Metabolic Interactions of Side-chain Extended and Unsaturated Vitamin D Analogs with Cytochrome P450 Enzymes: Integrating Theoretical and Experimental Approaches
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental
2.1.1. General Procedures
2.1.2. Mass Spectrometry and NMR Spectra
2.1.3. HPLC Chromatography and UV Spectroscopy
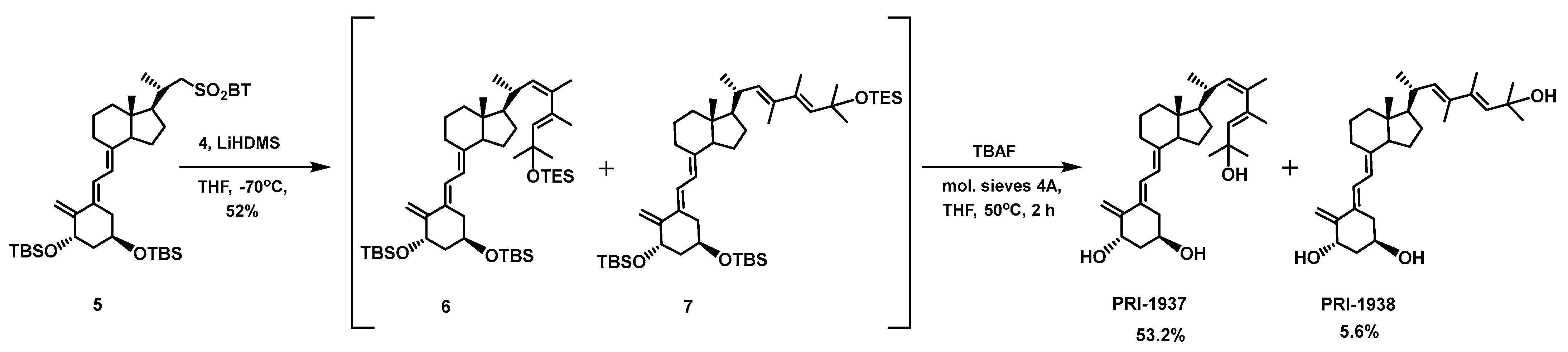
2.2. Syntheses
2.2.1. (E)-2,4-dimethylhex-3-ene-2,5-diol (2)
2.2.2. (E)-5-hydroxy-3,5-dimethylhex-3-en-2-one (3)
2.2.3. (E)-3,5-dimethyl-5-((triethylsilyl)oxy)hex-3-en-2-one (4)
2.2.4. (1S,3R,5E,7E,22Z,24E)-24-Dehydro-24a-homo-23-methyl-1,3-bis-(t-butyldimethylsilyl)-25-(triethylsilyl)ergocalciferol (6) and (1S,3R,5E,7E,22E,24E)-24-dehydro-24a-homo-23-methyl-1,3-bis-(t-butyldimethylsilyl)-25-(triethylsilyl)ergocalciferol (7)
2.2.5. (1S,3R,5E,7E,22Z,24E)-24-Dehydro-24a-homo-23-methyl-1,25-dihydroxyergocalciferol (PRI-1937) and (1S,3R,5E,7E,22E,24E)-24-dehydro-24a-homo-23-methyl-1,25-dihydroxyergocalciferol (PRI-1938)
2.2.6. (1S,3R,5Z,7E,22Z,24E)-24-Dehydro-24a-homo-23-methyl-1,25-dihydroxyergocalciferol (PRI-1927)
