Relationship of S100 Proteins with Neuroinflammation
Abstract
1. Introduction
2. Biology of S100 Proteins
2.1. Structural Features of S100 Proteins
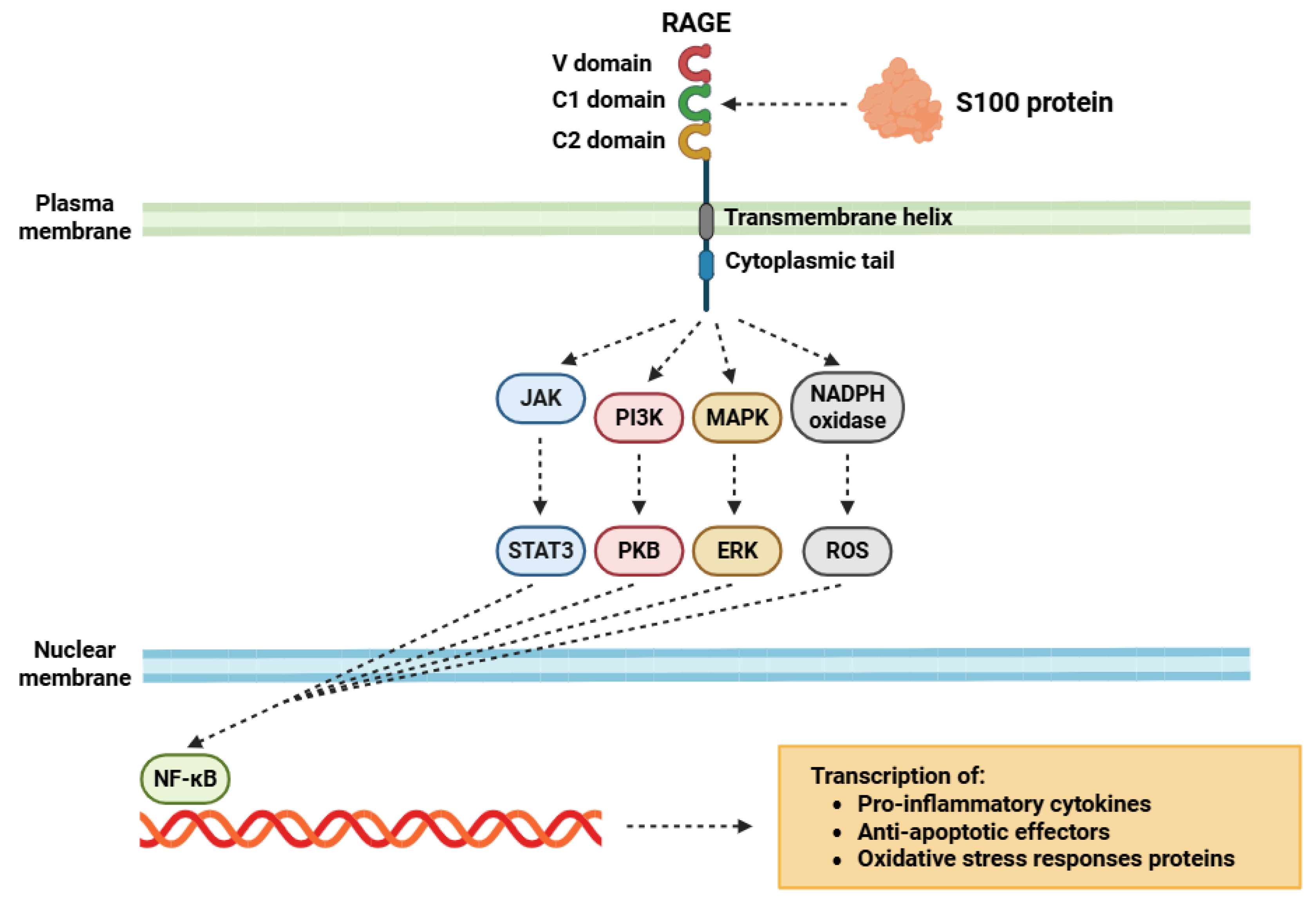
2.2. S100 Protein Receptors
2.3. Biological Functions of S100 Proteins
3. Role of S100 Proteins in Neuroinflammation
3.1. Contribution of S100 Proteins to Neuroinflammatory Mechanisms
3.2. Role of S100 Proteins in Alzheimer’s Disease
3.3. Role of S100 Proteins in Parkinson’s Disease
3.4. Role of S100 Proteins in Multiple Sclerosis
3.5. Role of S100 Proteins in Another Neuroinflammatory Diseases
4. Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| (NH4)2SO4 | Ammonium sulfate |
| AD | Alzheimer’s disease |
| APP | Amyloid precursor protein |
| Aβ | Beta-amyloid |
| BACE1 | β-site APP-cleaving enzyme 1 |
| BBB | Blood-brain barrier |
| Bcl-xL | B-cell lymphoma-extra large |
| C1q | Complement component 1q |
| Ca2+ | Calcium ion |
| CaMKII | Calcium/calmodulin-dependent protein kinase II |
| CCL2 | C-C motif chemokine ligand 2 |
| CD200 | Cluster of differentiation 200 |
| CK2 | Casein kinase 2 |
| CNS | Central nervous system |
| COX-2 | Cyclooxygenase 2 |
| CSF | Cerebrospinal fluid |
| CX3CL1 | Chemokine (C-X3-C motif) ligand 1 |
| CXCL10 | C-X-C motif chemokine ligand 10 |
| DAMP | Damage-associated molecular patterns |
| DIAPH1 | Diaphanous-related formin-1 |
| ECM | Extracellular matrix |
| EDC | Epidermal differentiation complex |
| EF-hand | Helix–loop–helix structural motif for Ca2+ binding |
| ERK | Extracellular signal-regulated kinase |
| ERK1/2 | Extracellular signal-regulated kinases 1 and 2 |
| FGF1 | Fibroblast growth factor 1 |
| GM-CSF | Granulocyte-macrophage colony-stimulating factor |
| HIF-1α | Hypoxia-Inducible Factor 1-alpha |
| ICAM-1 | Intercellular adhesion molecule 1 |
| IgG | Immunoglobulin G |
| IKK | IκB kinase |
| IL-1α | Interleukin 1 alpha |
| IL-1β | Interleukin 1 beta |
| IL-17 | Interleukin 17 |
| IL-6 | Interleukin 6 |
| IL-8 | Interleukin 8 |
| iNOS | Inducible nitric oxide synthase |
| JAK | Janus kinase |
| JNK | c-Jun N-terminal kinase |
| LTP | Long-term potentiation |
| MAPK | Mitogen-activated protein kinase |
| MMP | Matrix metalloproteinase |
| MS | Multiple sclerosis |
| MyD88 | Myeloid differentiation primary response 88 |
| NADPH | Nicotinamide adenine dinucleotide phosphate |
| NEMO | NF-κB essential modulator (regulatory subunit IKKγ) |
| NFAT | Nuclear factor of activated T-cell |
| NF-κB | Nuclear factor kappa-light-chain-enhancer of activated B cells |
| NLR | NOD-like receptors |
| NLRP3 | NOD-, LRR- and pyrin domain-containing protein 3 |
| NO | Nitric oxide |
| OPC | Oligodendrocyte precursor cell |
| p53 | Tumor protein p53 |
| PAMPs | Pathogen-associated molecular patterns |
| PD | Parkinson’s disease |
| PI3K | Phosphoinositide 3-kinase |
| PKB | Protein kinase B |
| PKC | Protein kinase C |
| PRR | Pattern recognition receptor |
| PTM | Post-translational modification |
| RAGE | Receptor for advanced glycation end-products |
| Ras-MAPK | Ras-mitogen-activated protein kinase |
| ROS | Reactive oxygen species |
| S100 | S100 protein family |
| SCF(βTrCP) | Skp, cullin, F-box containing complex (β-TrCP subunit) |
| SNpc | Substantia nigra pars compacta |
| sRAGE | Soluble receptor for advanced glycation end products |
| STAT3 | Signal transducer and activator of transcription 3 |
| TBI | Traumatic brain injury |
| Th1 | T helper type 1 cells |
| Th17 | T helper type 17 cells |
| TLR | Toll-like receptor |
| TLR4 | Toll-like receptor 4 |
| TNF-α | Tumor necrosis factor alpha |
| VCAM-1 | Vascular cell adhesion molecule 1 |
| VEGF | Vascular endothelial growth factor |
| Zn2+ | Zinc ion |
| τ | Tau protein |
References
- Kölliker-Frers, R.; Udovin, L.; Otero-Losada, M.; Kobiec, T.; Herrera, M.I.; Palacios, J.; Razzitte, G.; Capani, F. Neuroinflammation: An Integrating Overview of Reactive-Neuroimmune Cell Interactions in Health and Disease. Mediat. Inflamm. 2021, 2021, 9999146. [Google Scholar] [CrossRef]
- Ceulemans, A.G.; Zgavc, T.; Kooijman, R.; Hachimi-Idrissi, S.; Sarre, S.; Michotte, Y. The dual role of the neuroinflammatory response after ischemic stroke: Modulatory effects of hypothermia. J. Neuroinflamm. 2010, 7, 74. [Google Scholar] [CrossRef]
- Kim, M.E.; Lee, J.S. Mechanisms and Emerging Regulators of Neuroinflammation: Exploring New Therapeutic Strategies for Neurological Disorders. Curr. Issues Mol. Biol. 2024, 47, 8. [Google Scholar] [CrossRef]
- Shabab, T.; Khanabdali, R.; Moghadamtousi, S.Z.; Kadir, H.A.; Mohan, G. Neuroinflammation pathways: A general review. Int. J. Neurosci. 2017, 127, 624–633. [Google Scholar] [CrossRef]
- Di Vito, A.; Donato, G.; Tomassoni, D. Molecular and Cellular Mechanisms of Neuroinflammation. Biomed. Res. Int. 2017, 2017, 8417183. [Google Scholar] [CrossRef] [PubMed]
- Afridi, R.; Bhusal, A.; Tsuda, M.; Ryu, H.; Suk, K. Function of Glial Cells in Neuroinflammatory and Neuroimmunological Responses II. Cells 2023, 12, 1750. [Google Scholar] [CrossRef] [PubMed]
- Muzio, L.; Viotti, A.; Martino, G. Microglia in Neuroinflammation and Neurodegeneration: From Understanding to Therapy. Front. Neurosci. 2021, 15, 742065. [Google Scholar] [CrossRef] [PubMed]
- Figuera-Losada, M.; Rojas, C.; Slusher, B.S. Inhibition of microglia activation as a phenotypic assay in early drug discovery. J. Biomol. Screen 2014, 19, 17–31. [Google Scholar] [CrossRef]
- Gülke, E.; Gelderblom, M.; Magnus, T. Danger signals in stroke and their role on microglia activation after ischemia. Ther. Adv. Neurol. Disord. 2018, 11, 1756286418774254. [Google Scholar] [CrossRef]
- Smith, J.A.; Das, A.; Ray, S.K.; Banik, N.L. Role of pro-inflammatory cytokines released from microglia in neurodegenerative diseases. Brain Res. Bull. 2012, 87, 10–20. [Google Scholar] [CrossRef]
- Simpson, D.S.A.; Oliver, P.L. ROS Generation in Microglia: Understanding Oxidative Stress and Inflammation in Neurodegenerative Disease. Antioxidants 2020, 9, 743. [Google Scholar] [CrossRef]
- Liy, P.M.; Puzi, N.N.A.; Jose, S.; Vidyadaran, S. Nitric oxide modulation in neuroinflammation and the role of mesenchymal stem cells. Exp. Biol. Med. 2021, 246, 2399–2406. [Google Scholar] [CrossRef]
- Zhou, Y.; Cui, C.; Ma, X.; Luo, W.; Zheng, S.G.; Qiu, W. Nuclear Factor κB (NF-κB)-Mediated Inflammation in Multiple Sclerosis. Front. Immunol. 2020, 11, 391. [Google Scholar] [CrossRef]
- Ji, R.R.; Suter, M.R. p38 MAPK, microglial signaling, and neuropathic pain. Mol. Pain 2007, 3, 33. [Google Scholar] [CrossRef]
- Chen, Y.; Ye, X.; Escames, G.; Lei, W.; Zhang, X.; Li, M.; Jing, T.; Yao, Y.; Qiu, Z.; Wang, Z.; et al. The NLRP3 inflammasome: Contributions to inflammation-related diseases. Cell. Mol. Biol. Lett. 2023, 28, 51. [Google Scholar] [CrossRef]
- Fiebich, B.L.; Batista, C.R.A.; Saliba, S.W.; Yousif, N.M.; de Oliveira, A.C.P. Role of Microglia TLRs in Neurodegeneration. Front. Cell. Neurosci. 2018, 12, 329. [Google Scholar] [CrossRef]
