Significant Reduction of Chenodeoxycholic Acid and Glycochenodeoxycholic Acid in the Elderly with Severe COVID-19
Abstract
1. Introduction
2. Materials and Methods
2.1. Patient Inclusion and Sample Collection
2.2. Cell Culture, Viral Stimulation, and Bile Acid Intervention
2.3. LC-MS/MS Untargeted Metabolomics
2.4. LC-MS/MS Targeted Bile Acids Metabolomics
2.5. qPCR (Quantitative Real-Time PCR)
2.6. Cytokine Quantification
2.7. RNA Sequencing of THP-1 Cells
2.8. Statistical Analysis
3. Results
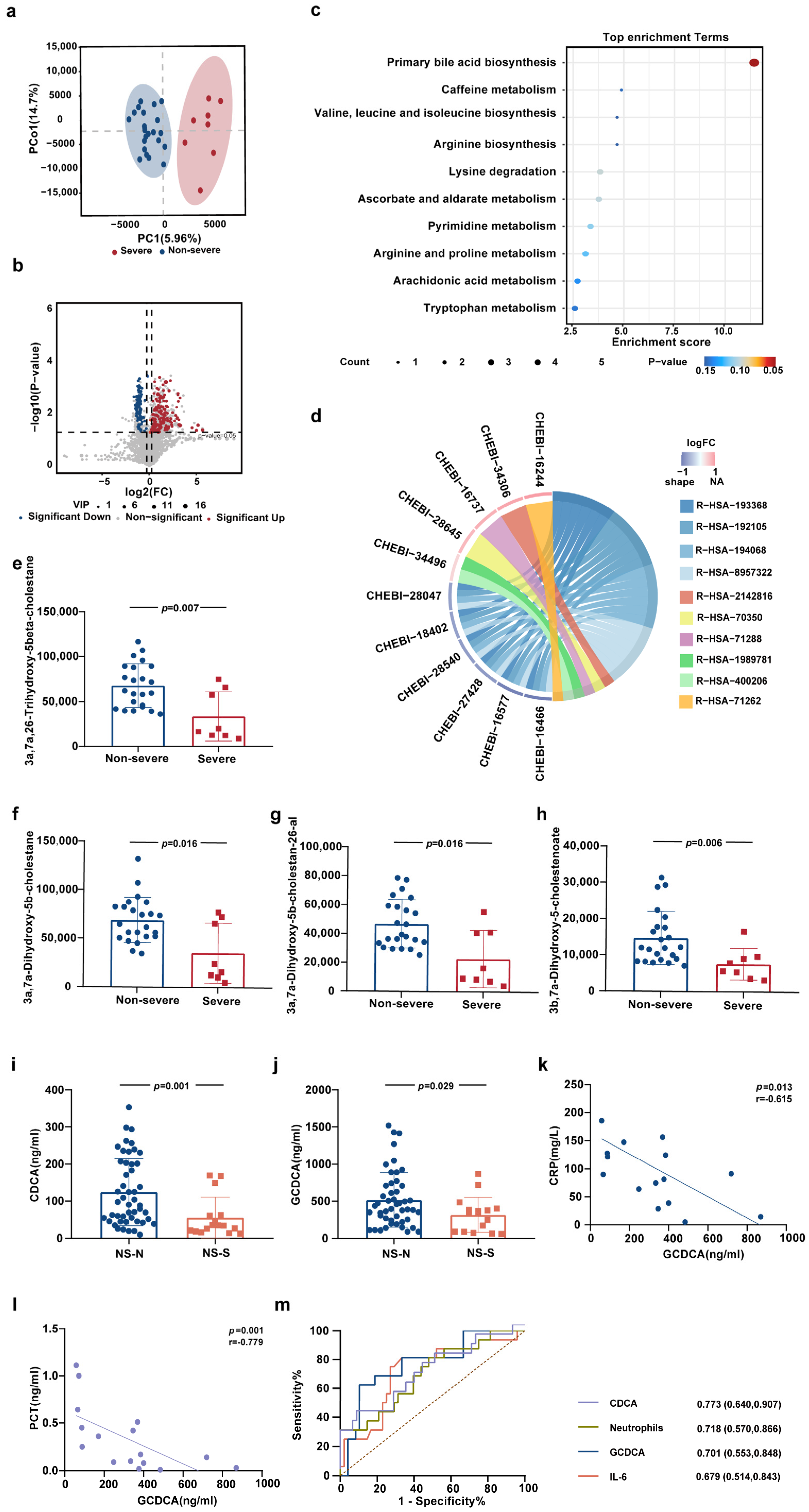
3.1. Alterations of Primary Bile Acids Biosynthesis in Peripheral Blood of Elderly Patients with Severe COVID-19
3.2. CDCA and GCDCA Were Significantly Downregulated in Elderly Patients with Severe COVID-19
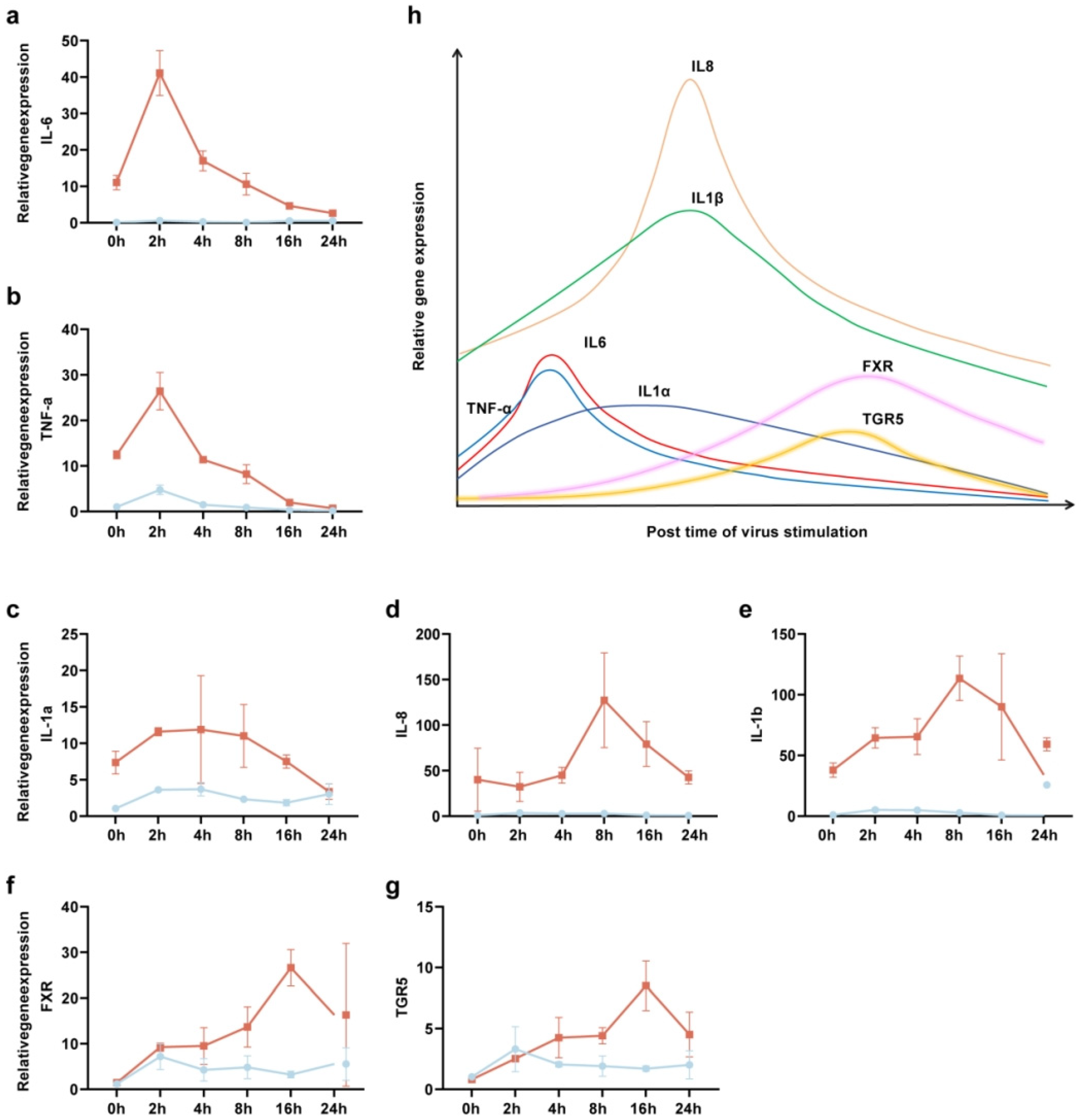
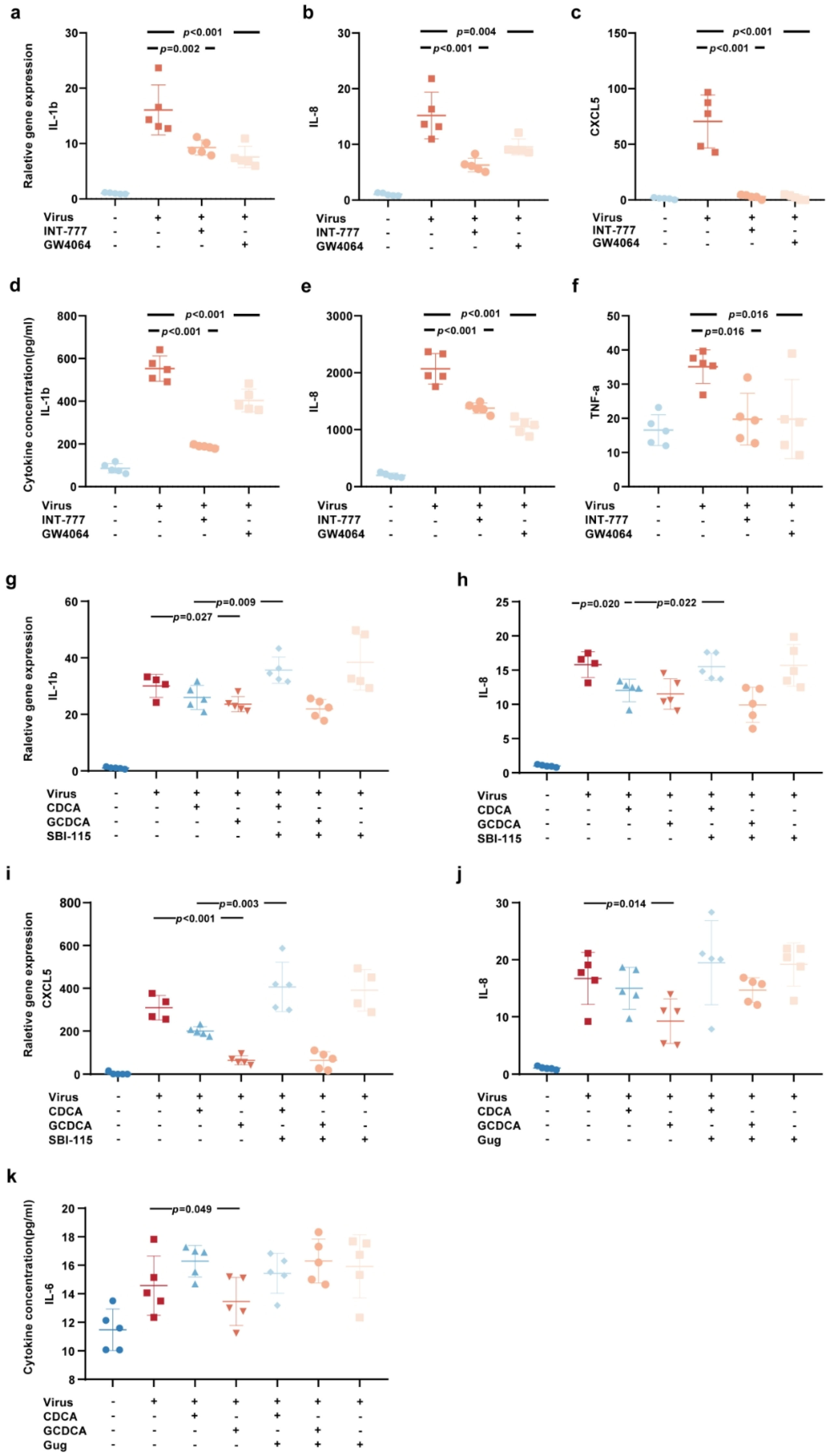
3.3. CDCA and GCDCA Inhibit the Inflammatory Response Induced by SARS-CoV-2 in THP-1 Cells
3.4. The Activation of Bile Acid Receptors FXR and TGR5 Regulate the Inflammatory Response Induced by SARS-CoV-2
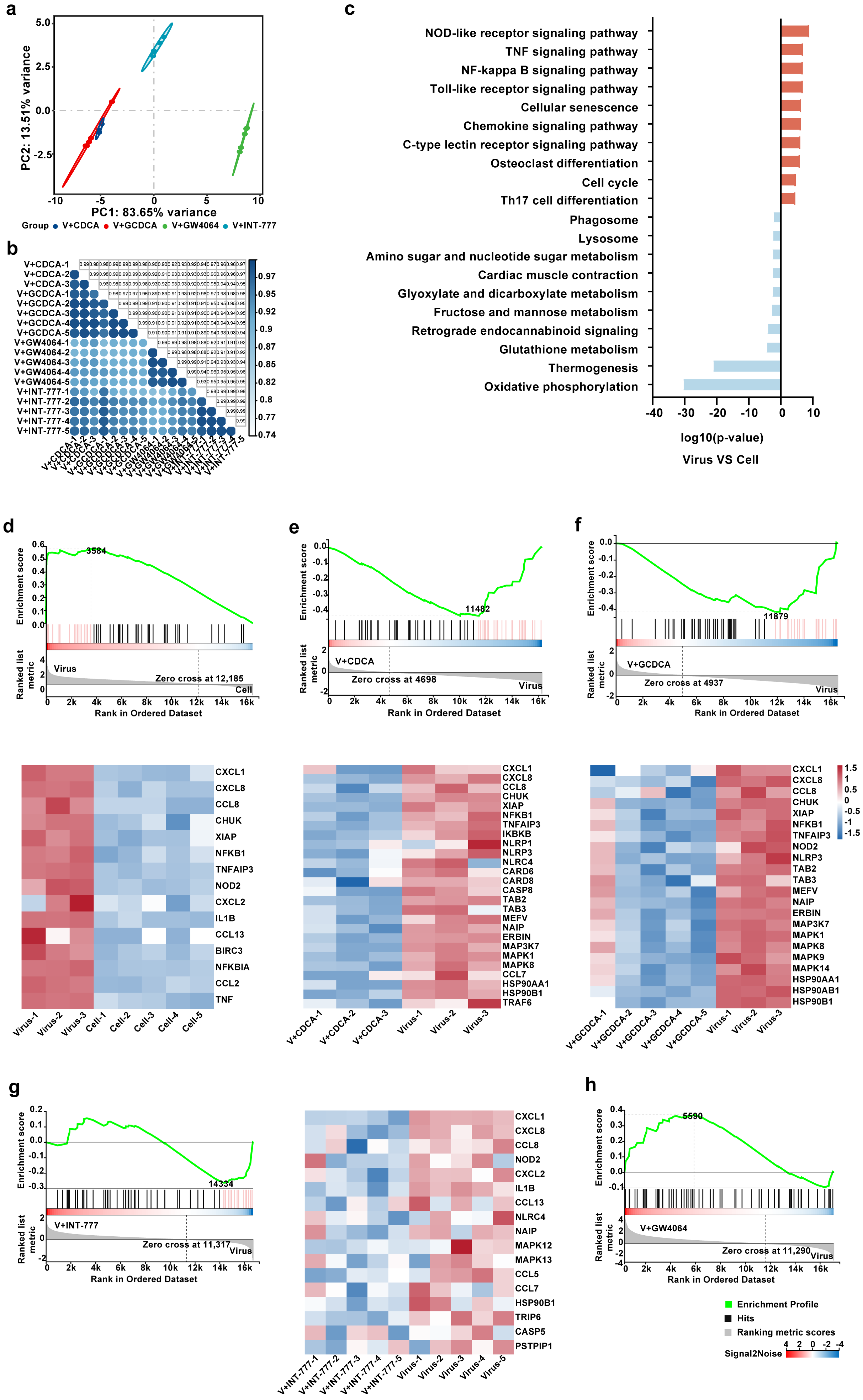
3.5. CDCA/GCDCA Might Suppress SARS-CoV-2-Induced Inflammatory Responses by Inhibiting the NOD-like Receptor Signaling Pathway
3.6. CDCA and GCDCA Predict the Progression of Severe COVID-19 in Elderly Patients
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- WHO. WHO Director-General’s Opening Remarks at the Media Briefing—5 May 2023. Available online: https://www.who.int/director-general/speeches/detail/who-director-general-s-opening-remarks-at-the-media-briefing---5-may-2023 (accessed on 5 May 2023).
- WHO. WHO COVID-19 Dashboard Data. Available online: https://data.who.int/dashboards/covid19/data (accessed on 9 November 2024).
- Pedreañez, A.; Mosquera-Sulbaran, J.; Muñoz, N. SARS-CoV-2 infection represents a high risk for the elderly: Analysis of pathogenesis. Arch. Virol. 2021, 166, 1565–1574. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Fang, X.; Cai, Z.; Wu, X.; Gao, X.; Min, J.; Wang, F. Comorbid Chronic Diseases and Acute Organ Injuries Are Strongly Correlated with Disease Severity and Mortality among COVID-19 Patients: A Systemic Review and Meta-Analysis. Research 2020, 2020, 2402961. [Google Scholar] [CrossRef]
