Phytocompounds in Precision Dermatology: COX-2 Inhibitors as a Therapeutic Target in Atopic-Prone Skin
Abstract
1. Introduction
2. Materials and Methods
2.1. COX-2 3D Structure Retrieval and Minimization
2.2. Compound Database Retrieval
2.3. Rule of Five Filtering
2.4. Virtual Screening
2.5. Induced-Fit Docking (IFD)
2.6. Visualization and Interaction Analysis
2.7. Molecular Dynamics Simulation Analysis
2.8. Post-Simulation Stability, Compactness, and Residual Fluctuation Analyses
2.9. Binding Free Energy Analysis
2.10. Pharmacokinetic Analysis of Control and Shortlisted Compounds
3. Results and Discussion
3.1. Virtual Screening of Phytocompounds Against COX-2
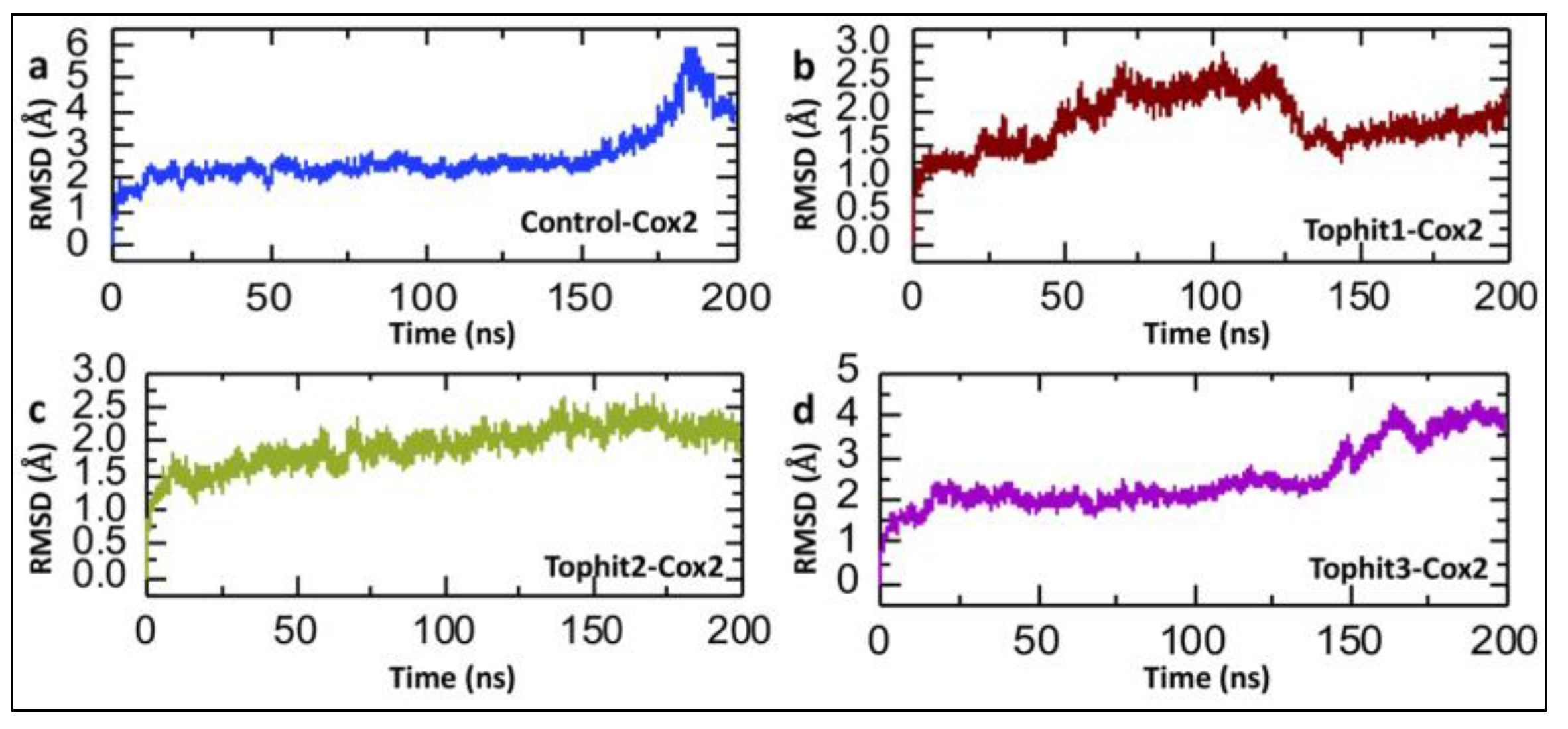
3.2. Molecular Dynamics Stability Analysis of Tophit-COX-2 Complexes
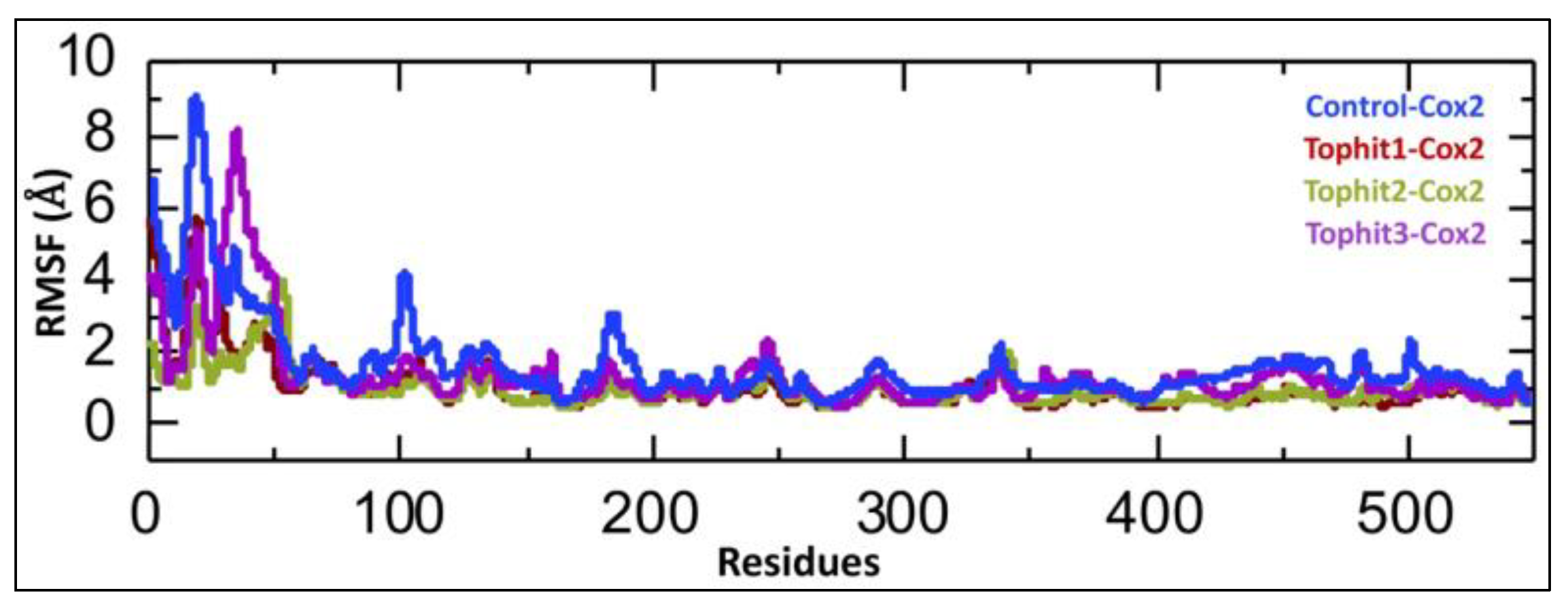
3.3. Dynamic Residual Fluctuation Analysis of Tophit-COX-2 Complexes
3.4. Post-Simulation Compactness Analysis of Tophit-COX-2 Complexes
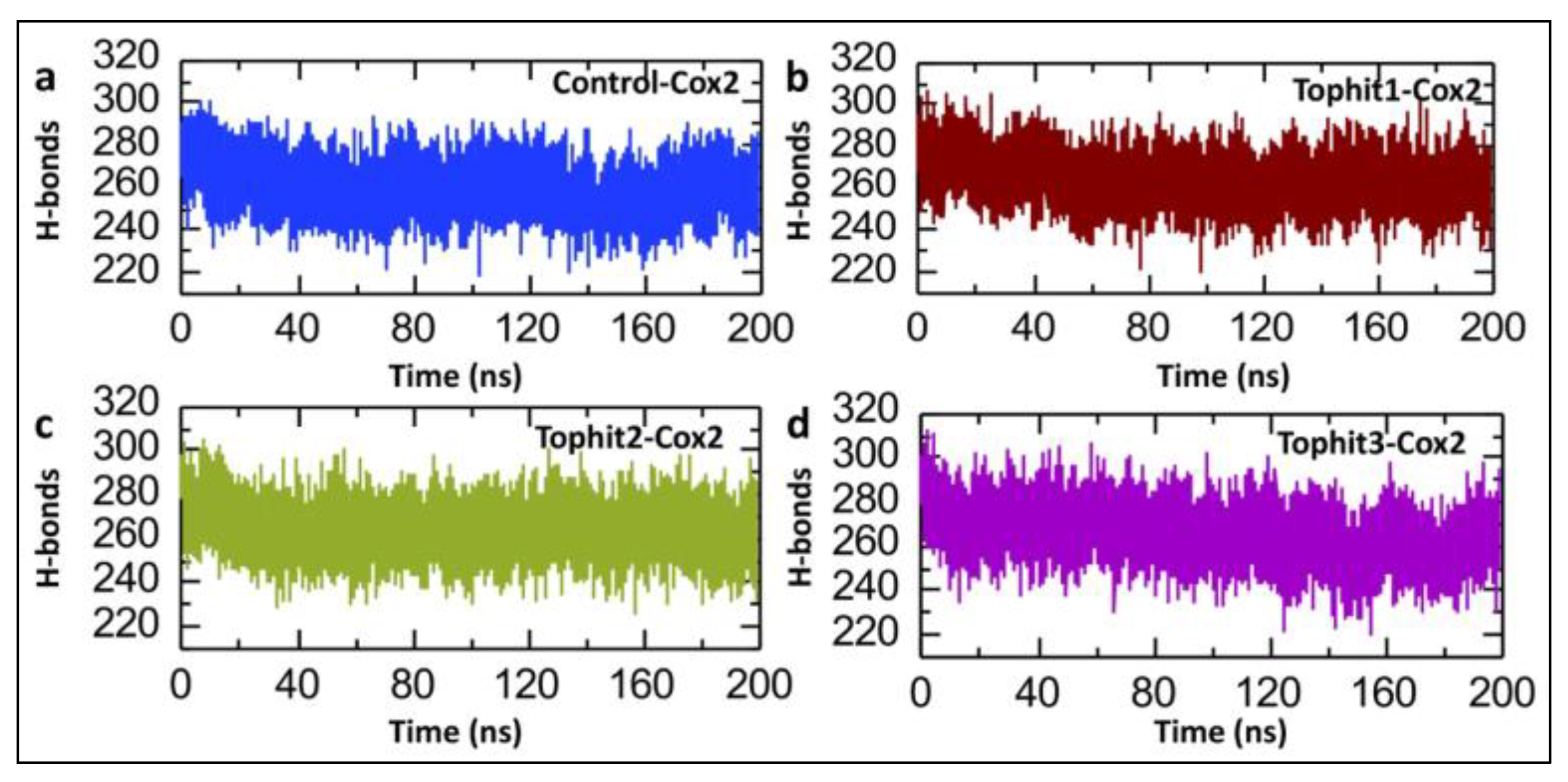
3.5. Post-Simulation Hydrogen Bond Analysis
3.6. Binding Free Energy Calculations of Control and Top Hit Compound-COX-2 Complexes
3.7. Pharmacokinetic Properties Analysis of Control and Lead Compounds
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Tian, J.; Zhang, D.; Yang, Y.; Huang, Y.; Wang, L.; Yao, X.; Lu, Q. Global epidemiology of atopic dermatitis: A comprehensive systematic analysis and modelling study. Br. J. Dermatol. 2023, 190, 55–61. [Google Scholar] [CrossRef]
- Flohr, C.; Mann, J. New insights into the epidemiology of childhood atopic dermatitis. Allergy 2014, 69, 3–16. [Google Scholar] [CrossRef] [PubMed]
- Egawa, G.; Kabashima, K. Barrier dysfunction in the skin allergy. Allergol. Int. 2018, 67, 3–11. [Google Scholar] [CrossRef]
- Kim, J.; Kim, B.E.; Ahn, K.; Leung, D.Y.M. Interactions Between Atopic Dermatitis and Staphylococcus aureus Infection: Clinical Implications. Allergy Asthma Immunol. Res. 2019, 11, 593–603. [Google Scholar] [CrossRef] [PubMed]
- Tamagawa-Mineoka, R.; Katoh, N. Atopic Dermatitis: Identification and Management of Complicating Factors. Int. J. Mol. Sci. 2020, 21, 2671. [Google Scholar] [CrossRef]
- Brown, S.J.; McLean, W.I. One remarkable molecule: Filaggrin. J. Investig. Dermatol. 2012, 132 Pt 2, 751–762. [Google Scholar] [CrossRef] [PubMed]
- Czarnowicki, T.; He, H.; Krueger, J.G.; Guttman-Yassky, E. Atopic dermatitis endotypes and implications for targeted therapeutics. J. Allergy Clin. Immunol. 2019, 143, 1–11. [Google Scholar] [CrossRef]