- Freeman, L.; Guo, H.; David, C.N.; Brickey, W.J.; Jha, S.; Ting, J.P. NLR members NLRC4 and NLRP3 mediate sterile inflammasome activation in microglia and astrocytes. J. Exp. Med. 2017, 214, 1351–1370. [Google Scholar] [CrossRef] [PubMed]
- Gradisnik, L.; Velnar, T. Astrocytes in the central nervous system and their functions in health and disease: A review. World J. Clin. Cases 2023, 11, 3385–3394. [Google Scholar] [CrossRef] [PubMed]
- Nutma, E.; van Gent, D.; Amor, S.; Peferoen, L.A.N. Astrocyte and Oligodendrocyte Cross-Talk in the Central Nervous System. Cells 2020, 9, 600. [Google Scholar] [CrossRef] [PubMed]
- Bouvier, D.S.; Fixemer, S.; Heurtaux, T.; Jeannelle, F.; Frauenknecht, K.B.M.; Mittelbronn, M. The Multifaceted Neurotoxicity of Astrocytes in Ageing and Age-Related Neurodegenerative Diseases: A Translational Perspective. Front. Physiol. 2022, 13, 814889. [Google Scholar] [CrossRef]
- Manu, D.R.; Slevin, M.; Barcutean, L.; Forro, T.; Boghitoiu, T.; Balasa, R. Astrocyte Involvement in Blood-Brain Barrier Function: A Critical Update Highlighting Novel, Complex, Neurovascular Interactions. Int. J. Mol Sci. 2023, 24, 17146. [Google Scholar] [CrossRef]
- Manich, G.; Recasens, M.; Valente, T.; Almolda, B.; González, B.; Castellano, B. Role of the CD200-CD200R Axis During Homeostasis and Neuroinflammation. Neuroscience 2019, 405, 118–136. [Google Scholar] [CrossRef]
- Cook, A.; Hippensteel, R.; Shimizu, S.; Nicolai, J.; Fatatis, A.; Meucci, O. Interactions between chemokines: Regulation of fractalkine/CX3CL1 homeostasis by SDF/CXCL12 in cortical neurons. J. Biol. Chem. 2010, 285, 10563–10571. [Google Scholar] [CrossRef] [PubMed]
- Ledonne, A.; Mercuri, N.B. On the Modulatory Roles of Neuregulins/ErbB Signaling on Synaptic Plasticity. Int. J. Mol. Sci. 2019, 21, 275. [Google Scholar] [CrossRef]
- Müller, L.; Di Benedetto, S.; Müller, V. From Homeostasis to Neuroinflammation: Insights into Cellular and Molecular Interactions and Network Dynamics. Cells 2025, 14, 54. [Google Scholar] [CrossRef]
- Bresnick, A.R. S100 proteins as therapeutic targets. Biophys. Rev. 2018, 10, 1617–1629. [Google Scholar] [CrossRef]
- Fritz, G.; Botelho, H.M.; Morozova-Roche, L.A.; Gomes, C.M. Natural and amyloid self-assembly of S100 proteins: Structural basis of functional diversity. FEBS J. 2010, 277, 4578–4590. [Google Scholar] [CrossRef] [PubMed]
- Zimmer, D.B.; Wright Sadosky, P.; Weber, D.J. Molecular mechanisms of S100-target protein interactions. Microsc. Res. Tech. 2003, 60, 552–559. [Google Scholar] [CrossRef]
- Singh, P.; Ali, S.A. Multifunctional Role of S100 Protein Family in the Immune System: An Update. Cells 2022, 11, 2274. [Google Scholar] [CrossRef]
- Heizmann, C.W. Ca2+-binding S100 proteins in the central nervous system. Neurochem. Res. 1999, 24, 1097–1100. [Google Scholar] [CrossRef] [PubMed]
- Donato, R.; Cannon, B.R.; Sorci, G.; Riuzzi, F.; Hsu, K.; Weber, D.J.; Geczy, C.L. Functions of S100 proteins. Curr. Mol. Med. 2013, 13, 24–57. [Google Scholar] [CrossRef]
- Chaturvedi, N.; Ahmad, K.; Yadav, B.S.; Lee, E.J.; Sonkar, S.C.; Marina, N.; Choi, I. Understanding Calcium-Dependent Conformational Changes in S100A1 Protein: A Combination of Molecular Dynamics and Gene Expression Study in Skeletal Muscle. Cells 2020, 9, 181. [Google Scholar] [CrossRef]
- Völkers, M.; Rohde, D.; Goodman, C.; Most, P. S100A1: A regulator of striated muscle sarcoplasmic reticulum Ca2+ handling, sarcomeric, and mitochondrial function. J. Biomed. Biotechnol. 2010, 2010, 178614. [Google Scholar] [CrossRef] [PubMed]
- Mueller, A.; Schäfer, B.W.; Ferrari, S.; Weibel, M.; Makek, M.; Höchli, M.; Heizmann, C.W. The calcium-binding protein S100A2 interacts with p53 and modulates its transcriptional activity. J. Biol. Chem. 2005, 280, 29186–29193. [Google Scholar] [CrossRef]
- Pan, S.C.; Li, C.Y.; Kuo, C.Y.; Kuo, Y.Z.; Fang, W.Y.; Huang, Y.H.; Hsieh, T.C.; Kao, H.Y.; Kuo, Y.; Kang, Y.R.; et al. The p53-S100A2 Positive Feedback Loop Negatively Regulates Epithelialization in Cutaneous Wound Healing. Sci. Rep. 2018, 8, 5458. [Google Scholar] [CrossRef] [PubMed]
- Guan, W.; Deng, Q.; Yu, X.L.; Yuan, Y.S.; Gao, J.; Li, J.J.; Zhou, L.; Xia, P.; Han, G.Y.; Han, W.; et al. Blockade of S100A3 activity inhibits murine hair growth. Genet. Mol. Res. 2015, 14, 13532–13544. [Google Scholar] [CrossRef]
- Li, Z.H.; Bresnick, A.R. The S100A4 metastasis factor regulates cellular motility via a direct interaction with myosin-IIA. Cancer Res. 2006, 66, 5173–5180. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Liu, L.; Qi, L.; Knifley, T.; Piecoro, D.W.; Rychahou, P.; Liu, J.; Mitov, M.I.; Martin, J.; Wang, C.; Wu, J.; et al. S100A4 alters metabolism and promotes invasion of lung cancer cells by up-regulating mitochondrial complex I protein NDUFS2. J. Biol. Chem. 2019, 294, 7516–7527. [Google Scholar] [CrossRef]
- Chan, W.Y.; Xia, C.L.; Dong, D.C.; Heizmann, C.W.; Yew, D.T. Differential expression of S100 proteins in the developing human hippocampus and temporal cortex. Microsc. Res. Tech. 2003, 60, 600–613. [Google Scholar] [CrossRef]
- Jurewicz, E.; Robaszkiewicz, K.; Moraczewska, J.; Filipek, A. Binding of S100A6 to actin and the actin-tropomyosin complex. Sci. Rep. 2020, 10, 12824. [Google Scholar] [CrossRef]
- Bhatt, T.; Bhosale, A.; Bajantri, B.; Mathapathi, M.S.; Rizvi, A.; Scita, G.; Majumdar, A.; Jamora, C. Sustained Secretion of the Antimicrobial Peptide S100A7 Is Dependent on the Downregulation of Caspase-8. Cell Rep. 2019, 29, 2546–2555.e4. [Google Scholar] [CrossRef] [PubMed]
- Emberley, E.D.; Alowami, S.; Snell, L.; Murphy, L.C.; Watson, P.H. S100A7 (psoriasin) expression is associated with aggressive features and alteration of Jab1 in ductal carcinoma in situ of the breast. Breast Cancer Res. 2004, 6, R308–R315. [Google Scholar] [CrossRef] [PubMed]
- Wilkie, T.; Verma, A.K.; Zhao, H.; Charan, M.; Ahirwar, D.K.; Kant, S.; Pancholi, V.; Mishra, S.; Ganju, R.K. Lipopolysaccharide from the commensal microbiota of the breast enhances cancer growth: Role of S100A7 and TLR4. Mol. Oncol. 2022, 16, 1508–1522. [Google Scholar] [CrossRef]
- Ma, L.; Sun, P.; Zhang, J.C.; Zhang, Q.; Yao, S.L. Proinflammatory effects of S100A8/A9 via TLR4 and RAGE signaling pathways in BV-2 microglial cells. Int. J. Mol. Med. 2017, 40, 31–38. [Google Scholar] [CrossRef]
- Skronska-Wasek, W.; Durlanik, S.; Le, H.Q.; Schroeder, V.; Kitt, K.; Garnett, J.P.; Pflanz, S. The antimicrobial peptide S100A8/A9 produced by airway epithelium functions as a potent and direct regulator of macrophage phenotype and function. Eur. Respir. J. 2022, 59, 2002732. [Google Scholar] [CrossRef]
- Morel, E.; Gruenberg, J. The p11/S100A10 light chain of annexin A2 is dispensable for annexin A2 association to endosomes and functions in endosomal transport. PLoS ONE 2007, 2, e1118. [Google Scholar] [CrossRef] [PubMed]
- Miller, V.A.; Madureira, P.A.; Kamaludin, A.A.; Komar, J.; Sharma, V.; Sahni, G.; Thelwell, C.; Longstaff, C.; Waisman, D.M. Mechanism of plasmin generation by S100A10. Thromb. Haemost. 2017, 117, 1058–1071. [Google Scholar] [CrossRef]