- Nagao, M.; Matsumura, Y.; Yamamoto, M.; Shinohara, K.; Noguchi, T.; Yukawa, S.; Tsuchido, Y.; Teraishi, H.; Inoue, H.; Ikeda, T. Incidence of and risk factors for suspected COVID-19 reinfection in Kyoto City: A population-based epidemiological study. Eur. J. Clin. Microbiol. Infect. Dis. 2023, 42, 973–979. [Google Scholar] [CrossRef]
- Zhou, F.; Yu, T.; Du, R.; Fan, G.; Liu, Y.; Liu, Z.; Xiang, J.; Wang, Y.; Song, B.; Gu, X.; et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: A retrospective cohort study. Lancet 2020, 395, 1054–1062. [Google Scholar] [CrossRef]
- Rathod, D.; Kargirwar, K.; Patel, M.; Kumar, V.; Shalia, K.; Singhal, P. Risk Factors associated with COVID-19 Patients in India: A Single Center Retrospective Cohort Study. J. Assoc. Physicians India 2023, 71, 11–12. [Google Scholar]
- Liu, B.; Spokes, P.; He, W.; Kaldor, J. High risk groups for severe COVID-19 in a whole of population cohort in Australia. BMC Infect. Dis. 2021, 21, 685. [Google Scholar] [CrossRef]
- Fiorucci, S.; Urbani, G.; Biagioli, M.; Sepe, V.; Distrutti, E.; Zampella, A. Bile acids and bile acid activated receptors in the treatment of COVID-19. Biochem. Pharmacol. 2024, 228, 115983. [Google Scholar] [CrossRef] [PubMed]
- Brevini, T.; Maes, M.; Webb, G.J.; John, B.V.; Fuchs, C.D.; Buescher, G.; Wang, L.; Griffiths, C.; Brown, M.L.; Scott, W.E., 3rd; et al. FXR inhibition may protect from SARS-CoV-2 infection by reducing ACE2. Nature 2023, 615, 134–142. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; Huang, S.; Yin, L. The cytokine storm and COVID-19. J. Med. Virol. 2021, 93, 250–256. [Google Scholar] [CrossRef]
- Smyth, J.S.; Truong, J.K.; Rao, A.; Lin, R.; Foulke-Abel, J.; Adorini, L.; Donowitz, M.; Dawson, P.A.; Keely, S.J. Farnesoid X receptor enhances epithelial ACE2 expression and inhibits virally induced IL-6 secretion: Implications for intestinal symptoms of SARS-CoV-2. Am. J. Physiol. Gastrointest. Liver Physiol. 2023, 325, G446–G452. [Google Scholar] [CrossRef]
