Single-Nucleus Transcriptomics Reveals Glial Metabolic–Immune Rewiring and Intercellular Signaling Disruption in Chronic Migraine
Abstract
1. Introduction
2. Materials and Methods
2.1. Establishment of a CM Rat Model
2.2. Assessment of Paw Withdrawal Mechanical Threshold (PWMT)
2.3. Assessment of Tail-Flick Test (TFT)
2.4. Single-Cell Suspension Preparation and Nuclei Isolation
2.5. snRNA-Seq Data Quality Control and Preprocessing
2.6. Cell Type Identification and Functional Annotation
2.7. Estimating Cellular Composition Preference Using Ro/e
2.8. Cell-Type-Specific Inflammation Score Analysis
2.9. Pathway Enrichment Analysis
2.10. Pseudotime Trajectory Analysis
2.11. Cell–Cell Communication Analysis
2.12. Statistical Analysis
3. Results
3.1. NTG Induces Progressive Hyperalgesia in a Rat Model of CM
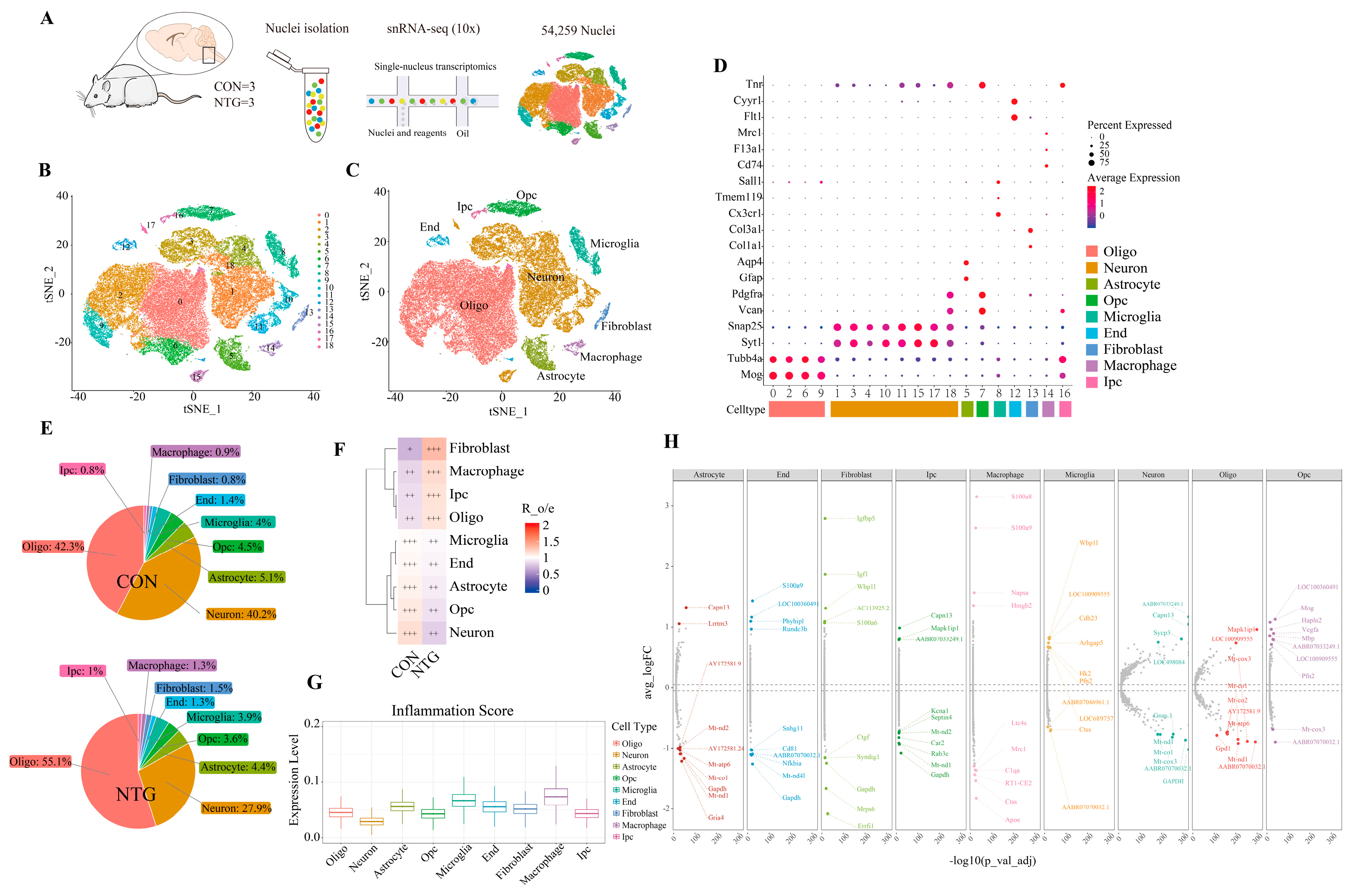
3.2. NTG Drives Cellular Remodeling and Neuroinflammatory Activation in the TNC
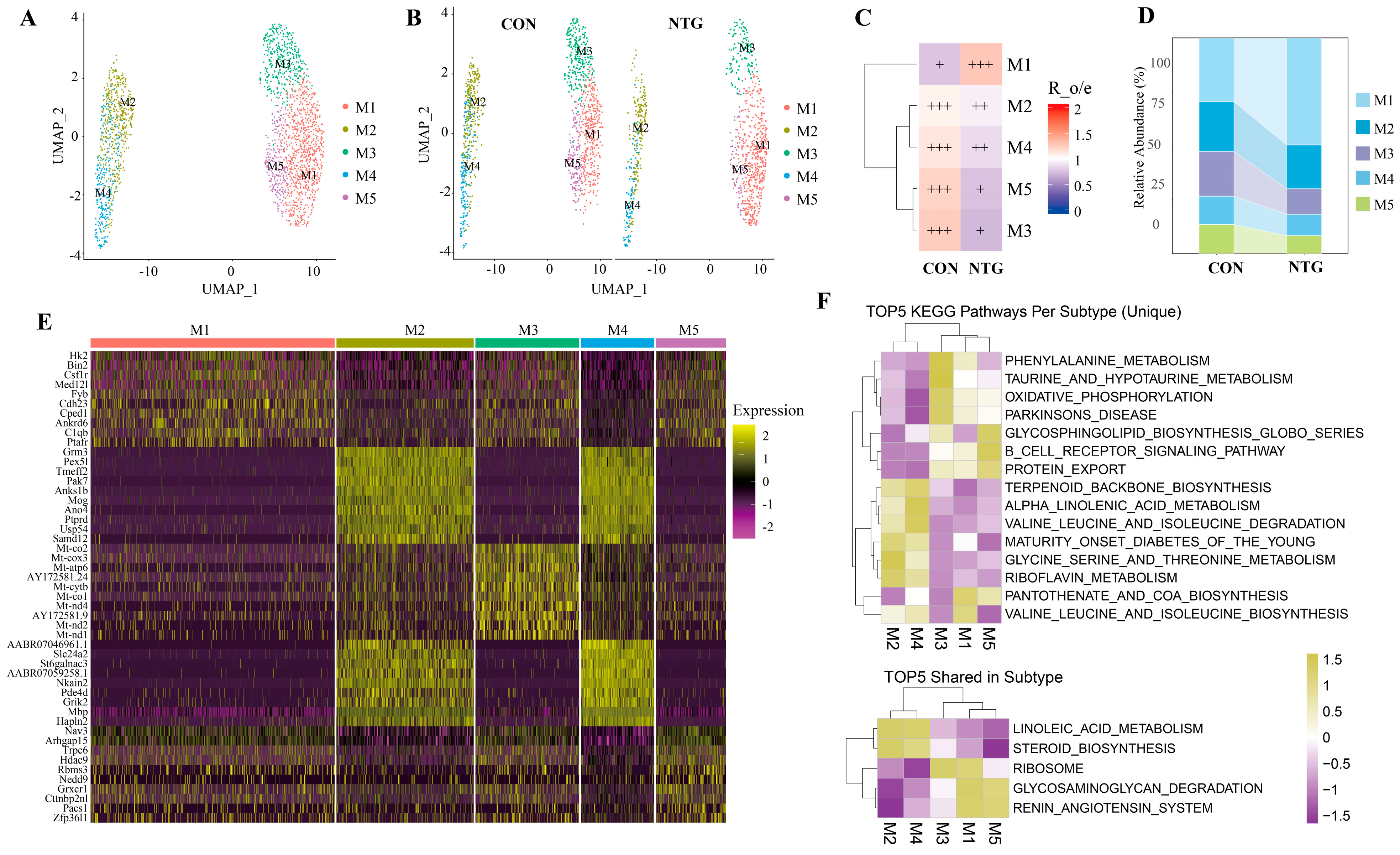
3.3. NTG-Mic: A Proinflammatory Microglial Subpopulation Enriched in Glycolysis and Complement Activation
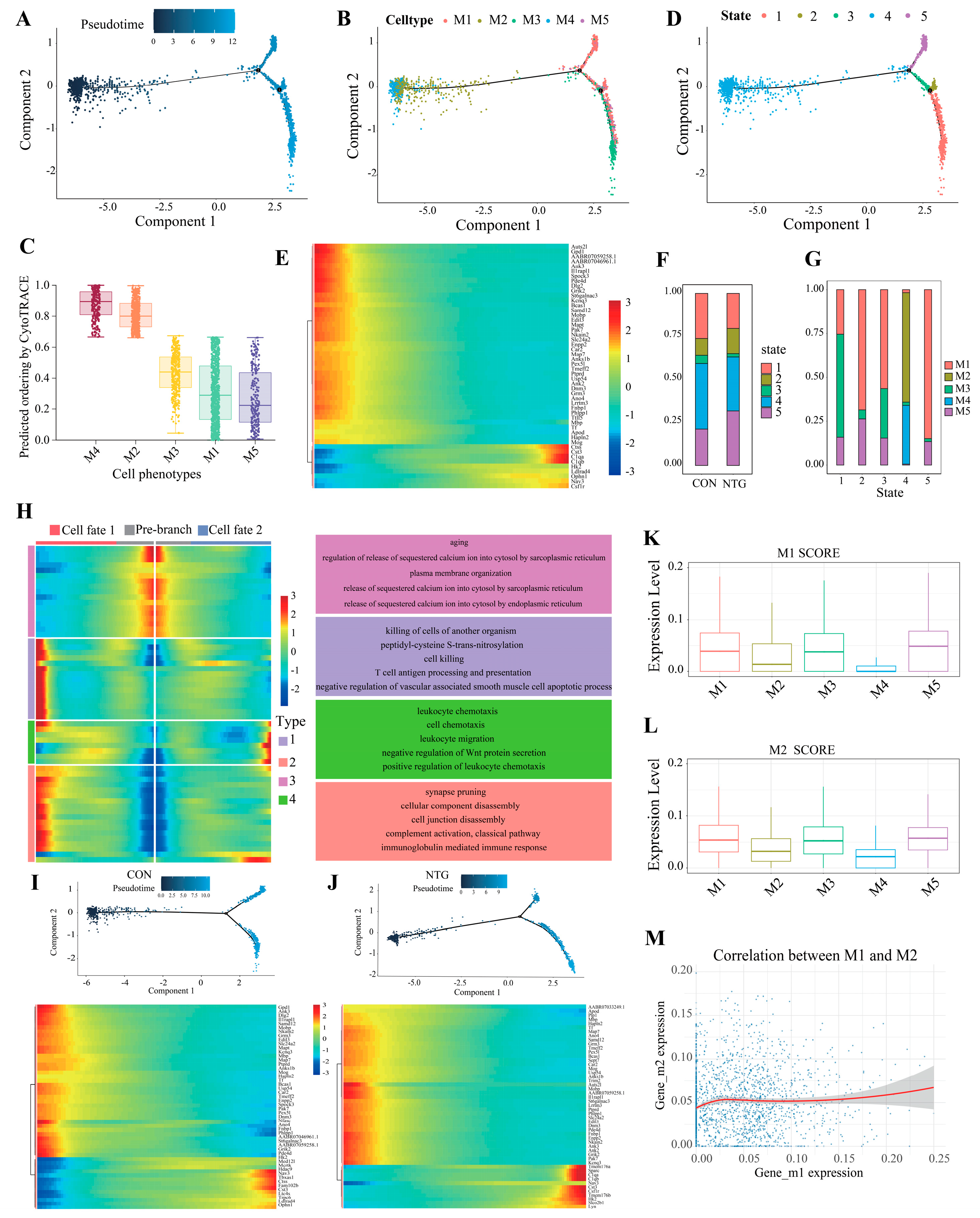
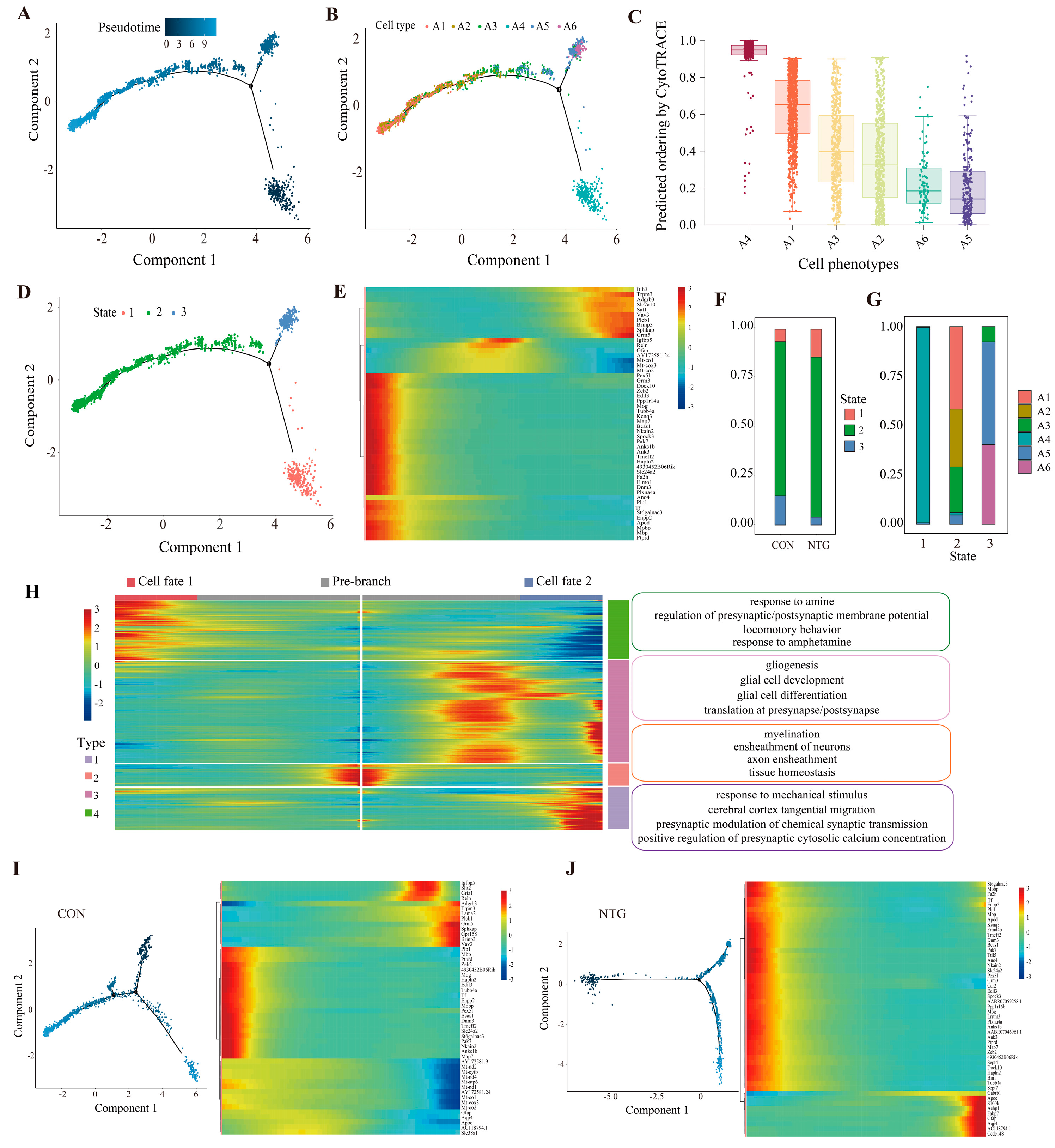
3.4. Pseudotime Analysis Reveals Microglial Divergence Toward Inflammatory Fates
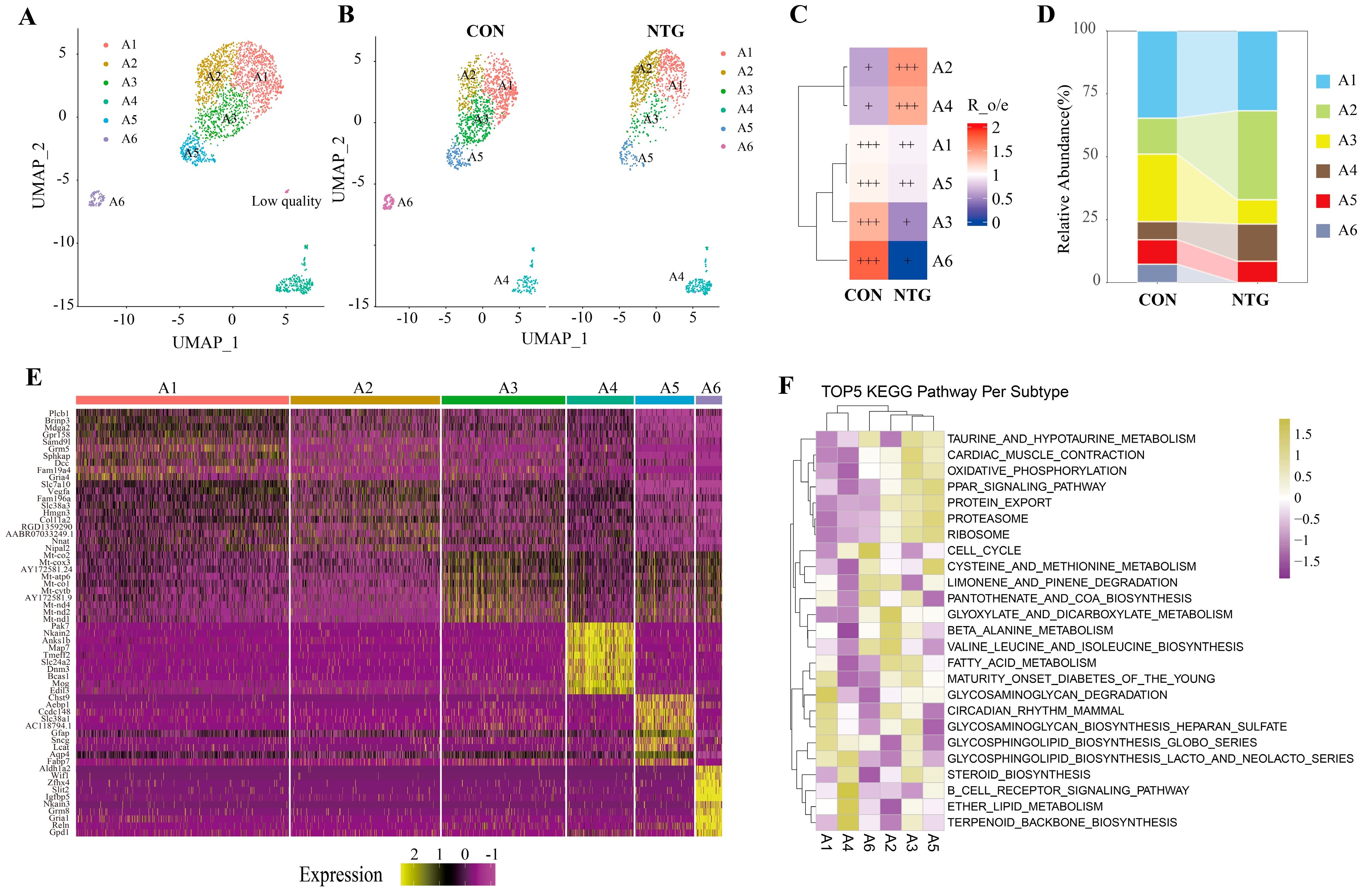
3.5. NTG-Asts: A Reactive Astrocyte Subpopulation with Dual Inflammatory and Metabolic Programs
3.6. Pseudotime Analysis Reveals Astrocytic Trajectories Toward Distinct Reactive States
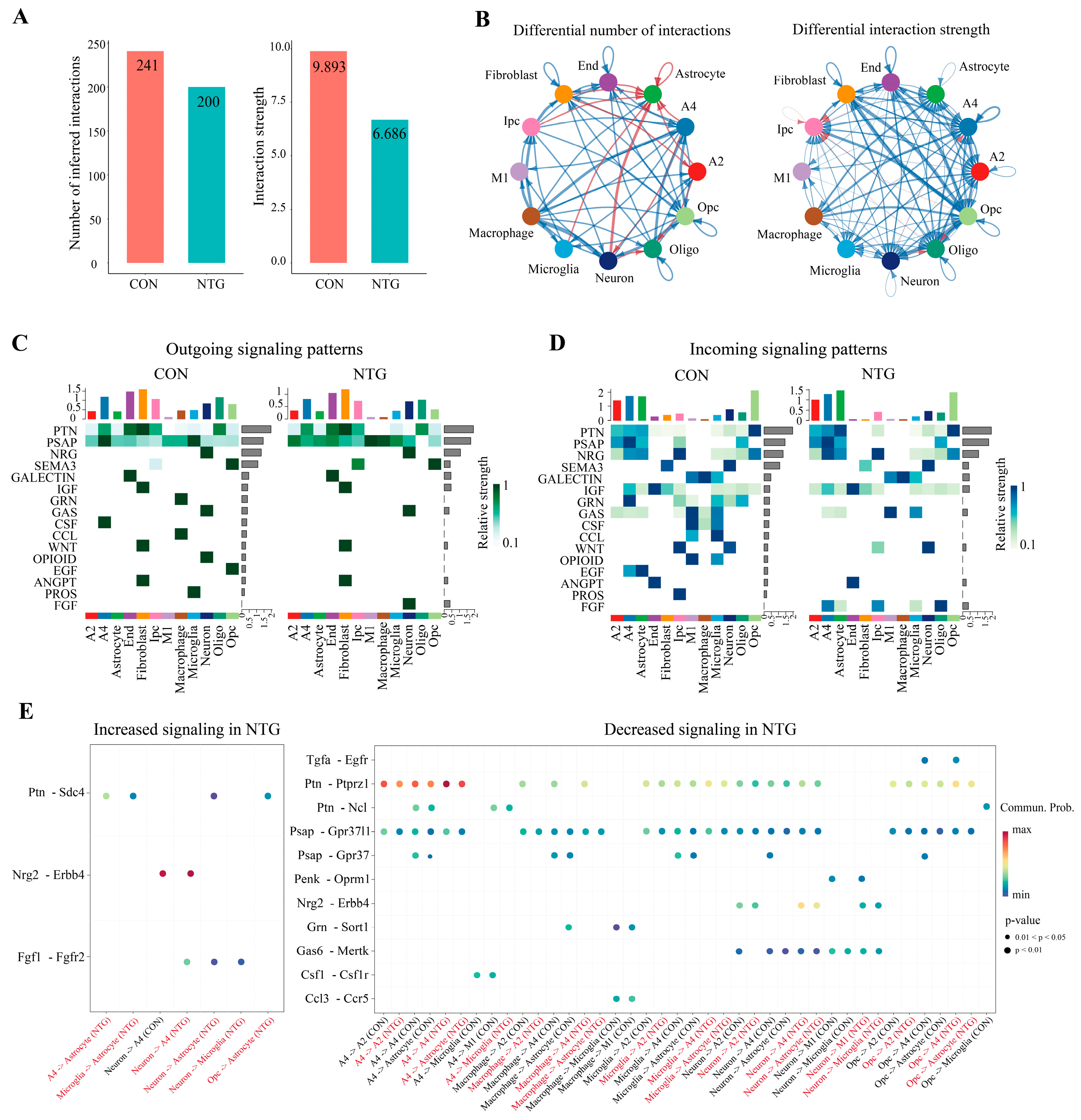
3.7. NTG Rewires Glial Communication Networks in the TNC
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| CM | Chronic migraine |
| CNS | Central nervous system |
| GO | Gene ontology |
| GSVA | Gene set variation analysis |
| KEGG | Kyoto encyclopedia of genes and genomes |
| NTG | Nitroglycerin |
| PPI | Protein–protein interaction |
| PWMT | Paw withdrawal mechanical threshold |
| TFT | Tail-flick test |
| TNC | Trigeminal nucleus caudalis |
| snRNA | Single-nucleus RNA sequencing |
References
- May, A.; Schulte, L.H. Chronic migraine: Risk factors, mechanisms and treatment. Nat. Rev. Neurol. 2016, 12, 455–464. [Google Scholar] [CrossRef] [PubMed]
