Signal Peptides: From Molecular Mechanisms to Applications in Protein and Vaccine Engineering
Abstract
1. Introduction
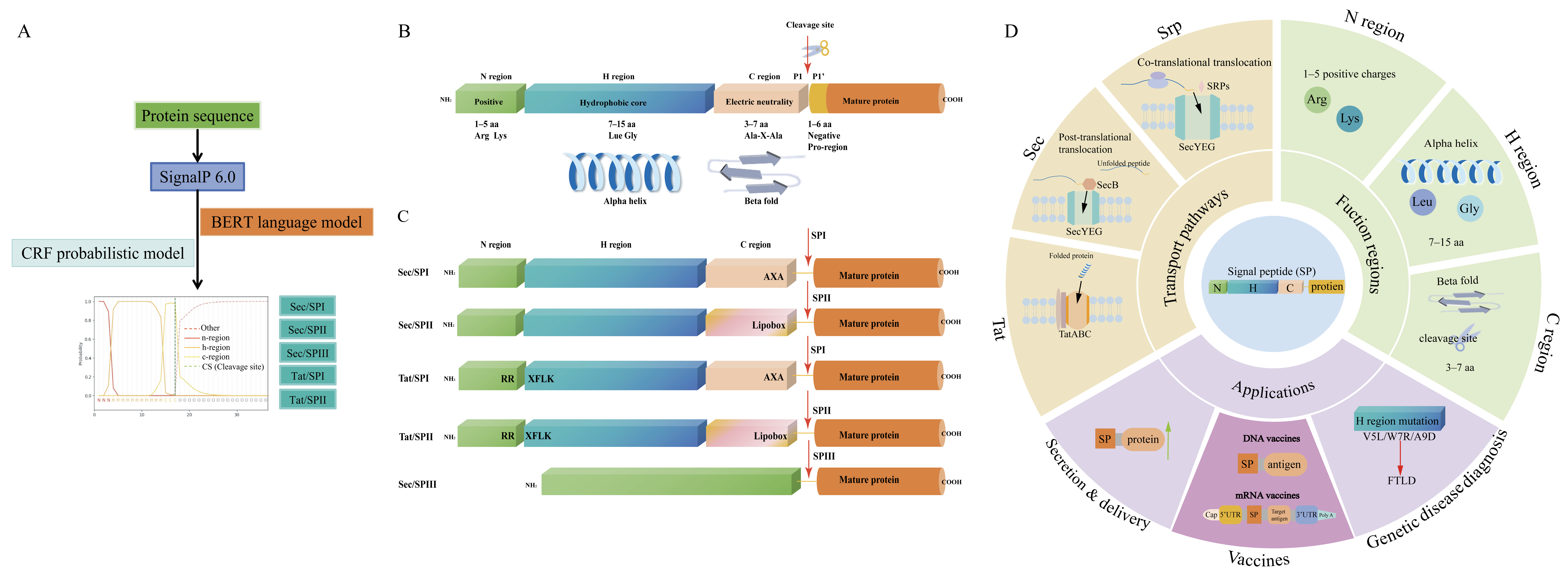
2. SP Structure
2.1. N-Region
2.1.1. The Role of Positive Charges in the N-Region
2.1.2. The Influence of Positive Charges in the N-Region on Translocation Efficiency
2.2. H-Region
2.2.1. Hydrophobicity and Transport Efficiency of the H-Region
2.2.2. Hydrophobicity and Transport Pathway of the H-Region
2.2.3. H-Region α-Helix and Transport Efficiency
2.2.4. H-Region Mutations in Hereditary Diseases
2.3. C-Region
2.3.1. Cleavage Motifs and Amino Acid Preferences in the C-Region
2.3.2. The Role of the Pro-Region in Protein Secretion
2.3.3. SPVs and C-Region Cleavage Errors
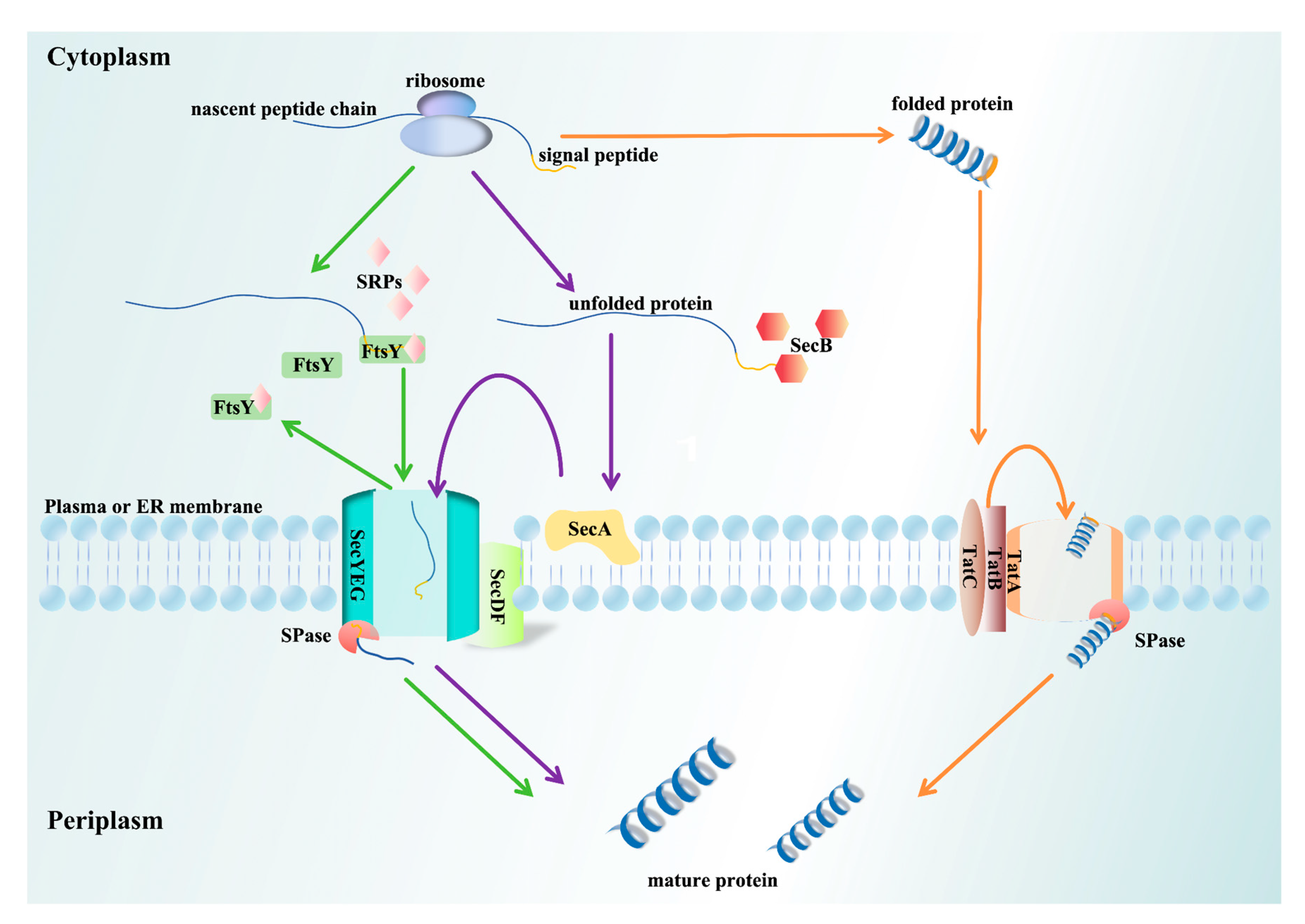
3. SP Transport Pathways
3.1. Srp Pathway
3.2. Sec Pathway
3.3. Tat Pathway
3.4. Uncanonical Transport Pathway
4. SP Applications
4.1. SPs in Prokaryotic Expression System
4.2. SPs in Eukaryotic Expression System
4.3. SPs in Vaccine Enhancement
4.3.1. SPs in Enhancing Subunit Vaccine Immunogenicity
4.3.2. SPs in Enhancing DNA Vaccine Immunogenicity
4.3.3. SPs in Enhancing Viral Vector Vaccine Immunogenicity
4.3.4. SPs in Enhancing mRNA Vaccine Immunogenicity
5. Concluding Remark
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Imai, K.; Nakai, K. Tools for the Recognition of Sorting Signals and the Prediction of Subcellular Localization of Proteins From Their Amino Acid Sequences. Front. Genet. 2020, 11, 607812. [Google Scholar] [CrossRef] [PubMed]
- Byun-McKay, S.A.; Geeta, R. Protein subcellular relocalization: A new perspective on the origin of novel genes. Trends Ecol. Evol. 2007, 22, 338–344. [Google Scholar] [CrossRef]
- Shimoji, M.; Figueroa, R.A.; Neve, E.; Maksel, D.; Imreh, G.; Morgenstern, R.; Hallberg, E. Molecular basis for the dual subcellular distribution of microsomal glutathione transferase 1. Biochim. Biophys. Acta Biomembr. 2017, 1859, 238–244. [Google Scholar] [CrossRef]
- Milstein, C.; Brownlee, G.G.; Harrison, T.M.; Mathews, M.B. A possible precursor of immunoglobulin light chains. Nat. New Biol. 1972, 239, 117–120. [Google Scholar] [CrossRef] [PubMed]
