Carboxy-Methylation of the Catalytic Subunit of Protein Phosphatase 2A (PP2Ac) Integrates Methionine Availability with Methionine Addicted Cancer Cell Proliferation
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Lines and Growth Conditions
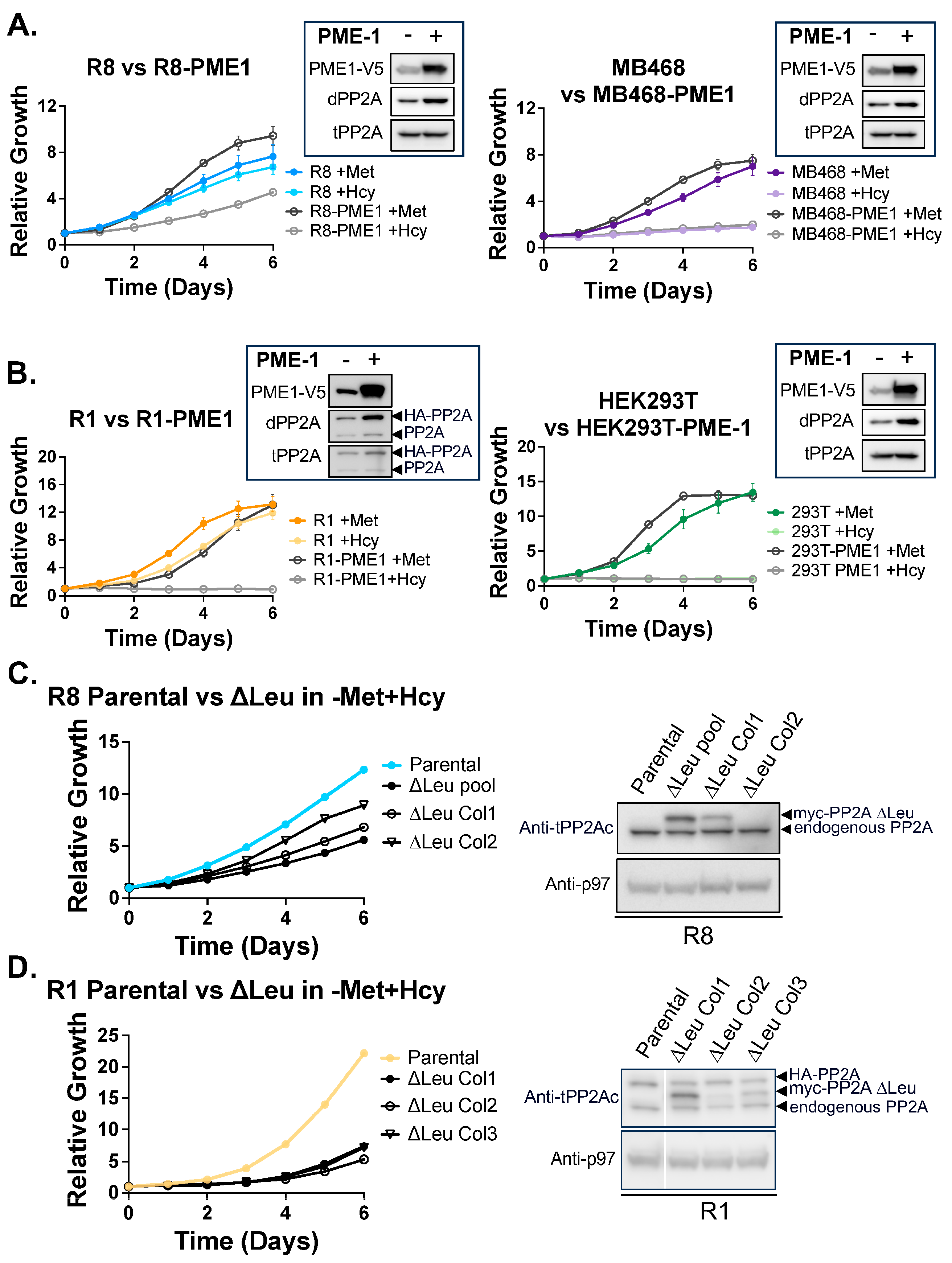
2.2. PME-1 Overexpression Cell Lines
2.3. ∆Leu309 Cell Lines
2.4. Growth Curves, Viability Assay, IC50 Determination
2.5. Protein Analysis
2.6. Cell Cycle Assays
2.7. Cell Death Assay
2.8. RNA Sequencing
2.9. Metabolite Measurements Using LC-MS
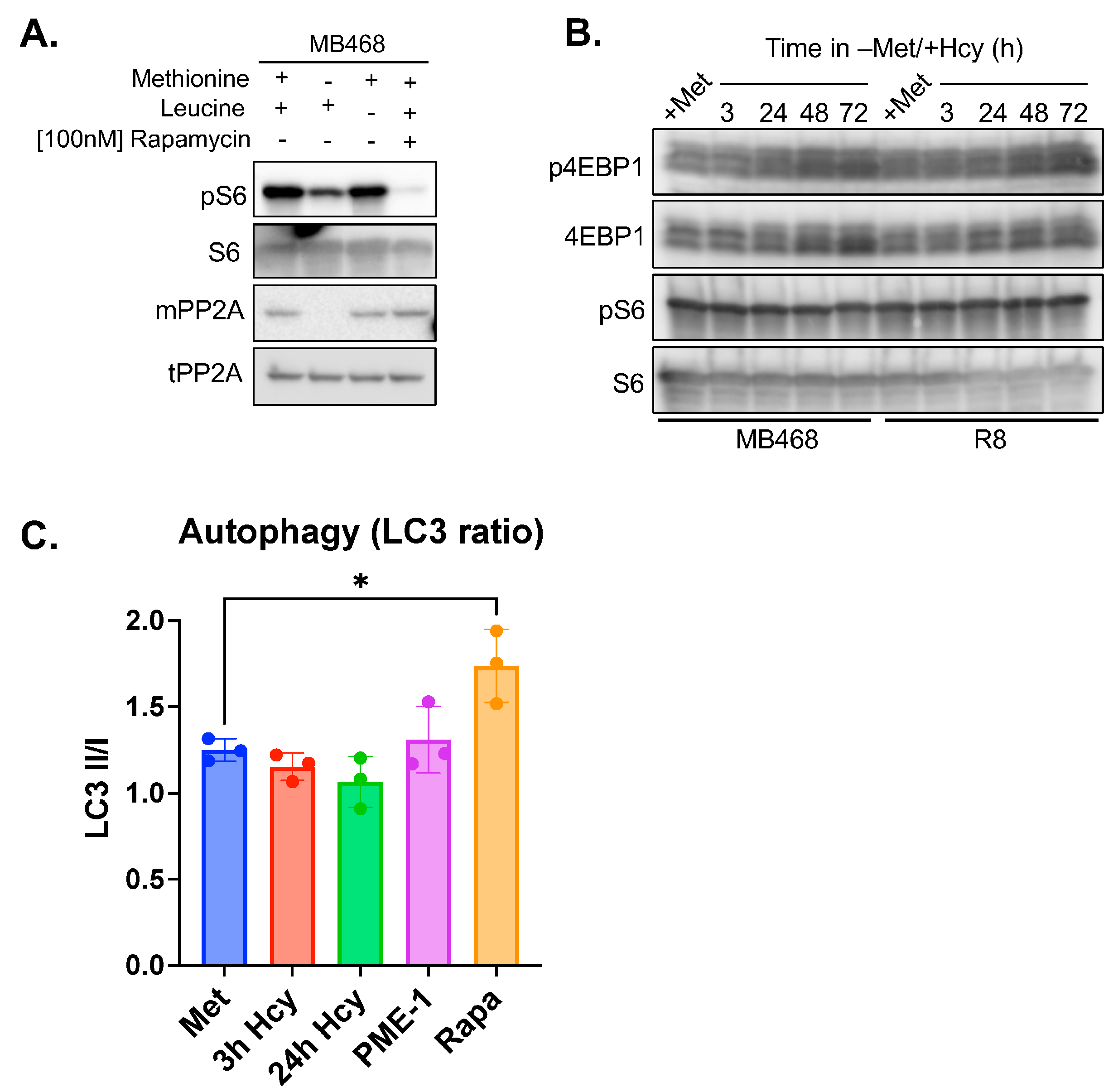
2.10. Autophagy
2.11. Activity Assay
2.12. Methyl-Proteome Profiling
Lysate Preparation
2.13. Mass Spectrometric Analysis for 3 Hr Met vs. Hcy Treatment
2.14. Identification of Peptides
2.15. Methyl False Discovery Estimation
2.16. Bioinformatic Analysis
3. Results
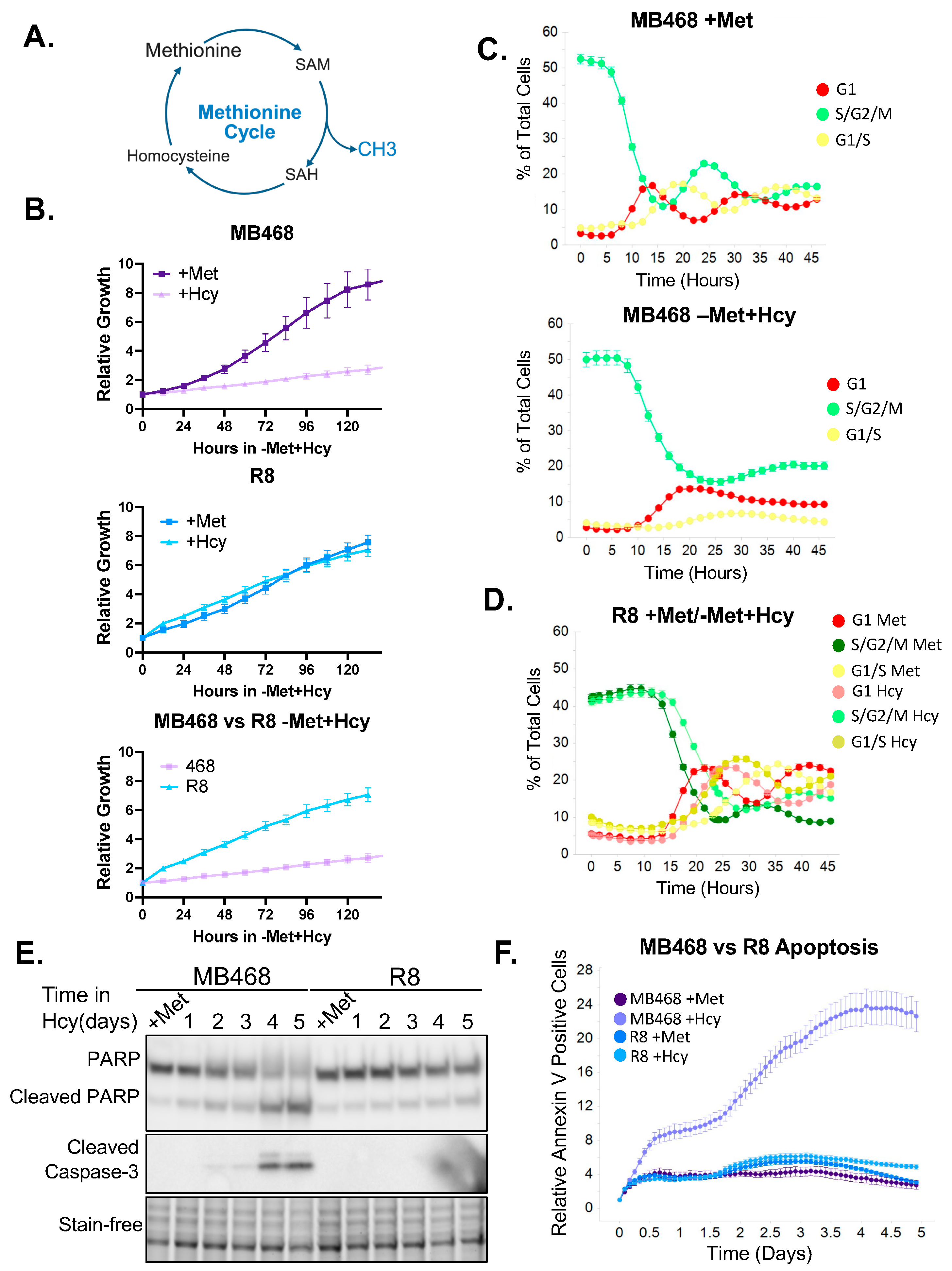
3.1. Characterization of the Cell Cycle and Survival Response of Methionine-Dependent and Independent Cells
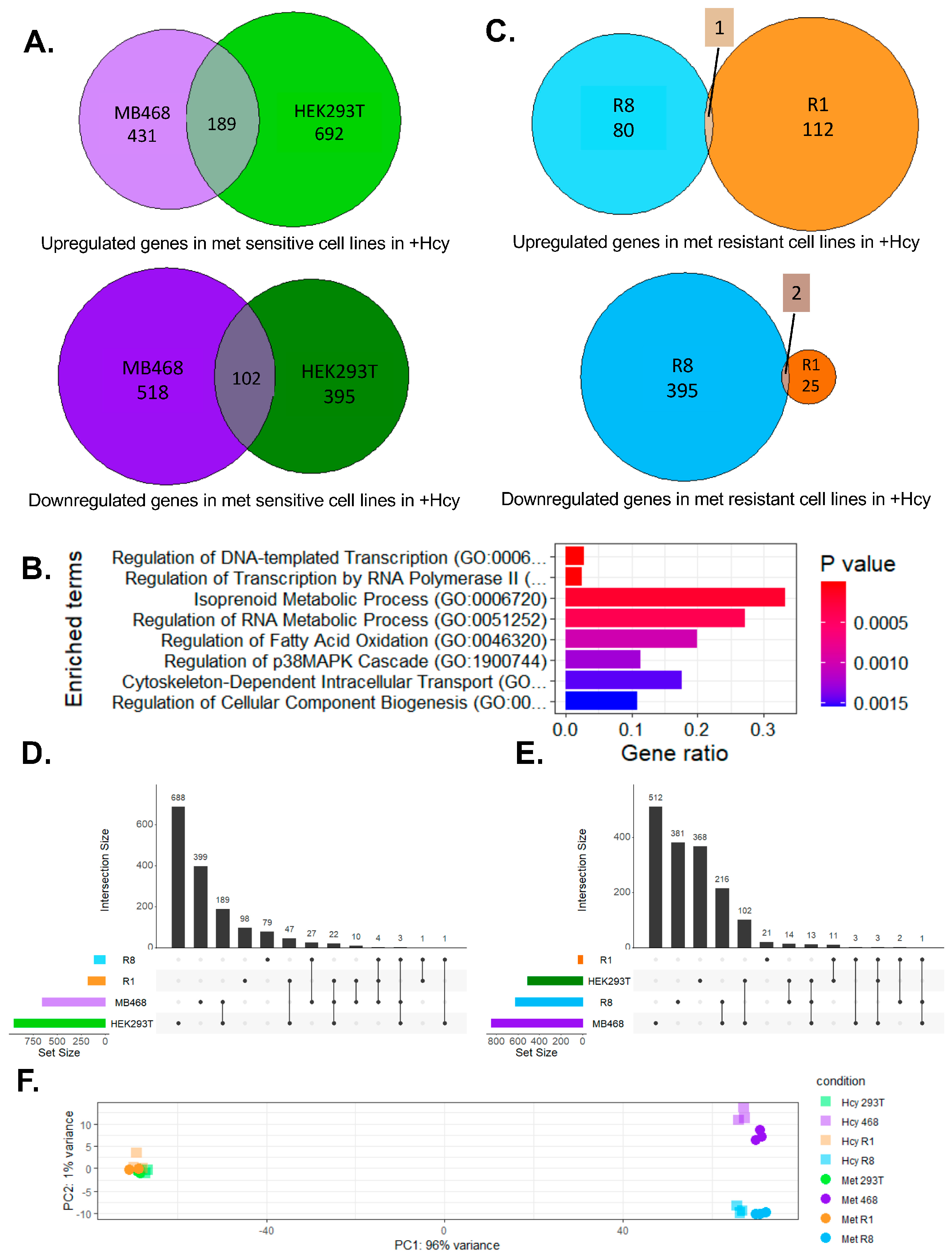
3.2. Methionine-Independent Cells Likely Employ Distinct Adaptive Mechanisms
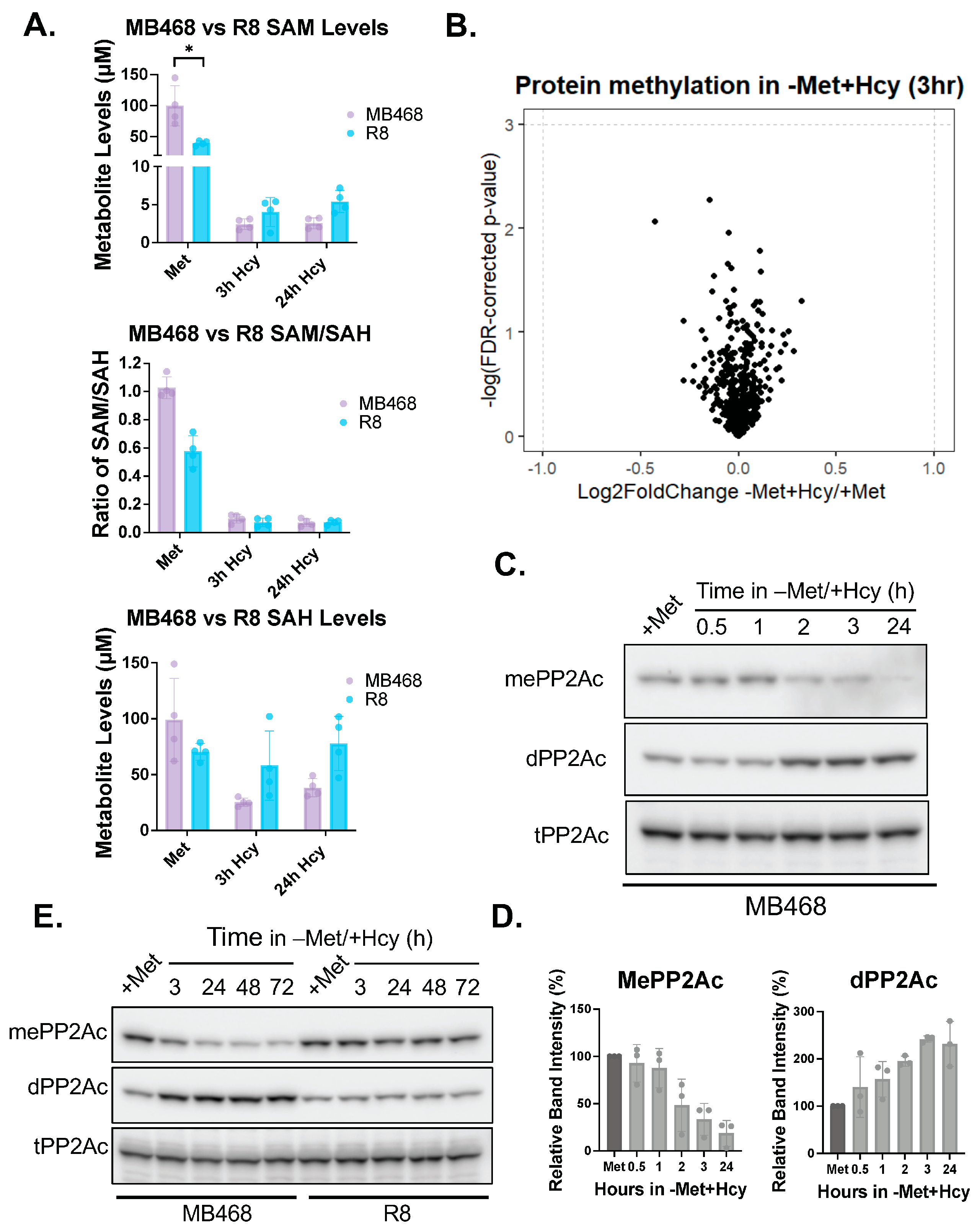
3.3. Carboxy-Methylation of the Catalytic PP2A Subunit Is Highly Responsive to Changes in Exogenous Methionine
3.4. Methionine-Dependence Is Not Mediated Through mTORC1 Signaling
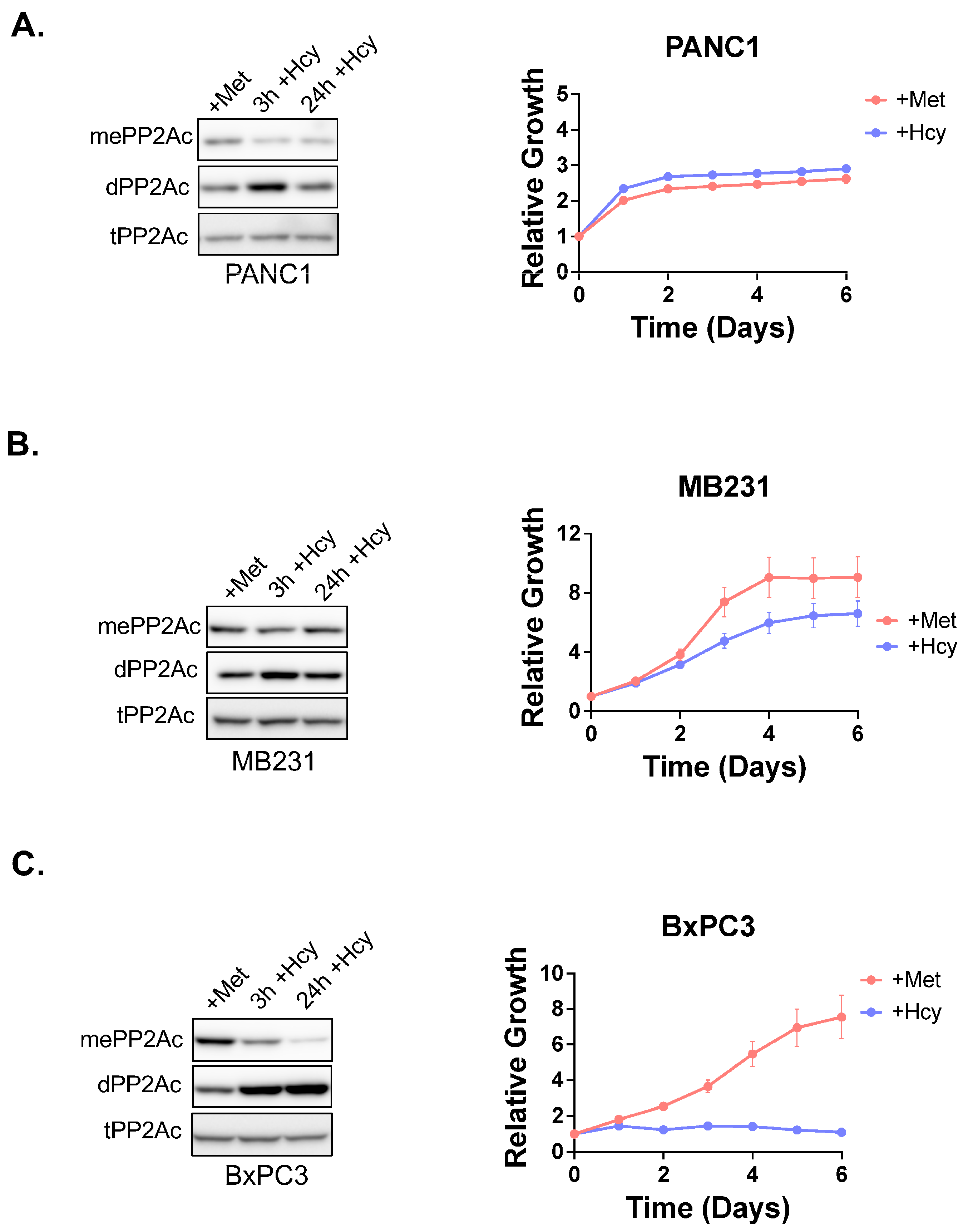
3.5. PP2Ac Methylation State Controls Methionine Dependence
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| PP2A | Protein Phosphatase 2A |
| SAM | S-Adenosyl-Methionine |
| -Met+Hcy | Methionine depleted media supplemented with homocysteine |
References
- Weindruch, R.; Walford, R.L.; Fligiel, S.; Guthrie, D. The retardation of aging in mice by dietary restriction: Longevity, cancer, immunity and lifetime energy intake. J. Nutr. 1986, 116, 641–654. [Google Scholar] [CrossRef]
- Lees, E.K.; Banks, R.; Cook, C.; Hill, S.; Morrice, N.; Grant, L.; Mody, N.; Delibegovic, M. Direct comparison of methionine restriction with leucine restriction on the metabolic health of C57BL/6J mice. Sci. Rep. 2017, 7, 9977. [Google Scholar] [CrossRef] [PubMed]
- Green, C.L.; Lamming, D.W.; Fontana, L. Molecular mechanisms of dietary restriction promoting health and longevity. Nat. Rev. Mol. Cell Biol. 2021, 23, 56. [Google Scholar] [CrossRef]