- Davey, G.E.; Murmann, P.; Hoechli, M.; Tanaka, T.; Heizmann, C.W. Calcium-dependent translocation of S100A11 requires tubulin filaments. Biochim. Biophys. Acta 2000, 1498, 220–232. [Google Scholar] [CrossRef]
- Shin, H.; Lee, J.; Kim, Y.; Jang, S.; Lee, Y.; Kim, S.; Lee, Y. Knockdown of BC200 RNA expression reduces cell migration and invasion by destabilizing mRNA for calcium-binding protein S100A11. RNA Biol. 2017, 14, 1418–1430. [Google Scholar] [CrossRef][Green Version]
- Zhang, M.X.; Gan, W.; Jing, C.Y.; Zheng, S.S.; Yi, Y.; Zhang, J.; Xu, X.; Lin, J.J.; Zhang, B.H.; Qiu, S.J. S100A11 promotes cell proliferation via P38/MAPK signaling pathway in intrahepatic cholangiocarcinoma. Mol. Carcinog. 2019, 58, 19–30. [Google Scholar] [CrossRef]
- Meijer, B.; Gearry, R.B.; Day, A.S. The role of S100A12 as a systemic marker of inflammation. Int. J. Inflamm. 2012, 2012, 907078. [Google Scholar] [CrossRef]
- Mikkelsen, S.E.; Novitskaya, V.; Kriajevska, M.; Berezin, V.; Bock, E.; Norrild, B.; Lukanidin, E. S100A12 protein is a strong inducer of neurite outgrowth from primary hippocampal neurons. J. Neurochem. 2001, 79, 767–776. [Google Scholar] [CrossRef]
- Landriscina, M.; Soldi, R.; Bagalá, C.; Micucci, I.; Bellum, S.; Tarantini, F.; Prudovsky, I.; Maciag, T. S100A13 participates in the release of fibroblast growth factor 1 in response to heat shock in vitro. J. Biol. Chem. 2001, 276, 22544–22552. [Google Scholar] [CrossRef]
- Mohan, S.K.; Yu, C. The IL1alpha-S100A13 heterotetrameric complex structure: A component in the non-classical pathway for interleukin 1alpha secretion. J. Biol. Chem. 2011, 286, 14608–14617. [Google Scholar] [CrossRef] [PubMed]
- Landriscina, M.; Schinzari, G.; Di Leonardo, G.; Quirino, M.; Cassano, A.; D’Argento, E.; Lauriola, L.; Scerrati, M.; Prudovsky, I.; Barone, C. S100A13, a new marker of angiogenesis in human astrocytic gliomas. J. Neurooncol. 2006, 80, 251–259. [Google Scholar] [CrossRef]
- Mandinova, A.; Soldi, R.; Graziani, I.; Bagala, C.; Bellum, S.; Landriscina, M.; Tarantini, F.; Prudovsky, I.; Maciag, T. S100A13 mediates the copper-dependent stress-induced release of IL-1alpha from both human U937 and murine NIH 3T3 cells. J. Cell Sci. 2003, 116, 2687–2696. [Google Scholar] [CrossRef] [PubMed]
- Zhao, F.T.; Jia, Z.S.; Yang, Q.; Song, L.; Jiang, X.J. S100A14 promotes the growth and metastasis of hepatocellular carcinoma. Asian Pac. J. Cancer Prev. 2013, 14, 3831–3836. [Google Scholar] [CrossRef]
- Jiang, S.; Zhu, Y.; Chen, Z.; Huang, Z.; Liu, B.; Xu, Y.; Li, Z.; Lin, Z.; Li, M. S100A14 inhibits cell growth and epithelial-mesenchymal transition (EMT) in prostate cancer through FAT1-mediated Hippo signaling pathway. Hum. Cell 2021, 34, 1215–1226. [Google Scholar] [CrossRef] [PubMed]
- Wolf, R.; Lewerenz, V.; Büchau, A.S.; Walz, M.; Ruzicka, T. Human S100A15 splice variants are differentially expressed in inflammatory skin diseases and regulated through Th1 cytokines and calcium. Exp. Dermatol. 2007, 16, 685–691. [Google Scholar] [CrossRef]
- Liu, Y.; Zhang, R.; Xin, J.; Sun, Y.; Li, J.; Wei, D.; Zhao, A.Z. Identification of S100A16 as a novel adipogenesis promoting factor in 3T3-L1 cells. Endocrinology 2011, 152, 903–911. [Google Scholar] [CrossRef]
- Xiang, Y.Y.; Liu, J.H.; Yi, X.; Luo, J.Y.; Yu, Y.; Yi, G.L. S100 A16 promotes the progression of osteosarcoma by activating the PI3 K/AKT signaling pathway through ANXA2. Sci. Rep. 2025, 15, 19962. [Google Scholar] [CrossRef] [PubMed]
- Seguella, L.; Capuano, R.; Pesce, M.; Annunziata, G.; Pesce, M.; de Conno, B.; Sarnelli, G.; Aurino, L.; Esposito, G. S100B Protein Stimulates Proliferation and Angiogenic Mediators Release through RAGE/pAkt/mTOR Pathway in Human Colon Adenocarcinoma Caco-2 Cells. Int. J. Mol. Sci. 2019, 20, 3240. [Google Scholar] [CrossRef]
- Lin, J.; Yang, Q.; Wilder, P.T.; Carrier, F.; Weber, D.J. The calcium-binding protein S100B down-regulates p53 and apoptosis in malignant melanoma. J. Biol. Chem. 2010, 285, 27487–27498. [Google Scholar] [CrossRef]
- Rezaei, O.; Pakdaman, H.; Gharehgozli, K.; Simani, L.; Vahedian-Azimi, A.; Asaadi, S.; Sahraei, Z.; Hajiesmaeili, M. S100 B: A new concept in neurocritical care. Iran J. Neurol. 2017, 16, 83–89. [Google Scholar] [PubMed]
- Hong, E.J.; Jeung, E.B. Biological significance of calbindin-D9k within duodenal epithelium. Int. J. Mol. Sci. 2013, 14, 23330–23340. [Google Scholar] [CrossRef]
- Cong, Y.; Cui, Y.; Wang, S.; Jiang, L.; Cao, J.; Zhu, S.; Birkin, E.; Lane, J.; Ruge, F.; Jiang, W.G.; et al. Calcium-Binding Protein S100P Promotes Tumor Progression but Enhances Chemosensitivity in Breast Cancer. Front. Oncol. 2020, 10, 566302. [Google Scholar] [CrossRef]
- Barry, S.; Chelala, C.; Lines, K.; Sunamura, M.; Wang, A.; Marelli-Berg, F.M.; Brennan, C.; Lemoine, N.R.; Crnogorac-Jurcevic, T. S100P is a metastasis-associated gene that facilitates transendothelial migration of pancreatic cancer cells. Clin. Exp. Metastasis 2013, 30, 251–264. [Google Scholar] [CrossRef] [PubMed]
- Gribenko, A.V.; Hopper, J.E.; Makhatadze, G.I. Molecular characterization and tissue distribution of a novel member of the S100 family of EF-hand proteins. Biochemistry 2001, 40, 15538–15548. [Google Scholar] [CrossRef]
- Gonzalez, L.L.; Garrie, K.; Turner, M.D. Role of S100 proteins in health and disease. Biochim. Biophys. Acta 2020, 1867, 118677. [Google Scholar] [CrossRef]
- Sreejit, G.; Flynn, M.C.; Patil, M.; Krishnamurthy, P.; Murphy, A.J.; Nagareddy, P.R. S100 family proteins in inflammation and beyond. Adv. Clin. Chem. 2020, 98, 173–231. [Google Scholar]
- Abdi, W.; Romasco, A.; Alkurdi, D.; Santacruz, E.; Okinedo, I.; Zhang, Y.; Kannan, S.; Shakiba, S.; Richmond, J.M. An overview of S100 proteins and their functions in skin homeostasis, interface dermatitis conditions and other skin pathologies. Exp. Dermatol. 2024, 33, e15158. [Google Scholar] [CrossRef]
- Zimmer, D.B.; Cornwall, E.H.; Reynolds, P.D.; Donald, C.M. S100A1 regulates neurite organization, tubulin levels, and proliferation in PC12 cells. J. Biol. Chem. 1998, 273, 4705–4711. [Google Scholar] [CrossRef]
- Wang, H.; Mao, X.; Ye, L.; Cheng, H.; Dai, X. The Role of the S100 Protein Family in Glioma. J. Cancer 2022, 13, 3022–3030. [Google Scholar] [CrossRef]
- Hernández-Ortega, K.; Canul-Euan, A.A.; Solis-Paredes, J.M.; Borboa-Olivares, H.; Reyes-Muñoz, E.; Estrada-Gutierrez, G.; Camacho-Arroyo, I. S100B actions on glial and neuronal cells in the developing brain: An overview. Front. Neurosci. 2024, 18, 1425525. [Google Scholar] [CrossRef] [PubMed]
- Santamaria-Kisiel, L.; Rintala-Dempsey, A.C.; Shaw, G.S. Calcium-dependent and -independent interactions of the S100 protein family. Biochem. J. 2006, 396, 201–214. [Google Scholar] [CrossRef]
- Hermann, A.; Donato, R.; Weiger, T.M.; Chazin, W.J. S100 calcium binding proteins and ion channels. Front. Pharmacol. 2012, 3, 67. [Google Scholar] [CrossRef]
- Xia, C.; Braunstein, Z.; Toomey, A.C.; Zhong, J.; Rao, X. S100 Proteins As an Important Regulator of Macrophage Inflammation. Front. Immunol. 2018, 8, 1908. [Google Scholar] [CrossRef]
