Enhancement of Protein–Protein Interactions by Destabilizing Mutations Revealed by HDX-MS
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Antibody Expression and Purification
2.3. DSC
2.4. ITC
2.5. HDX-MS
3. Results
3.1. Human Growth Hormone Variant (hGHv)
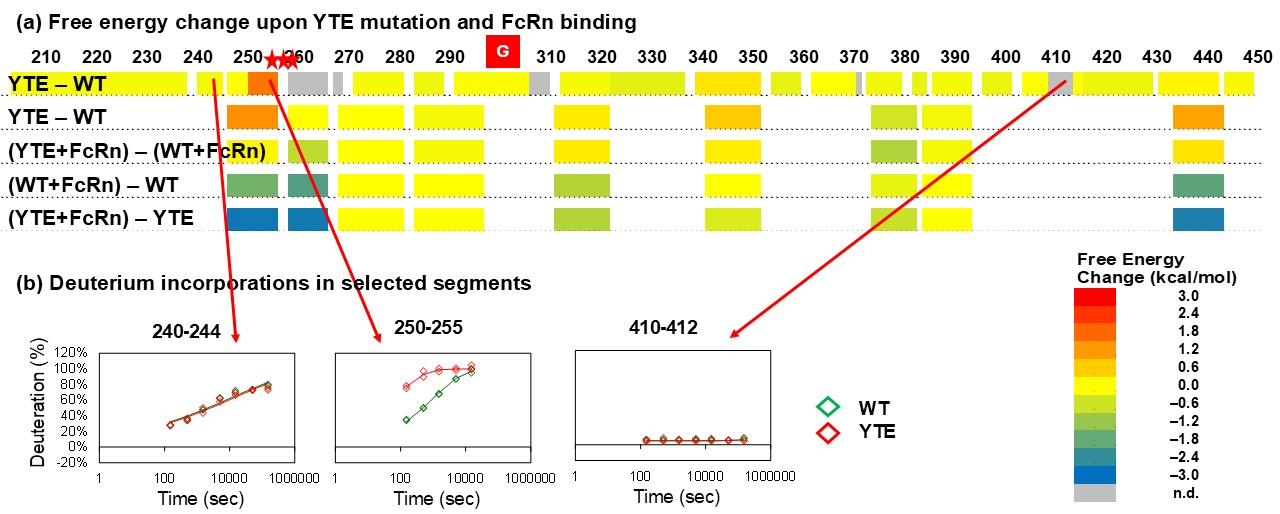
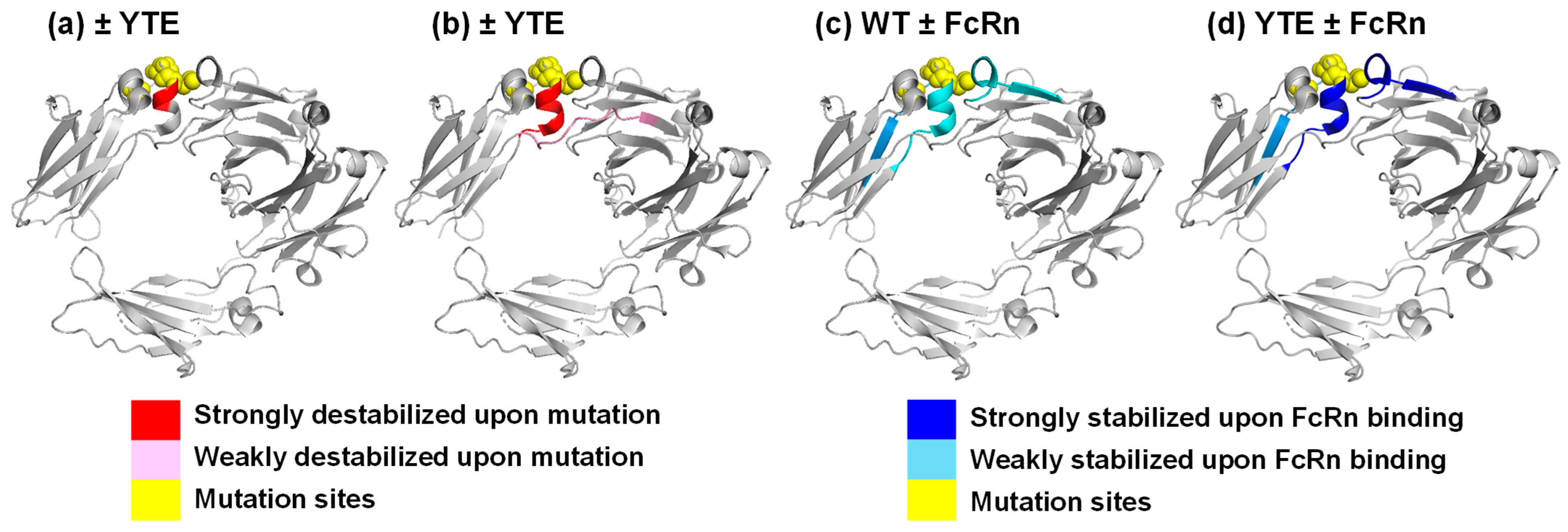
3.2. YTE Mutant
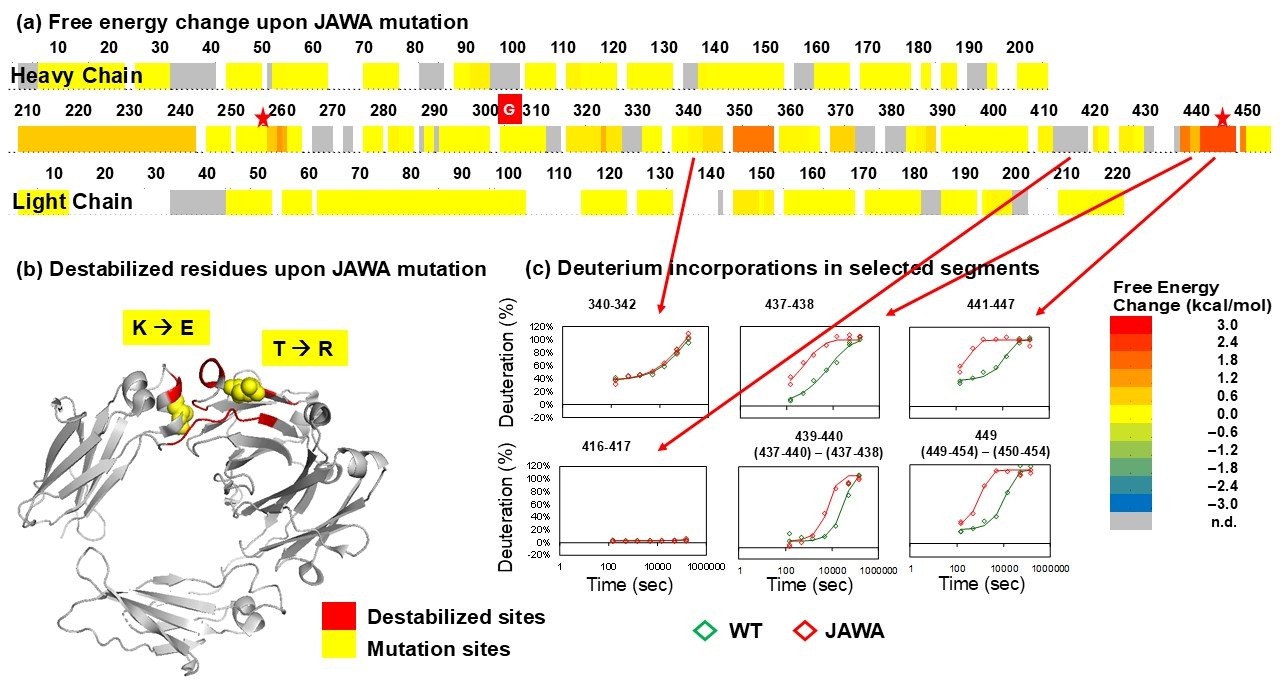
3.3. JAWA Mutant
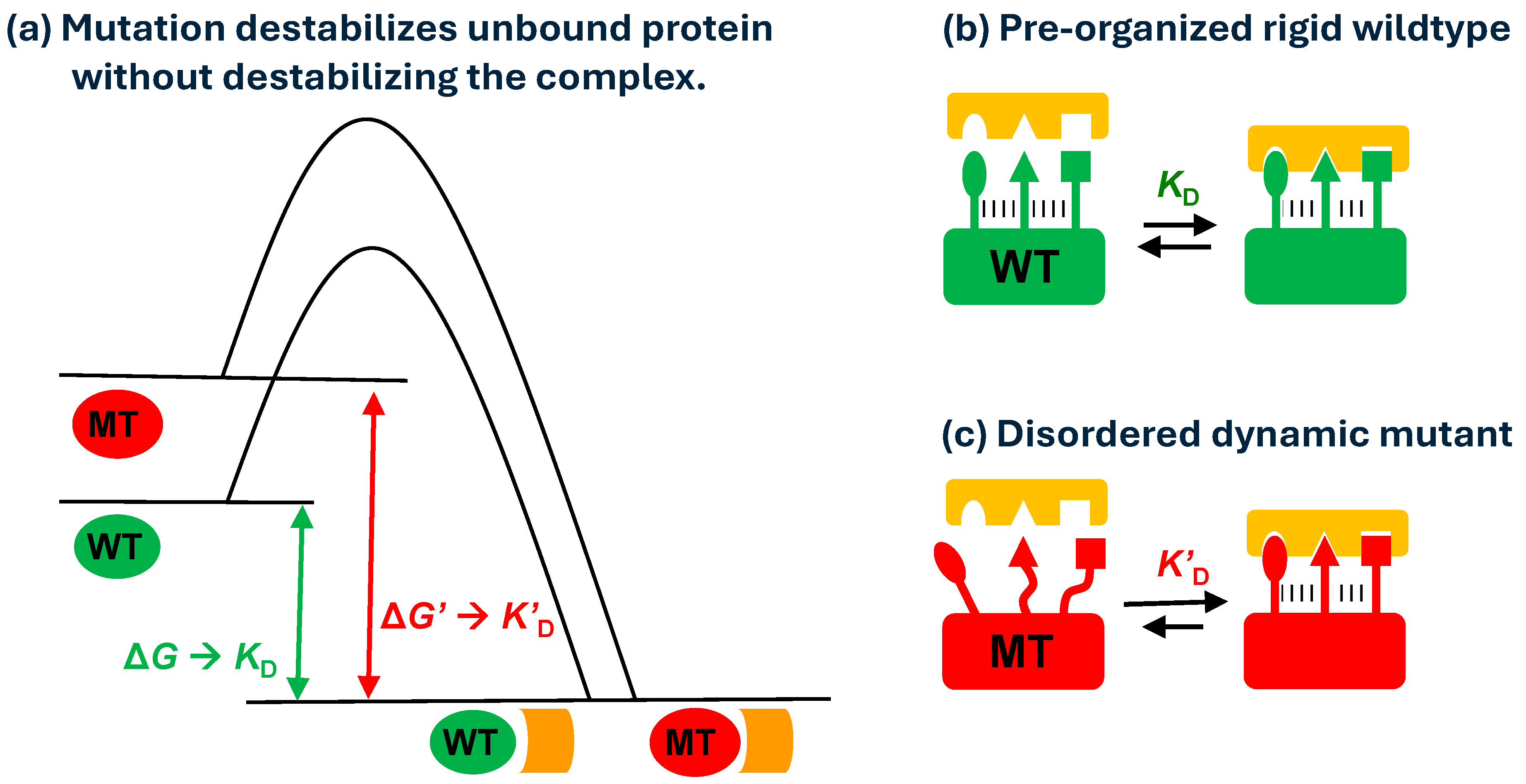
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Ebrahimi, S.B.; Samanta, D. Engineering protein-based therapeutics through structural and chemical design. Nat. Commun. 2023, 14, 2411–2421. [Google Scholar] [CrossRef] [PubMed]
- Frokjaer, S.; Otzen, D.E. Protein drug stability: A formulation challenge. Nat. Rev. Drug Discov. 2005, 4, 298–306. [Google Scholar] [CrossRef]
- Zhang, P.; Fang, S.; Tsao, C.; Liu, S.; Jain, P.; Sinclair, A.; Hung, H.-C.; Bai, T.; Wu, K.; Jiang, S. Zwitterionic gel encapsulation promotes protein stability, enhances pharmacokinetics, and reduces immunogenicity. Proc. Natl. Acad. Sci. USA 2015, 112, 12046–12051. [Google Scholar] [CrossRef]
- Pearce, K.H., Jr.; Cunningham, B.C.; Fuh, G.; Teeri, T.; Wells, J.A. Growth hormone binding affinity for its receptor surpasses the requirements for cellular activity. Biochemistry 1999, 38, 81–89. [Google Scholar] [CrossRef]
- Wang, X.; Mathieu, M.; Brezski, R.J. IgG Fc engineering to modulate antibody effector functions. Protein Cell 2018, 9, 63–73. [Google Scholar] [CrossRef]
- Vidal, L.S.; Isalan, M.; Heap, J.T.; Ledesma-Amaro, R. A primer to directed evolution: Current methodologies and future directions. RSC Chem. Biol. 2023, 4, 271–291. [Google Scholar] [CrossRef]