- Orentreich, N.; Matias, J.R.; DeFelice, A.; Zimmerman, J.A. Low methionine ingestion by rats extends life span. J. Nutr. 1993, 123, 269–274. [Google Scholar] [CrossRef]
- Parkhitko, A.A.; Wang, L.; Filine, E.; Jouandin, P.; Leshchiner, D.; Binari, R.; Asara, J.M.; Rabinowitz, J.D.; Perrimon, N. A genetic model of methionine restriction extends Drosophila health- and lifespan. Proc. Natl. Acad. Sci. USA 2021, 118, e2110387118. [Google Scholar] [CrossRef]
- Sugimura, T.; Birnbaum, S.M.; Winitz, M.; Greenstein, J.P. Quantitative nutritional studies with water-soluble, chemically defined diets. VIII. The forced feeding of diets each lacking in one essential amino acid. Arch. Biochem. Biophys. 1959, 81, 448–455. [Google Scholar] [CrossRef]
- Gao, X.; Sanderson, S.M.; Dai, Z.; Reid, M.A.; Cooper, D.E.; Lu, M.; John P Richie, J.; Ciccarella, A.; Calcagnotto, A.; Mikhael, P.G.; et al. Dietary methionine links nutrition and metabolism to the efficacy of cancer therapies. Nature 2019, 572, 397. [Google Scholar] [CrossRef]
- Guo, H.; Lishko, V.K.; Herrera, H.; Groce, A.; Kubota, T.; Hoffman, R.M. Therapeutic tumor-specific cell cycle block induced by methionine starvation In Vivo. Cancer Res. 1993, 53, 5676–5679. [Google Scholar]
- Poirson-Bichat, F.; Gonfalone, G.; Bras-Gonçalves, R.A.; Dutrillaux, B.; Poupon, M.F. Growth of methionine-dependent human prostate cancer (PC-3) is inhibited by ethionine combined with methionine starvation. Br. J. Cancer 1997, 75, 1605. [Google Scholar] [CrossRef]
- Kaiser, P. Methionine Dependence of Cancer. Biomolecules 2020, 10, 568. [Google Scholar] [CrossRef]
- Hoffman, R.M.; Erbe, R.W. High In Vivo rates of methionine biosynthesis in transformed human and malignant rat cells auxotrophic for methionine. Proc. Natl. Acad. Sci. USA 1976, 73, 1523. [Google Scholar] [CrossRef]
- Stern, P.H.; Wallace, C.D.; Hoffman, R.M. Altered methionine metabolism occurs in all members of a set of diverse human tumor cell lines. J. Cell. Physiol. 1984, 119, 29–34. [Google Scholar] [CrossRef]
- Chello, P.L.; Bertino, J.R. Dependence of 5-methyltetrahydrofolate utilization by L5178Y murine leukemia cells In Vitro on the presence of hydroxycobalamin and transcobalamin II. Cancer Res. 1973, 33, 1898–1904. [Google Scholar]