- Permyakov, S.E.; Denesyuk, A.I.; Denessiouk, K.A.; Permyakova, M.E.; Kazakov, A.S.; Ismailov, R.G.; Rastrygina, V.A.; Sokolov, A.S.; Permyakov, E.A. Monomeric state of S100P protein: Experimental and molecular dynamics study. Cell Calcium 2019, 80, 152–159. [Google Scholar] [CrossRef]
- Sivaraja, V.; Kumar, T.K.; Prudovsky, I.; Yu, C. Three-dimensional solution structure of a unique S100 protein. Biochem. Biophys. Res. Commun. 2005, 335, 1140–1148. [Google Scholar] [CrossRef] [PubMed]
- Gifford, J.L.; Walsh, M.P.; Vogel, H.J. Structures and metal-ion-binding properties of the Ca2+-binding helix-loop-helix EF-hand motifs. Biochem. J. 2007, 405, 199–221. [Google Scholar] [CrossRef] [PubMed]
- Yap, K.L.; Ames, J.B.; Swindells, M.B.; Ikura, M. Diversity of conformational states and changes within the EF-hand protein superfamily. Proteins 1999, 37, 499–507. [Google Scholar] [CrossRef]
- Denessiouk, K.; Permyakov, S.; Denesyuk, A.; Permyakov, E.; Johnson, M.S. Two structural motifs within canonical EF-hand calcium-binding domains identify five different classes of calcium buffers and sensors. PLoS ONE 2014, 9, e109287. [Google Scholar] [CrossRef]
- Sattar, Z.; Lora, A.; Jundi, B.; Railwah, C.; Geraghty, P. The S100 Protein Family as Players and Therapeutic Targets in Pulmonary Diseases. Pulm. Med. 2021, 2021, 5488591. [Google Scholar] [CrossRef]
- Moore, B.W. A soluble protein characteristic of the nervous system. Biochem. Biophys. Res. Commun. 1965, 19, 739–744. [Google Scholar] [CrossRef] [PubMed]
- Kiss, B.; Ecsédi, P.; Simon, M.; Nyitray, L. Isolation and Characterization of S100 Protein-Protein Complexes. In Calcium-Binding Proteins of the EF-Hand Superfamily: From Basics to Medical Applications; Human Press: New York, NY, USA, 2019; pp. 325–338. [Google Scholar]
- Zimmer, D.B.; Eubanks, J.O.; Ramakrishnan, D.; Criscitiello, M.F. Evolution of the S100 family of calcium sensor proteins. Cell Calcium 2013, 53, 170–179. [Google Scholar] [CrossRef]
- Sedaghat, F.; Notopoulos, A. S100 protein family and its application in clinical practice. Hippokratia 2008, 12, 198–204. [Google Scholar] [PubMed]
- Liu, M.; Wang, Y.; Miettinen, J.J.; Kumari, R.; Majumder, M.M.; Tierney, C.; Bazou, D.; Parsons, A.; Suvela, M.; Lievonen, J.; et al. S100 Calcium Binding Protein Family Members Associate With Poor Patient Outcome and Response to Proteasome Inhibition in Multiple Myeloma. Front. Cell Dev. Biol. 2021, 9, 723016. [Google Scholar] [CrossRef]
- Marenholz, I.; Volz, A.; Ziegler, A.; Davies, A.; Ragoussis, I.; Korge, B.P.; Mischke, D. Genetic analysis of the epidermal differentiation complex (EDC) on human chromosome 1q21: Chromosomal orientation, new markers, and a 6-Mb YAC contig. Genomics 1996, 37, 295–302. [Google Scholar] [CrossRef]
- Kizawa, K.; Takahara, H.; Unno, M.; Heizmann, C.W. S100 and S100 fused-type protein families in epidermal maturation with special focus on S100A3 in mammalian hair cuticles. Biochimie 2011, 93, 2038–2047. [Google Scholar] [CrossRef]
- Holthaus, K.B.; Sachslehner, A.P.; Steinbinder, J.; Eckhart, L. Epidermal Differentiation Genes of the Common Wall Lizard Encode Proteins with Extremely Biased Amino Acid Contents. Genes 2024, 15, 1136. [Google Scholar] [CrossRef]
- Shang, X.; Cheng, H.; Zhou, R. Chromosomal mapping, differential origin and evolution of the S100 gene family. Genet. Sel. Evol. 2008, 40, 449–464. [Google Scholar] [CrossRef] [PubMed]
- Cross, S.S.; Hamdy, F.C.; Deloulme, J.C.; Rehman, I. Expression of S100 proteins in normal human tissues and common cancers using tissue microarrays: S100A6, S100A8, S100A9 and S100A11 are all overexpressed in common cancers. Histopathology 2005, 46, 256–269. [Google Scholar] [CrossRef] [PubMed]
- Leśniak, W. Epigenetic regulation of S100 protein expression. Clin. Epigenetics 2011, 2, 77–83. [Google Scholar] [CrossRef][Green Version]
- Lindsey, J.C.; Lusher, M.E.; Anderton, J.A.; Gilbertson, R.J.; Ellison, D.W.; Clifford, S.C. Epigenetic deregulation of multiple S100 gene family members by differential hypomethylation and hypermethylation events in medulloblastoma. Br. J. Cancer 2007, 97, 267–274. [Google Scholar] [CrossRef]
- Mossel, D.M.; Moganti, K.; Riabov, V.; Weiss, C.; Kopf, S.; Cordero, J.; Dobreva, G.; Rots, M.G.; Klüter, H.; Harmsen, M.C.; et al. Epigenetic Regulation of S100A9 and S100A12 Expression in Monocyte-Macrophage System in Hyperglycemic Conditions. Front. Immunol. 2020, 11, 1071. [Google Scholar] [CrossRef]
- Hofmann, M.A.; Drury, S.; Fu, C.; Qu, W.; Taguchi, A.; Lu, Y.; Avila, C.; Kambham, N.; Bierhaus, A.; Nawroth, P.; et al. RAGE mediates a novel proinflammatory axis: A central cell surface receptor for S100/calgranulin polypeptides. Cell 1999, 97, 889–901. [Google Scholar] [CrossRef]
- Leclerc, E.; Vetter, S.W. The role of S100 proteins and their receptor RAGE in pancreatic cancer. Biochim. Biophys. Acta. 2015, 1852, 2706–2711. [Google Scholar] [CrossRef]
- Ray, R.; Juranek, J.K.; Rai, V. RAGE axis in neuroinflammation, neurodegeneration and its emerging role in the pathogenesis of amyotrophic lateral sclerosis. Neurosci. Biobehav. Rev. 2016, 62, 48–55. [Google Scholar] [CrossRef]
- Leclerc, E.; Fritz, G.; Vetter, S.W.; Heizmann, C.W. Binding of S100 proteins to RAGE: An update. Biochim. Biophys. Acta 2009, 1793, 993–1007. [Google Scholar] [CrossRef]
- Penumutchu, S.R.; Chou, R.H.; Yu, C. Structural insights into calcium-bound S100P and the V domain of the RAGE complex. PLoS ONE 2014, 9, e103947. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.; Andersen, G.R.; Yatime, L. Crystal structure of human S100A8 in complex with zinc and calcium. BMC Struct. Biol. 2016, 16, 8. [Google Scholar] [CrossRef]
- Koch, M.; Chitayat, S.; Dattilo, B.M.; Schiefner, A.; Diez, J.; Chazin, W.J.; Fritz, G. Structural basis for ligand recognition and activation of RAGE. Structure 2010, 18, 1342–1352. [Google Scholar] [CrossRef] [PubMed]
- Hudson, B.I.; Lippman, M.E. Targeting RAGE Signaling in Inflammatory Disease. Annu. Rev. Med. 2018, 69, 349–364. [Google Scholar] [CrossRef] [PubMed]
- Sparvero, L.J.; Asafu-Adjei, D.; Kang, R.; Tang, D.; Amin, N.; Im, J.; Rutledge, R.; Lin, B.; Amoscato, A.A.; Zeh, H.J.; et al. RAGE (Receptor for Advanced Glycation Endproducts), RAGE ligands, and their role in cancer and inflammation. J. Transl. Med. 2009, 7, 17. [Google Scholar] [CrossRef] [PubMed]
- Arivazhagan, L.; Popp, C.J.; Ruiz, H.H.; Wilson, R.A.; Manigrasso, M.B.; Shekhtman, A.; Ramasamy, R.; Sevick, M.A.; Schmidt, A.M. The RAGE/DIAPH1 axis: Mediator of obesity and proposed biomarker of human cardiometabolic disease. Cardiovasc. Res. 2024, 119, 2813–2824. [Google Scholar] [CrossRef]
- Ramasamy, R.; Shekhtman, A.; Schmidt, A.M. The RAGE/DIAPH1 Signaling Axis & Implications for the Pathogenesis of Diabetic Complications. Int. J. Mol. Sci. 2022, 23, 4579. [Google Scholar] [CrossRef]