- Horn, J.R.; Kraybill, B.; Petro, E.J.; Coales, S.J.; Morrow, J.A.; Hamuro, Y.; Kossiakoff, A.A. The role of protein dynamics in increasing the binding affinity of an engineered protein-protein interaction established by H/D exchange mass spectrometry. Biochemistry 2006, 45, 8488–8498. [Google Scholar] [CrossRef]
- Walters, B.T.; Jensen, P.F.; Larraillet, V.; Lin, K.; Patapoff, T.; Schlothauer, T.; Rand, K.D.; Zhang, J. Conformational Destabilization of Immunoglobulin G Increases the Low pH Binding Affinity with the Neonatal Fc Receptor. J. Biol. Chem. 2016, 291, 1817–1825. [Google Scholar] [CrossRef] [PubMed]
- Kouadio, J.L.; Horn, J.R.; Pal, G.; Kossiakoff, A.A. Shotgun alanine scanning shows that growth hormone can bind productively to its receptor through a drastically minimized interface. J. Biol. Chem. 2005, 280, 25524–25532. [Google Scholar] [CrossRef] [PubMed]
- Majumdar, R.; Esfandiary, R.; Bishop, S.M.; Samra, H.S.; Middaugh, C.R.; Volkin, D.B.; Weis, D.D. Correlations between changes in conformational dynamics and physical stability in a mutant IgG1 mAb engineered for extended serum half-life. MABS 2015, 7, 84–95. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Armstrong, A.A.; Tam, S.H.; McCarthy, S.G.; Luo, J.; Gilliland, G.L.; Chiu, M.L. Functional optimization of agonistic antibodies to OX40 receptor with novel Fc mutations to promote antibody multimerization. MABS 2017, 9, 1129–1142. [Google Scholar] [CrossRef]
- Zhang, Z.; Smith, D.L. Determination of amide hydrogen exchange by mass spectrometry: A new tool for protein structure elucidation. Protein Sci. 1993, 2, 522–531. [Google Scholar] [CrossRef] [PubMed]