2.3. Metabolic Resistance of Vitamin D Analogs to hCYP24A1- and hCYP3A4-Mediated Degradation
2.4. Theoretical Calculations
2.4.1. Preparation of Vitamin D3 Analogs and Structural Models of rCYP24A1 and hCYP3A4 Enzymes
2.4.2. Initial System Configurations: Molecular Docking
2.4.3. Interaction with Enzyme Models rCYP24A1 and hCYP3A4: Molecular Dynamics Simulations
2.4.4. Evaluating Binding Free Energy with Molecular Mechanics Poisson–Boltzmann Surface Area (MM/PBSA)
3. Results and Discussion
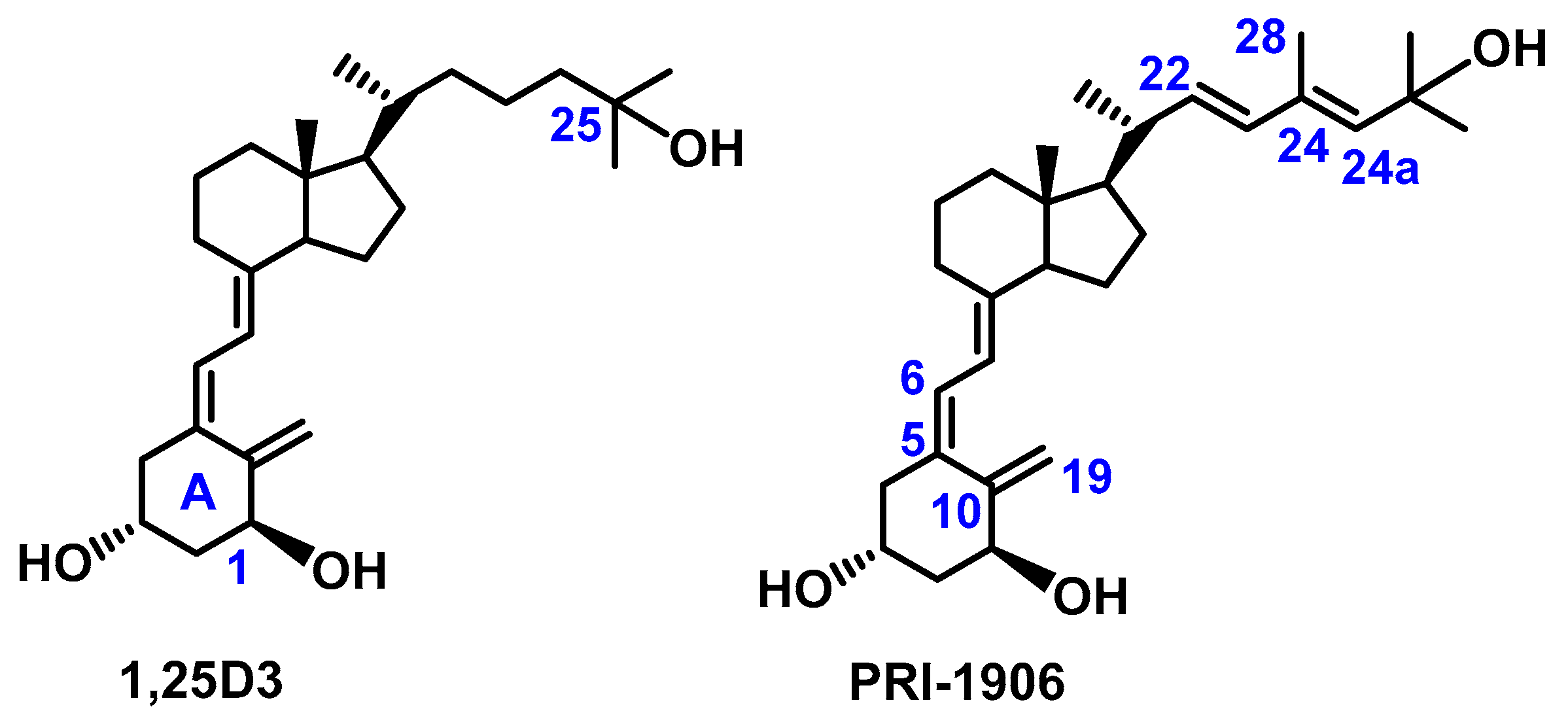
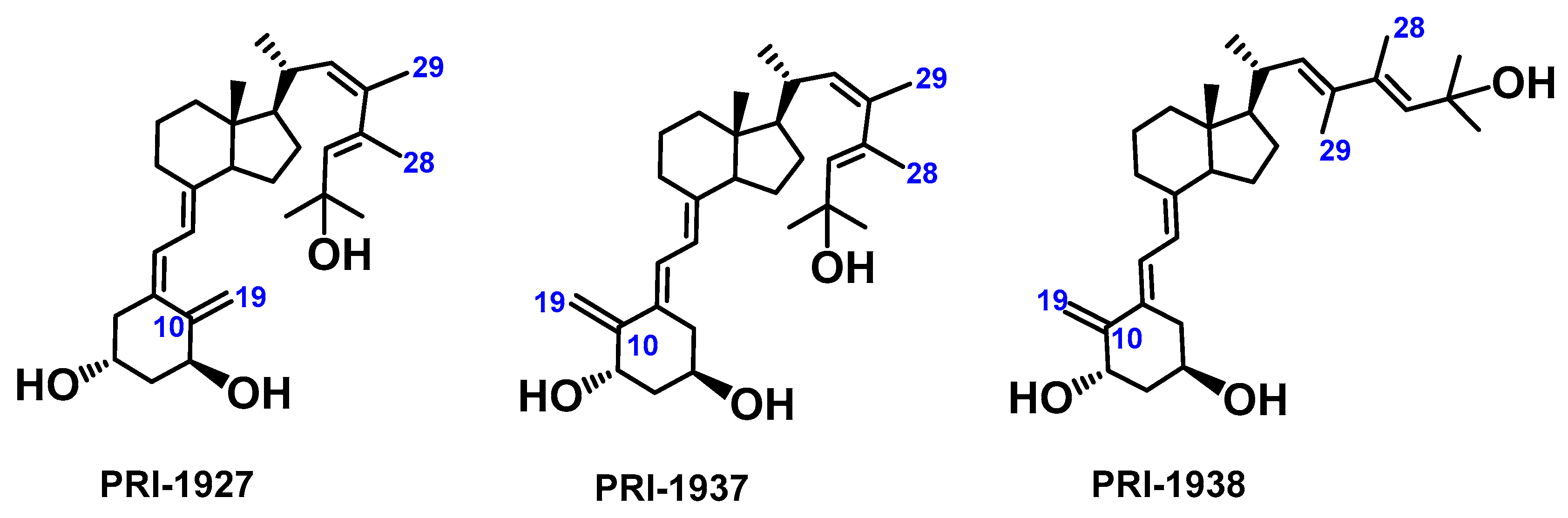
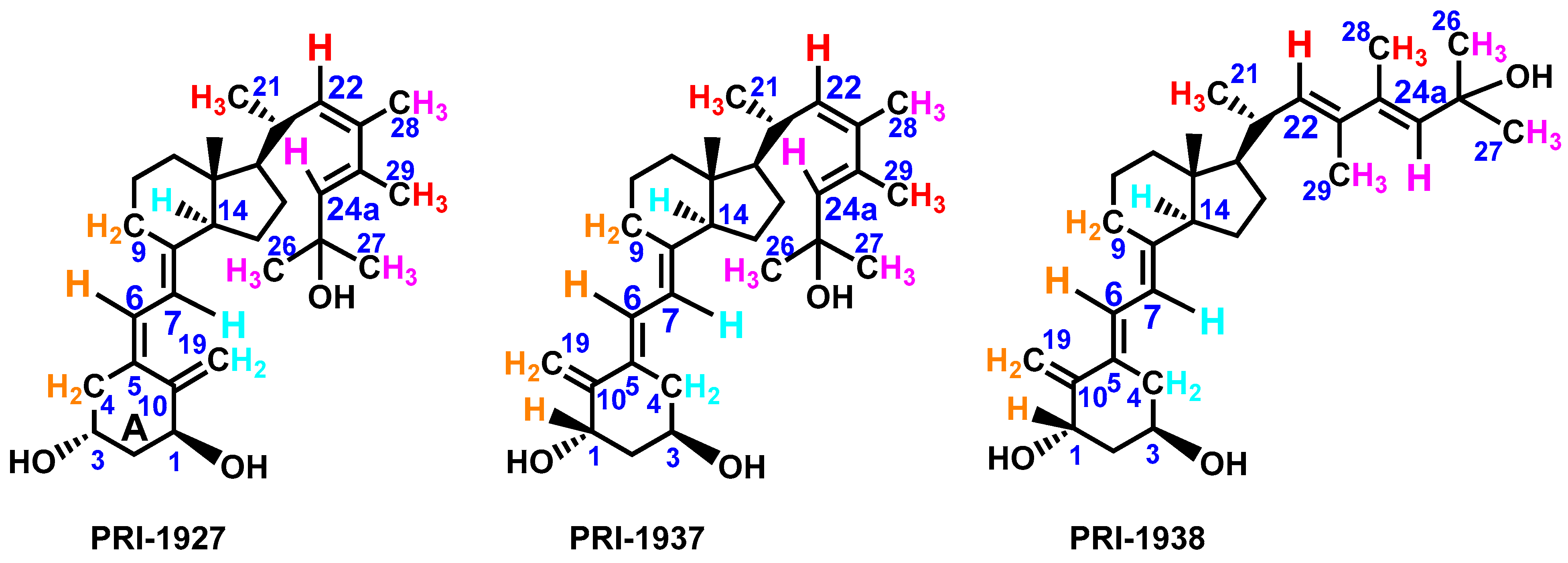
3.1. Synthesis of Analogs PRI-1927, PRI-1937, and PRI-1938
3.2. Structure Assignment of PRI-1927, PRI-1937, and PRI-1938 by TOF MS ES+ and 1H and 13C NMR
3.3. Metabolic Conversion of 1,25D3 and Its Analogs by Cytochrome CYP3A4
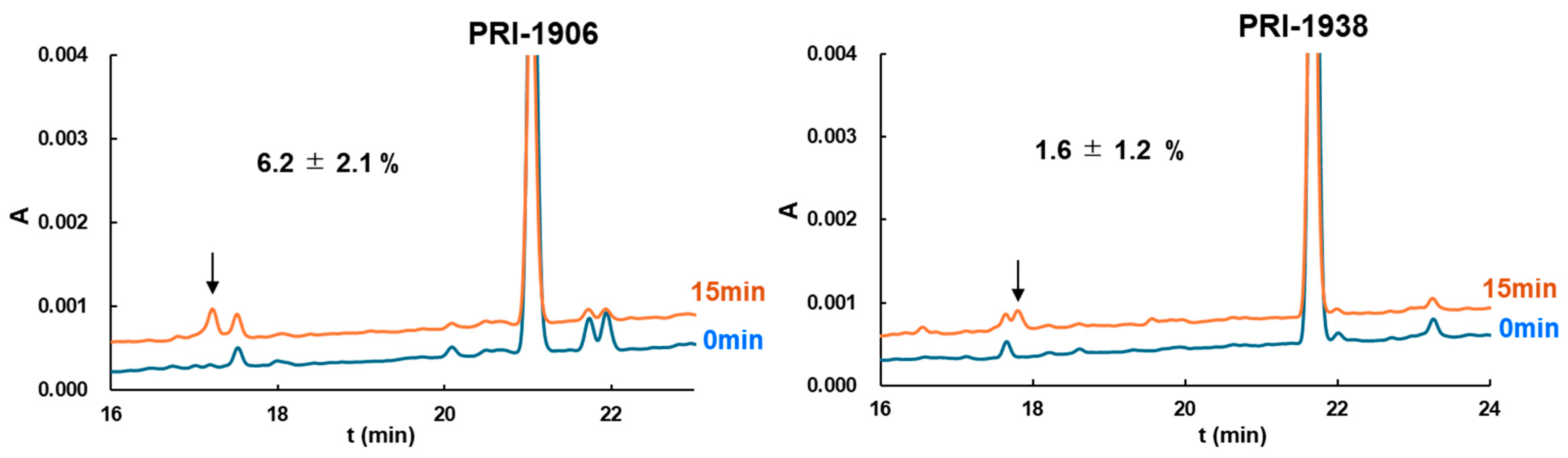
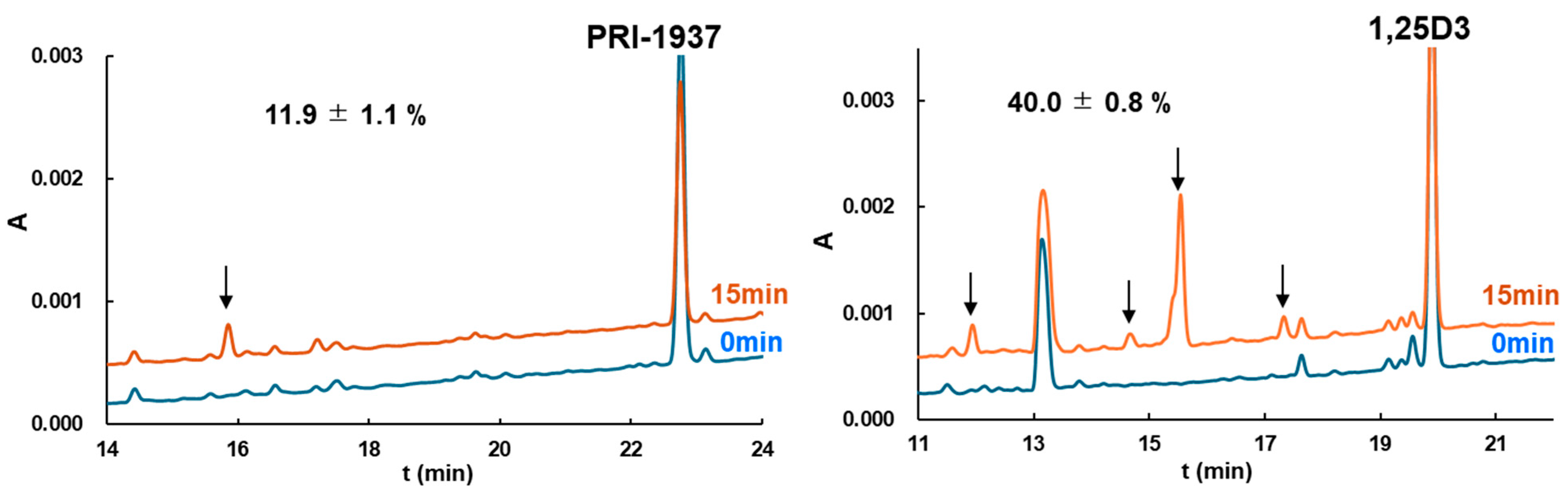
3.4. Metabolic Conversion of 1,25D3 and Its Analogs by Cytochrome CYP24A1
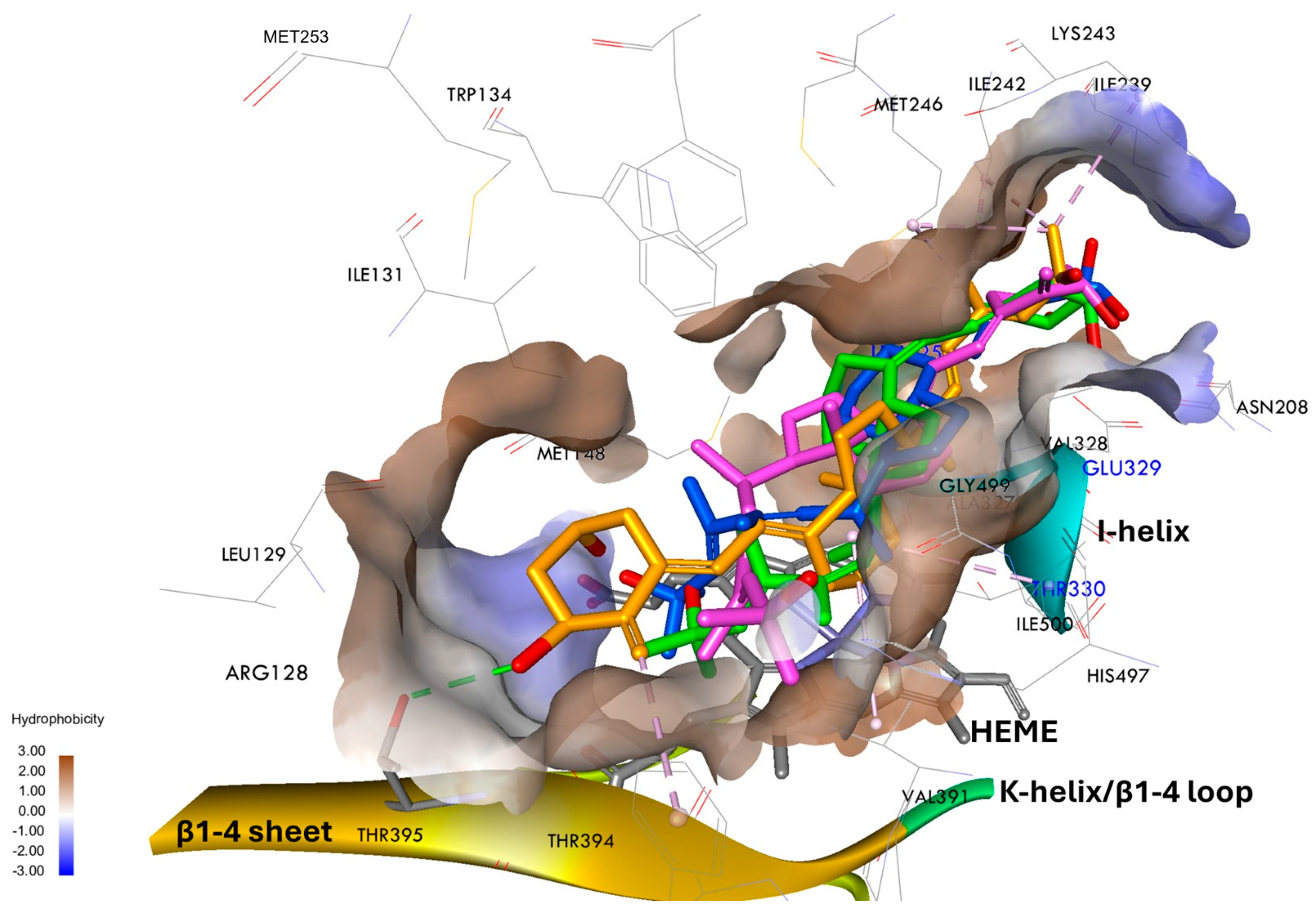
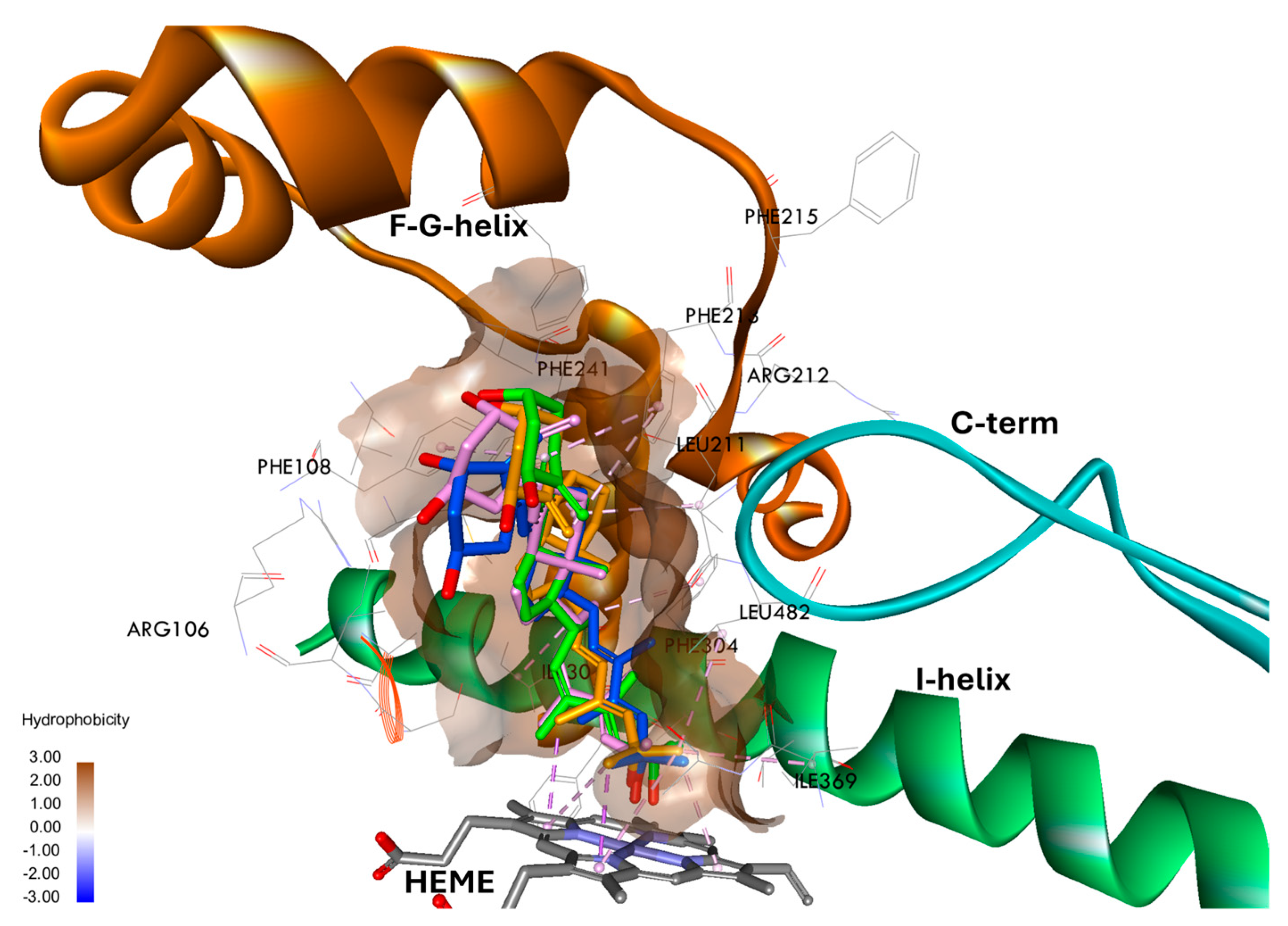