- Nagai, M.; Moriyama, M.; Ishii, C.; Mori, H.; Watanabe, H.; Nakahara, T.; Yamada, T.; Ishikawa, D.; Ishikawa, T.; Hirayama, A.; et al. High body temperature increases gut microbiota-dependent host resistance to influenza A virus and SARS-CoV-2 infection. Nat. Commun. 2023, 14, 3863. [Google Scholar] [CrossRef] [PubMed]
- National Health Commission of the People’s Republic of China. Notice on the Publication of the Diagnosis and Treatment Protocol for Novel Coronavirus Infection (Trial Version 10). Available online: https://www.gov.cn/zhengce/zhengceku/2023-01/06/content_5735343.htm (accessed on 5 January 2013).
- Asperges, E.; Albi, G.; Zuccaro, V.; Sambo, M.; Pieri, T.C.; Calia, M.; Colaneri, M.; Maiocchi, L.; Melazzini, F.; Lasagna, A.; et al. Dynamic NLR and PLR in Predicting COVID-19 Severity: A Retrospective Cohort Study. Infect. Dis. Ther. 2023, 12, 1625–1640. [Google Scholar] [CrossRef]
- Kosidło, J.W.; Wolszczak-Biedrzycka, B.; Matowicka-Karna, J.; Dymicka-Piekarska, V.; Dorf, J. Clinical Significance and Diagnostic Utility of NLR, LMR, PLR and SII in the Course of COVID-19: A Literature Review. J. Inflamm. Res. 2023, 16, 539–562. [Google Scholar] [CrossRef]
- Colaneri, M.; Genovese, C.; Fassio, F.; Canuti, M.; Giacomelli, A.; Ridolfo, A.L.; Asperges, E.; Albi, G.; Bruno, R.; Antinori, S.; et al. Prognostic Significance of NLR and PLR in COVID-19: A Multi-Cohort Validation Study. Infect. Dis. Ther. 2024, 13, 1147–1157. [Google Scholar] [CrossRef]
- Kim, Y.I.; Casel, M.A.B.; Kim, S.M.; Kim, S.G.; Park, S.J.; Kim, E.H.; Jeong, H.W.; Poo, H.; Choi, Y.K. Development of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) thermal inactivation method with preservation of diagnostic sensitivity. J. Microbiol. 2020, 58, 886–891. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; An, T.; Situ, B.; Hu, Y.; Ou, Z.; Li, Q.; He, X.; Zhang, Y.; Tian, P.; Sun, D.; et al. Heat inactivation of serum interferes with the immunoanalysis of antibodies to SARS-CoV-2. J. Clin. Lab. Anal. 2020, 34, e23411. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Li, F.; Yuan, B.; Li, W.; Gao, J.; Hu, X.; An, Y.; Liu, Z.; Li, Z.; Liu, H.; et al. Effects of a Serum Heating Procedure for Inactivating COVID-19 on Common Biochemical Tests. Clin. Lab. 2021, 67. [Google Scholar] [CrossRef]