- Hamuro, Y. Tutorial: Chemistry of Hydrogen/Deuterium Exchange Mass Spectrometry. J. Am. Soc. Mass Spectrom. 2021, 32, 133–151. [Google Scholar] [CrossRef]
- Chetty, P.S.; Mayne, L.; Lund-Katz, S.; Stranz, D.; Englander, S.W.; Phillips, M.C. Helical structure and stability in human apolipoprotein A-I by hydrogen exchange and mass spectrometry. Proc. Natl. Acad. Sci. USA 2009, 106, 19005–19010. [Google Scholar] [CrossRef]
- Bai, Y.; Milne, J.S.; Mayne, L.C.; Englander, S.W. Primary structure effects on peptide group hydrogen exchange. Proteins Struct. Funct. Genet. 1993, 17, 75–86. [Google Scholar] [CrossRef]
- Nguyen, D.; Mayne, L.; Phillips, M.C.; Englander, S.W. Reference Parameters for Protein Hydrogen Exchange Rates. J. Amer. Soc. Mass Spectrom. 2018, 29, 1936–1939. [Google Scholar] [CrossRef]
- Lowman, H.B.; Wells, J.A. Affinity maturation of human growth hormone by monovalent phage display. J. Mol. Biol. 1993, 234, 564–578. [Google Scholar] [CrossRef] [PubMed]
- Cunningham, B.C.; Wells, J.A. Comparison of a Structural and a Functional Epitope. J. Mol. Biol. 1993, 234, 554–563. [Google Scholar] [CrossRef]
- Clackson, T.; Wells, J.A. A hot spot of binding energy in a hormone–receptor interface. Science 1995, 267, 383–386. [Google Scholar] [CrossRef] [PubMed]
- Pal, G.; Kossiakoff, A.A.; Sidhu, S.S. The Functional Binding Epitope of a High Affinity Variant of Human Growth Hormone Mapped by Shotgun Alanine-scanning Mutagenesis: Insights into the Mechanisms Responsible for Improved Affinity. J. Mol. Biol. 2003, 332, 195–204. [Google Scholar] [CrossRef]