- Tóbon-Velasco, J.C.; Cuevas, E.; Torres-Ramos, M.A. Receptor for AGEs (RAGE) as mediator of NF-kB pathway activation in neuroinflammation and oxidative stress. CNS Neurol. Disord. Drug Targets 2014, 13, 1615–1626. [Google Scholar] [CrossRef]
- Hinz, M.; Scheidereit, C. The IκB kinase complex in NF-κB regulation and beyond. EMBO Rep. 2014, 15, 46–61. [Google Scholar] [CrossRef]
- Wang, X.; Peng, H.; Huang, Y.; Kong, W.; Cui, Q.; Du, J.; Jin, H. Post-translational Modifications of IκBα: The State of the Art. Front. Cell Dev. Biol. 2020, 8, 574706. [Google Scholar] [CrossRef]
- Kroll, M.; Margottin, F.; Kohl, A.; Renard, P.; Durand, H.; Concordet, J.P.; Bachelerie, F.; Arenzana-Seisdedos, F.; Benarous, R. Inducible degradation of IkappaBalpha by the proteasome requires interaction with the F-box protein h-betaTrCP. J. Biol. Chem. 1999, 274, 7941–7945. [Google Scholar] [CrossRef]
- Florio, T.J.; Lokareddy, R.K.; Yeggoni, D.P.; Sankhala, R.S.; Ott, C.A.; Gillilan, R.E.; Cingolani, G. Differential recognition of canonical NF-κB dimers by Importin α3. Nat. Commun. 2022, 13, 1207. [Google Scholar] [CrossRef]
- Wang, V.Y.; Huang, W.; Asagiri, M.; Spann, N.; Hoffmann, A.; Glass, C.; Ghosh, G. The transcriptional specificity of NF-κB dimers is coded within the κB DNA response elements. Cell Rep. 2012, 2, 824–839. [Google Scholar] [CrossRef] [PubMed]
- Mulero, M.C.; Wang, V.Y.; Huxford, T.; Ghosh, G. Genome reading by the NF-κB transcription factors. Nucleic Acids Res. 2019, 47, 9967–9989. [Google Scholar] [CrossRef]
- Liu, T.; Zhang, L.; Joo, D.; Sun, S.C. NF-κB signaling in inflammation. Signal Transduct. Target. Ther. 2017, 2, 17023. [Google Scholar] [CrossRef]
- Pan, Y.; Zhang, X.; Wang, Y.; Cai, L.; Ren, L.; Tang, L.; Wang, J.; Zhao, Y.; Wang, Y.; Liu, Q.; et al. Targeting JNK by a new curcumin analog to inhibit NF-kB-mediated expression of cell adhesion molecules attenuates renal macrophage infiltration and injury in diabetic mice. PLoS ONE 2013, 8, e79084. [Google Scholar] [CrossRef]
- Parrondo, R.; de las Pozas, A.; Reiner, T.; Rai, P.; Perez-Stable, C. NF-κB activation enhances cell death by antimitotic drugs in human prostate cancer cells. Mol. Cancer 2010, 9, 182. [Google Scholar] [CrossRef] [PubMed]
- Lingappan, K. NF-κB in Oxidative Stress. Curr. Opin. Toxicol. 2018, 7, 81–86. [Google Scholar] [CrossRef] [PubMed]
- Zglejc-Waszak, K.; Pomianowski, A.; Wojtkiewicz, J.; Banach, M.; Juranek, J.K. New insights into RAGE/Diaph1 interaction as a modulator of actin cytoskeleton dynamics in peripheral nervous system in long-term hyperglycaemia. Eur. J. Neurosci. 2023, 57, 1642–1656. [Google Scholar] [CrossRef]
- Zhu, P.; Ren, M.; Yang, C.; Hu, Y.X.; Ran, J.M.; Yan, L. Involvement of RAGE, MAPK and NF-κB pathways in AGEs-induced MMP-9 activation in HaCaT keratinocytes. Exp. Dermatol. 2012, 21, 123–129. [Google Scholar] [CrossRef] [PubMed]
- Bao, J.M.; He, M.Y.; Liu, Y.W.; Lu, Y.J.; Hong, Y.Q.; Luo, H.H.; Ren, Z.L.; Zhao, S.C.; Jiang, Y. AGE/RAGE/Akt pathway contributes to prostate cancer cell proliferation by promoting Rb phosphorylation and degradation. Am. J. Cancer Res. 2015, 5, 1741–1750. [Google Scholar] [CrossRef]
- Serban, A.I.; Stanca, L.; Geicu, O.I.; Dinischiotu, A. AGEs-Induced IL-6 Synthesis Precedes RAGE Up-Regulation in HEK 293 Cells: An Alternative Inflammatory Mechanism? Int. J. Mol. Sci. 2015, 16, 20100–20117. [Google Scholar] [CrossRef]
- Lim, S.Y.; Raftery, M.J.; Goyette, J.; Hsu, K.; Geczy, C.L. Oxidative modifications of S100 proteins: Functional regulation by redox. J. Leukoc. Biol. 2009, 86, 577–587. [Google Scholar] [CrossRef]
- Lee, J.M.; Hammarén, H.M.; Savitski, M.M.; Baek, S.H. Control of protein stability by post-translational modifications. Nat. Commun. 2023, 14, 201. [Google Scholar] [CrossRef]
- Seitz, A.; Busch, M.; Kroemer, J.; Schneider, A.; Simon, S.; Jungmann, A.; Katus, H.A.; Most, P.; Ritterhoff, J. S100A1’s single cysteine is an indispensable redox switch for the protection against diastolic calcium waves in cardiomyocytes. Am. J. Physiol. Heart Circ. Physiol. 2024, 327, H000. [Google Scholar] [CrossRef]
- Zaręba-Kozioł, M.; Burdukiewicz, M.; Wysłouch-Cieszyńska, A. Intracellular Protein S-Nitrosylation—A Cells Response to Extracellular S100B and RAGE Receptor. Biomolecules 2022, 12, 613. [Google Scholar] [CrossRef]
- Malik, P.; Kumar Mukherjee, T. Immunological methods for the determination of AGE-RAGE axis generated glutathionylated and carbonylated proteins as oxidative stress markers. Methods 2022, 203, 354–363. [Google Scholar] [CrossRef] [PubMed]
- Piras, S.; Furfaro, A.L.; Domenicotti, C.; Traverso, N.; Marinari, U.M.; Pronzato, M.A.; Nitti, M. RAGE Expression and ROS Generation in Neurons: Differentiation versus Damage. Oxidative Med. Cell. Longev. 2016, 2016, 9348651. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, F.; Umeda, Y.; Shimamoto, S.; Tsuchiya, M.; Tokumitsu, H.; Tokuda, M.; Kobayashi, R. S100 proteins modulate protein phosphatase 5 function: A link between CA2+ signal transduction and protein dephosphorylation. J. Biol. Chem. 2012, 287, 13787–13798. [Google Scholar] [CrossRef] [PubMed]
- Downs, C.A.; Kreiner, L.H.; Johnson, N.M.; Brown, L.A.; Helms, M.N. Receptor for advanced glycation end-products regulates lung fluid balance via protein kinase C-gp91phox signaling to epithelial sodium channels. Am. J. Respir. Cell Mol. Biol. 2015, 52, 75–87. [Google Scholar] [CrossRef]
- Coste, K.; Bruet, S.; Chollat-Namy, C.; Filhol, O.; Cochet, C.; Gallot, D.; Marceau, G.; Blanchon, L.; Sapin, V.; Belville, C. Characterization of RAGE and CK2 Expressions in Human Fetal Membranes. Int. J. Mol. Sci. 2023, 24, 4074. [Google Scholar] [CrossRef]
- Dong, W.; Yang, X.; Li, X.; Wei, S.; An, C.; Zhang, J.; Shi, X.; Dong, S. Investigation of N-Glycan Functions in Receptor for Advanced Glycation End Products V Domain through Chemical Glycoprotein Synthesis. J. Am. Chem. Soc. 2024, 146, 18270–18280. [Google Scholar] [CrossRef]
- Degani, G.; Barbiroli, A.; Magnelli, P.; Digiovanni, S.; Altomare, A.; Aldini, G.; Popolo, L. Insights into the effects of N-glycosylation on the characteristics of the VC1 domain of the human receptor for advanced glycation end products (RAGE) secreted by Pichia pastoris. Glycoconj. J. 2019, 36, 27–38. [Google Scholar] [CrossRef]
- Bertheloot, D.; Latz, E. HMGB1, IL-1α, IL-33 and S100 proteins: Dual-function alarmins. Cell. Mol. Immunol. 2017, 14, 43–64. [Google Scholar] [CrossRef] [PubMed]
- Barger, S.W.; Van Eldik, L.J. S100 beta stimulates calcium fluxes in glial and neuronal cells. J. Biol. Chem. 1992, 267, 9689–9694. [Google Scholar] [CrossRef] [PubMed]
- Lancaster, T.; Tabrizi, M.E.A.; Repici, M.; Gupta, J.; Gross, S.R. An Extracellular/Membrane-Bound S100P Pool Regulates Motility and Invasion of Human Extravillous Trophoblast Lines and Primary Cells. Biomolecules 2023, 13, 1231. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Van Eldik, L.J. S100 beta induces apoptotic cell death in cultured astrocytes via a nitric oxide-dependent pathway. Biochim. Biophys. Acta 1996, 1313, 239–245. [Google Scholar] [CrossRef] [PubMed]