- Jia, W.; Wei, M.; Rajani, C.; Zheng, X. Targeting the alternative bile acid synthetic pathway for metabolic diseases. Protein Cell 2021, 12, 411–425. [Google Scholar] [CrossRef]
- Chen, N.; Zhou, M.; Dong, X.; Qu, J.; Gong, F.; Han, Y.; Qiu, Y.; Wang, J.; Liu, Y.; Wei, Y.; et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: A descriptive study. Lancet 2020, 395, 507–513. [Google Scholar] [CrossRef]
- Guan, W.J.; Ni, Z.Y.; Hu, Y.; Liang, W.H.; Ou, C.Q.; He, J.X.; Liu, L.; Shan, H.; Lei, C.L.; Hui, D.S.C.; et al. Clinical Characteristics of Coronavirus Disease 2019 in China. N. Engl. J. Med. 2020, 382, 1708–1720. [Google Scholar] [CrossRef]
- Zhou, Z.; Ren, L.; Zhang, L.; Zhong, J.; Xiao, Y.; Jia, Z.; Guo, L.; Yang, J.; Wang, C.; Jiang, S.; et al. Heightened Innate Immune Responses in the Respiratory Tract of COVID-19 Patients. Cell Host Microbe 2020, 27, 883–890.e882. [Google Scholar] [CrossRef] [PubMed]
- Junqueira, C.; Crespo, Â.; Ranjbar, S.; de Lacerda, L.B.; Lewandrowski, M.; Ingber, J.; Parry, B.; Ravid, S.; Clark, S.; Schrimpf, M.R.; et al. FcγR-mediated SARS-CoV-2 infection of monocytes activates inflammation. Nature 2022, 606, 576–584. [Google Scholar] [CrossRef]
- Pence, B.D. Severe COVID-19 and aging: Are monocytes the key? Geroscience 2020, 42, 1051–1061. [Google Scholar] [CrossRef] [PubMed]
- Mencarelli, A.; Renga, B.; Distrutti, E.; Fiorucci, S. Antiatherosclerotic effect of farnesoid X receptor. Am. J. Physiol. Heart Circ. Physiol. 2009, 296, H272–H281. [Google Scholar] [CrossRef] [PubMed]
- Ichikawa, R.; Takayama, T.; Yoneno, K.; Kamada, N.; Kitazume, M.T.; Higuchi, H.; Matsuoka, K.; Watanabe, M.; Itoh, H.; Kanai, T.; et al. Bile acids induce monocyte differentiation toward interleukin-12 hypo-producing dendritic cells via a TGR5-dependent pathway. Immunology 2012, 136, 153–162. [Google Scholar] [CrossRef]
- Bizjak, D.A.; Stangl, M.; Börner, N.; Bösch, F.; Durner, J.; Drunin, G.; Buhl, J.L.; Abendroth, D. Kynurenine serves as useful biomarker in acute, Long- and Post-COVID-19 diagnostics. Front. Immunol. 2022, 13, 1004545. [Google Scholar] [CrossRef]