- Halpern, B.; Clark, B.; Hardy, D.; Halpern, R.; Smith, R. The effect of replacement of methionine by homocystine on survival of malignant and normal adult mammalian cells in culture. Proc. Natl. Acad. Sci. USA 1974, 71, 1133–1136. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, J.; Inubushi, S.; Han, Q.; Tashiro, Y.; Sugisawa, N.; Hamada, K.; Aoki, Y.; Miyake, K.; Matsuyama, R.; Bouvet, M.; et al. Linkage of methionine addiction, histone lysine hypermethylation, and malignancy. iScience 2022, 25, 104162. [Google Scholar] [CrossRef] [PubMed]
- Wanders, D.; Hobson, K.; Ji, X. Methionine Restriction and Cancer Biology. Nutrients 2020, 12, 684. [Google Scholar] [CrossRef] [PubMed]
- Mecham, J.; Rowitch, D.; Wallace, C.; Stern, P.; Hoffman, R. The metabolic defect of methionine dependence occurs frequently in human tumor cell lines. Biochem. Biophys. Res. Commun. 1983, 117, 429–434. [Google Scholar] [CrossRef]
- Booher, K.; Lin, D.-W.; Borrego, S.L.; Kaiser, P. Downregulation of Cdc6 and pre-replication complexes in response to methionine stress in breast cancer cells. Cell Cycle 2012, 11, 4414. [Google Scholar] [CrossRef]
- Wang, Z.; Yip, L.Y.; Lee, J.H.J.; Wu, Z.; Chew, H.Y.; Chong, P.K.W.; Teo, C.C.; Ang, H.Y.-K.; Peh, K.L.E.; Yuan, J.; et al. Methionine is a metabolic dependency of tumor-initiating cells. Nat. Med. 2019, 25, 825–837. [Google Scholar] [CrossRef]
- Stern, P.; Hoffman, R. Elevated overall rates of transmethylation in cell lines from diverse human tumors. Vitro 1984, 20, 663–670. [Google Scholar] [CrossRef]
- Coalson, D.W.; Mecham, J.O.; Stern, P.H.; Hoffman, R.M. Reduced availability of endogenously synthesized methionine for S-adenosylmethionine formation in methionine-dependent cancer cells. Proc. Natl. Acad. Sci. USA 1982, 79, 4248–4251. [Google Scholar] [CrossRef]
- Tisdale, M. Utilization of preformed and endogenously synthesized methionine by cells in tissue culture. Br. J. Cancer 1984, 49, 315–320. [Google Scholar] [CrossRef]
- Borrego, S.L.; Fahrmann, J.; Datta, R.; Stringari, C.; Grapov, D.; Zeller, M.; Chen, Y.; Wang, P.; Baldi, P.; Gratton, E.; et al. Metabolic changes associated with methionine stress sensitivity in MDA-MB-468 breast cancer cells. Cancer Metab. 2016, 4, 9. [Google Scholar] [CrossRef] [PubMed]