- Guttman-Yassky, E.; Krueger, J.G. Atopic dermatitis and psoriasis: Two different immune diseases or one spectrum? Curr. Opin. Immunol. 2017, 48, 68–73. [Google Scholar] [CrossRef]
- Brunner, P.M.; Pavel, A.B.; Khattri, S.; Leonard, A.; Malik, K.; Rose, S.; On, S.J.; Vekaria, A.S.; Traidl-Hoffmann, C.; Singer, G.K.; et al. Baseline IL-22 expression in patients with atopic dermatitis stratifies tissue responses to fezakinumab. J. Allergy Clin. Immunol. 2019, 143, 142–154. [Google Scholar] [CrossRef]
- Gittler, J.K.; Shemer, A.; Suárez-Fariñas, M.; Fuentes-Duculan, J.; Gulewicz, K.J.; Wang, C.Q.; Mitsui, H.; Cardinale, I.; de Guzman Strong, C.; Krueger, J.G.; et al. Progressive activation of T(H)2/T(H)22 cytokines and selective epidermal proteins characterizes acute and chronic atopic dermatitis. J. Allergy Clin. Immunol. 2012, 130, 1344–1354. [Google Scholar] [CrossRef]
- Ricciotti, E.; FitzGerald, G.A. Prostaglandins and inflammation. Arter. Thromb. Vasc. Biol. 2011, 31, 986–1000. [Google Scholar] [CrossRef][Green Version]
- Tang, L.; Li, X.L.; Deng, Z.X.; Xiao, Y.; Cheng, Y.H.; Li, J. Conjugated linoleic acid attenuates 2,4-dinitrofluorobenzene-induced atopic dermatitis in mice through dual inhibition of COX-2/5-LOX and TLR4/NF-κB signaling. J. Nutr. Biochem. 2020, 81, 108379. [Google Scholar] [CrossRef]
- Carola, C.; Salazar, A.; Rakers, C.; Himbert, F.; Do, Q.-T.; Bernard, P.; von Hagen, J.; Capasso, R. A Cornflower Extract Containing N-Feruloylserotonin Reduces Inflammation in Human Skin by Neutralizing CCL17 and CCL22 and Inhibiting COX-2 and 5-LOX. Mediat. Inflamm. 2021, 2021, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Liana, D.; Eurtivong, C.; Phanumartwiwath, A. Boesenbergia rotunda and Its Pinostrobin for Atopic Dermatitis: Dual 5-Lipoxygenase and Cyclooxygenase-2 Inhibitor and Its Mechanistic Study through Steady-State Kinetics and Molecular Modeling. Antioxidants 2024, 13, 74. [Google Scholar] [CrossRef] [PubMed]
- Atanasov, A.G.; Zotchev, S.B.; Dirsch, V.M.; International Natural Product Sciences Taskforce; Supuran, C.T. Natural products in drug discovery: Advances and opportunities. Nat. Rev. Drug Discov. 2021, 20, 200–216. [Google Scholar] [CrossRef]
- Chandel, P.; Rawal, R.K.; Kaur, R. Natural products and their derivatives as cyclooxygenase-2 inhibitors. Futur. Med. Chem. 2018, 10, 2471–2492. [Google Scholar] [CrossRef]
- Lionta, E.; Spyrou, G.; Vassilatis, D.K.; Cournia, Z. Structure-based virtual screening for drug discovery: Principles, applications and recent advances. Curr. Top. Med. Chem. 2014, 14, 1923–1938. [Google Scholar] [CrossRef]
