Role of Charge Density of Polycations in DNA Complexation and Condensation
Abstract
1. Introduction
2. Simulation Systems and Methods
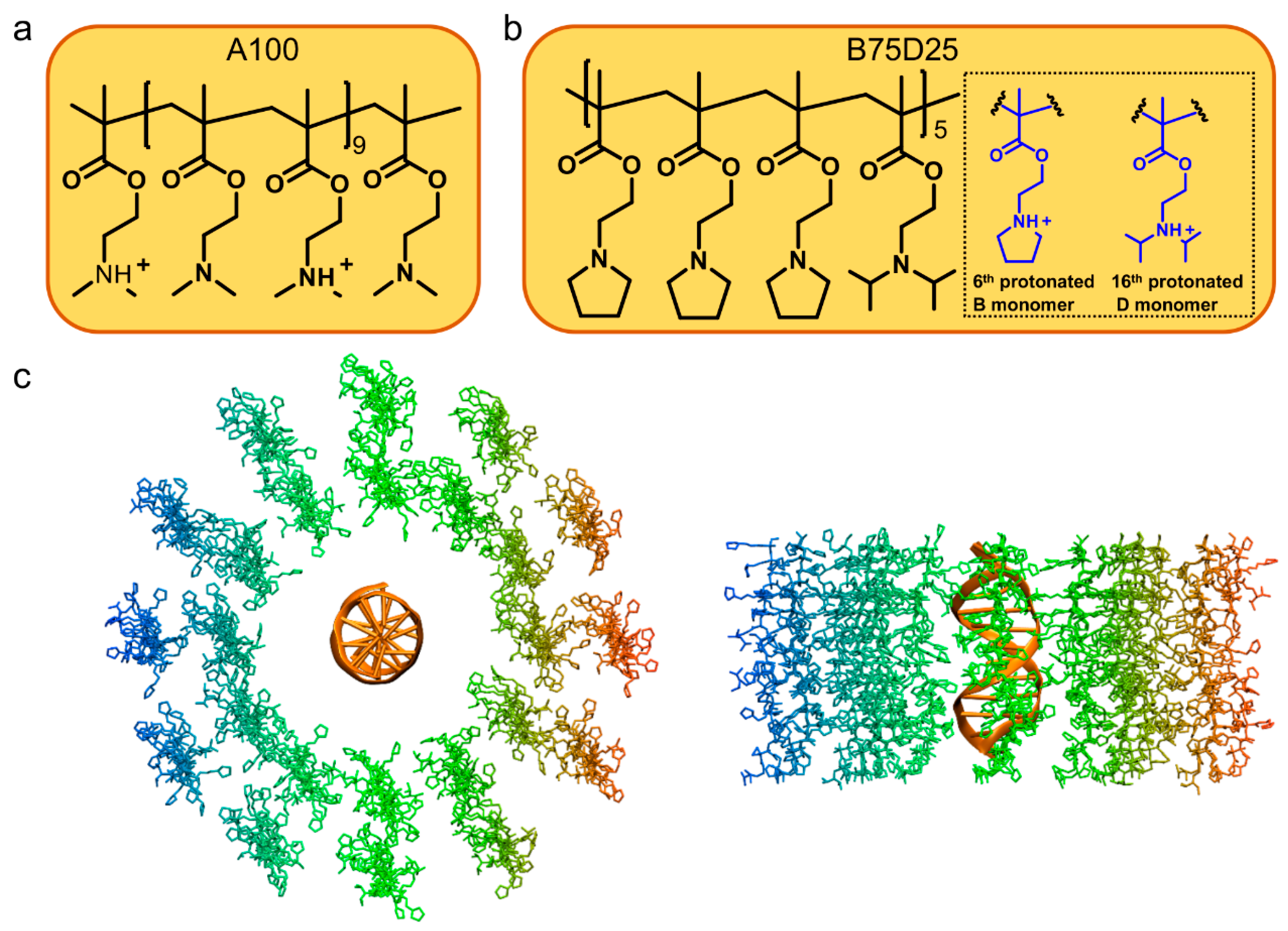
2.1. Simulation Systems
2.2. Simulation Methods and Structural Analysis
3. Results and Discussion
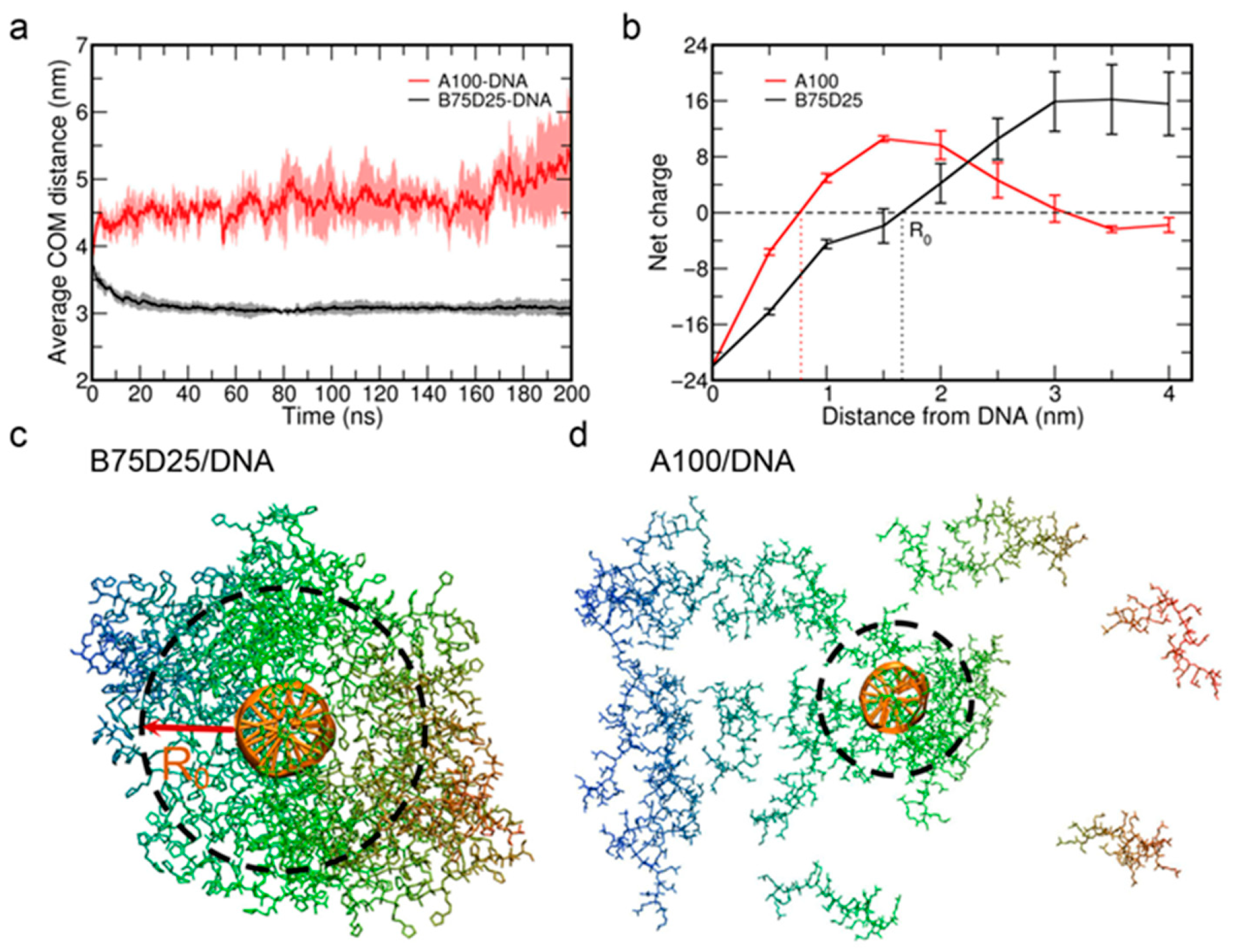
3.1. Formation of Polycation/DNA Complexes
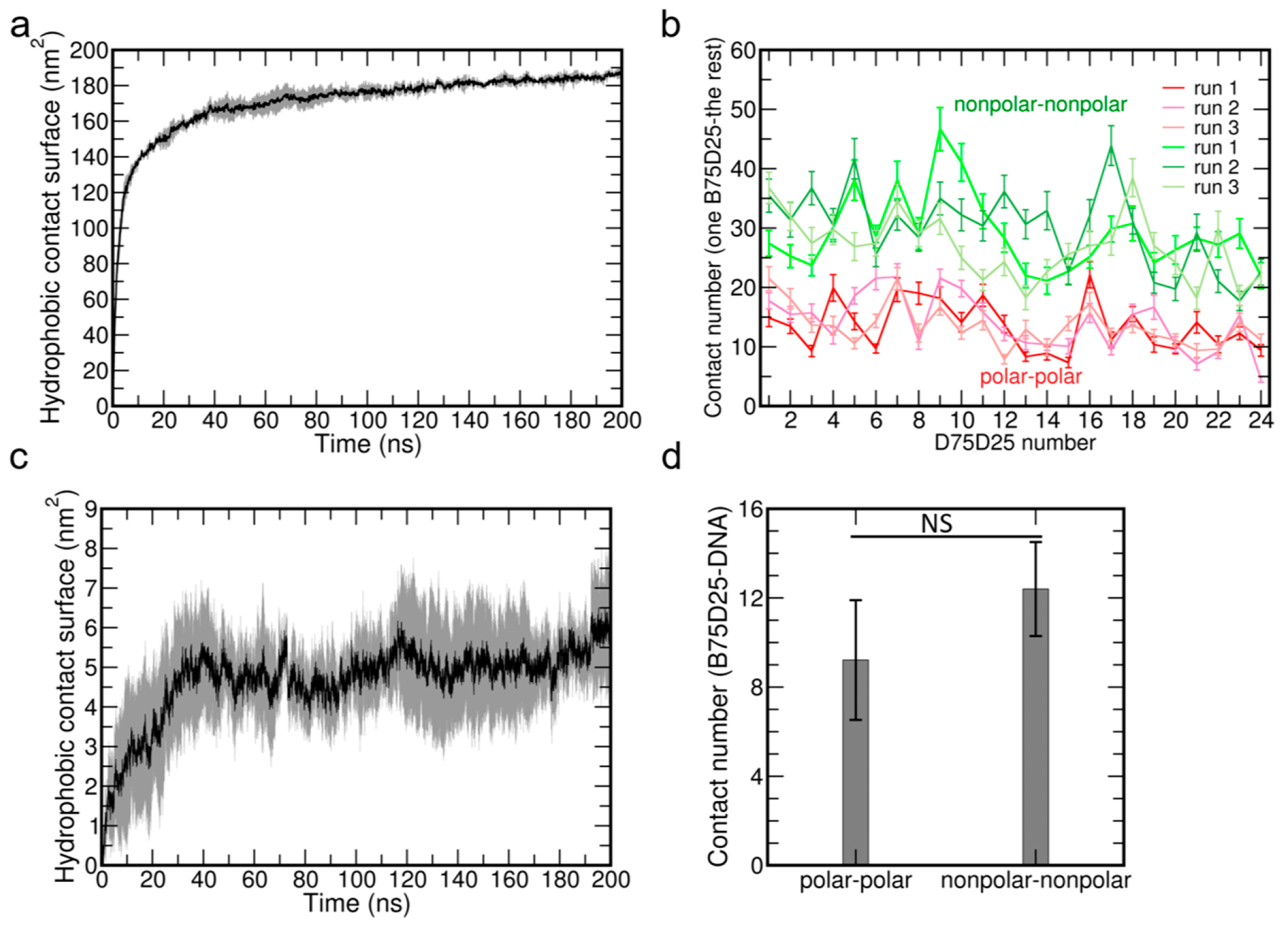
3.2. Hydrophobic Interactions Among B75D25 Polymers and Between B75D25s and DNA
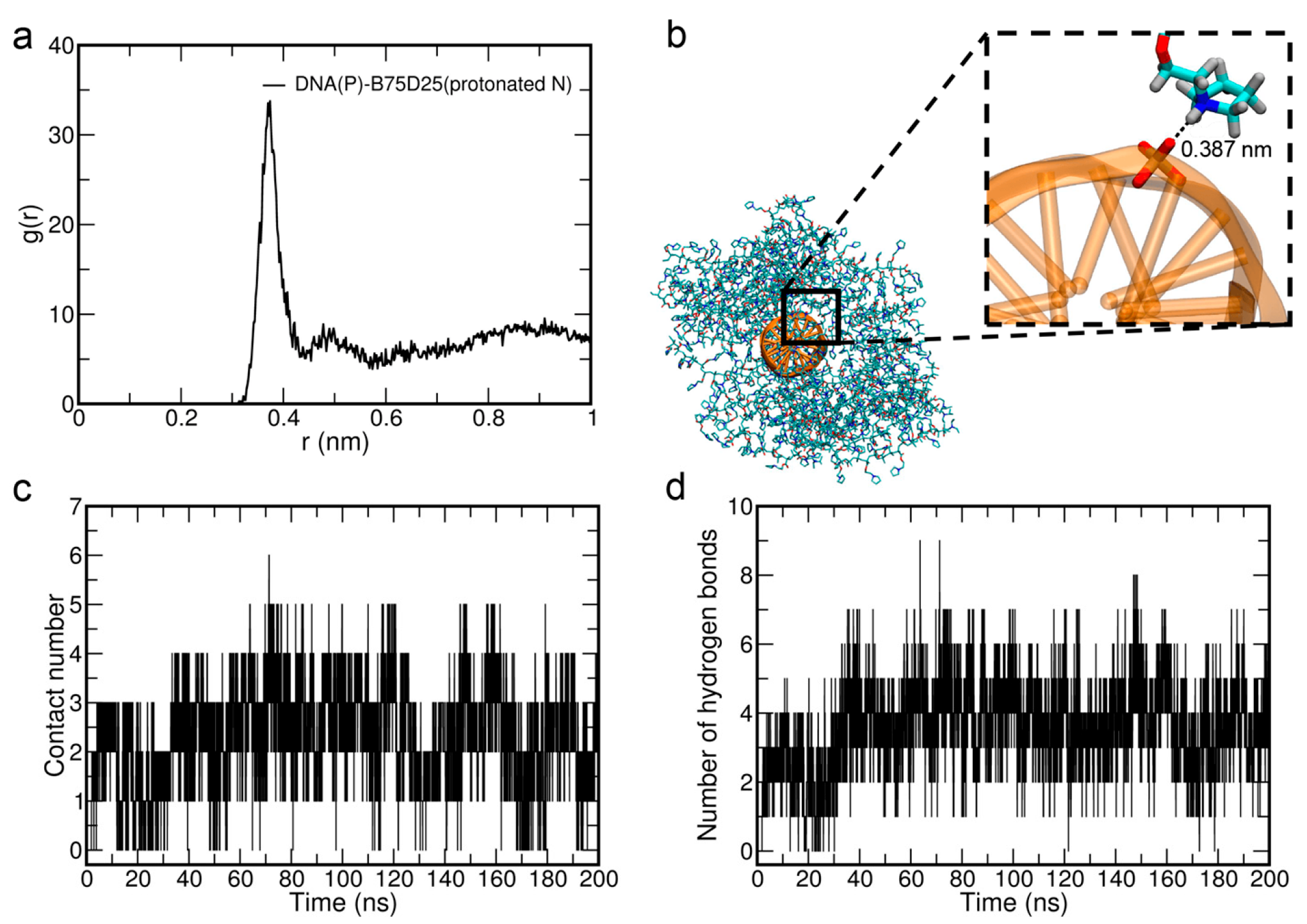
3.3. The Electrostatic Interactions Between B75D25s and DNA
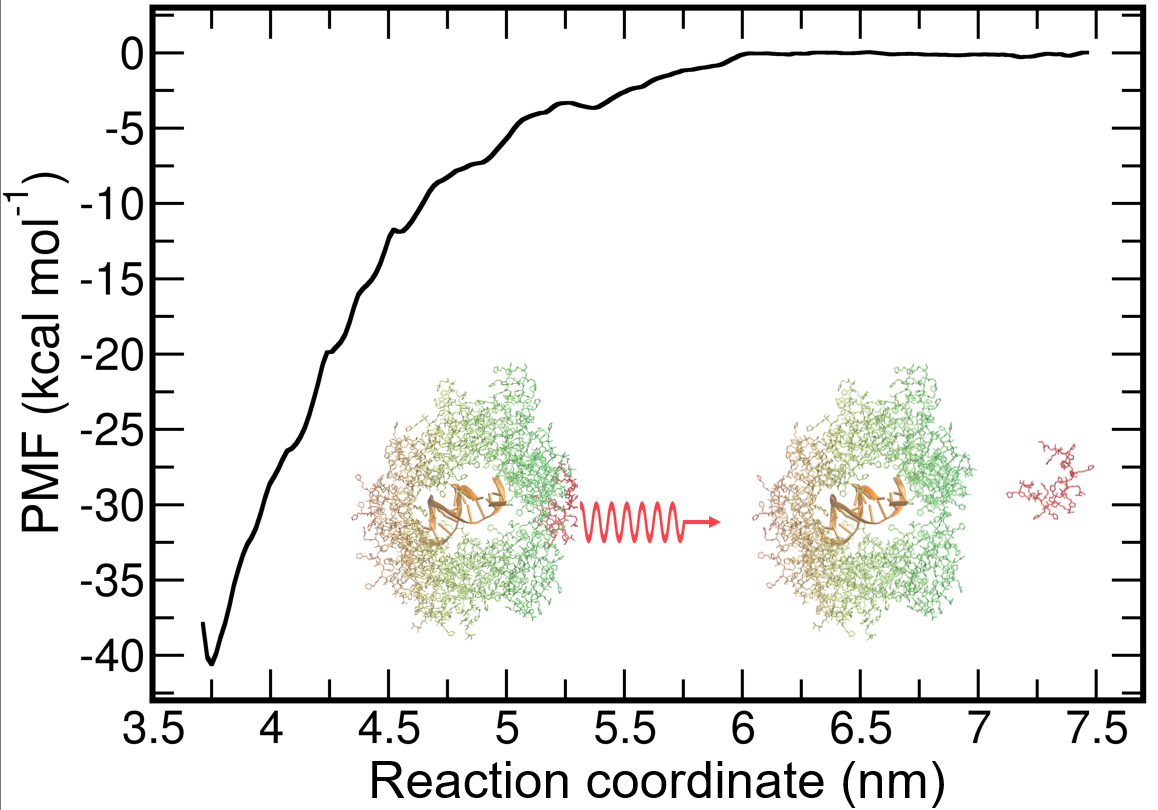
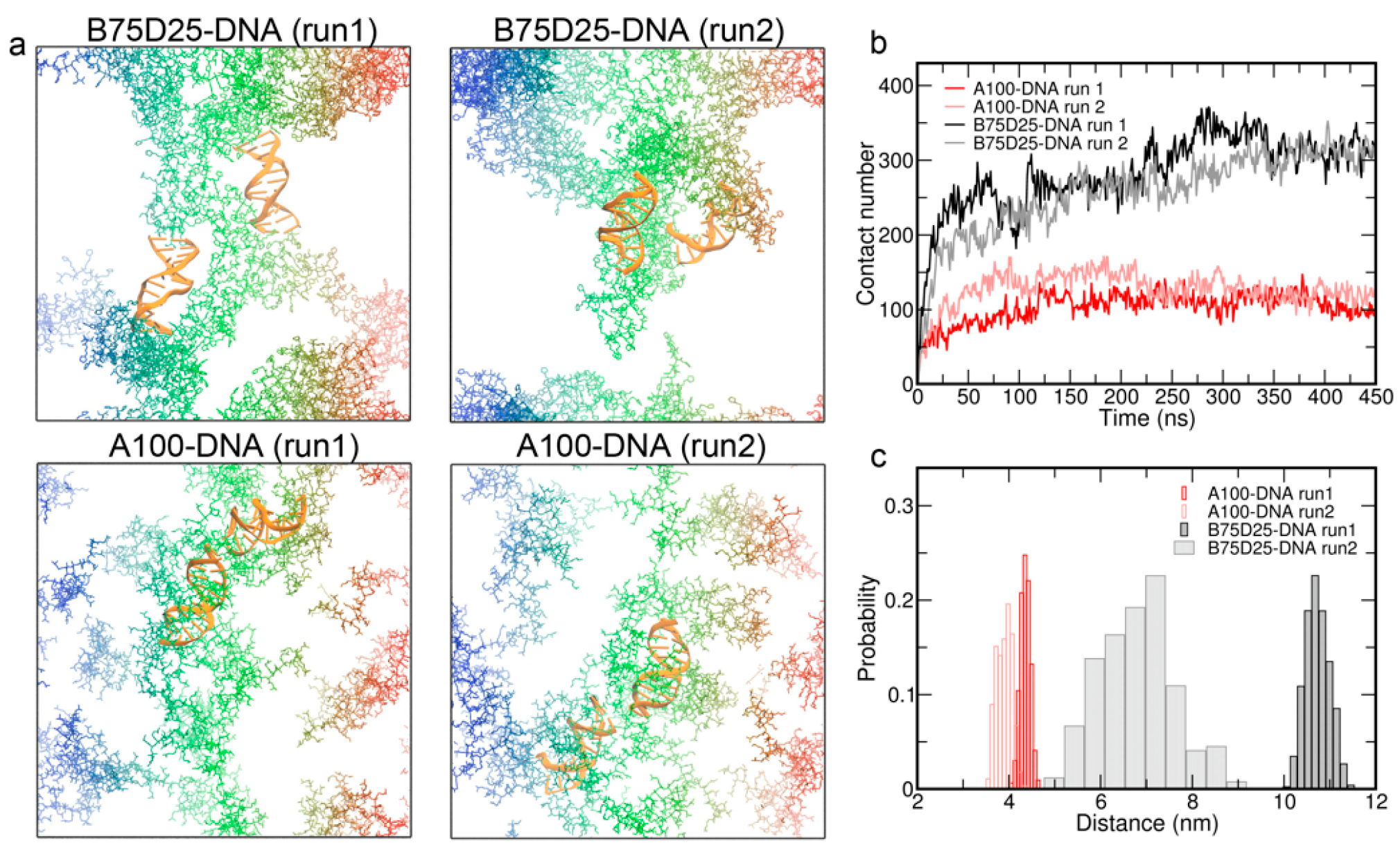
3.4. Simulations of Double DNA with Randomly Inserted Polycations
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yin, H.; Kanasty, R.L.; Eltoukhy, A.A.; Vegas, A.J.; Dorkin, J.R.; Anderson, D.G. Non-viral vectors for gene-based therapy. Nat. Rev. Genet. 2014, 15, 541–555. [Google Scholar] [CrossRef] [PubMed]
- Ibraheem, D.; Elaissari, A.; Fessi, H. Gene therapy and DNA delivery systems. Int. J. Pharm. 2014, 459, 70–83. [Google Scholar] [CrossRef]
- Zhou, Z.X.; Liu, X.R.; Zhu, D.C.; Wang, Y.; Zhang, Z.; Zhou, X.F.; Qiu, N.S.; Chen, X.S.; Shen, Y.Q. Nonviral cancer gene therapy: Delivery cascade and vector nanoproperty integration. Adv. Drug Deliv. Rev. 2017, 115, 115–154. [Google Scholar] [CrossRef] [PubMed]
- Ginn, S.L.; Amaya, A.K.; Alexander, I.E.; Edelstein, M.; Abedi, M.R. Gene therapy clinical trials worldwide to 2017: An update. J. Gene Med. 2018, 20, e3015. [Google Scholar] [CrossRef] [PubMed]