- Ashina, M.; Katsarava, Z.; Do, T.P.; Buse, D.C.; Pozo-Rosich, P.; Özge, A.; Krymchantowski, A.V.; Lebedeva, E.R.; Ravishankar, K.; Yu, S.; et al. Migraine: Epidemiology and systems of care. Lancet 2021, 397, 1485–1495. [Google Scholar] [CrossRef]
- Yeh, P.K.; An, Y.C.; Hung, K.S.; Yang, F.C. Influences of Genetic and Environmental Factors on Chronic Migraine: A Narrative Review. Curr. Pain Headache Rep. 2024, 28, 169–180. [Google Scholar] [CrossRef] [PubMed]
- Ha, W.S.; Chu, M.K. Altered immunity in migraine: A comprehensive scoping review. J. Headache Pain 2024, 25, 95. [Google Scholar] [CrossRef]
- Ashina, M.; Hansen, J.M.; Do, T.P.; Melo-Carrillo, A.; Burstein, R.; Moskowitz, M.A. Migraine and the trigeminovascular system-40 years and counting. Lancet Neurol. 2019, 18, 795–804. [Google Scholar] [CrossRef]
- Sacristan, C. Microglia and Astrocyte Crosstalk in Immunity. Trends Immunol. 2020, 41, 747–748. [Google Scholar] [CrossRef]
- Linnerbauer, M.; Wheeler, M.A.; Quintana, F.J. Astrocyte Crosstalk in CNS Inflammation. Neuron 2020, 108, 608–622. [Google Scholar] [CrossRef] [PubMed]
- Stratoulias, V.; Venero, J.L.; Tremblay, M.; Joseph, B. Microglial subtypes: Diversity within the microglial community. EMBO J. 2019, 38, e101997. [Google Scholar] [CrossRef]
- Sun, S.; Fan, Z.; Liu, X.; Wang, L.; Ge, Z. Microglia TREM1-mediated neuroinflammation contributes to central sensitization via the NF-kappaB pathway in a chronic migraine model. J. Headache Pain 2024, 25, 3. [Google Scholar] [CrossRef]
- Zhao, J.; Blaeser, A.S.; Levy, D. Astrocytes mediate migraine-related intracranial meningeal mechanical hypersensitivity. Pain 2021, 162, 2386–2396. [Google Scholar] [CrossRef]
- Wei, S.; Du, T.; Zhang, L.; Li, X.; Wang, Z.; Ning, Y.; Tang, Y.; Wu, X.; Han, J. A comprehensive exploration of astrocytes in migraine: A bibliometric and visual analysis. Eur. J. Med. Res. 2024, 29, 321. [Google Scholar] [CrossRef] [PubMed]
- Mathys, H.; Boix, C.A.; Akay, L.A.; Xia, Z.; Davila-Velderrain, J.; Ng, A.P.; Jiang, X.; Abdelhady, G.; Galani, K.; Mantero, J.; et al. Single-cell multiregion dissection of Alzheimer’s disease. Nature 2024, 632, 858–868. [Google Scholar] [CrossRef]
- Renthal, W. Localization of migraine susceptibility genes in human brain by single-cell RNA sequencing. Cephalalgia 2018, 38, 1976–1983. [Google Scholar] [CrossRef]