- Husseiny, M.M.A.E.; Nilsson, R. Methionine dependence in cancer cells due to lack of B12-dependent methionine synthase activity. Cancer Metab. 2025, 13, 34. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Garg, S.; Miousse, I.R. Rescue of Methionine Dependence by Cobalamin in a Human Colorectal Cancer Cell Line. Nutrients 2024, 16, 997. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, M.; Darnell, A.; Reilly, M.; Kunchok, T.; Joesch-Cohen, L.; Rosenberg, D.; Ali, A.; Rees, M.; Roth, J.; Lewis, C.; et al. Methionine synthase is essential for cancer cell proliferation in physiological folate environments. Nat. Metab. 2021, 3, 1500–1511. [Google Scholar] [CrossRef] [PubMed]
- Ghergurovich, J.; Xu, X.; Wang, J.; Yang, L.; Ryseck, R.; Wang, L.; Rabinowitz, J. Methionine synthase supports tumour tetrahydrofolate pools. Nat. Metab. 2021, 3, 1512–1520. [Google Scholar] [CrossRef]
- Hoffman, R.M.; Yano, S. Tumor-Specific S/G2-Phase Cell Cycle Arrest of Cancer Cells by Methionine Restriction. In Methionine Dependence of Cancer and Aging: Methods and Protocols; Hoffman, R.M., Ed.; Springer: New York, NY, USA, 2019; pp. 49–60. ISBN 978-1-4939-8796-2. [Google Scholar]
- Lin, D.; Chung, B.; Kaiser, P. S-adenosylmethionine limitation induces p38 mitogen-activated protein kinase and triggers cell cycle arrest in G1. J. Cell Sci. 2014, 127, 50–59. [Google Scholar] [CrossRef]
- Lauinger, L.; Kaiser, P. Sensing and Signaling of Methionine Metabolism. Metabolites 2021, 11, 83. [Google Scholar] [CrossRef]
- Laxman, S.; Sutter, B.; Wu, X.; Kumar, S.; Guo, X.; Trudgian, D.; Mirzaei, H.; Tu, B. Sulfur amino acids regulate translational capacity and metabolic homeostasis through modulation of tRNA thiolation. Cell 2013, 154, 416–429. [Google Scholar] [CrossRef]
- Kaiser, P.; Flick, K.; Wittenberg, C.; Reed, S.I. Regulation of Transcription by Ubiquitination without Proteolysis: Cdc34/SCFMet30-Mediated Inactivation of the Transcription Factor Met4. Cell 2000, 102, 303–314. [Google Scholar] [CrossRef]
- Patton, E.E.; Peyraud, C.; Rouillon, A.; Surdin-Kerjan, Y.; Tyers, M.; Thomas, D. SCFMet30-mediated control of the transcriptional activator Met4 is required for the G1–S transition. EMBO J. 2000, 19, 1613–1624. [Google Scholar] [CrossRef]
