 2020, 138, 149–155. [Google Scholar] [CrossRef]
- Boucher, E.; Plazy, C.; Richard, M.L.; Suau, A.; Mangin, I.; Cornet, M.; Aldebert, D.; Toussaint, B.; Hannani, D. Inulin prebiotic reinforces host cancer immunosurveillance via ɣδ T cell activation. Front. Immunol. 2023, 14, 1104224. [Google Scholar] [CrossRef]
- Gharaibeh, R.Z.; Jobin, C. Microbiota and cancer immunotherapy: In search of microbial signals. Gut 2019, 68, 385–388. [Google Scholar] [CrossRef]
- Kang, Y.-B.; Cai, Y. Faecal microbiota transplantation enhances efficacy of immune checkpoint inhibitors therapy against cancer. World J. Gastroenterol. 2021, 27, 5362–5375. [Google Scholar] [CrossRef]
- Lu, Y.; Yuan, X.; Wang, M.; He, Z.; Li, H.; Wang, J.; Li, Q. Gut microbiota influence immunotherapy responses: Mechanisms and therapeutic strategies. J. Hematol. Oncol. 2022, 15, 47. [Google Scholar] [CrossRef]
- McQuade, J.L.; Ologun, G.O.; Arora, R.; Wargo, J.A. Gut microbiome modulation via fecal microbiota transplant to augment immunotherapy in patients with melanoma or other cancers. Curr. Oncol. Rep. 2020, 22, 74. [Google Scholar] [CrossRef] [PubMed]
- Ali, H.; Khurana, S.; Ma, W.; Peng, Y.; Jiang, Z.-D.; DuPont, H.; Zhang, H.C.; Thomas, A.S.; Okhuysen, P.; Wang, Y. Safety and efficacy of fecal microbiota transplantation to treat and prevent recurrent Clostridioides difficile in cancer patients. J. Cancer 2021, 12, 6498–6506. [Google Scholar] [CrossRef]
- Subramanian, C.R.; Talluri, S.; Khan, S.U.; Katz, J.A.; Georgetson, M.; Sinh, P. Fecal Microbiota Transplantation for Recurrent Clostridium difficile Infection in Patients With Multiple Comorbidities: Long-Term Safety and Efficacy Results From a Tertiary Care Community Hospital. Gastroenterol. Res. 2020, 13, 138–145. [Google Scholar] [CrossRef] [PubMed]
- Hu, W.-T.; Zhang, Q.; Zhang, Z.; He, X.; Zhou, M.; Guo, Y.; Wang, X. Eosinophil and IFN-γ associated with immune-related adverse events as prognostic markers in patients with non-small cell lung cancer treated with immunotherapy. Front. Immunol. 2023, 14, 1112409. [Google Scholar] [CrossRef]
- Xu, X.; Ying, J. Gut microbiota and immunotherapy. Front. Microbiol. 2022, 13, 945887. [Google Scholar] [CrossRef]
- Woelk, C.H.; Snyder, A. Modulating gut microbiota to treat cancer. Science 2021, 371, 573–574. [Google Scholar] [CrossRef]
- Sun, J.Y.; Yin, T.L.; Zhou, J.; Xu, J.; Lu, X.J. Gut microbiome and cancer immunotherapy. J. Cell. Physiol. 2020, 235, 4082–4088. [Google Scholar] [CrossRef]
- Khan, M.A.W.; Ologun, G.; Arora, R.; McQuade, J.L.; Wargo, J.A. Gut microbiome modulates response to cancer immunotherapy. Dig. Dis. Sci. 2020, 65, 885–896. [Google Scholar] [CrossRef] [PubMed]
- Bae, J.; Park, K.; Kim, Y.-M. Commensal microbiota and cancer immunotherapy: Harnessing commensal bacteria for cancer therapy. Immune Netw. 2022, 22, e3. [Google Scholar] [CrossRef]
- Derosa, L.; Routy, B.; Fidelle, M.; Iebba, V.; Alla, L.; Pasolli, E.; Segata, N.; Desnoyer, A.; Pietrantonio, F.; Ferrere, G. Gut bacteria composition drives primary resistance to cancer immunotherapy in renal cell carcinoma patients. Eur. Urol. 2020, 78, 195–206. [Google Scholar] [CrossRef]
- Luke, J.J.; Piha-Paul, S.A.; Medina, T.; Verschraegen, C.F.; Varterasian, M.; Brennan, A.M.; Riese, R.J.; Sokolovska, A.; Strauss, J.; Hava, D.L. Phase I Study of SYNB1891, an Engineered E. coli Nissle Strain Expressing STING Agonist, with and without Atezolizumab in Advanced Malignancies. Clin. Cancer Res. 2023, 29, 2435–2444. [Google Scholar] [CrossRef] [PubMed]