- Vgontzas, A.; Renthal, W. Migraine-associated gene expression in cell types of the central and peripheral nervous system. Cephalalgia 2019, 40, 517–523. [Google Scholar] [CrossRef]
- Chen, L.; Li, Y.; Zhu, L.; Jin, H.; Kang, X.; Feng, Z. Single-cell RNA sequencing in the context of neuropathic pain: Progress, challenges, and prospects. Transl. Res. 2023, 251, 96–103. [Google Scholar] [CrossRef]
- Yang, L.; Xu, M.; Bhuiyan, S.A.; Li, J.; Zhao, J.; Cohrs, R.J.; Susterich, J.T.; Signorelli, S.; Green, U.; Stone, J.R.; et al. Human and mouse trigeminal ganglia cell atlas implicates multiple cell types in migraine. Neuron 2022, 110, 1806–1821.e1808. [Google Scholar] [CrossRef] [PubMed]
- Sureda-Gibert, P.; Romero-Reyes, M.; Akerman, S. Nitroglycerin as a model of migraine: Clinical and preclinical review. Neurobiol. Pain 2022, 12, 100105. [Google Scholar] [CrossRef] [PubMed]
- Demartini, C.; Greco, R.; Zanaboni, A.M.; Sances, G.; De Icco, R.; Borsook, D.; Tassorelli, C. Nitroglycerin as a comparative experimental model of migraine pain: From animal to human and back. Prog. Neurobiol. 2019, 177, 15–32. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, Y.; Tian, K.; Wang, Y.; Fan, X.; Pan, Q.; Qin, G.; Zhang, D.; Chen, L.; Zhou, J. Calcitonin gene-related peptide facilitates sensitization of the vestibular nucleus in a rat model of chronic migraine. J. Headache Pain 2020, 21, 72. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- McGinnis, C.S.; Murrow, L.M.; Gartner, Z.J. DoubletFinder: Doublet Detection in Single-Cell RNA Sequencing Data Using Artificial Nearest Neighbors. Cell Syst. 2019, 8, 329–337.e324. [Google Scholar] [CrossRef]
- Yang, S.; Corbett, S.E.; Koga, Y.; Wang, Z.; Johnson, W.E.; Yajima, M.; Campbell, J.D. Decontamination of ambient RNA in single-cell RNA-seq with DecontX. Genome Biol. 2020, 21, 57. [Google Scholar] [CrossRef]
- Hao, Y.; Hao, S.; Andersen-Nissen, E.; Mauck, W.M., 3rd; Zheng, S.; Butler, A.; Lee, M.J.; Wilk, A.J.; Darby, C.; Zager, M.; et al. Integrated analysis of multimodal single-cell data. Cell 2021, 184, 3573–3587.e3529. [Google Scholar] [CrossRef] [PubMed]