- Ru, J.; Li, P.; Wang, J.; Zhou, W.; Li, B.; Huang, C.; Li, P.; Guo, Z.; Tao, W.; Yang, Y.; et al. TCMSP: A database of systems pharmacology for drug discovery from herbal medicines. J. Cheminform. 2014, 6, 13. [Google Scholar] [CrossRef]
- Ntie-Kang, F.; Telukunta, K.K.; Döring, K.; Simoben, C.V.; Moumbock, A.F.A.; Malange, Y.I.; Njume, L.E.; Yong, J.N.; Sippl, W.; Günther, S. NANPDB: A Resource for Natural Products from Northern African Sources. J. Nat. Prod. 2017, 80, 2067–2076. [Google Scholar] [CrossRef]
- Lagorce, D.; Bouslama, L.; Becot, J.; Miteva, M.A.; Villoutreix, B.O. FAF-Drugs4: Free ADME-tox filtering computations for chemical biology and early stages drug discovery. Bioinformatics 2017, 33, 3658–3660. [Google Scholar] [CrossRef] [PubMed]
- Wang, E.; Sun, H.; Wang, J.; Wang, Z.; Liu, H.; Zhang, J.Z.H.; Hou, T. End-Point Binding Free Energy Calculation with MM/PBSA and MM/GBSA: Strategies and Applications in Drug Design. Chem. Rev. 2019, 119, 9478–9508. [Google Scholar] [CrossRef]
- Diallo, B.N.; Glenister, M.; Musyoka, T.M.; Lobb, K.; Bishop, Ö.T. SANCDB: An update on South African natural compounds and their readily available analogs. J. Chemin. 2021, 13, 1–14. [Google Scholar] [CrossRef]
- Sayaf, A.M.; Khalid, S.U.; Hameed, J.A.; Alshammari, A.; Khan, A.; Mohammad, A.; Alghamdi, S.; Wei, D.-Q.; Yeoh, K. Exploring the natural products chemical space through a molecular search to discover potential inhibitors that target the hypoxia-inducible factor (HIF) prolyl hydroxylase domain (PHD). Front. Pharmacol. 2023, 14, 1202128. [Google Scholar] [CrossRef] [PubMed]
- Ravindranath, P.A.; Forli, S.; Goodsell, D.S.; Olson, A.J.; Sanner, M.F.; Fetrow, J.S. AutoDockFR: Advances in Protein-Ligand Docking with Explicitly Specified Binding Site Flexibility. PLoS Comput. Biol. 2015, 11, e1004586. [Google Scholar] [CrossRef]
- Case, D.A.; Cheatham, T.E., 3rd; Darden, T.; Gohlke, H.; Luo, R.; Merz, K.M., Jr.; Onufriev, A.; Simmerling, C.; Wang, B.; Woods, R.J. The Amber biomolecular simulation programs. J. Comput. Chem. 2005, 26, 1668–1688. [Google Scholar] [CrossRef]
- Salomon-Ferrer, R.; Case, D.A.; Walker, R.C. An overview of the Amber biomolecular simulation package. Wiley Interdiscip. Rev. Comput. Mol. Sci. 2013, 3, 198–210. [Google Scholar] [CrossRef]
- Watowich, S.J.; Meyer, E.; Hagstrom, R.; Josephs, R. A stable, rapidly converging conjugate gradient method for energy minimization. J. Comput. Chem. 1988, 9, 650–661. [Google Scholar] [CrossRef]
- Meza, J.C. Steepest descent. Wiley Interdiscip. Rev. Comput. Stat. 2010, 2, 719–722. [Google Scholar] [CrossRef]