- Ghavami, S.; Eshragi, M.; Ande, S.R.; Chazin, W.J.; Klonisch, T.; Halayko, A.J.; McNeill, K.D.; Hashemi, M.; Kerkhoff, C.; Los, M. S100A8/A9 induces autophagy and apoptosis via ROS-mediated cross-talk between mitochondria and lysosomes that involves BNIP3. Cell Res. 2010, 20, 314–331. [Google Scholar] [CrossRef]
- Taneja, S.; Vetter, S.W.; Leclerc, E. Hypoxia and the Receptor for Advanced Glycation End Products (RAGE) Signaling in Cancer. Int. J. Mol. Sci. 2021, 22, 8153. [Google Scholar] [CrossRef]
- Fernandez-Fernandez, M.R.; Rutherford, T.J.; Fersht, A.R. Members of the S100 family bind p53 in two distinct ways. Protein Sci. 2008, 17, 1663–1670. [Google Scholar] [CrossRef]
- Ishijima, T.; Nakajima, K. Inflammatory cytokines TNFα, IL-1β, and IL-6 are induced in endotoxin- stimulated microglia through different signaling cascades. Sci. Prog. 2021, 104, 368504211054985. [Google Scholar] [CrossRef]
- Nam, A.R.; Kim, D.H.; Kim, M.J.; Lee, J.S.; Yang, S.J.; Kim, I.S. S100A8 Induces Secretion of MCP-1, IL-6, and IL-8 via TLR4 in Jurkat T Cells. Biomed. Sci. Lett. 2016, 22, 60–64. [Google Scholar] [CrossRef]
- Noack, M.; Miossec, P. Heterogeneous effects of S100 proteins during cell interactions between immune cells and stromal cells from synovium or skin. Clin. Exp. Immunol. 2023, 212, 276–284. [Google Scholar] [CrossRef]
- Kazakov, A.S.; Rastrygina, V.A.; Vologzhannikova, A.A.; Zemskova, M.Y.; Bobrova, L.A.; Deryusheva, E.I.; Permyakova, M.E.; Sokolov, A.S.; Litus, E.A.; Shevelyova, M.P.; et al. Recognition of granulocyte-macrophage colony-stimulating factor by specific S100 proteins. Cell Calcium 2024, 119, 102869. [Google Scholar] [CrossRef] [PubMed]
- Kushi, H.; Saito, T.; Makino, K.; Hayashi, N. L-8 is a key mediator of neuroinflammation in severe traumatic brain injuries. In Brain Edema XII; Springer: Vienna, Austria, 2003; pp. 347–350. [Google Scholar]
- Chen, J.; Liu, X.; Zhong, Y. Interleukin-17A: The Key Cytokine in Neurodegenerative Diseases. Front. Aging Neurosci. 2020, 12, 566922. [Google Scholar] [CrossRef]
- Croxford, A.L.; Spath, S.; Becher, B. GM-CSF in Neuroinflammation: Licensing Myeloid Cells for Tissue Damage. Trends Immunol. 2015, 36, 651–662. [Google Scholar] [CrossRef] [PubMed]
- Gross, S.R.; Sin, C.G.; Barraclough, R.; Rudland, P.S. Joining S100 proteins and migration: For better or for worse, in sickness and in health. Cell. Mol. Life Sci. 2014, 71, 1551–1579. [Google Scholar] [CrossRef]
- Bai, X.; Xu, P.C.; Chen, T.; Zhang, H.M.; Wu, S.J.; Yang, X.; Gao, S.; Jia, J.Y.; Jiang, J.Q.; Yan, T.K. The potential pathogenic roles of S100A8/A9 and S100A12 in patients with MPO-ANCA-positive vasculitis. BMC Immunol. 2022, 23, 42. [Google Scholar] [CrossRef] [PubMed]
- Yoshinouchi, T.; Ohtsuki, Y.; Ueda, R.; Sato, S.; Ueda, N. Myofibroblasts and S-100 protein positive cells in idiopathic pulmonary fibrosis and rheumatoid arthritis-associated interstitial pneumonia. Eur. Respir. J. 1999, 14, 579–584. [Google Scholar] [CrossRef]
- Lallyett, C.; Yeung, C.C.; Nielson, R.H.; Zeef, L.A.H.; Chapman-Jones, D.; Kjaer, M.; Kadler, K.E. Changes in S100 Proteins Identified in Healthy Skin following Electrical Stimulation: Relevance for Wound Healing. Adv. Ski. Wound Care 2018, 31, 322–327. [Google Scholar] [CrossRef]
- Cristóvão, J.S.; Gomes, C.M. S100 Proteins in Alzheimer’s Disease. Front. Neurosci. 2019, 13, 463. [Google Scholar] [CrossRef]
- Angelopoulou, E.; Paudel, Y.N.; Piperi, C. Emerging role of S100B protein implication in Parkinson’s disease pathogenesis. Cell. Mol. Life Sci. 2021, 78, 1445–1453. [Google Scholar] [CrossRef]
- Holzinger, D.; Foell, D.; Kessel, C. The role of S100 proteins in the pathogenesis and monitoring of autoinflammatory diseases. Mol. Cell. Pediatr. 2018, 5, 7. [Google Scholar] [CrossRef]
- Bresnick, A.R.; Weber, D.J.; Zimmer, D.B. S100 proteins in cancer. Nat. Rev. Cancer 2015, 15, 96–109. [Google Scholar] [CrossRef] [PubMed]
- Ackermann, G.E.; Marenholz, I.; Wolfer, D.P.; Chan, W.Y.; Schäfer, B.; Erne, P.; Heizmann, C.W. S100A1-deficient male mice exhibit increased exploratory activity and reduced anxiety-related responses. Biochim. Biophys. Acta 2006, 1763, 1307–1319. [Google Scholar] [CrossRef][Green Version]
- Filipek, A.; Leśniak, W. S100A6 and Its Brain Ligands in Neurodegenerative Disorders. Int. J. Mol. Sci. 2020, 21, 3979. [Google Scholar] [CrossRef]
- Tian, Q.; Li, Z.; Yan, Z.; Jiang, S.; Zhao, X.; Wang, L.; Li, M. Inflammatory role of S100A8/A9 in the central nervous system non-neoplastic diseases. Brain Res. Bull. 2024, 218, 111100. [Google Scholar] [CrossRef] [PubMed]
- Shepherd, C.E.; Goyette, J.; Utter, V.; Rahimi, F.; Yang, Z.; Geczy, C.L.; Halliday, G.M. Inflammatory S100A9 and S100A12 proteins in Alzheimer’s disease. Neurobiol. Aging 2006, 27, 1554–1563. [Google Scholar] [CrossRef] [PubMed]
- Steiner, J.; Bogerts, B.; Schroeter, M.L.; Bernstein, H.G. S100B protein in neurodegenerative disorders. Clin. Chem. Lab. Med. 2011, 49, 409–424. [Google Scholar] [CrossRef]
- Brozzi, F.; Arcuri, C.; Giambanco, I.; Donato, R. S100B Protein Regulates Astrocyte Shape and Migration via Interaction with Src Kinase: IMPLICATIONS FOR ASTROCYTE DEVELOPMENT, ACTIVATION, AND TUMOR GROWTH. J. Biol. Chem. 2009, 284, 8797–8811. [Google Scholar] [CrossRef]
- Hoyaux, D.; Boom, A.; Van den Bosch, L.; Belot, N.; Martin, J.J.; Heizmann, C.W.; Kiss, R.; Pochet, R. S100A6 overexpression within astrocytes associated with impaired axons from both ALS mouse model and human patients. J. Neuropathol. Exp. Neurol. 2002, 61, 736–744. [Google Scholar] [CrossRef]
- Wu, M.; Xu, L.; Wang, Y.; Zhou, N.; Zhen, F.; Zhang, Y.; Qu, X.; Fan, H.; Liu, S.; Chen, Y.; et al. S100A8/A9 induces microglia activation and promotes the apoptosis of oligodendrocyte precursor cells by activating the NF-κB signaling pathway. Brain Res. Bull. 2018, 143, 234–245. [Google Scholar] [CrossRef]
- Dong, N.; Wang, Y. MiR-30a Regulates S100A12-induced Retinal Microglial Activation and Inflammation by Targeting NLRP3. Curr. Eye Res. 2019, 44, 1236–1243. [Google Scholar] [CrossRef]
- Lisachev, P.D.; Shtark, M.B.; Sokolova, O.O.; Pustylnyak, V.O.; Salakhutdinova, M.Y.; Epstein, O.I. A Comparison of the Dynamics of S100B, S100A1, and S100A6 mRNA Expression in Hippocampal CA1 Area of Rats during Long-Term Potentiation and after Low-Frequency Stimulation. Cardiovasc. Psychiatry Neurol. 2010, 2010, 720958. [Google Scholar] [CrossRef]
- Rickmann, M.; Wolff, J.R. S100 protein expression in subpopulations of neurons of rat brain. Neuroscience 1995, 67, 977–991. [Google Scholar] [CrossRef]
- Leśniak, W.; Filipek, A. S100 Proteins—Intracellular and Extracellular Function in Norm and Pathology. Biomolecules 2024, 14, 432. [Google Scholar] [CrossRef] [PubMed]
- Donato, R.; Sorci, G.; Riuzzi, F.; Arcuri, C.; Bianchi, R.; Brozzi, F.; Tubaro, C.; Giambanco, I. S100B’s double life: Intracellular regulator and extracellular signal. Biochim. Biophys. Acta 2009, 1793, 1008–1022. [Google Scholar] [CrossRef]