3.5. Characterization of the Binding Sites for Vitamin D3 Analogs in the CYP24A1 and hCYP3A4 Models by Molecular Docking
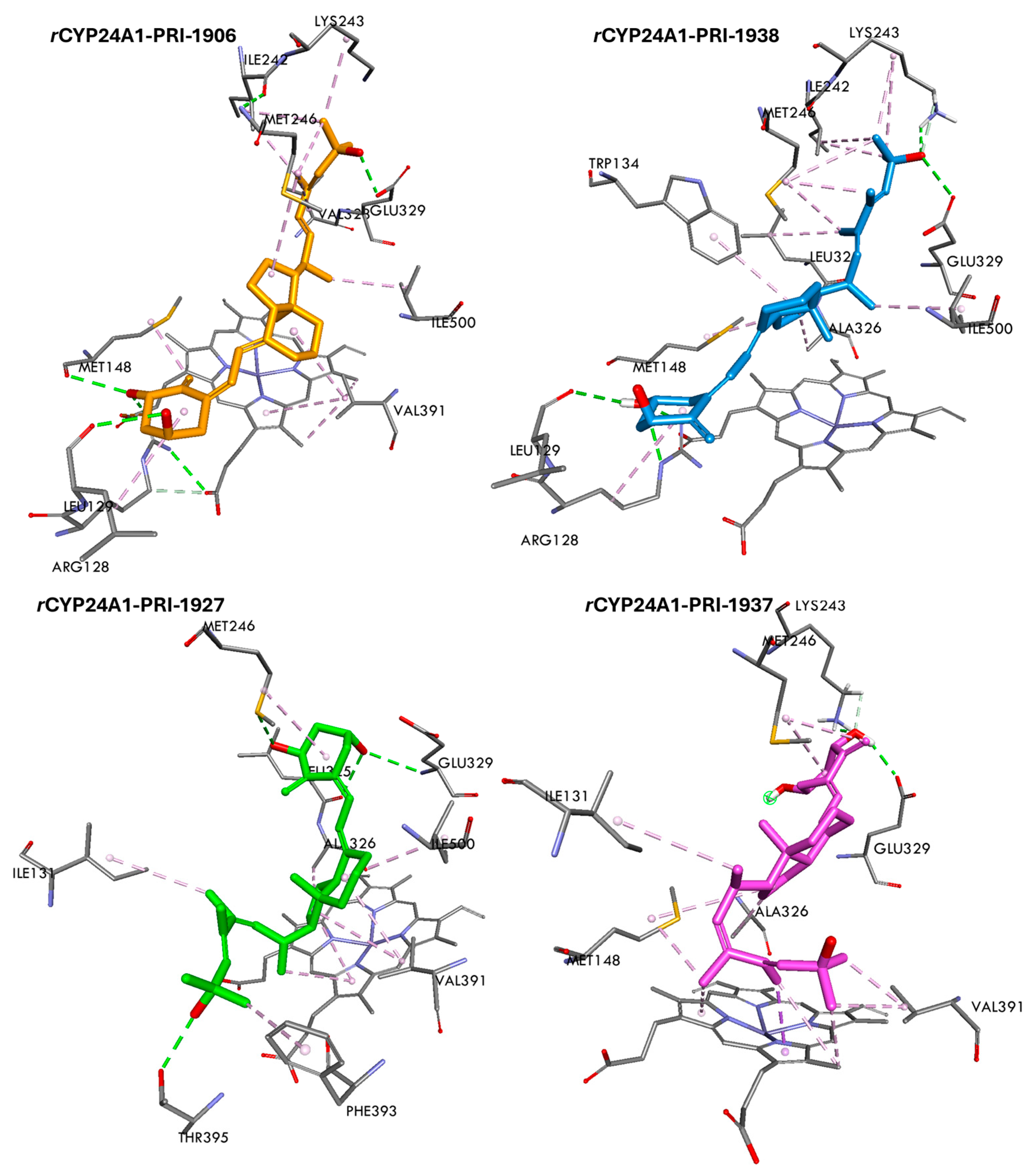
3.6. Binding Free Energy and Interactions of Vitamin D3 Analogs with rCYP24A1
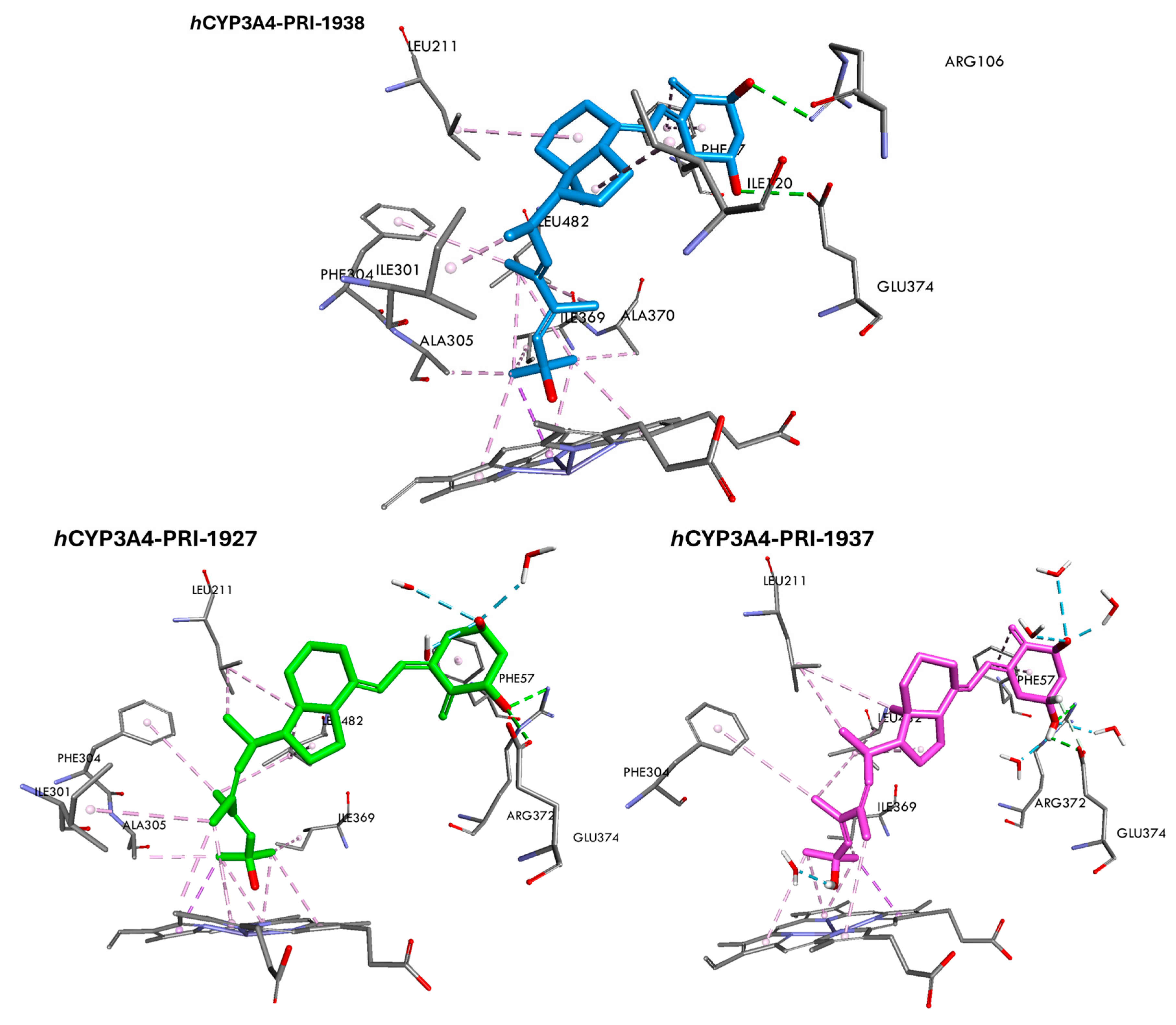
3.7. Binding Free Energy and Interactions of Vitamin D3 Analogs with hCYP3A4
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Uga, M.; Kaneko, I.; Shiozaki, Y.; Koike, M.; Tsugawa, N.; Jurutka, P.W.; Miyamoto, K.I.; Segawa, H. The Role of Intestinal Cytochrome P450s in Vitamin D Metabolism. Biomolecules 2024, 14, 717. [Google Scholar] [CrossRef]
- Saponaro, F.; Saba, A.; Zucchi, R. An Update on Vitamin D Metabolism. Int. J. Mol. Sci. 2020, 21, 6573. [Google Scholar] [CrossRef]
- Maestro, M.A.; Molnar, F.; Carlberg, C. Vitamin D and Its Synthetic Analogs. J. Med. Chem. 2019, 62, 6854–6875. [Google Scholar] [CrossRef]