- Delafiori, J.; Siciliano, R.F.; de Oliveira, A.N.; Nicolau, J.C.; Sales, G.M.; Dalçóquio, T.F.; Busanello, E.N.B.; Eguti, A.; de Oliveira, D.N.; Bertolin, A.J.; et al. Comparing plasma and skin imprint metabolic profiles in COVID-19 diagnosis and severity assessment. J. Mol. Med. 2024, 102, 183–195. [Google Scholar] [CrossRef]
- He, M.; Huang, Y.; Wang, Y.; Liu, J.; Han, M.; Xiao, Y.; Zhang, N.; Gui, H.; Qiu, H.; Cao, L.; et al. Metabolomics-based investigation of SARS-CoV-2 vaccination (Sinovac) reveals an immune-dependent metabolite biomarker. Front. Immunol. 2022, 13, 954801. [Google Scholar] [CrossRef]
- Porru, E.; Comito, R.; Interino, N.; Cerrato, A.; Contoli, M.; Rizzo, P.; Conti, M.; Campo, G.; Spadaro, S.; Caliceti, C.; et al. Sulfated Bile Acids in Serum as Potential Biomarkers of Disease Severity and Mortality in COVID-19. Cells 2024, 13, 1576. [Google Scholar] [CrossRef]
- Onoja, A.; von Gerichten, J.; Lewis, H.M.; Bailey, M.J.; Skene, D.J.; Geifman, N.; Spick, M. Meta-Analysis of COVID-19 Metabolomics Identifies Variations in Robustness of Biomarkers. Int. J. Mol. Sci. 2023, 24, 14371. [Google Scholar] [CrossRef]
- Xie, J.; Li, H.; Zhang, X.; Yang, T.; Yue, M.; Zhang, Y.; Chen, S.; Cui, N.; Yuan, C.; Li, J.; et al. Akkermansia muciniphila protects mice against an emerging tick-borne viral pathogen. Nat. Microbiol. 2023, 8, 91–106. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.L.; Takeda, K.; Sundrud, M.S. Emerging roles of bile acids in mucosal immunity and inflammation. Mucosal Immunol. 2019, 12, 851–861. [Google Scholar] [CrossRef]
- Hofmann, A.F.; Roda, A. Physicochemical properties of bile acids and their relationship to biological properties: An overview of the problem. J. Lipid Res. 1984, 25, 1477–1489. [Google Scholar] [CrossRef] [PubMed]
- Wagner, E.F.; Eferl, R. Fos/AP-1 proteins in bone and the immune system. Immunol. Rev. 2005, 208, 126–140. [Google Scholar] [CrossRef] [PubMed]