- Hu, C.; Li, T.; Xu, Y.; Zhang, X.; Li, F.; Bai, J.; Chen, J.; Jiang, W.; Yang, K.; Ou, Q.; et al. CellMarker 2.0: An updated database of manually curated cell markers in human/mouse and web tools based on scRNA-seq data. Nucleic Acids Res. 2022, 51, D870–D876. [Google Scholar] [CrossRef]
- Valihrach, L.; Matusova, Z.; Zucha, D.; Klassen, R.; Benesova, S.; Abaffy, P.; Kubista, M.; Anderova, M. Recent advances in deciphering oligodendrocyte heterogeneity with single-cell transcriptomics. Front. Cell. Neurosci. 2022, 16, 1025012. [Google Scholar] [CrossRef] [PubMed]
- Marques, S.; Zeisel, A.; Codeluppi, S.; van Bruggen, D.; Mendanha Falcão, A.; Xiao, L.; Li, H.; Häring, M.; Hochgerner, H.; Romanov, R.A.; et al. Oligodendrocyte heterogeneity in the mouse juvenile and adult central nervous system. Science 2016, 352, 1326–1329. [Google Scholar] [CrossRef]
- Cheng, S.; Li, Z.; Gao, R.; Xing, B.; Gao, Y.; Yang, Y.; Qin, S.; Zhang, L.; Ouyang, H.; Du, P.; et al. A pan-cancer single-cell transcriptional atlas of tumor infiltrating myeloid cells. Cell 2021, 184, 792–809.e723. [Google Scholar] [CrossRef]
- Trapnell, C.; Cacchiarelli, D.; Grimsby, J.; Pokharel, P.; Li, S.; Morse, M.; Lennon, N.J.; Livak, K.J.; Mikkelsen, T.S.; Rinn, J.L. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol. 2014, 32, 381–386. [Google Scholar] [CrossRef]
- Jin, S.; Guerrero-Juarez, C.F.; Zhang, L.; Chang, I.; Ramos, R.; Kuan, C.H.; Myung, P.; Plikus, M.V.; Nie, Q. Inference and analysis of cell-cell communication using CellChat. Nat. Commun. 2021, 12, 1088. [Google Scholar] [CrossRef]
- Jordão, M.J.C.; Sankowski, R.; Brendecke, S.M.; Sagar; Locatelli, G.; Tai, Y.H.; Tay, T.L.; Schramm, E.; Armbruster, S.; Hagemeyer, N.; et al. Single-cell profiling identifies myeloid cell subsets with distinct fates during neuroinflammation. Science 2019, 363, eaat7554. [Google Scholar] [CrossRef]
- Safaiyan, S.; Besson-Girard, S.; Kaya, T.; Cantuti-Castelvetri, L.; Liu, L.; Ji, H.; Schifferer, M.; Gouna, G.; Usifo, F.; Kannaiyan, N.; et al. White matter aging drives microglial diversity. Neuron 2021, 109, 1100–1117.e1110. [Google Scholar] [CrossRef] [PubMed]
- Gao, C.; Jiang, J.; Tan, Y.; Chen, S. Microglia in neurodegenerative diseases: Mechanism and potential therapeutic targets. Signal Transduct. Target. Ther. 2023, 8, 359. [Google Scholar] [CrossRef] [PubMed]