- Alshabrawy, A.K.; Cui, Y.; Sylvester, C.; Yang, D.; Petito, E.S.; Barratt, K.R.; Sawyer, R.K.; Heatlie, J.K.; Polara, R.; Sykes, M.J.; et al. Therapeutic Potential of a Novel Vitamin D(3) Oxime Analogue, VD1-6, with CYP24A1 Enzyme Inhibitory Activity and Negligible Vitamin D Receptor Binding. Biomolecules 2022, 12, 960. [Google Scholar] [CrossRef] [PubMed]
- Mizumoto, Y.; Sakamoto, R.; Nagata, A.; Sakane, S.; Kittaka, A.; Odagi, M.; Tera, M.; Nagasawa, K. Synthesis of C2-Alkoxy-Substituted 19-Nor Vitamin D(3) Derivatives: Stereoselectivity and Biological Activity. Biomolecules 2022, 12, 69. [Google Scholar] [CrossRef]
- Maekawa, K.; Ishizawa, M.; Ikawa, T.; Sajiki, H.; Matsumoto, T.; Tokiwa, H.; Makishima, M.; Yamada, S. Syntheses of 25-Adamantyl-25-alkyl-2-methylidene-1alpha,25-dihydroxyvitamin D(3) Derivatives with Structure-Function Studies of Antagonistic and Agonistic Active Vitamin D Analogs. Biomolecules 2023, 13, 1082. [Google Scholar] [CrossRef] [PubMed]
- Nachliely, M.; Trachtenberg, A.; Khalfin, B.; Nalbandyan, K.; Cohen-Lahav, M.; Yasuda, K.; Sakaki, T.; Kutner, A.; Danilenko, M. Dimethyl fumarate and vitamin D derivatives cooperatively enhance VDR and Nrf2 signaling in differentiating AML cells in vitro and inhibit leukemia progression in a xenograft mouse model. J. Steroid Biochem. Mol. Biol. 2019, 188, 8–16. [Google Scholar] [CrossRef]