- Morrison, C. $1-million price tag set for Glybera gene therapy. Nat. Biotechnol. 2015, 33, 217–218. [Google Scholar] [CrossRef]
- Thomas, C.E.; Ehrhardt, A.; Kay, M.A. Progress and problems with the use of viral vectors for gene therapy. Nat. Rev. Genet. 2003, 4, 346–358. [Google Scholar] [CrossRef]
- Bessis, N.; GarciaCozar, F.J.; Boissier, M.C. Immune responses to gene therapy vectors: Influence on vector function and effector mechanisms. Gene Ther. 2004, 11, S10–S17. [Google Scholar] [CrossRef]
- Cottard, V.; Valvason, C.; Falgarone, G.; Lutomski, D.; Boissier, M.C.; Bessis, N. Immune response against gene therapy vectors: Influence of synovial fluid on adeno-associated virus mediated gene transfer to chondrocytes. J. Clin. Immunol. 2004, 24, 162–169. [Google Scholar] [CrossRef] [PubMed]
- Baum, C.; Kustikova, O.; Modlich, U.; Li, Z.X.; Fehse, B. Mutagenesis and oncogenesis by chromosomal insertion of gene transfer vectors. Hum. Gene Ther. 2006, 17, 253–263. [Google Scholar] [CrossRef]
- Young, P. Treatment to cure: Advancing AAV gene therapy manufacture. Drug Discov. Today 2023, 28, 103610. [Google Scholar] [CrossRef]
- Wong, C.H.; Li, D.; Wang, N.; Gruber, J.; Lo, A.W.; Conti, R.M. The estimated annual financial impact of gene therapy in the United States. Gene Ther. 2023, 30, 761–773. [Google Scholar] [CrossRef] [PubMed]
- Ramamoorth, M.; Narvekar, A. Non viral vectors in gene therapy—An overview. J. Clin. Diagn. Res. 2015, 9, GE01–GE06. [Google Scholar] [CrossRef] [PubMed]