- Schiffer, C.; Ultsch, M.; Walsh, S.; Somers, W.; de Vos, A.M.; Kossiakoff, A.A. Structure of a phage display-derived variant of human growth hormone complexed to two copies of the extracellular domain of its receptor: Evidence for strong structural coupling between receptor binding sites. J. Mol. Biol. 2002, 316, 277–289. [Google Scholar] [CrossRef]
- Roopenian, D.C.; Akilesh, S. FcRn: The neonatal Fc receptor comes of age. Nat. Rev. 2007, 7, 715–725. [Google Scholar] [CrossRef]
- Dall’Acqua, W.F.; Woods, R.M.; Ward, E.S.; Palaszynski, S.R.; Patel, N.K.; Brewah, Y.A.; Wu, H.; Kiener, P.A.; Langermann, S. Increasing the affinity of a human IgG1 for the neonatal Fc receptor: Biological consequences. J. Immunol. 2002, 169, 5171–5180. [Google Scholar] [CrossRef]
- Robbie, G.; Criste, R.; Dall’Acqua, W.; Jensen, K.; Patel, N.; Losonsky, G.; Griffin, P.A. Novel investigational Fc modified humanized monoclonal antibody, motavizumab-YTE, has an extended half-life in healthy adults. A randomized study. Antimicrob. Agents Chemother. 2013, 57, 6147–6153. [Google Scholar] [CrossRef]
- Dall’Acqua, W.F.; Kiener, P.A.; Wu, H. Properties of human IgG1s engineered for enhanced binding to the neonatal Fc receptor (FcRn). J. Biol. Chem. 2006, 281, 23514–23524. [Google Scholar] [CrossRef] [PubMed]
- Oganesyan, V.; Damschroder, M.M.; Woods, R.M.; Cook, K.E.; Wu, H.; Dall’acqua, W.F. Structural characterization of a human Fc fragment engineered for extended serum half-life. Mol. Immunol. 2009, 46, 1750–1755. [Google Scholar] [CrossRef] [PubMed]
- Oganesyan, V.; Damschroder, M.M.; Cook, K.E.; Li, Q.; Gao, C.; Wu, H.; Dall’Acqua, W.F. Structural insights into neonatal Fc receptor based recycling mechanisms. J. Biol. Chem. 2014, 289, 7812–7824. [Google Scholar] [CrossRef] [PubMed]