- Su, N.; Flick, K.; Kaiser, P. The F-box protein Met30 is required for multiple steps in the budding yeast cell cycle. Mol. Cell. Biol. 2005, 25, 3875–3885. [Google Scholar] [CrossRef]
- Kaiser, P.; Sia, R.A.L.; Bardes, E.G.S.; Lew, D.J.; Reed, S.I. Cdc34 and the F-box protein Met30 are required for degradation of the Cdk-inhibitory kinase Swe1. Genes Dev. 1998, 12, 2587–2597. [Google Scholar] [CrossRef] [PubMed]
- Sutter, B.M.; Wu, X.; Laxman, S.; Tu, B.P. Methionine Inhibits Autophagy and Promotes Growth by Inducing the SAM-Responsive Methylation of PP2A. Cell 2013, 154, 403–415. [Google Scholar] [CrossRef] [PubMed]
- Jiang, C.; Liu, J.; He, S.; Xu, W.; Huang, R.; Pan, W.; Li, X.; Dai, X.; Guo, J.; Zhang, T.; et al. PRMT1 orchestrates with SAMTOR to govern mTORC1 methionine sensing via Arg-methylation of NPRL2. Cell Metab. 2023, 35, 2183. [Google Scholar] [CrossRef]
- Gu, X.; Orozco, J.M.; Saxton, R.A.; Condon, K.J.; Liu, G.Y.; Krawczyk, P.A.; Scaria, S.M.; Harper, J.W.; Gygi, S.P.; Sabatini, D.M. SAMTOR is an S-adenosylmethionine sensor for the mTORC1 pathway. Science 2017, 358, 813. [Google Scholar] [CrossRef] [PubMed]
- Lin, D.-W.; Carranza, F.G.; Borrego, S.; Lauinger, L.; Paula, L.D.d.; Pulipelli, H.R.; Andronicos, A.; Hertel, K.J.; Kaiser, P. Nutrient control of splice site selection contributes to methionine addiction of cancer. Mol. Metab. 2025, 93, 102103. [Google Scholar] [CrossRef]
- Zheng, H.; Qi, Y.; Hu, S.; Cao, X.; Xu, C.; Yin, Z.; Chen, X.; Li, Y.; Liu, W.; Li, J.; et al. Identification of Integrator-PP2A complex (INTAC), an RNA polymerase II phosphatase. Science 2020, 370, eabb5872. [Google Scholar] [CrossRef]
- Shi, Y. Serine/Threonine Phosphatases: Mechanism through Structure. Cell 2009, 139, 468–484. [Google Scholar] [CrossRef]
- Haesen, D.; Sents, W.; Lemaire, K.; Hoorne, Y.; Janssens, V. The Basic Biology of PP2A in Hematologic Cells and Malignancies. Front. Oncol. 2014, 4, 347. [Google Scholar] [CrossRef]