- Michetti, F.; Clementi, M.E.; Di Liddo, R.; Valeriani, F.; Ria, F.; Rende, M.; Di Sante, G.; Romano Spica, V. The S100B Protein: A Multifaceted Pathogenic Factor More Than a Biomarker. Int. J. Mol. Sci. 2023, 24, 9605. [Google Scholar] [CrossRef]
- Wright, N.T.; Cannon, B.R.; Zimmer, D.B.; Weber, D.J. S100A1: Structure, Function, and Therapeutic Potential. Curr. Chem. Biol. 2009, 3, 138–145. [Google Scholar] [PubMed]
- Donato, R.; Sorci, G.; Giambanco, I. S100A6 protein: Functional roles. Cell. Mol. Life Sci. 2017, 74, 2749–2760. [Google Scholar] [CrossRef] [PubMed]
- Viemann, D. S100-Alarmins Are Essential Pilots of Postnatal Innate Immune Adaptation. Front. Immunol. 2020, 11, 688. [Google Scholar] [CrossRef]
- Kleissner, M.; Sramko, M.; Kohoutek, J.; Kautzner, J.; Kettner, J. Serum S100 Protein Is a Reliable Predictor of Brain Injury After Out-of-Hospital Cardiac Arrest: A Cohort Study. Front. Cardiovasc. Med. 2021, 8, 624825. [Google Scholar] [CrossRef]
- Singh, A.K.; Asif, S.; Pandey, D.K.; Chaudhary, A.; Kapoor, V.; Verma, P.K. Biomarkers in Acute Traumatic Brain Injury: A Systematic Review and Meta-Analysis. Cureus 2024, 16, e63020. [Google Scholar] [CrossRef]
- Lopes, A.N.; Regner, A.; Simon, D. The Role of S100b Protein Biomarker in Brain Death: A Literature Review. Cureus 2024, 16, e62707. [Google Scholar] [CrossRef]
- Camponeschi, C.; De Carluccio, M.; Amadio, S.; Clementi, M.E.; Sampaolese, B.; Volonté, C.; Tredicine, M.; Romano Spica, V.; Di Liddo, R.; Ria, F.; et al. S100B Protein as a Therapeutic Target in Multiple Sclerosis: The S100B Inhibitor Arundic Acid Protects from Chronic Experimental Autoimmune Encephalomyelitis. Int. J. Mol. Sci. 2021, 22, 13558. [Google Scholar] [CrossRef]
- Migheli, A.; Cordera, S.; Bendotti, C.; Atzori, C.; Piva, R.; Schiffer, D. S-100β protein is upregulated in astrocytes and motor neurons in the spinal cord of patients with amyotrophic lateral sclerosis. Neurosci. Lett. 1999, 261, 25–28. [Google Scholar] [CrossRef]
- Hagmeyer, S.; Romão, M.A.; Cristóvão, J.S.; Vilella, A.; Zoli, M.; Gomes, C.M.; Grabrucker, A.M. Distribution and Relative Abundance of S100 Proteins in the Brain of the APP23 Alzheimer’s Disease Model Mice. Front. Neurosci. 2019, 13, 640. [Google Scholar] [CrossRef]
- Schuermans, S.; Kestens, C.; Marques, P.E. Systemic mechanisms of necrotic cell debris clearance. Cell Death Dis. 2024, 15, 557. [Google Scholar] [CrossRef] [PubMed]
- Heizmann, C.W. S100 proteins: Diagnostic and prognostic biomarkers in laboratory medicine. Biochim. Biophys. Acta 2019, 1866, 1197–1206. [Google Scholar] [CrossRef] [PubMed]
- Mori, T.; Koyama, N.; Arendash, G.W.; Horikoshi-Sakuraba, Y.; Tan, J.; Town, T. Overexpression of human S100B exacerbates cerebral amyloidosis and gliosis in the Tg2576 mouse model of Alzheimer’s disease. GLIA 2010, 58, 300–314. [Google Scholar] [CrossRef] [PubMed]
- Mrak, R.E.; Griffinbc, W.S. The role of activated astrocytes and of the neurotrophic cytokine S100B in the pathogenesis of Alzheimer’s disease. Neurobiol. Aging 2001, 22, 915–922. [Google Scholar] [CrossRef]
- Origlia, N.; Arancio, O.; Domenici, L.; Yan, S.S. MAPK, beta-amyloid and synaptic dysfunction: The role of RAGE. Expert Rev. Neurother. 2009, 9, 1635–1645. [Google Scholar] [CrossRef]
- Hampel, H.; Hardy, J.; Blennow, K.; Chen, C.; Perry, G.; Kim, S.H.; Villemagne, V.L.; Aisen, P.; Vendruscolo, M.; Iwatsubo, T.; et al. The Amyloid-β Pathway in Alzheimer’s Disease. Mol. Psychiatry 2021, 26, 5481–5503. [Google Scholar] [CrossRef]
- Esposito, G.; Scuderi, C.; Lu, J.; Savani, C.; De Filippis, D.; Iuvone, T.; Steardo, L., Jr.; Sheen, V.; Steardo, L. S100B induces tau protein hyperphosphorylation via Dickopff-1 up-regulation and disrupts the Wnt pathway in human neural stem cells. J. Cell. Mol. Med. 2008, 12, 914–927. [Google Scholar] [CrossRef]
- Dugger, B.N.; Whiteside, C.M.; Maarouf, C.L.; Walker, D.G.; Beach, T.G.; Sue, L.I.; Garcia, A.; Dunckley, T.; Meechoovet, B.; Reiman, E.M.; et al. The Presence of Select Tau Species in Human Peripheral Tissues and Their Relation to Alzheimer’s Disease. J. Alzheimers Dis. 2016, 51, 345–356. [Google Scholar] [CrossRef] [PubMed]
- Litus, E.A.; Shevelyova, M.P.; Vologzhannikova, A.A.; Deryusheva, E.I.; Machulin, A.V.; Nemashkalova, E.L.; Permyakova, M.E.; Sokolov, A.S.; Alikova, V.D.; Uversky, V.N.; et al. Binding of Pro-Inflammatory Proteins S100A8 or S100A9 to Amyloid-β Peptide Suppresses Its Fibrillation. Biomolecules 2025, 15, 431. [Google Scholar] [CrossRef]
- Lodeiro, M.; Puerta, E.; Ismail, M.A.; Rodriguez-Rodriguez, P.; Rönnbäck, A.; Codita, A.; Parrado-Fernandez, C.; Maioli, S.; Gil-Bea, F.; Merino-Serrais, P.; et al. Aggregation of the Inflammatory S100A8 Precedes Aβ Plaque Formation in Transgenic APP Mice: Positive Feedback for S100A8 and Aβ Productions. J. Gerontol. Ser. A Biomed. Sci. Med. Sci. 2017, 72, 319–328. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Klechikov, A.G.; Gharibyan, A.L.; Wärmländer, S.K.; Jarvet, J.; Zhao, L.; Jia, X.; Narayana, V.K.; Shankar, S.K.; Olofsson, A.; et al. The role of pro-inflammatory S100A9 in Alzheimer’s disease amyloid-neuroinflammatory cascade. Acta Neuropathol. 2014, 127, 507–522. [Google Scholar] [CrossRef] [PubMed]
- Sathe, K.; Maetzler, W.; Lang, J.D.; Mounsey, R.B.; Fleckenstein, C.; Martin, H.L.; Schulte, C.; Mustafa, S.; Synofzik, M.; Vukovic, Z.; et al. S100B is increased in Parkinson’s disease and ablation protects against MPTP-induced toxicity through the RAGE and TNF-α pathway. Brain 2012, 135, 3336–3347. [Google Scholar] [CrossRef]
- Reeve, A.K.; Ludtmann, M.H.; Angelova, P.R.; Simcox, E.M.; Horrocks, M.H.; Klenerman, D.; Gandhi, S.; Turnbull, D.M.; Abramov, A.Y. Aggregated α-synuclein and complex I deficiency: Exploration of their relationship in differentiated neurons. Cell Death Dis. 2015, 6, e1820. [Google Scholar] [CrossRef]
- Puspita, L.; Chung, S.Y.; Shim, J.W. Oxidative stress and cellular pathologies in Parkinson’s disease. Mol. Brain. 2017, 10, 53. [Google Scholar] [CrossRef]
- Radi, R. Peroxynitrite, a stealthy biological oxidant. J. Biol. Chem. 2013, 288, 26464–26472. [Google Scholar] [CrossRef]
- Chinta, S.J.; Andersen, J.K. Redox imbalance in Parkinson’s disease. Biochim. Biophys. Acta 2008, 1780, 1362–1367. [Google Scholar] [CrossRef]
- Fardell, C.; Zettergren, A.; Ran, C.; Carmine Belin, A.; Ekman, A.; Sydow, O.; Bäckman, L.; Holmberg, B.; Dizdar, N.; Söderkvist, P.; et al. S100B polymorphisms are associated with age of onset of Parkinson’s disease. BMC Med. Genet. 2018, 19, 42. [Google Scholar] [CrossRef]
- Zervides, K.A.; Jern, A.; Nystedt, J.; Gullstrand, B.; Nilsson, P.C.; Sundgren, P.C.; Bengtsson, A.A.; Jönsen, A. Serum S100A8/A9 concentrations are associated with neuropsychiatric involvement in systemic lupus erythematosus: A cross-sectional study. BMC Rheumatol. 2022, 6, 38. [Google Scholar] [CrossRef]
- Wang, S.; Song, R.; Wang, Z.; Jing, Z.; Wang, S.; Ma, J. S100A8/A9 in Inflammation. Front. Immunol. 2018, 9, 1298. [Google Scholar] [CrossRef]