- Kaushik, S.; He, H.; Dalbey, R.E. Bacterial Signal Peptides- Navigating the Journey of Proteins. Front. Physiol. 2022, 13, 933153. [Google Scholar] [CrossRef]
- Warren, G.; Dobberstein, B. Protein transfer across microsomal membranes reassembled from separated membrane components. Nature 1978, 273, 569–571. [Google Scholar] [CrossRef]
- Blobel, G. Intracellular protein topogenesis. Proc. Natl. Acad. Sci. USA 1980, 77, 1496–1500. [Google Scholar] [CrossRef]
- Parys, J.B.; Van Coppenolle, F. Sec61 complex/translocon: The role of an atypical ER Ca2+-leak channel in health and disease. Front. Physiol. 2022, 13, 991149. [Google Scholar] [CrossRef] [PubMed]
- Deshaies, R.J.; Schekman, R. A yeast mutant defective at an early stage in import of secretory protein precursors into the endoplasmic reticulum. J. Cell Biol. 1987, 105, 633–645. [Google Scholar] [CrossRef]
- Görlich, D.; Rapoport, T.A. Protein translocation into proteoliposomes reconstituted from purified components of the endoplasmic reticulum membrane. Cell 1993, 75, 615–630. [Google Scholar] [CrossRef]
- Liaci, A.M.; Förster, F. Take Me Home, Protein Roads: Structural Insights into Signal Peptide Interactions during ER Translocation. Int. J. Mol. Sci. 2021, 22, 11871. [Google Scholar] [CrossRef] [PubMed]
- Morrison, J.H.; Poeschla, E.M. The Feline Immunodeficiency Virus Envelope Signal Peptide Is a Tetherin Antagonizing Protein. Mbio 2023, 14, e0016123. [Google Scholar] [CrossRef] [PubMed]
- Hao, B.; Liu, L.; Liu, N.; Sun, L.; Fan, F.; Huang, J. The Bombyx mori Nucleopolyhedrovirus GP64 Retains the Transmembrane Helix of Signal Peptide to Contribute to Secretion across the Cytomembrane. Microbiol. Spectr. 2022, 10, e0191322. [Google Scholar] [CrossRef]
- Luo, Y.; Chen, Z.; Lian, S.; Ji, X.; Zhu, C.; Zhu, G.; Xia, P. The Love and Hate Relationship between T5SS and Other Secretion Systems in Bacteria. Int. J. Mol. Sci. 2023, 25, 281. [Google Scholar] [CrossRef]