- Fiorucci, S.; Cipriani, S.; Mencarelli, A.; Renga, B.; Distrutti, E.; Baldelli, F. Counter-regulatory role of bile acid activated receptors in immunity and inflammation. Curr. Mol. Med. 2010, 10, 579–595. [Google Scholar] [CrossRef]
- Wang, J.; Zhang, J.; Lin, X.; Wang, Y.; Wu, X.; Yang, F.; Gao, W.; Zhang, Y.; Sun, J.; Jiang, C.; et al. DCA-TGR5 signaling activation alleviates inflammatory response and improves cardiac function in myocardial infarction. J. Mol. Cell. Cardiol. 2021, 151, 3–14. [Google Scholar] [CrossRef]
- Zheng, C.; Wang, L.; Zou, T.; Lian, S.; Luo, J.; Lu, Y.; Hao, H.; Xu, Y.; Xiang, Y.; Zhang, X.; et al. Ileitis promotes MASLD progression via bile acid modulation and enhanced TGR5 signaling in ileal CD8(+) T cells. J. Hepatol. 2024, 80, 764–777. [Google Scholar] [CrossRef]
- Hu, X.; Yan, J.; Huang, L.; Araujo, C.; Peng, J.; Gao, L.; Liu, S.; Tang, J.; Zuo, G.; Zhang, J.H. INT-777 attenuates NLRP3-ASC inflammasome-mediated neuroinflammation via TGR5/cAMP/PKA signaling pathway after subarachnoid hemorrhage in rats. Brain Behav. Immun. 2021, 91, 587–600. [Google Scholar] [CrossRef]
- Fiorucci, S.; Urbani, G.; Distrutti, E. Bile Acids and SARS-CoV-2: Ursodeoxycholic Acid as a Potential Treatment of COVID-19. Recent Adv. Inflamm. Allergy Drug Discov. 2023, 17, 2–6. [Google Scholar] [CrossRef]
- Shi, Y.; Su, W.; Zhang, L.; Shi, C.; Zhou, J.; Wang, P.; Wang, H.; Shi, X.; Wei, S.; Wang, Q.; et al. TGR5 Regulates Macrophage Inflammation in Nonalcoholic Steatohepatitis by Modulating NLRP3 Inflammasome Activation. Front. Immunol. 2020, 11, 609060. [Google Scholar] [CrossRef]
- Guo, C.; Xie, S.; Chi, Z.; Zhang, J.; Liu, Y.; Zhang, L.; Zheng, M.; Zhang, X.; Xia, D.; Ke, Y.; et al. Bile Acids Control Inflammation and Metabolic Disorder through Inhibition of NLRP3 Inflammasome. Immunity 2016, 45, 802–816. [Google Scholar] [CrossRef] [PubMed]


| NS-N Group (n = 48) | NS-S Group (n = 16) | p-Value | |
|---|---|---|---|
| Personal information | |||
| Gender | |||
| Male | 30 (62.50%) | 14 (87.50%) | 0.119 |
| Female | 18 (37.50%) | 2 (12.50%) | |
| Age (year) * | 76.80 ± 10.93 | 83.49 ± 10.70 | 0.037 |
| Underlying comorbidities | |||
| Circulatory system diseases | 29 (60.42%) | 12 (75.00%) | 0.292 |