- Hardee, C.L.; Arevalo-Soliz, L.M.; Hornstein, B.D.; Zechiedrich, L. Advances in Non-Viral DNA Vectors for Gene Therapy. Genes 2017, 8, 65. [Google Scholar] [CrossRef]
- Wong, S.Y.; Pelet, J.M.; Putnam, D. Polymer systems for gene delivery-past, present, and future. Prog. Polym. Sci. 2007, 32, 799–837. [Google Scholar] [CrossRef]
- Thomas, M.; Klibanov, A.M. Non-viral gene therapy: Polycation-mediated DNA delivery. Appl. Microbiol. Biot. 2003, 62, 27–34. [Google Scholar] [CrossRef]
- Junquera, E.; Aicart, E. Recent progress in gene therapy to deliver nucleic acids with multivalent cationic vectors. Adv. Colloid. Interfac. 2016, 233, 161–175. [Google Scholar] [CrossRef]
- Kim, K.; Chen, W.C.W.; Heo, Y.; Wang, Y.D. Polycations and their biomedical applications. Prog. Polym. Sci. 2016, 60, 18–50. [Google Scholar] [CrossRef]
- Bruininks, B.M.; de Souza, P.C.T.; Marrink, S.J. Microscopic View on Non-viral Mediated Transfection. Biophys. J. 2018, 114, 602a. [Google Scholar] [CrossRef]
- Liu, X.; Xiang, J.J.; Zhu, D.C.; Jiang, L.M.; Zhou, Z.X.; Tang, J.B.; Liu, X.R.; Huang, Y.Z.; Shen, Y.Q. Fusogenic Reactive Oxygen Species Triggered Charge-Reversal Vector for Effective Gene Delivery. Adv. Mater. 2016, 28, 1743–1752. [Google Scholar] [CrossRef]
- Ahern, J.O.; Sigen, A.; Zhou, D.Z.; Gao, Y.S.; Lyu, J.; Meng, Z.; Cutlar, L.; Pierucci, L.; Wang, W.X. Brushlike Cationic Polymers with Low Charge Density for Gene Delivery. Biomacromolecules 2018, 19, 1410–1415. [Google Scholar] [CrossRef]