- Stanevich, V.; Jiang, L.; Satyshur, K.A.; Li, Y.; Jeffrey, P.D.; Li, Z.; Menden, P.; Semmelhack, M.F.; Xing, Y. The structural basis for tight control of PP2A methylation and function by LCMT-1. Mol. Cell 2011, 41, 331. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Du, X.; Moreno, C.; Green, R.; Ogris, E.; Feng, Q.; Chou, L.; McQuoid, M.; Pallas, D. Methylation of the protein phosphatase 2A catalytic subunit is essential for association of Balpha regulatory subunit but not SG2NA, striatin, or polyomavirus middle tumor antigen. Mol. Biol. Cell 2001, 12, 185–199. [Google Scholar] [CrossRef] [PubMed]
- Longin, S.; Zwaenepoel, K.; Louis, J.V.; Dilworth, S.; Goris, J.; Janssens, V. Selection of Protein Phosphatase 2A Regulatory Subunits Is Mediated by the C Terminus of the Catalytic Subunit. J. Biol. Chem. 2007, 282, 26971–26980. [Google Scholar] [CrossRef] [PubMed]
- Lin, D.; Chung, B.; Huang, J.; Wang, X.; Huang, L.; Kaiser, P. Microhomology-based CRISPR tagging tools for protein tracking, purification, and depletion. J. Biol. Chem. 2019, 294, 10877–10885. [Google Scholar] [CrossRef]
- Hoffman, R.M.; Jacobsen, S.J.; Erbe, R.W. Reversion to methionine independence in simian virus 40-transformed human and malignant rat fibroblasts is associated with altered ploidy and altered properties of transformation. Proc. Natl. Acad. Sci. USA 1979, 76, 1313–1317. [Google Scholar] [CrossRef]
- Borrego, S.L.; Lin, D.-W.; Kaiser, P. Isolation and Characterization of Methionine-Independent Clones from Methionine-Dependent Cancer Cells. Methods Mol. Biol. 2019, 1866, 37–48. [Google Scholar] [CrossRef]
- Borrego, S.; Fahrmann, J.; Hou, J.; Lin, D.-W.; Tromberg, B.J.; Fiehn, O.; Kaiser, P. Lipid remodeling in response to methionine stress in MDA-MBA-468 triple-negative breast cancer cells. J. Lipid Res. 2021, 62, 100056. [Google Scholar] [CrossRef]
- Bray, N.L.; Pimentel, H.; Melsted, P.; Pachter, L. Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 2016, 34, 525–527. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Wickham, H. ggplot2 Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016; Available online: https://www.scirp.org/reference/referencespapers?referenceid=3610100 (accessed on 26 May 2025).
- Larsson, J. Eulerr: Area-Proportional Euler and Venn Diagrams with Ellipses. 2016. Available online: https://cran.r-project.org/web/packages/eulerr/index.html (accessed on 28 June 2025).