- Fyta, M. Atomistic Methods in Computational Approaches in Physics; Morgan & Claypool Publishers: San Rafael, CA, USA, 2016. [Google Scholar]
- Salomon-Ferrer, R.; Götz, A.W.; Poole, D.; Le Grand, S.; Walker, R.C. Routine Microsecond Molecular Dynamics Simulations with AMBER on GPUs. 2. Explicit Solvent Particle Mesh Ewald. J. Chem. Theory Comput. 2013, 9, 3878–3888. [Google Scholar] [CrossRef]
- Roe, D.R.; Cheatham, T.E., 3rd. PTRAJ and CPPTRAJ: Software for Processing and Analysis of Molecular Dynamics Trajectory Data. J. Chem. Theory Comput. 2013, 9, 3084–3095. [Google Scholar] [CrossRef]
- Maiorov, V.N.; Crippen, G.M. Significance of root-mean-square deviation in comparing three-dimensional structures of globular proteins. J. Mol. Biol. 1994, 235, 625–634. [Google Scholar] [CrossRef]
- Cooper, A. Thermodynamic fluctuations in protein molecules. Proc. Natl. Acad. Sci. USA 1976, 73, 2740–2741. [Google Scholar] [CrossRef] [PubMed]
- Lobanov, M.I.; Bogatyreva, N.S.; Galzitskaia, O.V. Radius of gyration is indicator of compactness of protein structure. Mol. Biol. 2008, 42, 701–706. (In Russian) [Google Scholar] [CrossRef]
- Poli, G.; Granchi, C.; Rizzolio, F.; Tuccinardi, T. Application of MM-PBSA Methods in Virtual Screening. Molecules 2020, 25, 1971. [Google Scholar] [CrossRef] [PubMed]
- Pu, C.; Yan, G.; Shi, J.; Li, R. Assessing the performance of docking scoring function, FEP, MM-GBSA, and QM/MM-GBSA ap-proaches on a series of PLK1 inhibitors. Medchemcomm 2017, 8, 1452–1458. [Google Scholar] [CrossRef]
- Genheden, S.; Ryde, U. The MM/PBSA and MM/GBSA methods to estimate ligand-binding affinities. Expert. Opin. Drug Discov. 2015, 10, 449–461. [Google Scholar] [CrossRef]
- Pollastri, M.P. Overview on the Rule of Five. Curr. Protoc. Pharmacol. 2010, 49, 9.12.1–9.12.8. [Google Scholar] [CrossRef]
- Daina, A.; Michielin, O.; Zoete, V. SwissADME: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017, 7, 42717. [Google Scholar] [CrossRef]
- Pires, D.E.V.; Blundell, T.L.; Ascher, D.B. pkCSM: Predicting Small-Molecule Pharmacokinetic and Toxicity Properties Using Graph-Based Signatures. J. Med. Chem. 2015, 58, 4066–4072. [Google Scholar] [CrossRef]
- Lees, P.; Toutain, P.L.; Elliott, J.; Giraudel, J.M.; Pelligand, L.; King, J.N. Pharmacology, safety, efficacy and clinical uses of the COX-2 inhibitor robenacoxib. J. Vet. Pharmacol. Ther. 2022, 45, 325–351. [Google Scholar] [CrossRef] [PubMed]
- Osmaniye, D. Novel thiadiazol derivatives; design, synthesis, biological activity, molecular docking and molecular dynamics. J. Mol. Struct. 2023, 1272, 134171. [Google Scholar] [CrossRef]