- Sibilska-Kaminski, I.K.; Fabisiak, A.; Brzeminski, P.; Plum, L.A.; Sicinski, R.R.; DeLuca, H.F. Novel superagonist analogs of 2-methylene calcitriol: Design, molecular docking, synthesis and biological evaluation. Bioorg. Chem. 2022, 118, 105416. [Google Scholar] [CrossRef]
- Kawagoe, F.; Mototani, S.; Yasuda, K.; Mano, H.; Sakaki, T.; Kittaka, A. Stereoselective Synthesis of 24-Fluoro-25-Hydroxyvitamin D3 Analogues and Their Stability to hCYP24A1-Dependent Catabolism. Int. J. Mol. Sci. 2021, 22, 11863. [Google Scholar] [CrossRef] [PubMed]
- Chodynski, M.; Wietrzyk, J.; Marcinkowska, E.; Opolski, A.; Szelejewski, W.; Kutner, A. Synthesis and antiproliferative activity of side-chain unsaturated and homologated analogs of 1,25-dihydroxyvitamin D(2): (24E)-(1S)-24-Dehydro-24a-homo-1,25-dihydroxyergocalciferol and congeners. Steroids 2002, 67, 789–798. [Google Scholar] [CrossRef]
- Zolek, T.; Yasuda, K.; Brown, G.; Sakaki, T.; Kutner, A. In Silico Prediction of the Metabolic Resistance of Vitamin D Analogs against CYP3A4 Metabolizing Enzyme. Int. J. Mol. Sci. 2022, 23, 7845. [Google Scholar] [CrossRef]
- Norlin, M.; Wikvall, K. Enzymatic activation in vitamin D signaling—Past, present and future. Arch. Biochem. Biophys. 2023, 742, 109639. [Google Scholar] [CrossRef] [PubMed]
- Pineda-Lancheros, L.E.; Galvez-Navas, J.M.; Rojo-Tolosa, S.; Membrive-Jimenez, C.; Valverde-Merino, M.I.; Martinez-Martinez, F.; Sanchez-Martin, A.; Ramirez-Tortosa, M.; Perez-Ramirez, C.; Jimenez-Morales, A. Polymorphisms in VDR, CYP27B1, CYP2R1, GC and CYP24A1 Genes as Biomarkers of Survival in Non-Small Cell Lung Cancer: A Systematic Review. Nutrients 2023, 15, 1525. [Google Scholar] [CrossRef]
- Annalora, A.J.; Goodin, D.B.; Hong, W.X.; Zhang, Q.; Johnson, E.F.; Stout, C.D. Crystal structure of CYP24A1, a mitochondrial cytochrome P450 involved in vitamin D metabolism. J. Mol. Biol. 2010, 396, 441–451. [Google Scholar] [CrossRef] [PubMed]
- BIOVIA Discovery Studio, version 2022; Dassault Systèmes: San Diego, CA, USA, 2022.