- Wu, T.; Hu, E.; Xu, S.; Chen, M.; Guo, P.; Dai, Z.; Feng, T.; Zhou, L.; Tang, W.; Zhan, L.; et al. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovative 2021, 2, 100141. [Google Scholar] [CrossRef]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium—PubMed. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef]
- Conway, J.R.; Lex, A.; Gehlenborg, N. UpSetR: An R package for the visualization of intersecting sets and their properties. Bioinformatics 2017, 33, 2938. [Google Scholar] [CrossRef]
- Fellner, T.; Lackner, D.H.; Hombauer, H.; Piribauer, P.; Mudrak, I.; Zaragoza, K.; Juno, C.; Ogris, E. A novel and essential mechanism determining specificity and activity of protein phosphatase 2A (PP2A) In Vivo. Genes Dev. 2003, 17, 2138–2150. [Google Scholar] [CrossRef]
- Hartel, N.G.; Liu, C.Z.; Graham, N.A. Improved Discrimination of Asymmetric and Symmetric Arginine Dimethylation by Optimization of the Normalized Collision Energy in Liquid Chromatography–Mass Spectrometry Proteomics. J. Proteome Res. 2020, 19, 3123–3129. [Google Scholar] [CrossRef] [PubMed]
- Bruderer, R.; Bernhardt, O.M.; Gandhi, T.; Xuan, Y.; Sondermann, J.; Schmidt, M.; Gomez-Varela, D.; Reiter, L. Optimization of Experimental Parameters in Data-Independent Mass Spectrometry Significantly Increases Depth and Reproducibility of Results. Mol. Cell. Proteom. 2017, 16, 2296. [Google Scholar] [CrossRef] [PubMed]
- Demichev, V.; Messner, C.B.; Vernardis, S.I.; Lilley, K.S.; Ralser, M. DIA-NN: Neural networks and interference correction enable deep proteome coverage in high throughput. Nat. Methods 2020, 17, 41–44. [Google Scholar] [CrossRef] [PubMed]
- Elias, J.E.; Gygi, S.P. Target-Decoy Search Strategy for Mass Spectrometry-Based Proteomics. Methods Mol. Biol. 2010, 604, 55. [Google Scholar] [CrossRef]
- Shah, J.S.; Rai, S.N.; DeFilippis, A.P.; Hill, B.G.; Bhatnagar, A.; Brock, G.N. Distribution based nearest neighbor imputation for truncated high dimensional data with applications to pre-clinical and clinical metabolomics studies. BMC Bioinform. 2017, 18, 114. [Google Scholar] [CrossRef]
- Tyanova, S.; Cox, J. Perseus: A Bioinformatics Platform for Integrative Analysis of Proteomics Data in Cancer Research. In Cancer Systems Biology; Humana Press: New York, NY, USA, 2018; pp. 133–148. ISBN 978-1-4939-7493-1. [Google Scholar]
- Zhang, B.; Pirmoradian, M.; Zubarev, R.; Käll, L. Covariation of Peptide Abundances Accurately Reflects Protein Concentration Differences. Mol. Cell. Proteom. 2017, 16, 936–948. [Google Scholar] [CrossRef]
- Kohler, D.; Tsai, T.-H.; Verschueren, E.; Huang, T.; Hinkle, T.; Phu, L.; Choi, M.; Vitek, O. MSstatsPTM: Statistical Relative Quantification of Posttranslational Modifications in Bottom-Up Mass Spectrometry-Based Proteomics. Mol. Cell. Proteom. 2022, 22, 100477. [Google Scholar] [CrossRef] [PubMed]
- Aoki, Y.; Han, Q.; Tome, Y.; Yamamoto, J.; Kubota, Y.; Masaki, N.; Obara, K.; Hamada, K.; Wang, J.D.; Inubushi, S.; et al. Reversion of methionine addiction of osteosarcoma cells to methionine independence results in loss of malignancy, modulation of the epithelial-mesenchymal phenotype and alteration of histone-H3 lysine-methylation. Front. Oncol. 2022, 12, 1009548. [Google Scholar] [CrossRef] [PubMed]
- Sartorius AG. Live-Cell Imaging & Analysis. Sartorius. Available online: https://www.sartorius.com/en/products/live-cell-imaging-analysis (accessed on 19 August 2025).
- Ahuja, D.; Sáenz-Robles, M.T.; Pipas, J.M. SV40 large T antigen targets multiple cellular pathways to elicit cellular transformation. Oncogene 2005, 24, 7729–7745. [Google Scholar] [CrossRef] [PubMed]
- Graham, F.L.; Smiley, J.; Russell, W.C.; Nairn, R. Characteristics of a Human Cell Line Transformed by DNA from Human Adenovirus Type 5. J. Gen. Virol. 1977, 36, 59–72. [Google Scholar] [CrossRef]
- Hoffman, R.M.; Jacobsen, S.J. Reversible growth arrest in simian virus 40-transformed human fibroblasts. Proc. Natl. Acad. Sci. USA 1980, 77, 7306. [Google Scholar] [CrossRef]
- Yano, S.; Li, S.; Han, Q.; Tan, Y.; Bouvet, M.; Fujiwara, T.; Hoffman, R.M. Selective methioninase-induced trap of cancer cells in S/G2 phase visualized by FUCCI imaging confers chemosensitivity. Oncotarget 2014, 5, 8729. [Google Scholar] [CrossRef]
- Tan, Y.; Xu, M.; Hoffman, R.M. Broad Selective Efficacy of Recombinant Methioninase and Polyethylene Glycol-modified Recombinant Methioninase on Cancer Cells In Vitro. Anticancer Res. 2010, 30, 1041–1046. [Google Scholar]
- Hartel, N.G.; Chew, B.; Qin, J.; Xu, J.; Graham, N.A. Deep Protein Methylation Profiling by Combined Chemical and Immunoaffinity Approaches Reveals Novel PRMT1 Targets. Mol. Cell. Proteom. 2019, 18, 2149–2164. [Google Scholar] [CrossRef]
- Thomas, D.; Kuras, L.; Barbey, R.; Cherest, H.; Blaiseau, P.; Surdin-Kerjan, Y. Met30p, a yeast transcriptional inhibitor that responds to S-adenosylmethionine, is an essential protein with WD40 repeats. Mol. Cell. Biol. 1995, 15, 6526–6534. [Google Scholar] [CrossRef]
- Kim, S.G.; Buel, G.R.; Blenis, J. Nutrient Regulation of the mTOR Complex 1 Signaling Pathway. Mol. Cells 2013, 35, 463. [Google Scholar] [CrossRef]
- Singh, G.; Akcakanat, A.; Sharma, C.; Luyimbazi, D.; Naff, K.A.; Meric-Bernstam, F. Effect of Leucine Restriction on Akt/mTOR Signaling in Breast Cancer Cell Lines In Vitro and In Vivo. Nutr. Cancer 2011, 63, 264. [Google Scholar] [CrossRef] [PubMed]
- Khayati, K.; Antikainen, H.; Bonder, E.; Weber, G.; Kruger, W.; Jakubowski, H.; Dobrowolski, R. The amino acid metabolite homocysteine activates mTORC1 to inhibit autophagy and form abnormal proteins in human neurons and mice. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2017, 31, 598–609. [Google Scholar] [CrossRef] [PubMed]