- Teplyakov, A.; Zhao, Y.; Malia, T.J.; Obmolova, G.; Gilliland, G.L. IgG2 Fc structure and the dynamic features of the IgG CH2-CH3 interface. Mol. Immunol. 2013, 56, 131–139. [Google Scholar] [CrossRef]
- van Kampen, M.D.; Wilt, L.H.A.M.K.-D.; van Egmond, M.L.; Reinders-Blankert, P.; van den Bremer, E.T.J.; Wang, G.; Heck, A.J.R.; Parren, P.W.H.I.; Beurskens, F.J.; Schuurman, J.; et al. Biophysical Characterization and Stability of Modified IgG1 Antibodies with Different Hexamerization Propensities. J. Pharm. Sci. 2022, 111, 1587–1598. [Google Scholar] [CrossRef]
- Hughes-Jones, N.C.; Gardner, B. Reaction between the isolated globular sub-units of the complement component C1q and IgG-complexes. Mol. Immunol. 1979, 16, 697–701. [Google Scholar] [CrossRef]
- Hamuro, Y. Interpretation of Hydrogen/Deuterium Exchange Mass Spectrometry. J. Am. Soc. Mass Spectrom. 2024, 35, 819–828. [Google Scholar] [CrossRef] [PubMed]
- Ultsch, M.; Somers, W.; Kossiakoff, A.A.; de Vos, A.M. The crystal structure of affinity matured human growth hormone at 2 Å resolution. J. Mol. Biol. 1994, 236, 286–299. [Google Scholar] [CrossRef] [PubMed]

| Protein | Chemical Stability | DSC | ITC | HDX-MS –Ligands | HDX-MS +Ligands |
|---|---|---|---|---|---|
| (a) hGHv | [7] | ----- | [9] | [7] | [7] |
| (b) IgG-YTE | ----- | [10] | current | [8], current | [8] |
| (c) IgGB-JAWA | ----- | current | ----- | current | ----- |
| Protein | Chemical Stability | DSC | ||
|---|---|---|---|---|
| CH2 | CH3 | Fab | ||
| (a) hGHwt *1 | 11.6 kcal/mol | ----- | ----- | ----- |