- Zhou, J.B.; Liu, J.; Cheng, C.J.; Patel, T.R.; Weller, C.E.; Piepmeier, J.M.; Jiang, Z.Z.; Saltzman, W.M. Biodegradable poly(amine-co-ester) terpolymers for targeted gene delivery. Nat. Mater. 2012, 11, 82–90. [Google Scholar] [CrossRef] [PubMed]
- McMillan, R.B.; Bediako, H.; Devenica, L.M.; Velasquez, A.; Hardy, I.P.; Ma, Y.E.; Roscoe, D.M.; Carter, A.R. Protamine folds DNA into flowers and loop stacks. Biophys. J. 2023, 122, 4288–4302. [Google Scholar] [CrossRef] [PubMed]
- Postigo, A.; Marcuello, C.; Verstraeten, W.; Sarasa, S.; Walther, T.; Lostao, A.; Göpfrich, K.; del Barrio, J.; Hernández-Ainsa, S. Folding and Functionalizing DNA Origami: A Versatile Approach Using a Reactive Polyamine. J. Am. Chem. Soc. 2025, 147, 3919–3924. [Google Scholar] [CrossRef]
- Zhang, Z.; Qiu, N.; Wu, S.; Liu, X.; Zhou, Z.; Tang, J.; Liu, Y.; Zhou, R.; Shen, Y. Dose-Independent Transfection of Hydrophobized Polyplexes. Adv. Mater. 2021, 33, e2102219. [Google Scholar] [CrossRef] [PubMed]
- Ziebarth, J.; Wang, Y.M. Molecular Dynamics Simulations of DNA-Polycation Complex Formation. Biophys. J. 2009, 97, 1971–1983. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.; Tang, T.; Uludag, H.; Cuervo, J.E. Molecular dynamics simulations of DNA/PEI complexes: Effect of PEI branching and protonation state. Biophys. J. 2011, 100, 2754–2763. [Google Scholar] [CrossRef]
- Ziebarth, J.D.; Kennetz, D.R.; Walker, N.J.; Wang, Y. Structural Comparisons of PEI/DNA and PEI/siRNA Complexes Revealed with Molecular Dynamics Simulations. J. Phys. Chem. B 2017, 121, 1941–1952. [Google Scholar] [CrossRef]