| Endocrine system diseases | 21 (43.75%) | 6 (37.50%) | 0.661 |
| Digestive system diseases | 11 (22.92%) | 2 (12.50%) | 0.590 |
| Nervous system diseases | 12 (25.00%) | 5 (31.25%) | 0.870 |
| Urinary system diseases | 7 (14.58%) | 3 (18.75%) | 1.000 |
| Respiratory system diseases | 5 (10.42%) | 0 (0.00%) | 0.420 |
| Musculoskeletal system | 5 (10.42%) | 3 (18.75%) | 0.663 |
| Others | 5 (10.42%) | 0 (0.00%) | 0.420 |
| Living habits | |||
| Allergies | 4 (8.33%) | 1 (6.25%) | 1.000 |
| Smoking | 3 (6.25%) | 0 (0.00%) | 0.733 |
| Drinking | 4 (8.33%) | 1 (6.25%) | 1.000 |
| Clinical information | |||
| Vaccination | |||
| Unvaccinated | 12 (25.00%) | 4 (25.00%) | 0.986 |
| Vaccinated | 17 (35.42%) | 6 (37.50%) | |
| Unclear | 19 (39.58%) | 6 (37.50%) | |
| Time from the onset to sample collection (day) | 5.50 (3.00, 8.50) | 6.00 (2.00, 8.50) | 0.749 |
| Time of positive nucleic acid (day) | 5.00 (3.00, 8.00) | 10.00 (4.00, 15.00) | 0.092 |
| Length of hospital stay (day) *** | 7.00 (5.00, 10.00) | 16.00 (12.00, 20.50) | <0.001 |
| Onset symptoms | |||
| Fever | 39 (81.25%) | 12 (75.00%) | 0.858 |
| Cough | 35 (72.92%) | 13 (81.25%) | 0.739 |
| Expectoration | 28 (58.33%) | 10 (62.50%) | 0.769 |
| Sore throat | 15 (31.25%) | 4 (25.00%) | 0.874 |
| Others | 15 (31.25%) | 4 (25.00%) | 0.874 |
| Immune-related parameters | |||
| Leukocyte (109/L) | 4.23 (3.61, 6.58) | 5.30 (3.69, 6.50) | 0.603 |
| Neutrophils (%) * | 67.06 ± 14.99 | 78.49 ± 15.27 | 0.011 |
| Lymphocytes (%) ** | 22.42 ± 12.00 | 12.69 ± 9.78 | 0.005 |
| Monocytes (%) | 8.60 (5.55, 12.65) | 6.05 (3.00, 8.70) | 0.061 |
| Neutrophils (109/L) | 2.73 (2.07, 5.03) | 4.18 (3.42, 4.93) | 0.195 |
| Lymphocytes (109/L) * | 1.07 ± 0.61 | 0.66 ± 0.49 | 0.018 |
| Monocytes (109)/L) | 0.45 (0.30, 0.56) | 0.30 (0.12, 0.53) | 0.239 |
| PCT (ng/mL) * | 0.05 (0.03, 0.09) | 0.12 (0.06, 0.44) | 0.032 |
| CRP (mg/L) | 20.90 (7.00, 45.25) | 30.20 (10.20, 62.60) | 0.522 |
| IL-6 (pg/mL) * | 13.91 (5.05, 39.67) | 32.03 (13.23, 123.80) | 0.034 |
| NLR * | 2.95 (1.88, 6.87) | 5.22 (3.83, 7.84) | 0.038 |
| PLR | 151.27 (122.81, 258.50) | 168.83 (98.40, 317.46) | 0.762 |