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Petersson, G.A.; Nakatsuji, H.; et al. GaussView, version 5.0.; Gaussian, Inc.: Wallingford, CT, USA, 2016. [Google Scholar]
- Breneman, C.M.; Wiberg, K.B. Determining Atom-Centered Monopoles from Molecular Electrostatic Potentials—The Need for High Sampling Density in Formamide Conformational-Analysis. J. Comput. Chem. 1990, 11, 361–373. [Google Scholar] [CrossRef]
- Prosser, D.E.; Kaufmann, M.; O’Leary, B.; Byford, V.; Jones, G. Single A326G mutation converts human CYP24A1 from 25-OH-D3-24-hydroxylase into -23-hydroxylase, generating 1alpha,25-(OH)2D3-26,23-lactone. Proc. Natl. Acad. Sci. USA 2007, 104, 12673–12678. [Google Scholar] [CrossRef]
- Jones, G.; Willett, P.; Glen, R.C. Molecular recognition of receptor sites using a genetic algorithm with a description of desolvation. J. Mol. Biol. 1995, 245, 43–53. [Google Scholar] [CrossRef]
- Jones, G.; Willett, P.; Glen, R.C.; Leach, A.R.; Taylor, R. Development and validation of a genetic algorithm for flexible docking. J. Mol. Biol. 1997, 267, 727–748. [Google Scholar] [CrossRef]
- GOLD, version 2024.3.0; User Guide; CCDC Software Ltd.: Cambrige, UK, 2024.
- Grayson, P.; Tajkhorshid, E.; Schulten, K. Mechanisms of selectivity in channels and enzymes studied with interactive molecular dynamics. Biophys. J. 2003, 85, 36–48. [Google Scholar] [CrossRef]
- Brooks, B.R.; Brooks, C.L., 3rd; Mackerell, A.D., Jr.; Nilsson, L.; Petrella, R.J.; Roux, B.; Won, Y.; Archontis, G.; Bartels, C.; Boresch, S.; et al. CHARMM: The biomolecular simulation program. J. Comput. Chem. 2009, 30, 1545–1614. [Google Scholar] [CrossRef]
- Guvench, O.; MacKerell, A.D., Jr. Comparison of protein force fields for molecular dynamics simulations. Methods Mol. Biol. 2008, 443, 63–88. [Google Scholar] [CrossRef]
- Jorgensen, W.L.; Chandrasekhar, J.; Madura, J.D.; Impey, R.W.; Klein, M.L. Comparison of Simple Potential Functions for Simulating Liquid Water. J. Chem. Phys. 1983, 79, 926–935. [Google Scholar] [CrossRef]
- Berendsen, H.J.C.; Postma, J.P.M.; Vangunsteren, W.F.; Dinola, A.; Haak, J.R. Molecular-Dynamics with Coupling to an External Bath. J. Chem. Phys. 1984, 81, 3684–3690. [Google Scholar] [CrossRef]
- Essmann, U.; Perera, L.; Berkowitz, M.L.; Darden, T.; Lee, H.; Pedersen, L.G. A Smooth Particle Mesh Ewald Method. J. Chem. Phys. 1995, 103, 8577–8593. [Google Scholar] [CrossRef]
- Sagui, C.; Darden, T.A. Molecular dynamics simulations of biomolecules: Long-range electrostatic effects. Annu. Rev. Biophys. Biomol. Struct. 1999, 28, 155–179. [Google Scholar] [CrossRef]
- Kollman, P.A.; Massova, I.; Reyes, C.; Kuhn, B.; Huo, S.; Chong, L.; Lee, M.; Lee, T.; Duan, Y.; Wang, W.; et al. Calculating structures and free energies of complex molecules: Combining molecular mechanics and continuum models. Acc. Chem. Res. 2000, 33, 889–897. [Google Scholar] [CrossRef] [PubMed]
- Verma, J.; Subbarao, N. In silico identification of small molecule protein-protein interaction inhibitors: Targeting hotspot regions at the interface of MXRA8 and CHIKV envelope protein. J. Biomol. Struct. Dyn. 2023, 41, 3349–3367. [Google Scholar] [CrossRef] [PubMed]
- Varughese, J.K.; J, K.; S, S.K.; Francis, D.; L, J.L.K.; G, A.T. Identification of some dietary flavonoids as potential inhibitors of TMPRSS2 through protein-ligand interaction studies and binding free energy calculations. Struct. Chem. 2022, 33, 1489–1502. [Google Scholar] [CrossRef] [PubMed]