- Niazi, S.K.; Mariam, Z. Computer-Aided Drug Design and Drug Discovery: A Prospective Analysis. Pharmaceuticals 2023, 17, 22. [Google Scholar] [CrossRef]
- Kim, K.; Choi, J.H.; Oh, J.; Park, J.Y.; Kim, Y.M.; Moon, J.H.; Park, J.H.; Cho, J.Y. New 8-C-p-Hydroxylbenzylflavonol Glycosides from Pumpkin (Cucurbita moschata Duch.) Tendril and Their Osteoclast Differentiation Inhibitory Activities. Molecules 2020, 25, 2077. [Google Scholar] [CrossRef] [PubMed]
- Abdissa, N.; Heydenreich, M.; Midiwo, J.O.; Ndakala, A.; Majer, Z.; Neumann, B.; Stammler, H.-G.; Sewald, N.; Yenesew, A. A xanthone and a phenylanthraquinone from the roots of Bulbine frutescens, and the revision of six seco-anthraquinones into xanthones. Phytochem. Lett. 2014, 9, 67–73. [Google Scholar] [CrossRef]
- Adem, F.A.; Kuete, V.; Mbaveng, A.T.; Heydenreich, M.; Ndakala, A.; Irungu, B.; Efferth, T.; Yenesew, A. Cytotoxic benzylbenzofuran derivatives from Dorstenia kameruniana. Fitoterapia 2018, 128, 26–30. [Google Scholar] [CrossRef] [PubMed]
- Makangara, J.; Jonker, S.; Nkunya, M. A novel phenanthrenolide and C-benzyl dihydrochalcones from uvaria puguensis. Nat. Prod. Lett. 2002, 16, 267–272. [Google Scholar] [CrossRef]
- Elhassan, G.O.M.; Adhikari, A.; Yousuf, S.; Rahman, H.; Khalid, A.; Omer, H.; Fun, H.-K.; Jahan, H.; Choudhary, M.I.; Yagi, S. Phytochemistry and antiglycation activity of Aloe sinkatana Reynolds. Phytochem. Lett. 2012, 5, 725–728. [Google Scholar] [CrossRef]
- Ola, A.R.B.; Thomy, D.; Lai, D.; Brötz-Oesterhelt, H.; Proksch, P. Inducing secondary metabolite production by the endophytic fungus Fusarium tricinctum through coculture with Bacillus subtilis. J. Nat. Prod. 2013, 76, 2094–2099. [Google Scholar] [CrossRef]
- Salo-Ahen, O.M.H.; Alanko, I.; Bhadane, R.; Bonvin, A.M.J.J.; Honorato, R.V.; Hossain, S.; Juffer, A.H.; Kabedev, A.; Lahtela-Kakkonen, M.; Larsen, A.S.; et al. Molecular Dynamics Simulations in Drug Discovery and Pharmaceutical Development. Processes 2021, 9, 71. [Google Scholar] [CrossRef]
- Manandhar, S.; Sankhe, R.; Priya, K.; Hari, G.; Kumar, B.H.; Mehta, C.H.; Nayak, U.Y.; Pai, K.S.R. Molecular dynamics and structure-based virtual screening and identification of natural compounds as Wnt signaling modulators: Possible therapeutics for Alzheimer’s disease. Mol. Divers. 2022, 26, 2793–2811. [Google Scholar] [CrossRef] [PubMed]
- Terefe, E.M.; Ghosh, A. Molecular Docking, Validation, Dynamics Simulations, and Pharmacokinetic Prediction of Phytochem-icals Isolated From Croton dichogamus Against the HIV-1 Reverse Transcriptase. Bioinform. Biol. Insights 2022, 16, 11779322221125605. [Google Scholar] [CrossRef]