- Kondinskaia, D.A.; Kostritskii, A.Y.; Nesterenko, A.M.; Antipina, A.Y.; Gurtovenko, A.A. Atomic-Scale Molecular Dynamics Simulations of DNA-Polycation Complexes: Two Distinct Binding Patterns. J. Phys. Chem. B 2016, 120, 6546–6554. [Google Scholar] [CrossRef]
- Li, W.; Nordenskiold, L.; Zhou, R.; Mu, Y. Conformation-dependent DNA attraction. Nanoscale 2014, 6, 7085–7092. [Google Scholar] [CrossRef]
- Dans, P.D.; Danilane, L.; Ivani, I.; Drasata, T.; Lankas, F.; Hospital, A.; Walther, J.; Pujagut, R.I.; Battistini, F.; Gelpi, J.L.; et al. Long-timescale dynamics of the Drew-Dickerson dodecamer. Nucleic Acids Res. 2016, 44, 4052–4066. [Google Scholar] [CrossRef] [PubMed]
- Drew, H.R.; Wing, R.M.; Takano, T.; Broka, C.; Tanaka, S.; Itakura, K.; Dickerson, R.E. Structure of a B-DNA Dodecamer—Conformation and Dynamics. Proc. Natl. Acad. Sci. USA 1981, 78, 2179–2183. [Google Scholar] [CrossRef]
- Baker, N.A.; Sept, D.; Joseph, S.; Holst, M.J.; McCammon, J.A. Electrostatics of nanosystems: Application to microtubules and the ribosome. Proc. Natl. Acad. Sci. USA 2001, 98, 10037–10041. [Google Scholar] [CrossRef]
- Bayly, C.I.; Cieplak, P.; Cornell, W.D.; Kollman, P.A. A Well-Behaved Electrostatic Potential Based Method Using Charge Restraints for Deriving Atomic Charges—The Resp Model. J. Phys. Chem. 1993, 97, 10269–10280. [Google Scholar] [CrossRef]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Petersson, G.A.; Nakatsuji, H.; et al. Gaussian 16 Rev. A.01; Gaussian, Inc.: Wallingford, CT, USA, 2016. [Google Scholar]