- Jarzynski, C. Nonequilibrium equality for free energy differences. Phys. Rev. Lett. 1997, 78, 2690–2693. [Google Scholar] [CrossRef]
- El Hassab, M.A.; Shoun, A.A.; Al-Rashood, S.T.; Al-Warhi, T.; Eldehna, W.M. Identification of a New Potential SARS-COV-2 RNA-Dependent RNA Polymerase Inhibitor via Combining Fragment-Based Drug Design, Docking, Molecular Dynamics, and MM-PBSA Calculations. Front. Chem. 2020, 8, 584894. [Google Scholar] [CrossRef]
- Milczarek, M.; Chodynski, M.; Filip-Psurska, B.; Martowicz, A.; Krupa, M.; Krajewski, K.; Kutner, A.; Wietrzyk, J. Synthesis and Biological Activity of Diastereomeric and Geometric Analogs of Calcipotriol, PRI-2202 and PRI-2205, Against Human HL-60 Leukemia and MCF-7 Breast Cancer Cells. Cancers 2013, 5, 1355–1378. [Google Scholar] [CrossRef]
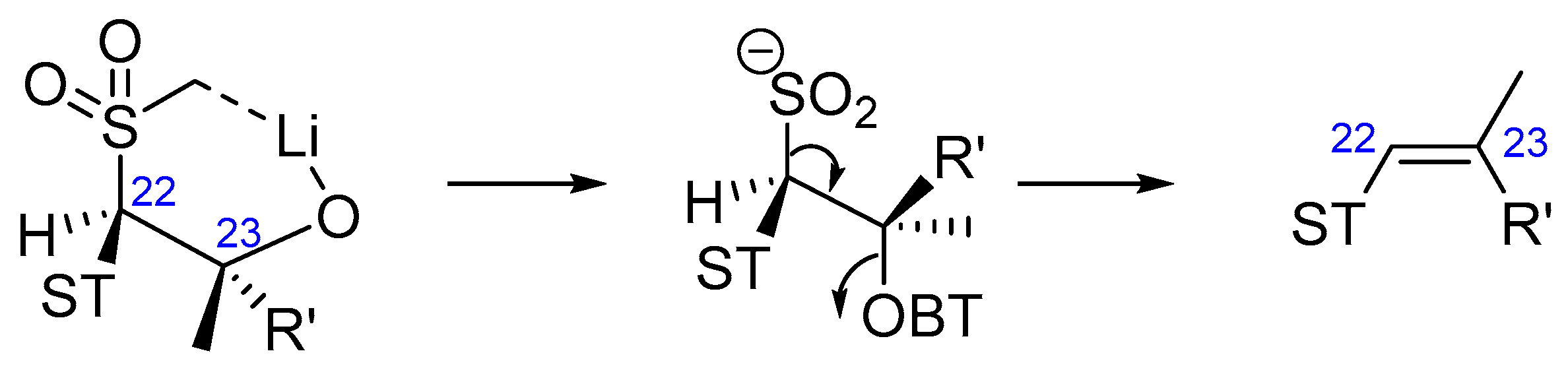
- Blakemore, P.R. The modified Julia olefination: Alkene synthesis via the condensation of metallated heteroarylalkylsulfones with carbonyl compounds. J. Chem. Soc. Perkin Trans. 1 2002, 1, 2563–2585. [Google Scholar] [CrossRef]
- Colovos, C.; Yeates, T.O. Verification of protein structures: Patterns of nonbonded atomic interactions. Protein Sci. 1993, 2, 1511–1519. [Google Scholar] [CrossRef] [PubMed]
- Bowie, J.U.; Luthy, R.; Eisenberg, D. A method to identify protein sequences that fold into a known three-dimensional structure. Science 1991, 253, 164–170. [Google Scholar] [CrossRef] [PubMed]

| Vitamin D Analogs | CYP24A1 | CYP3A4 | ||
|---|---|---|---|---|
| Metabolic Conversion by hCYP24A1 [% ± SD] | rCYP24A1 ΔGbind [kcal/mol] | Metabolic Conversion by hCYP3A4 [% ± SD] | hCYP3A4 ΔGbind[kcal/mol] | |
| PRI-1927 | 14.2 ± 0.8 | −56.97 | 16.7 ± 1.3 | −51.80 |
| PRI-1937 | 11.9 ± 1.1 | −74.37 | 16.3 ± 3.7 | −70.61 |
| PRI-1938 | 5.2 ± 1.3 | −124.54 | 1.6 ± 1.2 | −86.06 |
| PRI-1906 | 4.4 ± 0.7 | −93.49 | 6.2 ± 2.1 | −75.80 |
| 1,25D3 | 40.0 ± 0.8 | −12.91 | 14.9 ± 3.8 | −28.93 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Żołek, T.; Kadam, M.; Nadkarni, S.; Yasuda, K.; Chodyński, M.; Krajewski, K.; Michalak, O.; Tobiasz, J.; Kubiszewski, M.; Sakaki, T.; et al. Metabolic Interactions of Side-chain Extended and Unsaturated Vitamin D Analogs with Cytochrome P450 Enzymes: Integrating Theoretical and Experimental Approaches. Biomolecules 2025, 15, 1222. https://doi.org/10.3390/biom15091222
Żołek T, Kadam M, Nadkarni S, Yasuda K, Chodyński M, Krajewski K, Michalak O, Tobiasz J, Kubiszewski M, Sakaki T, et al. Metabolic Interactions of Side-chain Extended and Unsaturated Vitamin D Analogs with Cytochrome P450 Enzymes: Integrating Theoretical and Experimental Approaches. Biomolecules. 2025; 15(9):1222. https://doi.org/10.3390/biom15091222
Chicago/Turabian StyleŻołek, Teresa, Mayur Kadam, Sharmin Nadkarni, Kaori Yasuda, Michał Chodyński, Krzysztof Krajewski, Olga Michalak, Joanna Tobiasz, Marek Kubiszewski, Toshiyuki Sakaki, and et al. 2025. "Metabolic Interactions of Side-chain Extended and Unsaturated Vitamin D Analogs with Cytochrome P450 Enzymes: Integrating Theoretical and Experimental Approaches" Biomolecules 15, no. 9: 1222. https://doi.org/10.3390/biom15091222
APA StyleŻołek, T., Kadam, M., Nadkarni, S., Yasuda, K., Chodyński, M., Krajewski, K., Michalak, O., Tobiasz, J., Kubiszewski, M., Sakaki, T., & Kutner, A. (2025). Metabolic Interactions of Side-chain Extended and Unsaturated Vitamin D Analogs with Cytochrome P450 Enzymes: Integrating Theoretical and Experimental Approaches. Biomolecules, 15(9), 1222. https://doi.org/10.3390/biom15091222