- Kaur, A.; Westermarck, J. Regulation of protein phosphatase 2A (PP2A) tumor suppressor function by PME-1. Biochem. Soc. Trans. 2016, 44, 1683–1693. [Google Scholar] [CrossRef] [PubMed]
- Ogris, E.; Du, X.; Nelson, K.; Mak, E.; Yu, X.; Lane, W.; Pallas, D. A protein phosphatase methylesterase (PME-1) is one of several novel proteins stably associating with two inactive mutants of protein phosphatase 2A. J. Biol. Chem. 1999, 274, 14382–14391. [Google Scholar] [CrossRef]
- Longin, S.; Goris, J. 11 Reversible methylation of protein phosphatase 2A. Enzymes 2006, 24, 303–324. [Google Scholar] [CrossRef]
- Lyons, S.P.; Greiner, E.C.; Cressey, L.E.; Adamo, M.E.; Kettenbach, A.N. Regulation of PP2A, PP4, and PP6 holoenzyme assembly by carboxyl-terminal methylation. Sci. Rep. 2021, 11, 23031. [Google Scholar] [CrossRef]
- Lauinger, L.; Andronicos, A.; Flick, K.; Yu, C.; Durairaj, G.; Huang, L.; Kaiser, P. Cadmium binding by the F-box domain induces p97-mediated SCF complex disassembly to activate stress response programs. Nat. Commun. 2024, 15, 3894. [Google Scholar] [CrossRef]
- Liu, G.Y.; Sabatini, D.M. mTOR at the nexus of nutrition, growth, ageing and disease. Nat. Rev. Mol. Cell Biol. 2020, 21, 183. [Google Scholar] [CrossRef]
- Kim, J.; Guan, K.-L. mTOR as a central hub of nutrient signalling and cell growth. Nat. Cell Biol. 2019, 21, 63–71. [Google Scholar] [CrossRef]
- Tang, X.; Zhang, Y.; Wang, G.; Zhang, C.; Wang, F.; Shi, J.; Zhang, T.; Ding, J. Molecular mechanism of S-adenosylmethionine sensing by SAMTOR in mTORC1 signaling. Sci. Adv. 2022, 8, eabn3868. [Google Scholar] [CrossRef]
- Hu, H.; Luo, C.; Zheng, Y.G. Transient Kinetics Define a Complete Kinetic Model for Protein Arginine Methyltransferase 1. J. Biol. Chem. 2016, 291, 26722. [Google Scholar] [CrossRef] [PubMed]
- Qian, K.; Hu, H.; Xu, H.; Zheng, Y.G. Detection of PRMT1 inhibitors with stopped flow fluorescence. Signal Transduct. Target. Ther. 2018, 3, 6. [Google Scholar] [CrossRef] [PubMed]
- Frohner, I.; Mudrak, I.; Kronlachner, S.; Schüchner, S.; Ogris, E. Antibodies recognizing the C terminus of PP2A catalytic subunit are unsuitable for evaluating PP2A activity and holoenzyme composition. Sci. Signal. 2020, 13. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, J.; Han, Q.; Inubushi, S.; Sugisawa, N.; Hamda, K.; Nishino, H.; Miyake, K.; Kumamoto, T.; Matsuyama, R.; Bouvet, M.; et al. Histone methylation status of H3K4me3 and H3K9me3 under methionine restriction is unstable in methionine-addicted cancer cells, but stable in normal cells. Biochem. Biophys. Res. Commun. 2020, 533, 1034–1038. [Google Scholar] [CrossRef]
- Chen, Z.; Gu, X.; Chen, H.; Zhang, H.; Liu, J.; Yang, X.; Cai, Y.; Zhang, M.; Yan, L.; Yang, Y.; et al. RIPK1 senses S-adenosylmethionine scarcity to drive cell death and inflammation. Cell Metab. 2025, 37, 1732–1749. [Google Scholar] [CrossRef]
- Ji, M.; Xu, Q.; Li, X. Dietary methionine restriction in cancer development and antitumor immunity. Trends Endocrinol. Metab. 2024, 35, 400–412. [Google Scholar] [CrossRef]
- Epner, D.; Morrow, S.; Wilcox, M.; Houghton, J. Nutrient intake and nutritional indexes in adults with metastatic cancer on a phase I clinical trial of dietary methionine restriction. Nutr. Cancer 2002, 42, 158–166. [Google Scholar] [CrossRef]
- Thivat, E.; Farges, M.; Bacin, F.; D’Incan, M.; Mouret-Reynier, M.; Cellarier, E.; Madelmont, J.; Vasson, M.; Chollet, P.; Durando, X. Phase II trial of the association of a methionine-free diet with cystemustine therapy in melanoma and glioma. Anticancer Res. 2009, 29, 5235–5240. [Google Scholar]
- Goseki, N.; Yamazaki, S.; Shimojyu, K.; Kando, F.; Maruyama, M.; Endo, M.; Koike, M.; Takahashi, H. Synergistic effect of methionine-depleting total parenteral nutrition with 5-fluorouracil on human gastric cancer: A randomized, prospective clinical trial. Jpn. J. Cancer Res. GANN 1995, 86, 484–489. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Andronicos, A.; Yoneda, K.C.; Lin, D.-W.; Law, F.V.; Bae, H.; Basirattalab, A.; Graham, N.A.; Jang, C.; Kaiser, P. Carboxy-Methylation of the Catalytic Subunit of Protein Phosphatase 2A (PP2Ac) Integrates Methionine Availability with Methionine Addicted Cancer Cell Proliferation. Biomolecules 2025, 15, 1210. https://doi.org/10.3390/biom15091210
Andronicos A, Yoneda KC, Lin D-W, Law FV, Bae H, Basirattalab A, Graham NA, Jang C, Kaiser P. Carboxy-Methylation of the Catalytic Subunit of Protein Phosphatase 2A (PP2Ac) Integrates Methionine Availability with Methionine Addicted Cancer Cell Proliferation. Biomolecules. 2025; 15(9):1210. https://doi.org/10.3390/biom15091210
Chicago/Turabian StyleAndronicos, Anna, Kiku C. Yoneda, Da-Wei Lin, Fiona V. Law, Hosung Bae, Ali Basirattalab, Nicholas A. Graham, Cholsoon Jang, and Peter Kaiser. 2025. "Carboxy-Methylation of the Catalytic Subunit of Protein Phosphatase 2A (PP2Ac) Integrates Methionine Availability with Methionine Addicted Cancer Cell Proliferation" Biomolecules 15, no. 9: 1210. https://doi.org/10.3390/biom15091210
APA StyleAndronicos, A., Yoneda, K. C., Lin, D.-W., Law, F. V., Bae, H., Basirattalab, A., Graham, N. A., Jang, C., & Kaiser, P. (2025). Carboxy-Methylation of the Catalytic Subunit of Protein Phosphatase 2A (PP2Ac) Integrates Methionine Availability with Methionine Addicted Cancer Cell Proliferation. Biomolecules, 15(9), 1210. https://doi.org/10.3390/biom15091210