- Riese, R.; Luke, J.; Lewis, K.; Janku, F.; Piha-Paul, S.; Verschraegen, C.; Brennan, A.; Armstrong, M.; Varterasian, M.; Sokolovska, A. 500 SYNB1891, a bacterium engineered to produce a STING agonist, demonstrates target engagement in humans following intratumoral injection. J. Immunother. Cancer 2021, 9, A1–A1054. [Google Scholar] [CrossRef]
- Luo, K.; Li, N.; Ye, W.; Gao, H.; Luo, X.; Cheng, B. Activation of stimulation of interferon genes (STING) signal and cancer immunotherapy. Molecules 2022, 27, 4638. [Google Scholar] [CrossRef]
- Canale, F.P.; Basso, C.; Antonini, G.; Perotti, M.; Li, N.; Sokolovska, A.; Neumann, J.; James, M.J.; Geiger, S.; Jin, W. Metabolic modulation of tumours with engineered bacteria for immunotherapy. Nature 2021, 598, 662–666. [Google Scholar] [CrossRef]
- Spencer, K.R.; Wang, J.; Silk, A.W.; Ganesan, S.; Kaufman, H.L.; Mehnert, J.M. Biomarkers for immunotherapy: Current developments and challenges. Am. Soc. Clin. Oncol. Educ. Book 2016, 36, e493–e503. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.-R.; Wu, X.-L.; Sun, Y.-L. Therapeutic targets and biomarkers of tumor immunotherapy: Response versus non-response. Signal Transduct. Target. Ther. 2022, 7, 331. [Google Scholar] [CrossRef] [PubMed]
- Bai, R.; Lv, Z.; Xu, D.; Cui, J. Predictive biomarkers for cancer immunotherapy with immune checkpoint inhibitors. Biomark. Res. 2020, 8, 34. [Google Scholar] [CrossRef]
- Lommen, K.; Odeh, S.; de Theije, C.C.; Smits, K.M. Biobanking in molecular biomarker research for the early detection of cancer. Cancers 2020, 12, 776. [Google Scholar] [CrossRef]
- Elinav, E.; Garrett, W.S.; Trinchieri, G.; Wargo, J. The cancer microbiome. Nat. Rev. Cancer 2019, 19, 371–376. [Google Scholar] [CrossRef]
- Kit, O.I.; Timofeeva, S.V.; Sitkovskaya, A.O.; Novikova, I.A.; Kolesnikov, E.N. The biobank of the National Medical Research Centre for Oncology as a resource for research in the field of personalized medicine: A review. J. Mod. Oncol. 2022, 24, 6–11. [Google Scholar] [CrossRef]
- Liu, C.; Du, M.-X.; Abuduaini, R.; Yu, H.-Y.; Li, D.-H.; Wang, Y.-J.; Zhou, N.; Jiang, M.-Z.; Niu, P.-X.; Han, S.-S. Enlightening the taxonomy darkness of human gut microbiomes with a cultured biobank. Microbiome 2021, 9, 119. [Google Scholar] [CrossRef]
- Ma, L.; Guo, H.; Zhao, Y.; Liu, Z.; Wang, C.; Bu, J.; Sun, T.; Wei, J. Liquid biopsy in cancer: Current status, challenges and future prospects. Signal Transduct. Target. Ther. 2024, 9, 336. [Google Scholar] [CrossRef]
- Jee, J.; Brannon, A.R.; Singh, R.; Derkach, A.; Fong, C.; Lee, A.; Gray, L.; Pichotta, K.; Luthra, A.; Diosdado, M. DNA liquid biopsy-based prediction of cancer-associated venous thromboembolism. Nat. Med. 2024, 30, 2499–2507. [Google Scholar] [CrossRef]
- Mahuron, K.M.; Fong, Y. Applications of liquid biopsy for surgical patients with cancer: A review. JAMA Surg. 2024, 159, 96–103. [Google Scholar] [CrossRef] [PubMed]
- Poore, G.D.; Kopylova, E.; Zhu, Q.; Carpenter, C.; Fraraccio, S.; Wandro, S.; Kosciolek, T.; Janssen, S.; Metcalf, J.; Song, S.J. Microbiome analyses of blood and tissues suggest cancer diagnostic approach. Nature 2020, 579, 567–574. [Google Scholar] [CrossRef]
- Im, Y.; Tsui, D.; Diaz, L.; Wan, J. Next-generation liquid biopsies: Embracing data science in oncology. Trends Cancer 2021, 7, 283–292. [Google Scholar] [CrossRef]
- Piombino, C.; Mastrolia, I.; Omarini, C.; Candini, O.; Dominici, M.; Piacentini, F.; Toss, A. The role of exosomes in breast cancer diagnosis. Biomedicines 2021, 9, 312. [Google Scholar] [CrossRef]