- Galindo-Murillo, R.; Robertson, J.C.; Zgarbova, M.; Sponer, J.; Otyepka, M.; Jurecka, P.; Cheatham, T.E. Assessing the Current State of Amber Force Field Modifications for DNA. J. Chem. Theory Comput. 2016, 12, 4114–4127. [Google Scholar] [CrossRef]
- Jorgensen, W.L.; Chandrasekhar, J.; Madura, J.D.; Impey, R.W.; Klein, M.L. Comparison of Simple Potential Functions for Simulating Liquid Water. J. Chem. Phys. 1983, 79, 926–935. [Google Scholar] [CrossRef]
- Abraham, M.J.; Murtola, T.; Schulz, R.; Páll, S.; Smith, J.C.; Hess, B.; Lindahl, E. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. Softwarex 2015, s1–s2, 19–25. [Google Scholar] [CrossRef]
- Bussi, G.; Donadio, D.; Parrinello, M. Canonical sampling through velocity rescaling. J. Chem. Phys. 2007, 126, 014101. [Google Scholar] [CrossRef]
- Parrinello, M.; Rahman, A. Polymorphic Transitions in Single-Crystals—A New Molecular-Dynamics Method. J. Appl. Phys. 1981, 52, 7182–7190. [Google Scholar] [CrossRef]
- Hess, B.; Bekker, H.; Berendsen, H.J.C.; Fraaije, J.G.E.M. LINCS: A linear constraint solver for molecular simulations. J. Comput. Chem. 1997, 18, 1463–1472. [Google Scholar] [CrossRef]
- Darden, T.; York, D.; Pedersen, L. Particle mesh Ewald: An N⋅log(N) method for Ewald sums in large systems. J. Chem. Phys. 1993, 98, 10089–10092. [Google Scholar] [CrossRef]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]
- Torrie, G.M.; Valleau, J.P. Nonphysical sampling distributions in Monte Carlo free-energy estimation: Umbrella sampling. J. Comput. Phys. 1977, 23, 187–199. [Google Scholar] [CrossRef]