- Srinivasan, K.; Friedman, B.A.; Etxeberria, A.; Huntley, M.A.; van der Brug, M.P.; Foreman, O.; Paw, J.S.; Modrusan, Z.; Beach, T.G.; Serrano, G.E.; et al. Alzheimer’s Patient Microglia Exhibit Enhanced Aging and Unique Transcriptional Activation. Cell Rep. 2020, 31, 107843. [Google Scholar] [CrossRef]
- Heller, D.T.; Kolson, D.R.; Brandebura, A.N.; Amick, E.M.; Wan, J.; Ramadan, J.; Holcomb, P.S.; Liu, S.; Deerinck, T.J.; Ellisman, M.H.; et al. Astrocyte ensheathment of calyx-forming axons of the auditory brainstem precedes accelerated expression of myelin genes and myelination by oligodendrocytes. J. Comp. Neurol. 2024, 532, e25552. [Google Scholar] [CrossRef]
- Alghamdi, B.; Fern, R. Phenotype overlap in glial cell populations: Astroglia, oligodendroglia and NG-2(+) cells. Front. Neuroanat. 2015, 9, 49. [Google Scholar] [CrossRef]
- Cropper, H.C.; Conway, C.M.; Wyche, W.; Pradhan, A.A. Glial activation in pain and emotional processing regions in the nitroglycerin mouse model of chronic migraine. Headache: J. Head Face Pain 2024, 64, 973–982. [Google Scholar] [CrossRef]
- He, W.; Long, T.; Pan, Q.; Zhang, S.; Zhang, Y.; Zhang, D.; Qin, G.; Chen, L.; Zhou, J. Microglial NLRP3 inflammasome activation mediates IL-1β release and contributes to central sensitization in a recurrent nitroglycerin-induced migraine model. J. Neuroinflamm. 2019, 16, 78. [Google Scholar] [CrossRef] [PubMed]
- Hasel, P.; Dando, O.; Jiwaji, Z.; Baxter, P.; Todd, A.C.; Heron, S.; Márkus, N.M.; McQueen, J.; Hampton, D.W.; Torvell, M.; et al. Neurons and neuronal activity control gene expression in astrocytes to regulate their development and metabolism. Nat. Commun. 2017, 8, 15132. [Google Scholar] [CrossRef]
- Hu, X.; Leak, R.K.; Shi, Y.; Suenaga, J.; Gao, Y.; Zheng, P.; Chen, J. Microglial and macrophage polarization—New prospects for brain repair. Nat. Rev. Neurol. 2015, 11, 56–64. [Google Scholar] [CrossRef]
- Lawrence, J.M.; Schardien, K.; Wigdahl, B.; Nonnemacher, M.R. Roles of neuropathology-associated reactive astrocytes: A systematic review. Acta Neuropathol. Commun. 2023, 11, 42. [Google Scholar] [CrossRef]