- Bornot, A.; Etchebest, C.; de Brevern, A.G. Predicting protein flexibility through the prediction of local structures. Proteins 2011, 79, 839–852. [Google Scholar] [CrossRef] [PubMed]
- Bagewadi, Z.K.; Khan, T.Y.; Gangadharappa, B.; Kamalapurkar, A.; Shamsudeen, S.M.; Yaraguppi, D.A. Molecular dynamics and simulation analysis against superoxide dismutase (SOD) target of Micrococcus luteus with secondary metabolites from Bacillus licheniformis recognized by genome mining approach. Saudi J. Biol. Sci. 2023, 30, 103753. [Google Scholar] [CrossRef] [PubMed]
- Suleman, M.; Sayaf, A.M.; Aftab, S.; Alissa, M.; Alghamdi, A.; Alghamdi, S.A.; Alshehri, M.A.; Yeoh, K.K.; Crovella, S.; Shaito, A.A. Medicinal Phytocompounds as Potential Inhibitors of p300-HIF1α Interaction: A Structure-Based Screening and Molecular Dynamics Simulation Study. Pharmaceuticals 2025, 18, 602. [Google Scholar] [CrossRef]
- Chen, D.; Oezguen, N.; Urvil, P.; Ferguson, C.; Dann, S.M.; Savidge, T.C. Regulation of protein-ligand binding affinity by hydrogen bond pairing. Sci. Adv. 2016, 2, e1501240. [Google Scholar] [CrossRef]
- Suleman, M.; Murshed, A.; Sayaf, A.M.; Khan, A.; Khan, S.A.; Tricarico, P.M.; Moltrasio, C.; Agouni, A.; Yeoh, K.K.; Marzano, A.V.; et al. Exploring global natural product databases for NLRP3 inhibition: Unveiling novel combinatorial therapeutic strategy for hidradenitis suppurativa. J. Infect. Public Health 2025, 18, 102697. [Google Scholar] [CrossRef]
- Li, Y.; Meng, Q.; Yang, M.; Liu, D.; Hou, X.; Tang, L.; Wang, X.; Lyu, Y.; Chen, X.; Liu, K.; et al. Current trends in drug metabolism and pharmacokinetics. Acta Pharm. Sin. B 2019, 9, 1113–1144. [Google Scholar] [CrossRef]
- Reichel, A.; Lienau, P. Pharmacokinetics in Drug Discovery: An Exposure-Centred Approach to Optimising and Predicting Drug Efficacy and Safety. Handb. Exp. Pharmacol. 2016, 232, 235–260. [Google Scholar]
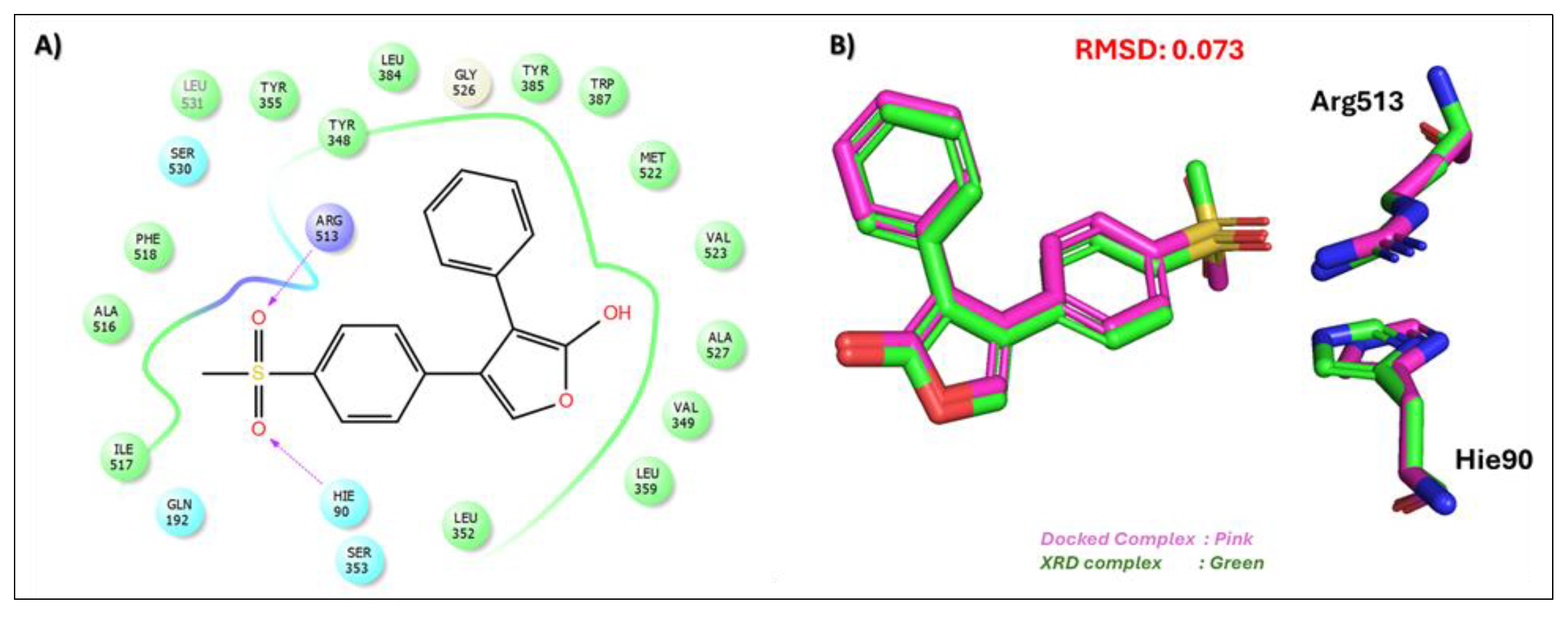
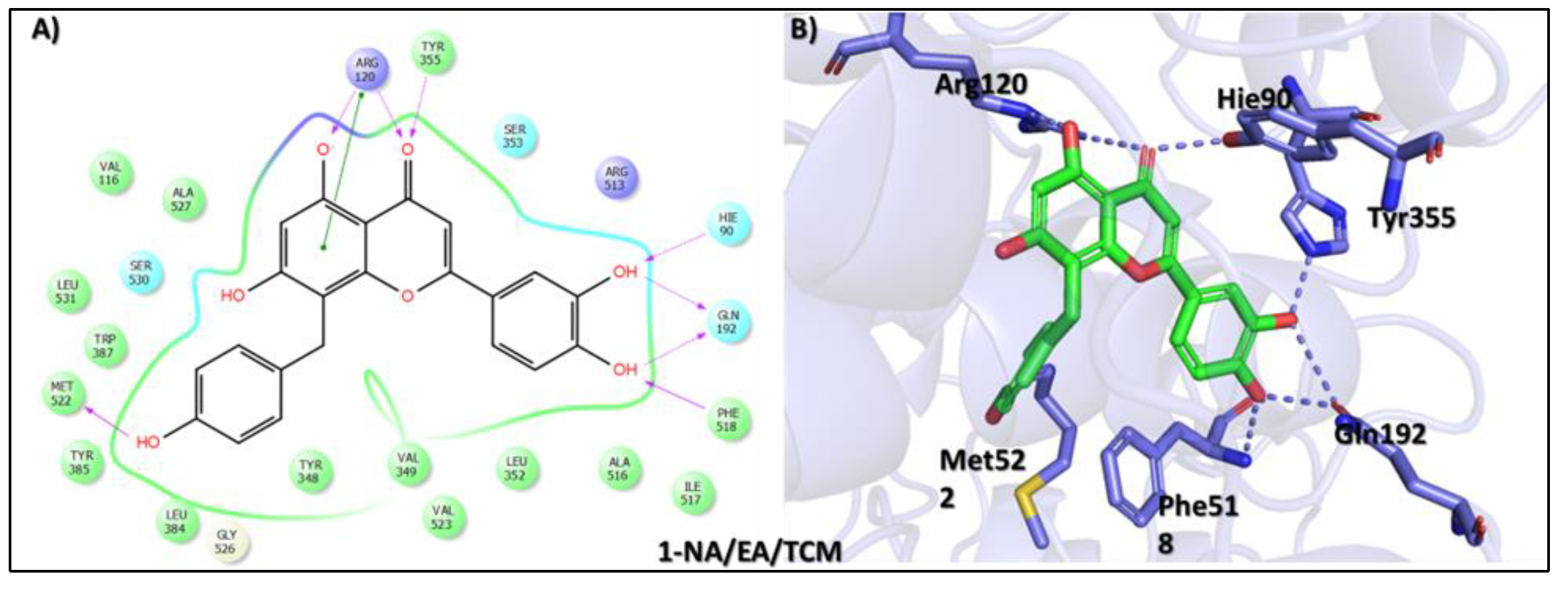
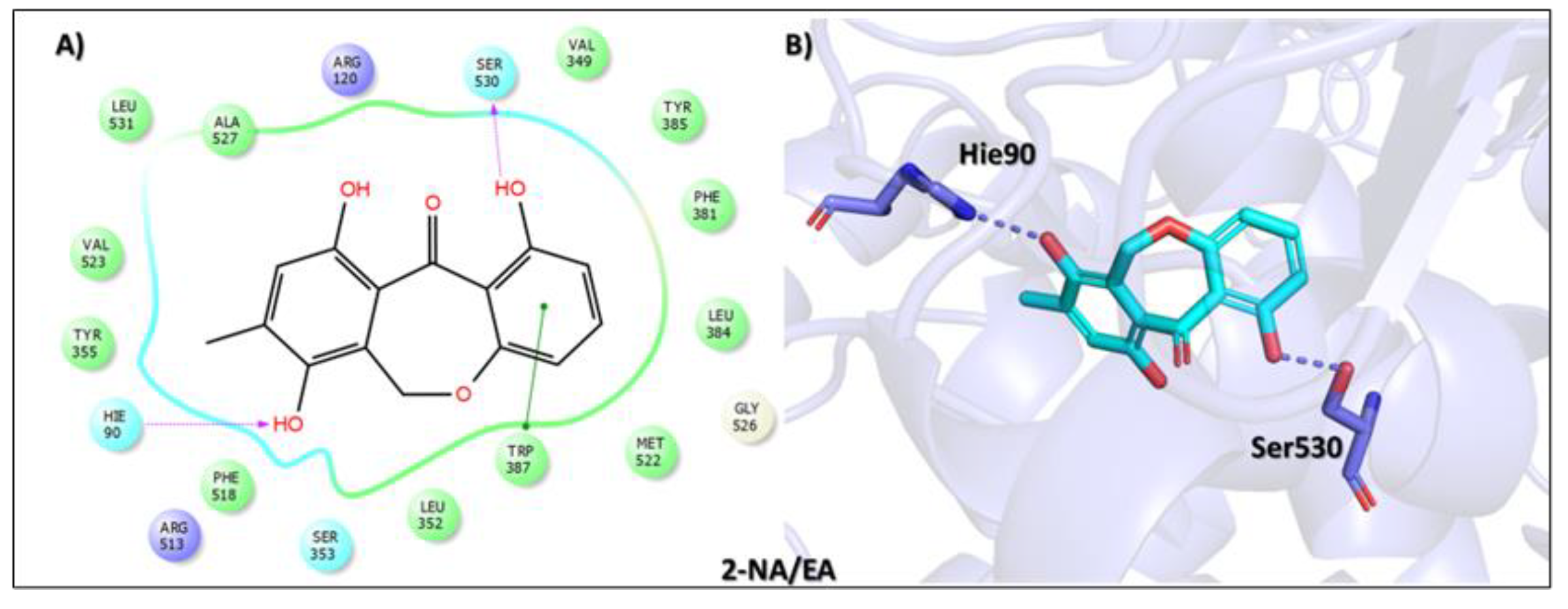

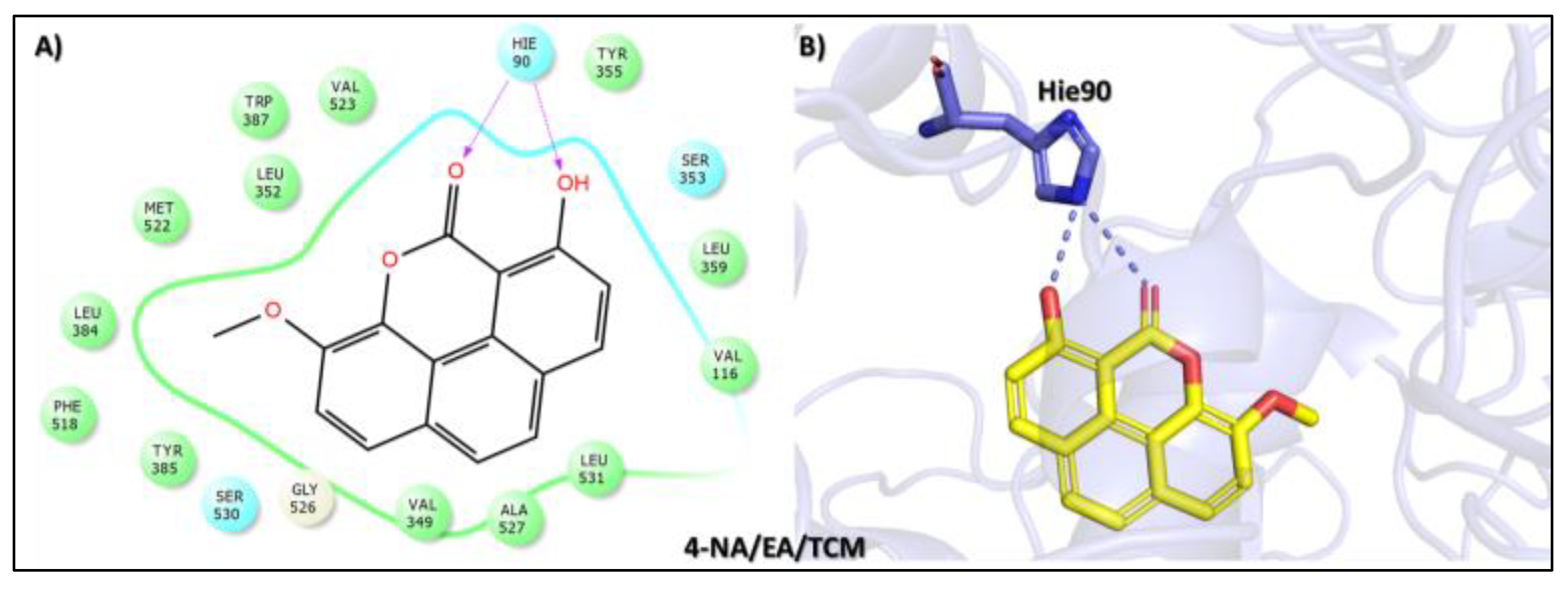


| Compound Name, Code, and Structure | Docking Score | Interacting Atom/FG | Interacting Residues | Interaction Nature |
|---|---|---|---|---|
Control (Rofecoxib (RCX)) [42]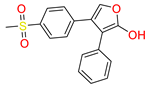 | −7.305 | O(Methylsulfonyl) | Arg513 | HB |
| O(Methylsulfonyl) | Hie90 | HB | ||
Tophit 1 (8-C-p-hydroxybenzylluteolin) [44]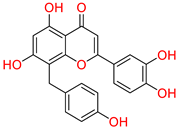 | −16.528 | OH(Resorcinol) | Arg120 | HB |