- Kumar, S.; Bouzida, D.; Swendsen, R.H.; Kollman, P.A.; Rosenberg, J.M. The Weighted Histogram Analysis Method for Free-Energy Calculations on Biomolecules.1. The Method. J. Comput. Chem. 1992, 13, 1011–1021. [Google Scholar] [CrossRef]
- Friesner, R.A.; Banks, J.L.; Murphy, R.B.; Halgren, T.A.; Klicic, J.J.; Mainz, D.T.; Repasky, M.P.; Knoll, E.H.; Shelley, M.; Perry, J.K.; et al. Glide: A New Approach for Rapid, Accurate Docking and Scoring. 1. Method and Assessment of Docking Accuracy. J. Med. Chem. 2004, 47, 1739–1749. [Google Scholar] [CrossRef] [PubMed]
- Kondinskaia, D.A.; Gurtovenko, A.A. Supramolecular complexes of DNA with cationic polymers: The effect of polymer concentration. Polymer 2018, 142, 277–284. [Google Scholar] [CrossRef]
- Semenyuk, P.I.; Zhiryakova, M.V.; Izumrudov, V.A. Supercharged Polyplexes: Full-Atom Molecular Dynamics Simulations and Experimental Study. Macromolecules 2018, 51, 5450–5459. [Google Scholar] [CrossRef]
- Shen, J.W.; Li, J.C.; Zhao, Z.N.; Zhang, L.; Peng, G.T.; Liang, L.J. Molecular dynamics study on the mechanism of polynucleotide encapsulation by chitosan. Sci. Rep. 2017, 7, 5050. [Google Scholar] [CrossRef]
- Ma, X.; Wang, Y.; Zhao, T.; Li, Y.; Su, L.C.; Wang, Z.; Huang, G.; Sumer, B.D.; Gao, J. Ultra-Ph-Sensitive Nanoprobe Library with Broad Ph Tunability and Fluorescence Emissions. J. Am. Chem. Soc. 2014, 136, 11085–11092. [Google Scholar] [CrossRef]