- Shao, F.; Wang, X.; Wu, H.; Wu, Q.; Zhang, J. Microglia and Neuroinflammation: Crucial Pathological Mechanisms in Traumatic Brain Injury-Induced Neurodegeneration. Front. Aging Neurosci. 2022, 14, 825086. [Google Scholar] [CrossRef]
- Batiuk, M.Y.; Martirosyan, A.; Wahis, J.; de Vin, F.; Marneffe, C.; Kusserow, C.; Koeppen, J.; Viana, J.F.; Oliveira, J.F.; Voet, T.; et al. Identification of region-specific astrocyte subtypes at single cell resolution. Nat. Commun. 2020, 11, 1220. [Google Scholar] [CrossRef]
- Li, D.; Yang, K.; Li, J.; Xu, X.; Gong, L.; Yue, S.; Wei, H.; Yue, Z.; Wu, Y.; Yin, S. Single-cell sequencing reveals glial cell involvement in development of neuropathic pain via myelin sheath lesion formation in the spinal cord. J. Neuroinflamm. 2024, 21, 213. [Google Scholar] [CrossRef]
- Li, H.; Liu, P.; Zhang, B.; Yuan, Z.; Guo, M.; Zou, X.; Qian, Y.; Deng, S.; Zhu, L.; Cao, X.; et al. Acute ischemia induces spatially and transcriptionally distinct microglial subclusters. Genome Med. 2023, 15, 109. [Google Scholar] [CrossRef] [PubMed]
- Schain, A.J.; Melo-Carrillo, A.; Borsook, D.; Grutzendler, J.; Strassman, A.M.; Burstein, R. Activation of pial and dural macrophages and dendritic cells by cortical spreading depression. Ann. Neurol. 2018, 83, 508–521. [Google Scholar] [CrossRef] [PubMed]
- Khan, S.; Amin, F.M.; Fliedner, F.P.; Christensen, C.E.; Tolnai, D.; Younis, S.; Olinger, A.C.R.; Birgens, H.; Daldrup-Link, H.; Kjær, A.; et al. Investigating macrophage-mediated inflammation in migraine using ultrasmall superparamagnetic iron oxide-enhanced 3T magnetic resonance imaging. Cephalalgia 2019, 39, 1407–1420. [Google Scholar] [CrossRef] [PubMed]
- Stevens, B.; Allen, N.J.; Vazquez, L.E.; Howell, G.R.; Christopherson, K.S.; Nouri, N.; Micheva, K.D.; Mehalow, A.K.; Huberman, A.D.; Stafford, B.; et al. The Classical Complement Cascade Mediates CNS Synapse Elimination. Cell 2007, 131, 1164–1178. [Google Scholar] [CrossRef]
- Baik, S.H.; Kang, S.; Lee, W.; Choi, H.; Chung, S.; Kim, J.I.; Mook-Jung, I. A Breakdown in Metabolic Reprogramming Causes Microglia Dysfunction in Alzheimer’s Disease. Cell Metab. 2019, 30, 493–507.e496. [Google Scholar] [CrossRef]