| Resorcinol ring | Arg120 | Pi-Pi | ||
| O(Dihydropyran-4-one) | Arg120 | HB | ||
| O(Dihydropyran-4-one) | Try355 | HB | ||
| OH(Phenol) | Met522 | HB | ||
| OH(Pyrocatechol) | Hie90 | HB | ||
| OH(Pyrocatechol) | Gln192 | HB | ||
| OH(Pyrocatechol) | Gln192 | HB | ||
| OH(Pyrocatechol) | Phe518 | HB | ||
Tophit 2 (Eptosphaerin D) [45]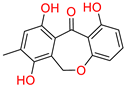 | −10.879 | OH(Methylhydroquinone) | Hie90 | HB |
| OH(Phenol) | Ser530 | HB | ||
| Phenol ring | Trp387 | Pi-Pi | ||
Tophit 3 (2-(p-hydroxybenzyl)-7-methoxybenzofuran-6-ol) [46]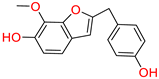 | −9.760 | OH(Phenol) | Met522 | HB |
| Phenol ring | Trp387 | Pi-Pi | ||
| Phenol ring | Phe518 | Pi-Pi | ||
| OH(2-methoxyphenol) | Arg513 | HB | ||
| 2-methoxyphenol | Tyr355 | Pi-Pi | ||
Tophit 4 (Puguenolide) [47]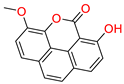 | −9.752 | O(Oxan-2-one) | Hie90 | HB |
| OH(Phenol) | Hie90 | HB | ||
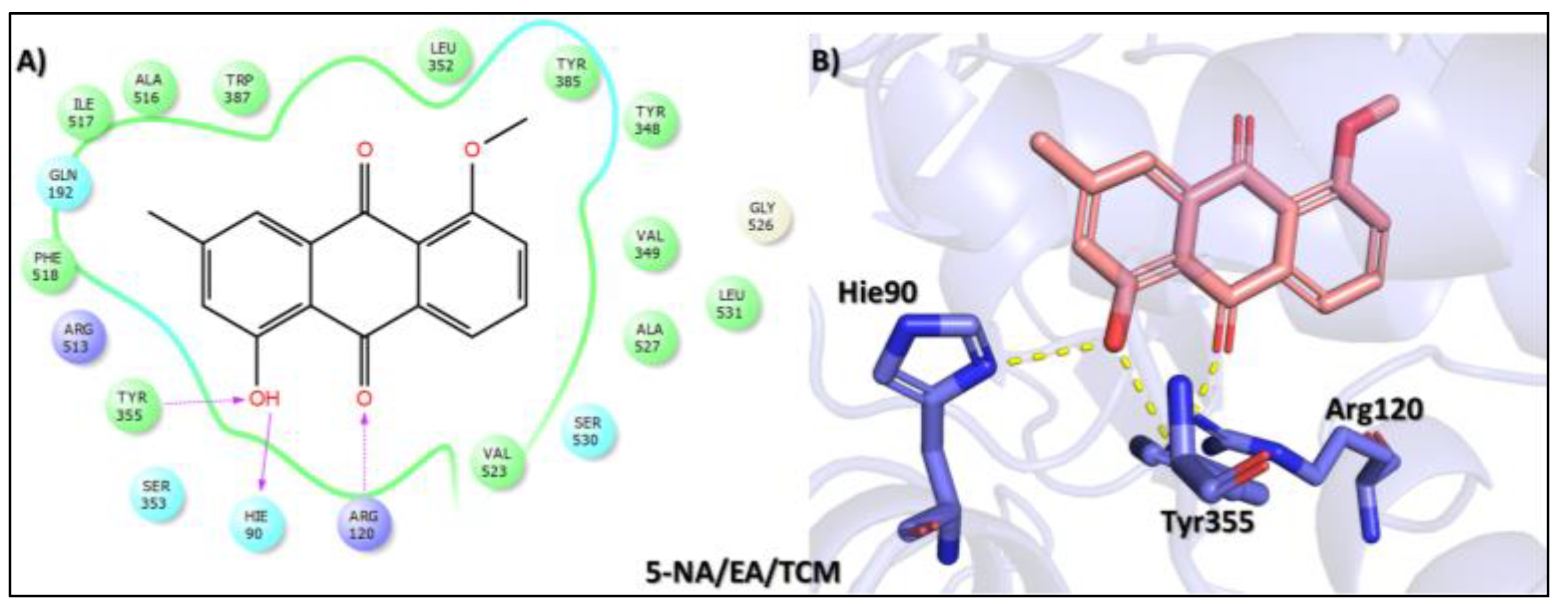
Tophit 5 (1-hydroxy-5-methoxy-3-methyl-9,10 dihydroanthracene 9,10-dione) [48]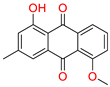 | −8.742 | OH(Methylphenol) | Arg120 | |
| OH(Methylphenol) | Hie90 | |||
| CO(Cyclohexane-1,4-dione) | Tyr355 | |||
Tophit 6 (Macrocarpon C) [49]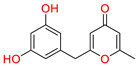 | −8.098 | CO(Methylpyran-4-one) | Arg513 | HB |
| OH(Resorcinol) | Met522 | HB |
| MM/GBSA | ||||
|---|---|---|---|---|
| Parameters | Tophit1-COX-2 | Tophit2-COX-2 | Tophit3-COX-2 | Control-COX-2 |
| ΔEvdw | −54.4187 ± 0.31 | 2.4877 ± 0.12 | −44.3097 ± 0.02 | −44.7286 ± 0.23 |
| ΔEele | −185.7878 ± 1.20 | −30.518 ± 0.03 | −4.1429 ± 0.13 | −9.7443 ± 0.29 |
| EGB | 195.9706 ± 1.10 | −10.8805 ± 0.02 | 10.6177 ± 0.12 | 57.3348 ± 0.28 |
| ESURF | −6.0761 ± 0.01 | 2.3955 ± 0.00 | −4.5258 ± 0.01 | −5.9503 ± 0.01 |
| Delta G Gas | −240.2065 ± 1.25 | −28.0303 ± 0.14 | −48.4525 ± 0.21 | −54.473 ± 0.32 |
| Delta G Solv | 189.8945 ± 1.10 | −8.485 ± 0.02 | 6.0919 ± 0.12 | 51.3845 ± 0.28 |
| ∆G Total | −50.312 xB1; 0.34 | −36.5153 ± 0.14 | −42.3606 ± 0.20 | −3.0885 ± 0.32 |
| Drugs ID | Molecular Weight | Hydrogen Acceptors | Hydrogen Donors | Consensus Log P | Lipinski’s Rule | |
|---|---|---|---|---|---|---|
| Results | Violation | |||||
| Control-COX-2 | 314.362 | 4 | 1 | 3.72 | Yes | 0 |
| Tophit1-COX-2 | 391.355 | 7 | 4 | 2.94 | Yes | 0 |
| Tophit2-COX-2 | 272.256 | 5 | 3 | 2.23 | Yes | 0 |
| Tophit3-COX-2 | 270.284 | 4 | 2 | 3.4434 | Yes | 0 |
| Properties | Control-COX-2 | Tophit1-COX-2 | Tophit2-COX-2 | Tophit3-COX-2 |
|---|---|---|---|---|