- Liu, M.; Allegood, J.; Zhu, X.; Seo, J.; Gebre, A.K.; Boudyguina, E.; Cheng, D.; Chuang, C.C.; Shelness, G.S.; Spiegel, S.; et al. Uncleaved ApoM signal peptide is required for formation of large ApoM/sphingosine 1-phosphate (S1P)-enriched HDL particles. J. Biol. Chem. 2015, 290, 7861–7870. [Google Scholar] [CrossRef] [PubMed]
- Peng, C.; Shi, C.; Cao, X.; Li, Y.; Liu, F.; Lu, F. Factors Influencing Recombinant Protein Secretion Efficiency in Gram-Positive Bacteria: Signal Peptide and Beyond. Front. Bioeng. Biotechnol. 2019, 7, 139. [Google Scholar] [CrossRef]
- Teufel, F.; Almagro Armenteros, J.J.; Johansen, A.R.; Gíslason, M.H.; Pihl, S.I.; Tsirigos, K.D.; Winther, O.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 6.0 predicts all five types of signal peptides using protein language models. Nat. Biotechnol. 2022, 40, 1023–1025. [Google Scholar] [CrossRef]
- Liu, C.; Angius, F.; Pol, A.; Mesman, R.A.; Versantvoort, W.; Op den Camp, H.J.M. Identification and characterization of an abundant lipoprotein from Methylacidiphilum fumariolicum SolV. Arch. Microbiol. 2023, 205, 261. [Google Scholar] [CrossRef]
- Craig, L.; Forest, K.T.; Maier, B. Type IV pili: Dynamics, biophysics and functional consequences. Nat. Rev. Microbiol. 2019, 17, 429–440. [Google Scholar] [CrossRef]
- Aledo, J.C. Methionine in proteins: The Cinderella of the proteinogenic amino acids. Protein Sci. A Publ. Protein Soc. 2019, 28, 1785–1796. [Google Scholar] [CrossRef]
- Petrychenko, V.; Yi, S.H.; Liedtke, D.; Peng, B.Z.; Rodnina, M.V.; Fischer, N. Structural basis for translational control by the human 48S initiation complex. Nat. Struct. Mol. Biol. 2025, 32, 62–72. [Google Scholar] [CrossRef] [PubMed]
- Lahry, K.; Datta, M.; Varshney, U. Genetic analysis of translation initiation in bacteria: An initiator tRNA-centric view. Mol. Microbiol. 2024, 122, 772–788. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.S.; Kim, D.; Hwang, C.S. Where Does N-Formylmethionine Come from? What for? Where Is It Going? What is the origin of N-formylmethionine in eukaryotic cells? Mol. Cells 2022, 45, 109–111. [Google Scholar] [CrossRef]
- Frain, K.M.; Robinson, C.; van Dijl, J.M. Transport of Folded Proteins by the Tat System. Protein J. 2019, 38, 377–388. [Google Scholar] [CrossRef]