- Zamboni, M.; Llorens-Bobadilla, E.; Magnusson, J.P.; Frisén, J. A Widespread Neurogenic Potential of Neocortical Astrocytes Is Induced by Injury. Cell Stem Cell 2020, 27, 605–617.e605. [Google Scholar] [CrossRef]
- Matusova, Z.; Hol, E.M.; Pekny, M.; Kubista, M.; Valihrach, L. Reactive astrogliosis in the era of single-cell transcriptomics. Front. Cell. Neurosci. 2023, 17, 1173200. [Google Scholar] [CrossRef]
- Liddelow, S.A.; Guttenplan, K.A.; Clarke, L.E.; Bennett, F.C.; Bohlen, C.J.; Schirmer, L.; Bennett, M.L.; Münch, A.E.; Chung, W.-S.; Peterson, T.C.; et al. Neurotoxic reactive astrocytes are induced by activated microglia. Nature 2017, 541, 481–487. [Google Scholar] [CrossRef]
- Xiang, C.; Li, H.; Tang, W. Targeting CSF-1R represents an effective strategy in modulating inflammatory diseases. Pharmacol. Res. 2023, 187, 106566. [Google Scholar] [CrossRef] [PubMed]
- Jian, J.; Konopka, J.; Liu, C. Insights into the role of progranulin in immunity, infection, and inflammation. J. Leukoc. Biol. 2013, 93, 199–208. [Google Scholar] [CrossRef]
- Wang, Y.; Szretter, K.J.; Vermi, W.; Gilfillan, S.; Rossini, C.; Cella, M.; Barrow, A.D.; Diamond, M.S.; Colonna, M. IL-34 is a tissue-restricted ligand of CSF1R required for the development of Langerhans cells and microglia. Nat. Immunol. 2012, 13, 753–760. [Google Scholar] [CrossRef] [PubMed]
- Nandi, S.; Gokhan, S.; Dai, X.-M.; Wei, S.; Enikolopov, G.; Lin, H.; Mehler, M.F.; Stanley, E.R. The CSF-1 receptor ligands IL-34 and CSF-1 exhibit distinct developmental brain expression patterns and regulate neural progenitor cell maintenance and maturation. Dev. Biol. 2012, 367, 100–113. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.; Xu, Y.; Guo, P.; Chen, Y.J.; Zhou, J.; Xia, M.; Tan, B.; Liu, X.; Feng, H.; Chen, Y. CCL5/CCR5-mediated peripheral inflammation exacerbates blood–brain barrier disruption after intracerebral hemorrhage in mice. J. Transl. Med. 2023, 21, 196. [Google Scholar] [CrossRef]
- Zeng, Z.; Lan, T.; Wei, Y.; Wei, X. CCL5/CCR5 axis in human diseases and related treatments. Genes. Dis. 2022, 9, 12–27. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Li, H.; Li, Y.; Xu, H.; Nazir, F.H.; Jiang, W.; Zheng, L.; Tang, C. Astrocyte-derived PTN alleviates deficits in hippocampal neurogenesis and cognition in models of multiple sclerosis. Stem Cell Rep. 2025, 20, 102383. [Google Scholar] [CrossRef]