| LMR | 2.16 (1.72, 3.85) | 2.27 (0.61, 2.90) | 0.351 |
| SII | 415.66 (282.17, 1167.54) | 645.50 (465.05, 1924.34) | 0.225 |
| Variants | Univariate Logistic Regression | Multivariate Logistic Regression | ||
|---|---|---|---|---|
| p-Value | OR (95%CI) | p-Value | OR (95%CI) | |
| Age (years) (60–74) | ||||
| 75–90 | 0.610 | 1.437 (0.357, 5.795) | ||
| >90 | 0.121 | 3.067 (0.745, 12.625) | ||
| Neutrophils (%) | 0.016 | 1.054 (1.010, 1.100) | 0.018 | 1.082 (1.014, 1.155) |
| Lymphocytes (%) | 0.009 | 0.922 (0.868, 0.980) | ||
| Lymphocytes (109/L) | 0.021 | 0.208 (0.055, 0.785) | ||
| PCT (ng/mL) | 0.182 | 3.682 (0.543, 24.991) | ||
| IL-6 (pg/mL) | 0.033 | 3.889 (1.119, 13.516) | 0.028 | 1.026 (1.003, 1.050) |
| CDCA (ng/mL) | 0.015 | 0.986 (0.975, 0.997) | 0.018 | 0.978 (0.960, 0.996) |
| GCDCA (ng/mL) | 0.058 | 0.998 (0.995, 1.000) | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, S.; Xu, W.; Tu, B.; Xiao, Z.; Li, X.; Huang, L.; Yuan, X.; Luo, S.; Zhou, J.; Yang, X.; et al. Significant Reduction of Chenodeoxycholic Acid and Glycochenodeoxycholic Acid in the Elderly with Severe COVID-19. Biomolecules 2025, 15, 943. https://doi.org/10.3390/biom15070943
Liu S, Xu W, Tu B, Xiao Z, Li X, Huang L, Yuan X, Luo S, Zhou J, Yang X, et al. Significant Reduction of Chenodeoxycholic Acid and Glycochenodeoxycholic Acid in the Elderly with Severe COVID-19. Biomolecules. 2025; 15(7):943. https://doi.org/10.3390/biom15070943
Chicago/Turabian StyleLiu, Shiyang, Wen Xu, Bo Tu, Zhiqing Xiao, Xue Li, Lei Huang, Xin Yuan, Shengdong Luo, Juanjuan Zhou, Xinxin Yang, and et al. 2025. "Significant Reduction of Chenodeoxycholic Acid and Glycochenodeoxycholic Acid in the Elderly with Severe COVID-19" Biomolecules 15, no. 7: 943. https://doi.org/10.3390/biom15070943
APA StyleLiu, S., Xu, W., Tu, B., Xiao, Z., Li, X., Huang, L., Yuan, X., Luo, S., Zhou, J., Yang, X., Yang, J., Chang, D., Chen, W., & Wang, F.-S. (2025). Significant Reduction of Chenodeoxycholic Acid and Glycochenodeoxycholic Acid in the Elderly with Severe COVID-19. Biomolecules, 15(7), 943. https://doi.org/10.3390/biom15070943