- Welsh, J.A.; Goberdhan, D.C.I.; O’Driscoll, L.; Buzas, E.I.; Blenkiron, C.; Bussolati, B.; Cai, H.; Di Vizio, D.; Driedonks, T.A.P.; Erdbrügger, U.; et al. Minimal information for studies of extracellular vesicles (MISEV2023): From basic to advanced approaches. J. Extracell. Vesicles 2024, 13, e12404. [Google Scholar] [CrossRef] [PubMed]
- Raposo, G.; Stahl, P.D. Extracellular vesicles: A new communication paradigm? Nat. Rev. Mol. Cell Biol. 2019, 20, 509–510. [Google Scholar] [CrossRef] [PubMed]
- Ellis, G.I.; Riley, J.L. How to kill T(reg) cells for immunotherapy. Nat. Cancer 2020, 1, 1134–1135. [Google Scholar] [CrossRef]
- Zhang, K.K.; Reddy, N.; Janis, J.E. Office-based Plastic Surgery-Evidence-based Clinical and Administrative Guidelines. Plast. Reconstr. Surg. Glob. Open 2022, 10, e4634. [Google Scholar] [CrossRef] [PubMed]
- Salehi, M.; Sharifi, M. Exosomal miRNAs as novel cancer biomarkers: Challenges and opportunities. J. Cell. Physiol. 2018, 233, 6370–6380. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Zhang, L.; Montgomery, K.C.; Jiang, L.; Lyon, C.J.; Hu, T.Y. Advanced technologies for molecular diagnosis of cancer: State of pre-clinical tumor-derived exosome liquid biopsies. Mater. Today Bio 2023, 18, 100538. [Google Scholar] [CrossRef]
- Lee, Y.; Ni, J.; Beretov, J.; Wasinger, V.C.; Graham, P.; Li, Y. Recent advances of small extracellular vesicle biomarkers in breast cancer diagnosis and prognosis. Mol. Cancer 2023, 22, 33. [Google Scholar] [CrossRef]
- Tamura, T.; Yoshioka, Y.; Sakamoto, S.; Ichikawa, T.; Ochiya, T. Extracellular vesicles as a promising biomarker resource in liquid biopsy for cancer. Extracell. Vesicles Circ. Nucleic Acids 2021, 2, 148–174. [Google Scholar] [CrossRef]
- Ho, H.-Y.; Chung, K.-S.; Kan, C.-M.; Wong, S.-C. Liquid biopsy in the clinical management of cancers. Int. J. Mol. Sci. 2024, 25, 8594. [Google Scholar] [CrossRef]
- Febbo, P.G.; Allo, M.; Alme, E.B.; Cuyun Carter, G.; Dumanois, R.; Essig, A.; Kiernan, E.; Kubler, C.B.; Martin, N.; Popescu, M.C. Recommendations for the equitable and widespread implementation of liquid biopsy for cancer care. JCO Precis. Oncol. 2024, 8, e2300382. [Google Scholar] [CrossRef]
- Xiao, L.; Zhang, F.; Zhao, F. Large-scale microbiome data integration enables robust biomarker identification. Nat. Comput. Sci. 2022, 2, 307–316. [Google Scholar] [CrossRef]
- Heinken, A.; Basile, A.; Hertel, J.; Thinnes, C.; Thiele, I. Genome-scale metabolic modeling of the human microbiome in the era of personalized medicine. Annu. Rev. Microbiol. 2021, 75, 199–222. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; He, Y.; Yang, F.; Chen, K. Current and emerging applications of liquid biopsy in pan-cancer. Transl. Oncol. 2023, 34, 101720. [Google Scholar] [CrossRef]
- Tang, Q.; Jin, G.; Wang, G.; Liu, T.; Liu, X.; Wang, B.; Cao, H. Current sampling methods for gut microbiota: A call for more precise devices. Front. Cell. Infect. Microbiol. 2020, 10, 151. [Google Scholar] [CrossRef]
- Butterfield, L.H.; Najjar, Y.G. Immunotherapy combination approaches: Mechanisms, biomarkers and clinical observations. Nat. Rev. Immunol. 2024, 24, 399–416. [Google Scholar] [CrossRef]
- Yang, M.; Cui, M.; Sun, Y.; Liu, S.; Jiang, W. Mechanisms, combination therapy, and biomarkers in cancer immunotherapy resistance. Cell Commun. Signal. 2024, 22, 338. [Google Scholar] [CrossRef]
- Wisdom, A.J.; Barker, C.A.; Chang, J.Y.; Demaria, S.; Formenti, S.; Grassberger, C.; Gregucci, F.; Hoppe, B.S.; Kirsch, D.G.; Marciscano, A.E. The next chapter in immunotherapy and radiation combination therapy–cancer-specific perspectives. Int. J. Radiat. Oncol. Biol. Phys. 2024, 118, 1404–1421. [Google Scholar] [CrossRef]