- Xie, Y.; Su, N.; Yang, J.; Tan, Q.; Huang, S.; Jin, M.; Ni, Z.; Zhang, B.; Zhang, D.; Luo, F.; et al. FGF/FGFR signaling in health and disease. Signal Transduct. Target. Ther. 2020, 5, 181. [Google Scholar] [CrossRef]
- Vila-Pueyo, M.; Gliga, O.; Gallardo, V.J.; Pozo-Rosich, P. The Role of Glial Cells in Different Phases of Migraine: Lessons from Preclinical Studies. Int. J. Mol. Sci. 2023, 24, 12553. [Google Scholar] [CrossRef]
- Noseda, R.; Burstein, R. Migraine pathophysiology: Anatomy of the trigeminovascular pathway and associated neurological symptoms, CSD, sensitization and modulation of pain. Pain 2013, 154 (Suppl. S1), S44–S53. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Simoes, R.; Guo, T.; Cao, Y.Q. Neuroimmune interactions in the development and chronification of migraine headache. Trends Neurosci. 2024, 47, 819–833. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hu, S.; Tang, Z.; Sun, S.; Liu, L.; Wang, Y.; Xu, L.; Yuan, J.; Chen, Y.; Sun, M.; Zhao, L. Single-Nucleus Transcriptomics Reveals Glial Metabolic–Immune Rewiring and Intercellular Signaling Disruption in Chronic Migraine. Biomolecules 2025, 15, 942. https://doi.org/10.3390/biom15070942
Hu S, Tang Z, Sun S, Liu L, Wang Y, Xu L, Yuan J, Chen Y, Sun M, Zhao L. Single-Nucleus Transcriptomics Reveals Glial Metabolic–Immune Rewiring and Intercellular Signaling Disruption in Chronic Migraine. Biomolecules. 2025; 15(7):942. https://doi.org/10.3390/biom15070942
Chicago/Turabian StyleHu, Shuangyuan, Zili Tang, Shiqi Sun, Lu Liu, Yuyan Wang, Longyao Xu, Jing Yuan, Ying Chen, Mingsheng Sun, and Ling Zhao. 2025. "Single-Nucleus Transcriptomics Reveals Glial Metabolic–Immune Rewiring and Intercellular Signaling Disruption in Chronic Migraine" Biomolecules 15, no. 7: 942. https://doi.org/10.3390/biom15070942
APA StyleHu, S., Tang, Z., Sun, S., Liu, L., Wang, Y., Xu, L., Yuan, J., Chen, Y., Sun, M., & Zhao, L. (2025). Single-Nucleus Transcriptomics Reveals Glial Metabolic–Immune Rewiring and Intercellular Signaling Disruption in Chronic Migraine. Biomolecules, 15(7), 942. https://doi.org/10.3390/biom15070942