| (b) hGHv *1 | 8.0 kcal/mol | ----- | ----- | ----- |
| (c) IgGwt *2 | ----- | 70 °C | 83 °C | 87 °C |
| (d) IgG-YTE *2 | ----- | 64 °C | 83 °C | 87 °C |
| (e) IgGBwt | ----- | 73 °C | 83 °C | 77 °C |
| (f) IgGB-JAWA | ----- | 71 °C | 75 °C | 77 °C |
| Protein–Protein Interaction | SPR KD (nM) | ΔG (kcal/mol) | ΔH (kcal/mol) | −TΔS (kcal/mol) |
|---|---|---|---|---|
| (a) hGHwt + hGHbp *1 | <0.01 | −12 | −9 | −3 |
| (b) hGHv + hGHbp *1 | ~1.2 | −15 | −36 | 21 |
| (c) IgGAwt + FcRn | 5 *2 | −8.4 | −5.5 | −3.0 |
| (d) IgGA-YTE + FcRn | 0.4 *2 | −9.4 | −12.0 | 2.6 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hamuro, Y.; Armstrong, A.; Branson, J.; Wu, S.-J.; Huang, R.Y.-C.; Jacobs, S. Enhancement of Protein–Protein Interactions by Destabilizing Mutations Revealed by HDX-MS. Biomolecules 2025, 15, 1201. https://doi.org/10.3390/biom15081201
Hamuro Y, Armstrong A, Branson J, Wu S-J, Huang RY-C, Jacobs S. Enhancement of Protein–Protein Interactions by Destabilizing Mutations Revealed by HDX-MS. Biomolecules. 2025; 15(8):1201. https://doi.org/10.3390/biom15081201
Chicago/Turabian StyleHamuro, Yoshitomo, Anthony Armstrong, Jeffrey Branson, Sheng-Jiun Wu, Richard Y.-C. Huang, and Steven Jacobs. 2025. "Enhancement of Protein–Protein Interactions by Destabilizing Mutations Revealed by HDX-MS" Biomolecules 15, no. 8: 1201. https://doi.org/10.3390/biom15081201
APA StyleHamuro, Y., Armstrong, A., Branson, J., Wu, S.-J., Huang, R. Y.-C., & Jacobs, S. (2025). Enhancement of Protein–Protein Interactions by Destabilizing Mutations Revealed by HDX-MS. Biomolecules, 15(8), 1201. https://doi.org/10.3390/biom15081201