- Tang, F.; Ding, A.; Xu, Y.; Ye, Y.; Li, L.; Xie, R.; Huang, W. Gene and photothermal combination therapy: Principle, materials, and amplified anticancer intervention. Small 2024, 20, 2307078. [Google Scholar] [CrossRef]
- Willsmore, Z.N.; Coumbe, B.G.; Crescioli, S.; Reci, S.; Gupta, A.; Harris, R.J.; Chenoweth, A.; Chauhan, J.; Bax, H.J.; McCraw, A. Combined anti-PD-1 and anti-CTLA-4 checkpoint blockade: Treatment of melanoma and immune mechanisms of action. Eur. J. Immunol. 2021, 51, 544–556. [Google Scholar] [CrossRef]
- Váraljai, R.; Zimmer, L.; Al-Matary, Y.; Kaptein, P.; Albrecht, L.J.; Shannan, B.; Brase, J.C.; Gusenleitner, D.; Amaral, T.; Wyss, N. Interleukin 17 signaling supports clinical benefit of dual CTLA-4 and PD-1 checkpoint inhibition in melanoma. Nat. Cancer 2023, 4, 1292–1308. [Google Scholar] [CrossRef]
- Seidel, J.A.; Otsuka, A.; Kabashima, K. Anti-PD-1 and anti-CTLA-4 therapies in cancer: Mechanisms of action, efficacy, and limitations. Front. Oncol. 2018, 8, 86. [Google Scholar] [CrossRef] [PubMed]
- Ciernikova, S.; Sevcikova, A.; Drgona, L.; Mego, M. Modulating the gut microbiota by probiotics, prebiotics, postbiotics, and fecal microbiota transplantation: An emerging trend in cancer patient care. Biochim. Biophys. Acta Rev. Cancer 2023, 1878, 188990. [Google Scholar] [CrossRef] [PubMed]
- Kaźmierczak-Siedlecka, K.; Daca, A.; Fic, M.; van de Wetering, T.; Folwarski, M.; Makarewicz, W. Therapeutic methods of gut microbiota modification in colorectal cancer management–fecal microbiota transplantation, prebiotics, probiotics, and synbiotics. Gut Microbes 2020, 11, 1518–1530. [Google Scholar] [CrossRef] [PubMed]
- Daillère, R.; Vétizou, M.; Waldschmitt, N.; Yamazaki, T.; Isnard, C.; Poirier-Colame, V.; Duong, C.P.; Flament, C.; Lepage, P.; Roberti, M.P. Enterococcus hirae and Barnesiella intestinihominis facilitate cyclophosphamide-induced therapeutic immunomodulatory effects. Immunity 2016, 45, 931–943. [Google Scholar] [CrossRef]
- Zhang, X.; Cai, X.; Zheng, H. Anti-PD1 or anti-PD-L1 antibodies alone or in combination with chemotherapy first-line treatment of advanced non-small cell lung cancer. Thorac. Cancer 2022, 13, 1104–1105. [Google Scholar] [CrossRef]
- Li, L.; Chen, L.; Yan, L.; Guo, Y.; Li, F.; Fan, M.; Lan, M.; Lai, X.; Zhou, J.; Huang, Y. Initial analysis of the synergy of programmed cell death-1 (PD-1) inhibitor and concurrent chemoradiotherapy treatment for recurrent/metastatic head and neck squamous cell carcinoma patients. Radiat. Oncol. 2023, 18, 109. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Lin, J.; Yang, X.; Long, J.; Bai, Y.; Yang, X.; Mao, Y.; Sang, X.; Seery, S.; Zhao, H. Combination regimens with PD-1/PD-L1 immune checkpoint inhibitors for gastrointestinal malignancies. J. Hematol. Oncol. 2019, 12, 42. [Google Scholar] [CrossRef] [PubMed]
- Page, D.B.; Bear, H.; Prabhakaran, S.; Gatti-Mays, M.E.; Thomas, A.; Cobain, E.; McArthur, H.; Balko, J.M.; Gameiro, S.R.; Nanda, R. Two may be better than one: PD-1/PD-L1 blockade combination approaches in metastatic breast cancer. NPJ Breast Cancer 2019, 5, 34. [Google Scholar] [CrossRef] [PubMed]
- Shi, M.; Chen, Y.; Ji, D. The implications from the interplay of neoadjuvant chemoradiotherapy and the immune microenvironment in rectal cancer. Future Oncol. 2022, 18, 3229–3244. [Google Scholar] [CrossRef]
- Takamori, S.; Toyokawa, G.; Takada, K.; Shoji, F.; Okamoto, T.; Maehara, Y. Combination Therapy of Radiotherapy and Anti-PD-1/PD-L1 Treatment in Non-Small-cell Lung Cancer: A Mini-review. Clin. Lung Cancer 2018, 19, 12–16. [Google Scholar] [CrossRef]