- Nielsen, H. Predicting Secretory Proteins with SignalP. Methods Mol. Biol. 2017, 1611, 59–73. [Google Scholar] [CrossRef]
- Van Puyenbroeck, V.; Pauwels, E.; Provinciael, B.; Bell, T.W.; Schols, D.; Kalies, K.U.; Hartmann, E.; Vermeire, K. Preprotein signature for full susceptibility to the co-translational translocation inhibitor cyclotriazadisulfonamide. Traffic 2020, 21, 250–264. [Google Scholar] [CrossRef]
- Jeon, E.J.; Lee, S.M.; Hong, H.S.; Jeong, K.J. Design of fully synthetic signal peptide library and its use for enhanced secretory production of recombinant proteins in Corynebacterium glutamicum. Microb. Cell Factories 2024, 23, 252. [Google Scholar] [CrossRef] [PubMed]
- Low, K.O.; Muhammad Mahadi, N.; Md Illias, R. Optimisation of signal peptide for recombinant protein secretion in bacterial hosts. Appl. Microbiol. Biotechnol. 2013, 97, 3811–3826. [Google Scholar] [CrossRef]
- Kajava, A.V.; Zolov, S.N.; Kalinin, A.E.; Nesmeyanova, M.A. The net charge of the first 18 residues of the mature sequence affects protein translocation across the cytoplasmic membrane of gram-negative bacteria. J. Bacteriol. 2000, 182, 2163–2169. [Google Scholar] [CrossRef]
- Musik, J.E.; Zalucki, Y.M.; Beacham, I.R.; Jennings, M.P. The role of signal sequence proximal residues in the mature region of bacterial secreted proteins in E. coli. Biochim. Et Biophys. Acta Biomembr. 2022, 1864, 184000. [Google Scholar] [CrossRef]
- Gu, Y.; Xu, X.; Wu, Y.; Niu, T.; Liu, Y.; Li, J.; Du, G.; Liu, L. Advances and prospects of Bacillus subtilis cellular factories: From rational design to industrial applications. Metab. Eng. 2018, 50, 109–121. [Google Scholar] [CrossRef] [PubMed]
- Cui, W.; Han, L.; Suo, F.; Liu, Z.; Zhou, L.; Zhou, Z. Exploitation of Bacillus subtilis as a robust workhorse for production of heterologous proteins and beyond. World J. Microbiol. Biotechnol. 2018, 34, 145. [Google Scholar] [CrossRef] [PubMed]