- De Carluccio, M.; Di Sante, G.; Clementi, M.E.; Ruggirello, M.; Stabile, A.M.; Pistilli, A.; Marini, S.; Romano Spica, V.; Rende, M.; Ria, F.; et al. Effect on Different Glial Cell Types of S100B Modulation in Multiple Sclerosis Experimental Models. Int. J. Mol. Sci. 2025, 26, 5948. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, S.; Linington, C.; Zipp, F.; Sotgiu, S.; de Waal Malefyt, R.; Wekerle, H.; Hohlfeld, R. Multiple sclerosis: Comparison of the human T-cell response to S100 beta and myelin basic protein reveals parallels to rat experimental autoimmune panencephalitis. Brain 1997, 120, 1437–1445. [Google Scholar] [CrossRef]
- Lovett-Racke, A.E.; Yang, Y.; Racke, M.K. Th1 versus Th17: Are T cell cytokines relevant in multiple sclerosis? Biochim. Biophys. Acta 2011, 1812, 246–251. [Google Scholar] [CrossRef] [PubMed]
- Süssmuth, S.D.; Tumani, H.; Ecker, D.; Ludolph, A.C. Amyotrophic lateral sclerosis: Disease stage related changes of tau protein and S100 beta in cerebrospinal fluid and creatine kinase in serum. Neurosci. Lett. 2003, 353, 57–60. [Google Scholar] [CrossRef] [PubMed]
- Juranek, J.K.; Daffu, G.K.; Wojtkiewicz, J.; Lacomis, D.; Kofler, J.; Schmidt, A.M. Receptor for Advanced Glycation End Products and its Inflammatory Ligands are Upregulated in Amyotrophic Lateral Sclerosis. Front. Cell. Neurosci. 2015, 9, 485. [Google Scholar] [CrossRef]
- Kamo, H.; Haebara, H.; Akiguchi, I.; Kameyama, M.; Kimura, H.; McGeer, P.L. A distinctive distribution of reactive astroglia in the precentral cortex in amyotrophic lateral sclerosis. Acta Neuropathol. 1987, 74, 33–38. [Google Scholar] [CrossRef]
- Dıaz-Amarilla, P.; Olivera-Bravo, S.; Trias, E.; Cragnolini, A.; MartınezPalma, L.; Cassina, P.; Beckman, J.; Barbeito, L. Phenotypically aberrant astrocytes that promote motoneuron damage in a model of inherited amyotrophic lateral sclerosis. Proc. Natl. Acad. Sci. USA 2011, 108, 18126–18131. [Google Scholar] [CrossRef]
- Serrano, A.; Donno, C.; Giannetti, S.; Peric, M.; Andjus, P.; D’Ambrosi, N.; Michetti, F. The astrocytic S100B protein with its receptor RAGE is aberrantly expressed in SOD1G93A models, and its inhibition decreases the expression of proinflammatory genes. Mediat. Inflamm. 2017, 2017, 1626204. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Ferreira, A.; Van Eldik, L.J. S100β induces neuronal cell death through nitric oxide release from astrocytes. J. Neurochem. 1997, 69, 2294–2301. [Google Scholar] [CrossRef]
- Koh, S.X.; Lee, J.K. S100B as a marker for brain damage and blood-brain barrier disruption following exercise. Sports Med. 2014, 44, 369–385. [Google Scholar] [CrossRef] [PubMed]
- Steinruecke, M.; Lonergan, R.M.; Selvaraj, B.T.; Chandran, S.; Diaz-Castro, B.; Stavrou, M. Blood-CNS barrier dysfunction in amyotrophic lateral sclerosis: Proposed mechanisms and clinical implications. J. Cereb. Blood Flow Metab. 2023, 43, 642–654. [Google Scholar] [CrossRef] [PubMed]
- Thelin, E.P.; Nelson, D.W.; Bellander, B.M. A review of the clinical utility of serum S100B protein levels in the assessment of traumatic brain injury. Acta Neurochir. 2017, 159, 209–225. [Google Scholar] [CrossRef]
- Janigro, D.; Mondello, S.; Posti, J.P.; Unden, J. GFAP and S100B: What You Always Wanted to Know and Never Dared to Ask. Front. Neurol. 2022, 13, 835597. [Google Scholar] [CrossRef]
- Oris, C.; Kahouadji, S.; Durif, J.; Bouvier, D.; Sapin, V. S100B, Actor and Biomarker of Mild Traumatic Brain Injury. Int. J. Mol. Sci. 2023, 24, 6602. [Google Scholar] [CrossRef]
- Dmytriyeva, O.; Pankratova, S.; Owczarek, S.; Sonn, K.; Soroka, V.; Ridley, C.M.; Marsolais, A.; Lopez-Hoyos, M.; Ambartsumian, N.; Lukanidin, E.; et al. The metastasis-promoting S100A4 protein confers neuroprotection in brain injury. Nat. Commun. 2012, 3, 1197. [Google Scholar] [CrossRef]
- Fang, B.; Liang, M.; Yang, G.; Ye, Y.; Xu, H.; He, X.; Huang, J.H. Expression of S100A6 in rat hippocampus after traumatic brain injury due to lateral head acceleration. Int. J. Mol. Sci. 2014, 15, 6378–6390. [Google Scholar] [CrossRef] [PubMed]
- He, G.Y.; Zhao, C.H.; Wu, D.G.; Cheng, H.; Sun, L.A.; Zhang, D.L.; Yang, X.J.; Fan, X.R.; Di, G.F.; Jiang, X.C. S100A8 Promotes Inflammation via Toll-Like Receptor 4 After Experimental Traumatic Brain Injury. Front. Neurosci. 2021, 14, 616559. [Google Scholar] [CrossRef] [PubMed]


| S100 Protein Member | Chromosomal Location | Functions | References |
|---|---|---|---|
| S100A1 | 1q21.3 | Modulates contractility | [32] |
| Regulates Ca2+ handling in heart and skeletal muscle | [33] | ||
| S100A2 | 1q21.3 | Involved in p53-mediated cell cycle arrest and tumor suppression | [34] |
| Negatively impacts tissue repair | [35] | ||
| S100A3 | 1q21.3 | Involved in hair shaft formation | [36] |
| S100A4 | 1q21.3 | Promotes cell motility, invasion, and metastasis | [37,38] |
| S100A5 | 1q21.3 | Modulation of neuronal activity | [39] |
| S100A6 | 1q21.3 | Regulates cytoskeletal dynamics and proliferation | [40] |
| (Calcyclin) | |||
| S100A7 | 1q21.3 | Antimicrobial peptide | [41] |
| (Psoriasin) | Its overexpression is linked to psoriasis and breast cancer progression | [42,43] | |
| S100A8/9 | 1q21.3 | Pro-inflammatory and antimicrobial roles | [44,45] |
| (Calprotectin) | |||
| S100A10 | 1q21.3 | Regulates membrane trafficking and plasminogen activation | [46,47] |
| S100A11 | 1q21.3 | Involved in cell proliferation, motility, and Ca2+ signal transduction | [48,49,50] |
| S100A12 | 1q21.3 | Associated with inflammatory diseases | [51] |
| (Calgranulin C) | Inducer of neurite growth | [52] | |
| S100A13 | 1q21.3 | Mediates non-classical secretion of FGF1 and IL-1α | [53,54] |
| Involved in angiogenesis and cellular stress responses | [55,56] | ||
| S100A14 | 1q21.3 | Influences cell proliferation and apoptosis: dual role in cancer | [57,58] |
| S100A15 | 1q21.3 | Involved in skin immune response | [59] |
| S100A16 | 1q21.3 | Implicated in adipocyte differentiation and tumor progression | [60,61] |
| S100B | 21q22.3 | Regulates cell proliferation and apoptosis | [62,63] |
| Marker of CNS injury | [64] | ||
| S100G | Xp22.2 | Involved in intestinal Ca2+ absorption | [65] |
| (Calbindin-D9k) | |||
| S100P | 4p16.1 | Promotes tumor progresion and metastasis | [66,67] |
| (Placental S100) | |||
| S100Z | 5q13.3 | Interacts with S100P | [68] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
García-Domínguez, M. Relationship of S100 Proteins with Neuroinflammation. Biomolecules 2025, 15, 1125. https://doi.org/10.3390/biom15081125
García-Domínguez M. Relationship of S100 Proteins with Neuroinflammation. Biomolecules. 2025; 15(8):1125. https://doi.org/10.3390/biom15081125
Chicago/Turabian StyleGarcía-Domínguez, Mario. 2025. "Relationship of S100 Proteins with Neuroinflammation" Biomolecules 15, no. 8: 1125. https://doi.org/10.3390/biom15081125
APA StyleGarcía-Domínguez, M. (2025). Relationship of S100 Proteins with Neuroinflammation. Biomolecules, 15(8), 1125. https://doi.org/10.3390/biom15081125