- Wang, Y.; Deng, W.; Li, N.; Neri, S.; Sharma, A.; Jiang, W.; Lin, S.H. Combining immunotherapy and radiotherapy for cancer treatment: Current challenges and future directions. Front. Pharmacol. 2018, 9, 185. [Google Scholar] [CrossRef]
- Yu, S.; Wang, Y.; He, P.; Shao, B.; Liu, F.; Xiang, Z.; Yang, T.; Zeng, Y.; He, T.; Ma, J. Effective combinations of immunotherapy and radiotherapy for cancer treatment. Front. Oncol. 2022, 12, 809304. [Google Scholar] [CrossRef]
- Spaas, M.; Lievens, Y. Is the combination of immunotherapy and radiotherapy in non-small cell lung cancer a feasible and effective approach? Front. Med. 2019, 6, 244. [Google Scholar] [CrossRef]
- Sardaro, A.; Ferrari, C.; Carbonara, R.; Altini, C.; Lavelli, V.; Rubini, G. Synergism between immunotherapy and radiotherapy in esophageal cancer: An overview of current knowledge and future perspectives. Cancer Biother. Radiopharm. 2021, 36, 123–132. [Google Scholar] [CrossRef]
- Thorsson, V.; Gibbs, D.L.; Brown, S.D.; Wolf, D.; Bortone, D.S.; Ou Yang, T.H.; Porta-Pardo, E.; Gao, G.F.; Plaisier, C.L.; Eddy, J.A.; et al. The Immune Landscape of Cancer. Immunity 2018, 48, 812–830.e814. [Google Scholar] [CrossRef]
- Sharma, P.; Siddiqui, B.A.; Anandhan, S.; Yadav, S.S.; Subudhi, S.K.; Gao, J.; Goswami, S.; Allison, J.P. The Next Decade of Immune Checkpoint Therapy. Cancer Discov. 2021, 11, 838–857. [Google Scholar] [CrossRef]
- Mokhtari, R.B.; Sampath, D.; Eversole, P.; Yu Lin, M.O.; Bosykh, D.A.; Boopathy, G.T.K.; Sivakumar, A.; Wang, C.C.; Kumar, R.; Sheng, J.Y.P.; et al. An Agrin-YAP/TAZ Rigidity Sensing Module Drives EGFR-Addicted Lung Tumorigenesis. Adv. Sci. 2025, 12, e2413443. [Google Scholar] [CrossRef]
- Mokhtari, R.B.; Ashayeri, N.; Baghaie, L.; Sambi, M.; Satari, K.; Baluch, N.; Bosykh, D.A.; Szewczuk, M.R.; Chakraborty, S. The Hippo Pathway Effectors YAP/TAZ-TEAD Oncoproteins as Emerging Therapeutic Targets in the Tumor Microenvironment. Cancers 2023, 15, 3468. [Google Scholar] [CrossRef]
- Chakraborty, S.; Sampath, D.; Yu Lin, M.O.; Bilton, M.; Huang, C.K.; Nai, M.H.; Njah, K.; Goy, P.A.; Wang, C.C.; Guccione, E.; et al. Agrin-Matrix Metalloproteinase-12 axis confers a mechanically competent microenvironment in skin wound healing. Nat. Commun. 2021, 12, 6349. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, S.; Njah, K.; Hong, W. Agrin Mediates Angiogenesis in the Tumor Microenvironment. Trends Cancer 2020, 6, 81–85. [Google Scholar] [CrossRef]
- Chakraborty, S.; Hong, W. Oncogenetic engagement with mechanosensing. Nat. Mater. 2020, 19, 707–709. [Google Scholar] [CrossRef] [PubMed]
- Njah, K.; Chakraborty, S.; Qiu, B.; Arumugam, S.; Raju, A.; Pobbati, A.V.; Lakshmanan, M.; Tergaonkar, V.; Thibault, G.; Wang, X.; et al. A Role of Agrin in Maintaining the Stability of Vascular Endothelial Growth Factor Receptor-2 during Tumor Angiogenesis. Cell Rep. 2019, 28, 949–965.E7. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, S.; Hong, W. Linking Extracellular Matrix Agrin to the Hippo Pathway in Liver Cancer and Beyond. Cancers 2018, 10, 45. [Google Scholar] [CrossRef]
- Chakraborty, S.; Njah, K.; Pobbati, A.V.; Lim, Y.B.; Raju, A.; Lakshmanan, M.; Tergaonkar, V.; Lim, C.T.; Hong, W. Agrin as a Mechanotransduction Signal Regulating YAP through the Hippo Pathway. Cell Rep. 2017, 18, 2464–2479. [Google Scholar] [CrossRef]
- Chakraborty, S.; Lakshmanan, M.; Swa, H.L.; Chen, J.; Zhang, X.; Ong, Y.S.; Loo, L.S.; Akincilar, S.C.; Gunaratne, J.; Tergaonkar, V.; et al. An oncogenic role of Agrin in regulating focal adhesion integrity in hepatocellular carcinoma. Nat. Commun. 2015, 6, 6184. [Google Scholar] [CrossRef]