| Absorption | ||||
| Water solubility log S | −4.663 | −3.605 | −3.237 | −3.246 |
| Caco-2 permeability × 10−6 | 0.847 | −0.135 | 0.84 | 1.053 |
| Human intestinal absorption (%) | 93.2 | 83.276 | 95.145 | 91.873 |
| Distribution | ||||
| VDss (human) | −0.372 | −1.276 | 0.13 | 0.318 |
| BBB permeability | No | No | No | Yes |
| CNS permeability | −1.881 | −2.398 | −2.209 | −2.052 |
| Subcellular localization | Mitochondria | Mitochondria | Mitochondria | Mitochondria |
| Metabolism | ||||
| CYP2D6 substrate | No | No | No | No |
| CYP3A4 substrate | Yes | No | No | No |
| CYP1A2 inhibitor | Yes | Yes | Yes | Yes |
| CYP2C19 inhibitor | Yes | Yes | Yes | Yes |
| CYP3A4 inhibitor | Yes | Yes | No | Yes |
| Excretion | ||||
| Total clearance | 0.768 | 0.353 | 0.061 | 0.359 |
| Renal OCT2 substrate | No | No | No | No |
| Toxicity | ||||
| AMES toxicity | No | Yes | Yes | No |
| Skin sensitization | No | Yes | No | No |
| Hepatotoxicity | No | No | No | No |
| Carcinogenic | No | No | No | No |
| Respiratory diseases | Safe | Toxic | Safe | Safe |
| Max. tolerated dose (log mg/kg/day) | 0.201 | 0.428 | 0.539 | 0.55 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Suleman, M.; Sayaf, A.M.; Moltrasio, C.; Tricarico, P.M.; Giambuzzi, F.; Rimondi, E.; Melloni, E.; Secchiero, P.; Marcuzzi, A.; Marzano, A.V.; et al. Phytocompounds in Precision Dermatology: COX-2 Inhibitors as a Therapeutic Target in Atopic-Prone Skin. Biomolecules 2025, 15, 998. https://doi.org/10.3390/biom15070998
Suleman M, Sayaf AM, Moltrasio C, Tricarico PM, Giambuzzi F, Rimondi E, Melloni E, Secchiero P, Marcuzzi A, Marzano AV, et al. Phytocompounds in Precision Dermatology: COX-2 Inhibitors as a Therapeutic Target in Atopic-Prone Skin. Biomolecules. 2025; 15(7):998. https://doi.org/10.3390/biom15070998
Chicago/Turabian StyleSuleman, Muhammad, Abrar Mohammad Sayaf, Chiara Moltrasio, Paola Maura Tricarico, Francesco Giambuzzi, Erika Rimondi, Elisabetta Melloni, Paola Secchiero, Annalisa Marcuzzi, Angelo Valerio Marzano, and et al. 2025. "Phytocompounds in Precision Dermatology: COX-2 Inhibitors as a Therapeutic Target in Atopic-Prone Skin" Biomolecules 15, no. 7: 998. https://doi.org/10.3390/biom15070998
APA StyleSuleman, M., Sayaf, A. M., Moltrasio, C., Tricarico, P. M., Giambuzzi, F., Rimondi, E., Melloni, E., Secchiero, P., Marcuzzi, A., Marzano, A. V., & Crovella, S. (2025). Phytocompounds in Precision Dermatology: COX-2 Inhibitors as a Therapeutic Target in Atopic-Prone Skin. Biomolecules, 15(7), 998. https://doi.org/10.3390/biom15070998