- Lee, H.; Son, S.H.; Sharma, R.; Won, Y.Y. A Discussion of the Ph-Dependent Protonation Behaviors of Poly(2-(Dimethylamino)Ethyl Methacrylate) (Pdmaema) and Poly(Ethylenimine-Ran-2-Ethyl-2-Oxazoline) (P(Ei-R-Eoz)). J. Phys. Chem. B 2011, 115, 844–860. [Google Scholar] [CrossRef]
| Distance from DNA (nm) | A100/DNA | B75D25/DNA |
|---|---|---|
| 0 | −22.0 (0) | −22.0 (0) |
| 0.5 | −14.2 (0.420) | −5.63 (0.450) |
| 1.0 | −4.50 (0.648) | 4.97 (0.634) |
| 1.5 | −1.90 (2.45) | 10.6 (0.419) |
| 2.0 | 4.20 (2.82) | 9.67 (2.05) |
| 2.5 | 10.5 (2.95) | 4.63 (2.51) |
| 3.0 | 15.9 (4.26) | 0.567 (1.92) |
| 3.5 | 16.2 (4.98) | −2.37 (0.450) |
| 4.0 | 15.6 (4.53) | −1.77 (1.05) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, J.; Jiang, Y.; Zhang, D.; Li, J.; Shen, Y.; Zhou, R. Role of Charge Density of Polycations in DNA Complexation and Condensation. Biomolecules 2025, 15, 983. https://doi.org/10.3390/biom15070983
Huang J, Jiang Y, Zhang D, Li J, Shen Y, Zhou R. Role of Charge Density of Polycations in DNA Complexation and Condensation. Biomolecules. 2025; 15(7):983. https://doi.org/10.3390/biom15070983
Chicago/Turabian StyleHuang, Jianxiang, Yangwei Jiang, Dong Zhang, Jingyuan Li, Youqing Shen, and Ruhong Zhou. 2025. "Role of Charge Density of Polycations in DNA Complexation and Condensation" Biomolecules 15, no. 7: 983. https://doi.org/10.3390/biom15070983
APA StyleHuang, J., Jiang, Y., Zhang, D., Li, J., Shen, Y., & Zhou, R. (2025). Role of Charge Density of Polycations in DNA Complexation and Condensation. Biomolecules, 15(7), 983. https://doi.org/10.3390/biom15070983