- Yang, Y.; Chen, X.; Pan, J.; Ning, H.; Zhang, Y.; Bo, Y.; Ren, X.; Li, J.; Qin, S.; Wang, D. Pan-cancer single-cell dissection reveals phenotypically distinct B cell subtypes. Cell 2024, 187, 4790–4811.e22. [Google Scholar] [CrossRef]
- Savage, S.R.; Yi, X.; Lei, J.T.; Wen, B.; Zhao, H.; Liao, Y.; Jaehnig, E.J.; Somes, L.K.; Shafer, P.W.; Lee, T.D. Pan-cancer proteogenomics expands the landscape of therapeutic targets. Cell 2024, 187, 4389–4407.e15. [Google Scholar] [CrossRef]
- Aaltonen, L.A.; Abascal, F.; Abeshouse, A.; Aburatani, H.; Adams, D.J.; Agrawal, N.; Ahn, K.S.; Ahn, S.-M.; Aikata, H.; Akbani, R.; et al. Pan-cancer analysis of whole genomes. Nature 2020, 578, 82–93. [Google Scholar] [CrossRef]
- Hegde, P.S.; Chen, D.S. Top 10 Challenges in Cancer Immunotherapy. Immunity 2020, 52, 17–35. [Google Scholar] [CrossRef]
- Stadtmauer, E.A.; Fraietta, J.A.; Davis, M.M.; Cohen, A.D.; Weber, K.L.; Lancaster, E.; Mangan, P.A.; Kulikovskaya, I.; Gupta, M.; Chen, F.; et al. CRISPR-engineered T cells in patients with refractory cancer. Science 2020, 367, eaba7365. [Google Scholar] [CrossRef] [PubMed]
- Wahida, A.; Buschhorn, L.; Frohling, S.; Jost, P.J.; Schneeweiss, A.; Lichter, P.; Kurzrock, R. The coming decade in precision oncology: Six riddles. Nat. Rev. Cancer 2023, 23, 43–54. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Edil, B.H.; Li, M. Combination therapies for cancer: Challenges and opportunities. BMC Med. 2023, 21, 171. [Google Scholar] [CrossRef] [PubMed]
- Schupack, D.A.; Mars, R.A.; Voelker, D.H.; Abeykoon, J.P.; Kashyap, P.C. The promise of the gut microbiome as part of individualized treatment strategies. Nat. Rev. Gastroenterol. Hepatol. 2022, 19, 7–25. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Cai, X.; Yan, C. Opportunities and challenges in combining immunotherapy and radiotherapy in esophageal cancer. J. Cancer Res. Clin. Oncol. 2023, 149, 18253–18270. [Google Scholar] [CrossRef] [PubMed]
- Whiteside, T.L.; Demaria, S.; Rodriguez-Ruiz, M.E.; Zarour, H.M.; Melero, I. Emerging opportunities and challenges in cancer immunotherapy. Clin. Cancer Res. 2016, 22, 1845–1855. [Google Scholar] [CrossRef] [PubMed]
- Paver, E.C.; Morey, A.L. Biomarkers and biomarker validation: A pathologist’s guide to getting it right. Pathology 2024, 56, 147–157. [Google Scholar] [CrossRef] [PubMed]
- Robles, J.; Prakash, A.; Vizcaíno, J.A.; Casal, J.I. Integrated meta-analysis of colorectal cancer public proteomic datasets for biomarker discovery and validation. PLoS Comput. Biol. 2024, 20, e1011828. [Google Scholar] [CrossRef]
- Huang, Z.; Deng, C.; Ma, C.; He, G.; Tao, J.; Zhang, L.; Hu, X.; Mo, Y.; Qiu, L.; Zhang, N.; et al. Identification and validation of the surface proteins FIBG, PDGF-β, and TGF-β on serum extracellular vesicles for non-invasive detection of colorectal cancer: Experimental study. Int. J. Surg. 2024, 110, 4672–4687. [Google Scholar] [CrossRef]
- Novielli, P.; Romano, D.; Magarelli, M.; Bitonto, P.D.; Diacono, D.; Chiatante, A.; Lopalco, G.; Sabella, D.; Venerito, V.; Filannino, P.; et al. Explainable artificial intelligence for microbiome data analysis in colorectal cancer biomarker identification. Front. Microbiol. 2024, 15, 1348974. [Google Scholar] [CrossRef]
- Tabib, N.S.S.; Madgwick, M.; Sudhakar, P.; Verstockt, B.; Korcsmaros, T.; Vermeire, S. Big data in IBD: Big progress for clinical practice. Gut 2020, 69, 1520–1532. [Google Scholar] [CrossRef]
- Cammarota, G.; Ianiro, G.; Ahern, A.; Carbone, C.; Temko, A.; Claesson, M.J.; Gasbarrini, A.; Tortora, G. Gut microbiome, big data and machine learning to promote precision medicine for cancer. Nat. Rev. Gastroenterol. Hepatol. 2020, 17, 635–648. [Google Scholar] [CrossRef]
- Chen, L.; Wang, D.; Garmaeva, S.; Kurilshikov, A.; Vila, A.V.; Gacesa, R.; Sinha, T.; Segal, E.; Weersma, R.K.; Wijmenga, C. The long-term genetic stability and individual specificity of the human gut microbiome. Cell 2021, 184, 2302–2315.e12. [Google Scholar] [CrossRef]
- Vandeputte, D.; De Commer, L.; Tito, R.Y.; Kathagen, G.; Sabino, J.; Vermeire, S.; Faust, K.; Raes, J. Temporal variability in quantitative human gut microbiome profiles and implications for clinical research. Nat. Commun. 2021, 12, 6740. [Google Scholar] [CrossRef]
- McKean, W.B.; Moser, J.C.; Rimm, D.; Hu-Lieskovan, S. Biomarkers in Precision Cancer Immunotherapy: Promise and Challenges. Am. Soc. Clin. Oncol. Book 2020, 40, e275–e291. [Google Scholar] [CrossRef]
- Bernicker, E. Next-generation sequencing and immunotherapy biomarkers: A medical oncology perspective. Arch. Pathol. Lab. Med. 2016, 140, 245–248. [Google Scholar] [CrossRef]
- Zhang, W.; Shi, J.; Wang, Y.; Zhou, H.; Zhang, Z.; Han, Z.; Li, G.; Yang, B.; Cao, G.; Ke, Y. Next-generation sequencing-guided molecular-targeted therapy and immunotherapy for biliary tract cancers. Cancer Immunol. Immunother. 2021, 70, 1001–1014. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Späth, S.S.; Marjani, S.L.; Zhang, W.; Pan, X. Characterization of cancer genomic heterogeneity by next-generation sequencing advances precision medicine in cancer treatment. Precis. Clin. Med. 2018, 1, 29–48. [Google Scholar] [CrossRef]
- Waldman, A.D.; Fritz, J.M.; Lenardo, M.J. A guide to cancer immunotherapy: From T cell basic science to clinical practice. Nat. Rev. Immunol. 2020, 20, 651–668. [Google Scholar] [CrossRef] [PubMed]
- Said, S.S.; Ibrahim, W.N. Cancer resistance to immunotherapy: Comprehensive insights with future perspectives. Pharmaceutics 2023, 15, 1143. [Google Scholar] [CrossRef]
- Attili, I.; Tarantino, P.; Passaro, A.; Stati, V.; Curigliano, G.; de Marinis, F. Strategies to overcome resistance to immune checkpoint blockade in lung cancer. Lung Cancer 2021, 154, 151–160. [Google Scholar] [CrossRef] [PubMed]
- Hirsch, F.R.; McElhinny, A.; Stanforth, D.; Ranger-Moore, J.; Jansson, M.; Kulangara, K.; Richardson, W.; Towne, P.; Hanks, D.; Vennapusa, B. PD-L1 immunohistochemistry assays for lung cancer: Results from phase 1 of the blueprint PD-L1 IHC assay comparison project. J. Thorac. Oncol. 2017, 12, 208–222. [Google Scholar] [CrossRef]
- McLaughlin, J.; Han, G.; Schalper, K.A.; Carvajal-Hausdorf, D.; Pelekanou, V.; Rehman, J.; Velcheti, V.; Herbst, R.; LoRusso, P.; Rimm, D.L. Quantitative assessment of the heterogeneity of PD-L1 expression in non–small-cell lung cancer. JAMA Oncol. 2016, 2, 46–54. [Google Scholar] [CrossRef]
- Yang, Q.; Chen, M.; Gu, J.; Niu, K.; Zhao, X.; Zheng, L.; Xu, Z.; Yu, Y.; Li, F.; Meng, L. Novel biomarkers of dynamic blood PD-L1 expression for immune checkpoint inhibitors in advanced non-small-cell lung cancer patients. Front. Immunol. 2021, 12, 665133. [Google Scholar] [CrossRef]
- Hinterleitner, C.; Strähle, J.; Malenke, E.; Hinterleitner, M.; Henning, M.; Seehawer, M.; Bilich, T.; Heitmann, J.; Lutz, M.; Mattern, S.; et al. Platelet PD-L1 reflects collective intratumoral PD-L1 expression and predicts immunotherapy response in non-small cell lung cancer. Nat. Commun. 2021, 12, 7005. [Google Scholar] [CrossRef] [PubMed]
- Kao, C.J.; Charmsaz, S.; Alden, S.L.; Brancati, M.; Li, H.L.; Balaji, A.; Munjal, K.; Howe, K.; Mitchell, S.; Leatherman, J.; et al. Immune-related events in individuals with solid tumors on immunotherapy associate with Th17 and Th2 signatures. J. Clin. Investig. 2024, 134, e176567. [Google Scholar] [CrossRef] [PubMed]
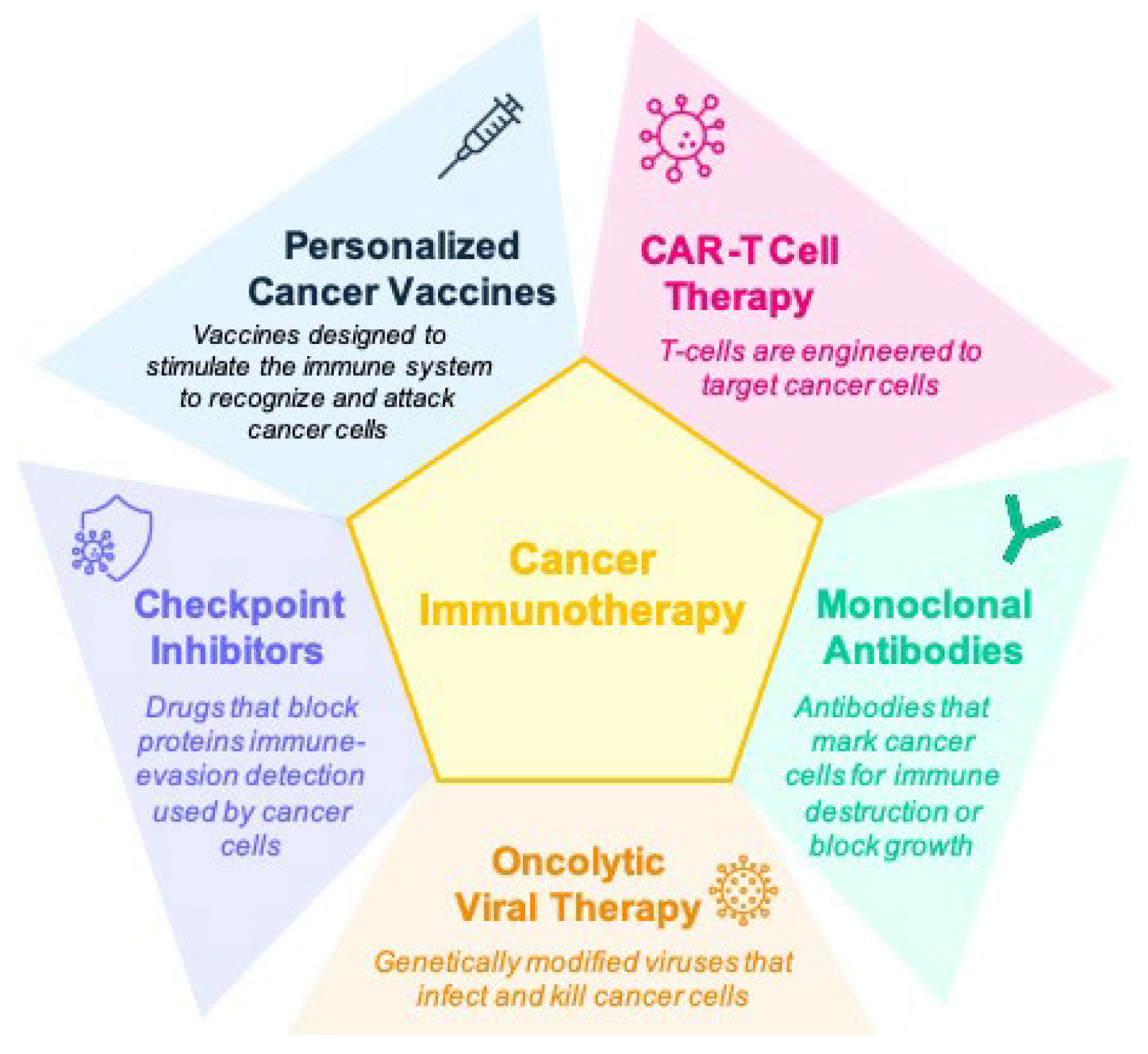
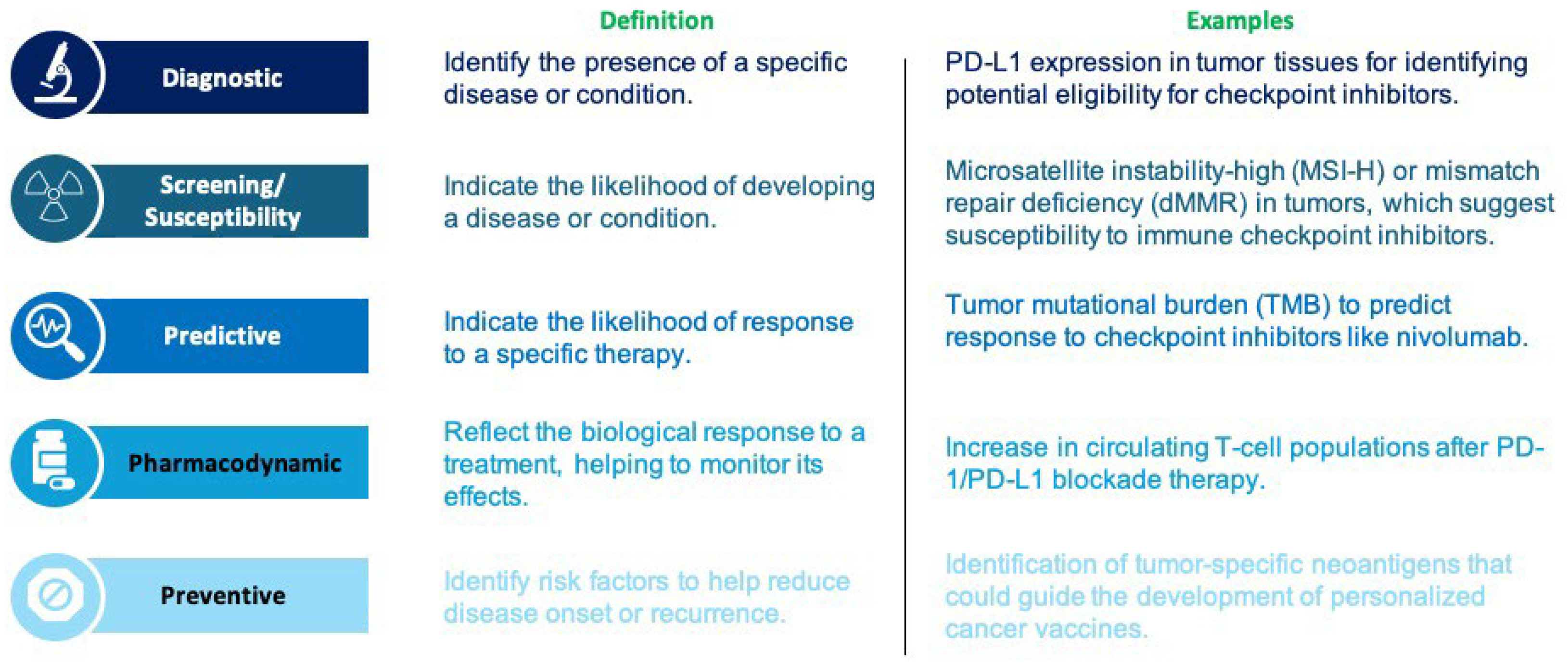

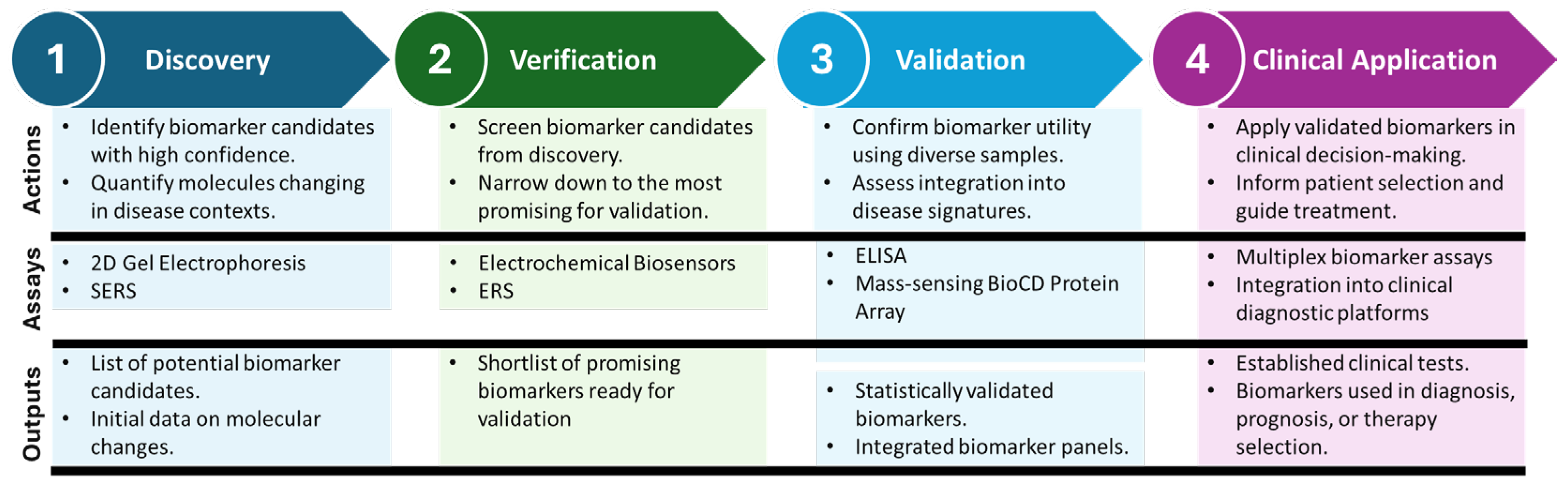
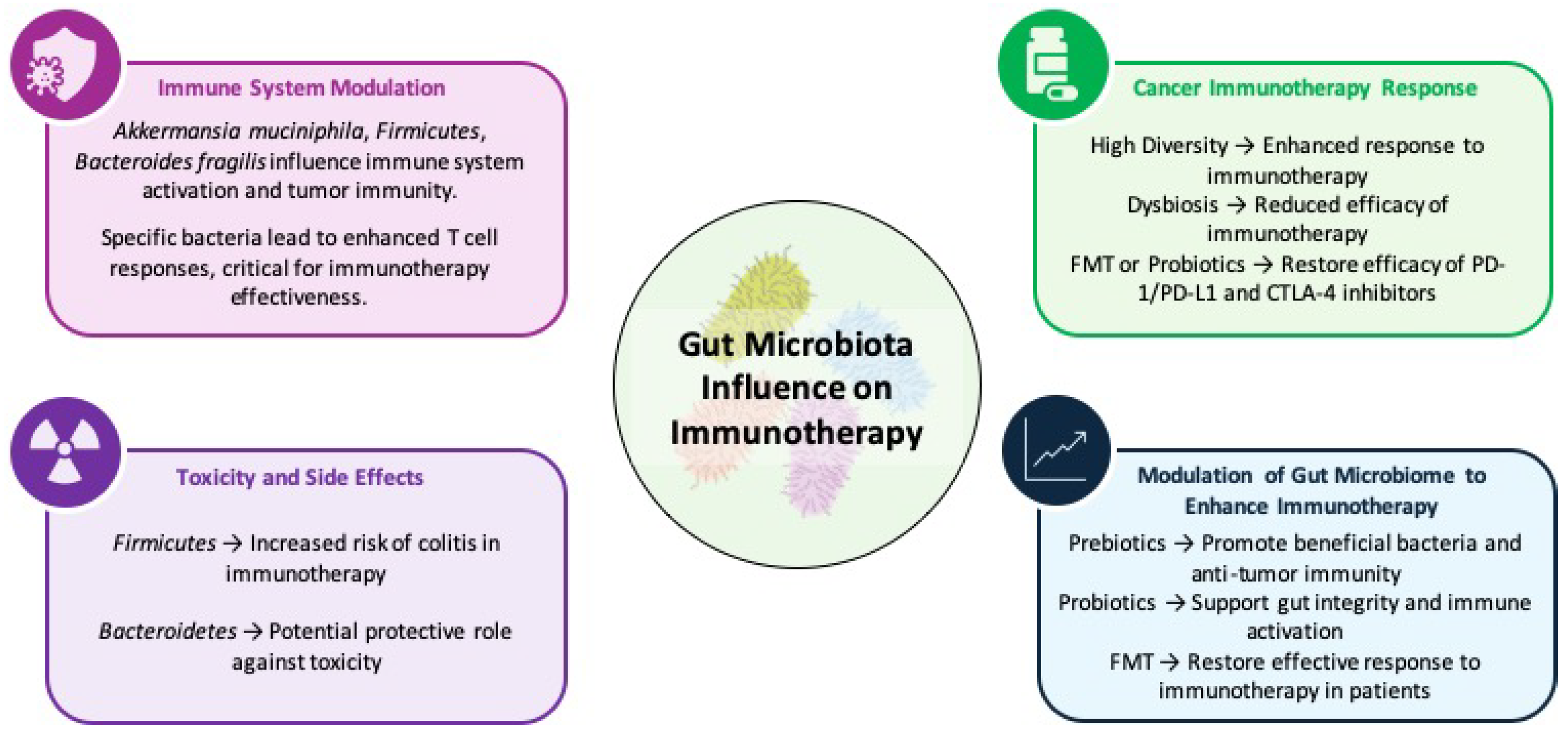
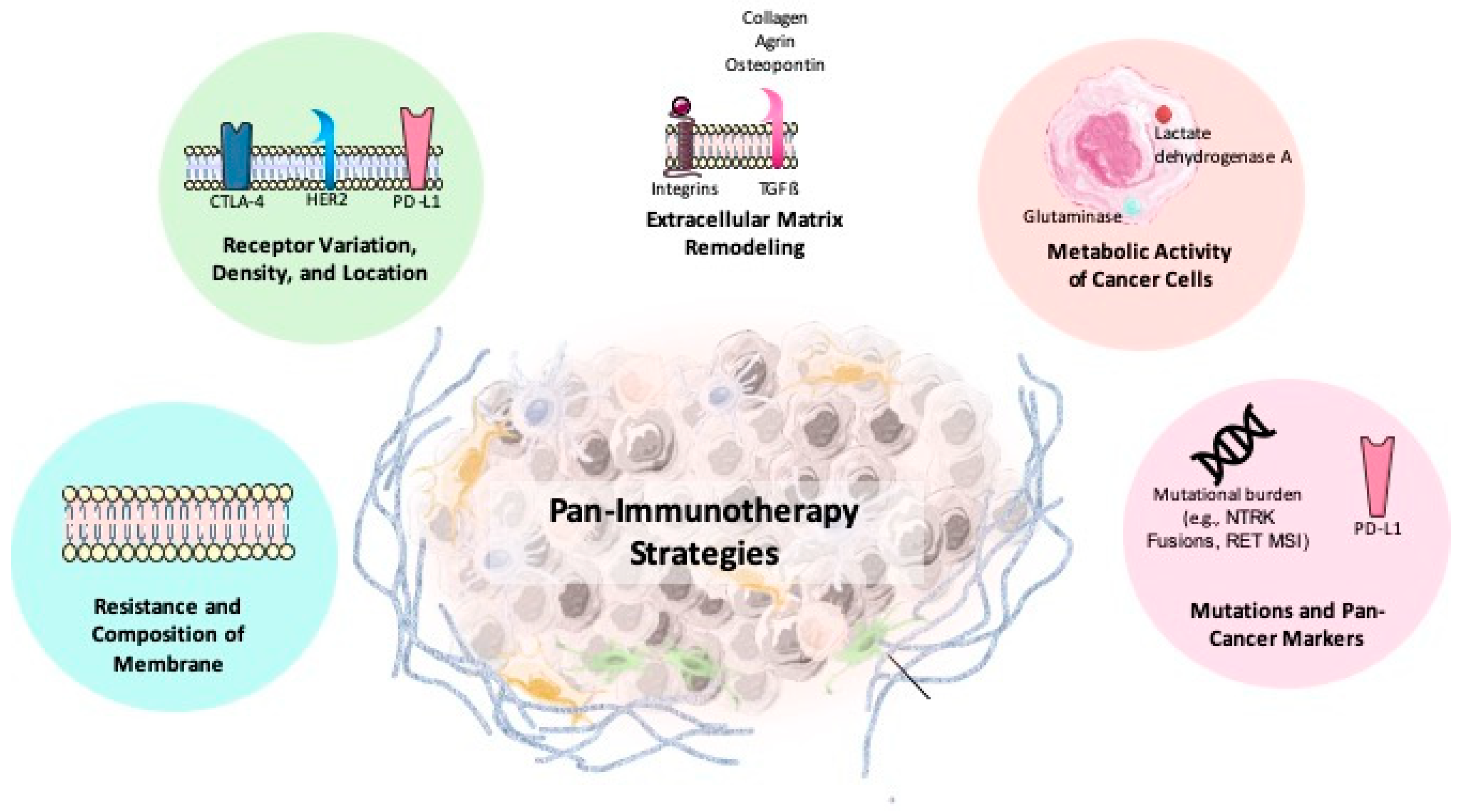
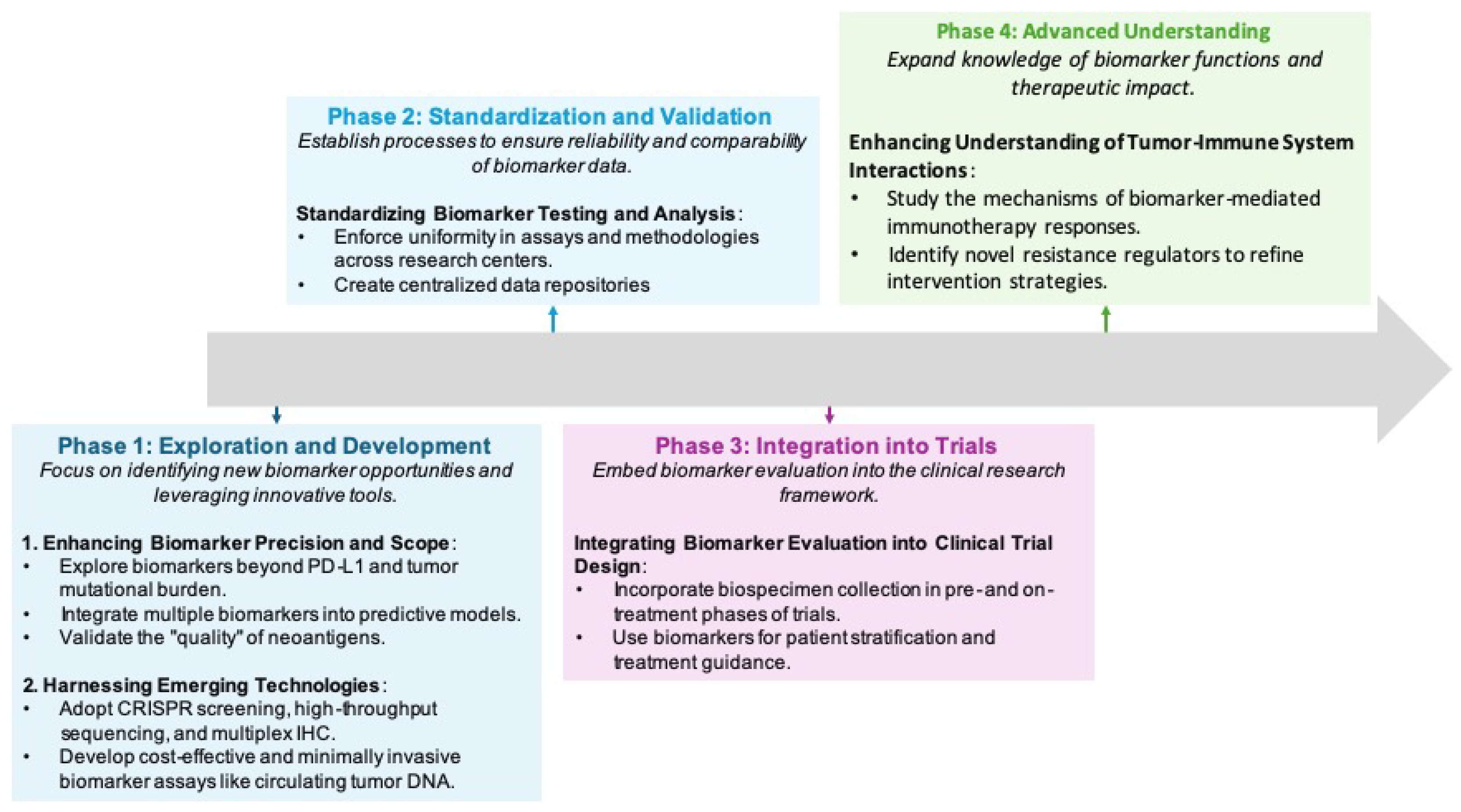
| Organ System | Possible irAEs |
|---|---|
| Dermatologic | Rash, pruritus, vitiligo, dermatitis |
| Endocrine | Hypothyroidism, hyperthyroidism, adrenal insufficiency, diabetes |
| Pulmonary | Pneumonitis, dyspnea, cough |
| Cardiovascular | Myocarditis, pericarditis, arrhythmias |
| Hepatic | Hepatitis, elevated liver enzymes |
| Musculoskeletal | Arthritis, myositis, joint pain |
| Renal | Nephritis, acute kidney injury |
| Neurological | Peripheral neuropathy, encephalitis, myasthenia gravis |
| Hematologic | Anemia, thrombocytopenia, neutropenia |
| Cancer Type | Diagnostic Biomarker | Description/Role |
|---|---|---|
| Non-Small-Cell Lung Cancer | Gut microbiota profiles (e.g., Bacillus, Bifidobacterium, Faecalibacterium) | Specific gut microbiota compositions can distinguish between NSCLC patients and healthy controls. Diagnostic models using bacterial abundance data have shown high accuracy. |
| Ovarian, Breast Cancer | CD39 expression | High CD39 expression on tumor-infiltrating lymphocytes (TILs) correlates with an immunosuppressive tumor environment and serves as a diagnostic marker. |
| Esophageal Squamous Cell Carcinoma | CD39-expressing T cells (combined with a clinical nomogram) | High levels of CD39+ T cells serve as a diagnostic and prognostic biomarker. A panel combining these cells with clinical indicators improves survival and prognosis predictions. |
| Pharmacodynamic Biomarker | Cancer Type | Application |
|---|---|---|
| 18F-fluoroestradiol (PET imaging) | Metastatic or recurrent breast cancer (ER-positive tumors) |
|
| Phosphorylated AKT | Solid tumors and lymphoma |
|
| IFN-γ and CXCL9 | Head and neck squamous cell carcinoma, non-small cell lung cancer |
|
| IL-8 | Head and neck squamous cell carcinoma, non-small cell lung cancer |
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mokhtari, R.B.; Sambi, M.; Shekari, F.; Satari, K.; Ghafoury, R.; Ashayeri, N.; Eversole, P.; Baluch, N.; Harless, W.W.; Muscarella, L.A.; et al. A Comprehensive Oncological Biomarker Framework Guiding Precision Medicine. Biomolecules 2025, 15, 1304. https://doi.org/10.3390/biom15091304
Mokhtari RB, Sambi M, Shekari F, Satari K, Ghafoury R, Ashayeri N, Eversole P, Baluch N, Harless WW, Muscarella LA, et al. A Comprehensive Oncological Biomarker Framework Guiding Precision Medicine. Biomolecules. 2025; 15(9):1304. https://doi.org/10.3390/biom15091304
Chicago/Turabian StyleMokhtari, Reza Bayat, Manpreet Sambi, Faezeh Shekari, Kosar Satari, Roya Ghafoury, Neda Ashayeri, Paige Eversole, Narges Baluch, William W. Harless, Lucia Anna Muscarella, and et al. 2025. "A Comprehensive Oncological Biomarker Framework Guiding Precision Medicine" Biomolecules 15, no. 9: 1304. https://doi.org/10.3390/biom15091304
APA StyleMokhtari, R. B., Sambi, M., Shekari, F., Satari, K., Ghafoury, R., Ashayeri, N., Eversole, P., Baluch, N., Harless, W. W., Muscarella, L. A., Yeger, H., Das, B., Szewczuk, M. R., & Chakraborty, S. (2025). A Comprehensive Oncological Biomarker Framework Guiding Precision Medicine. Biomolecules, 15(9), 1304. https://doi.org/10.3390/biom15091304