- Fu, L.L.; Xu, Z.R.; Shuai, J.B.; Hu, C.X.; Dai, W.; Li, W.F. High-level secretion of a chimeric thermostable lichenase from Bacillus subtilis by screening of site-mutated signal peptides with structural alterations. Curr. Microbiol. 2008, 56, 287–292. [Google Scholar] [CrossRef]
- Caspers, M.; Brockmeier, U.; Degering, C.; Eggert, T.; Freudl, R. Improvement of Sec-dependent secretion of a heterologous model protein in Bacillus subtilis by saturation mutagenesis of the N-domain of the AmyE signal peptide. Appl. Microbiol. Biotechnol. 2010, 86, 1877–1885. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.; Sun, J.; Li, X.; Xiong, Y.; Wang, H.; Shu, H.; Zhu, R.; Liu, Q.; Huang, Y.; Madley, R.; et al. Positive charge in the n-region of the signal peptide contributes to efficient post-translational translocation of small secretory preproteins. J. Biol. Chem. 2018, 293, 1899–1907. [Google Scholar] [CrossRef]
- Lee, E.; Shrestha, K.L.; Kang, S.; Ramakrishnan, N.; Kwon, Y. Cell-Based Sensors for the Detection of EGF and EGF-Stimulated Ca2+ Signaling. Biosensors 2023, 13, 383. [Google Scholar] [CrossRef]
- Avendaño-Monsalve, M.C.; Mendoza-Martínez, A.E.; Ponce-Rojas, J.C.; Poot-Hernández, A.C.; Rincón-Heredia, R.; Funes, S. Positively charged amino acids at the N terminus of select mitochondrial proteins mediate early recognition by import proteins αβ’-NAC and Sam37. J. Biol. Chem. 2022, 298, 101984. [Google Scholar] [CrossRef]
- Liaci, A.M.; Steigenberger, B.; Telles de Souza, P.C.; Tamara, S.; Gröllers-Mulderij, M.; Ogrissek, P.; Marrink, S.J.; Scheltema, R.A.; Förster, F. Structure of the human signal peptidase complex reveals the determinants for signal peptide cleavage. Mol. Cell 2021, 81, 3934–3948.e11. [Google Scholar] [CrossRef]
- Ibrahim, A.M.; Missiakas, D. A novel polysaccharide in the envelope of S. aureus influences the septal secretion of preproteins with a YSIRK/GXXS motif. J. Bacteriol. 2025, 207, e0047824. [Google Scholar] [CrossRef]
- Zhang, R.; Jia, Y.; Scaffidi, S.J.; Madsen, J.J.; Yu, W. Signal peptidase SpsB coordinates staphylococcal cell cycle, surface protein septal trafficking, and LTA synthesis. Mbio 2025, 16, e0267324. [Google Scholar] [CrossRef] [PubMed]
- Ulfig, A.; Fröbel, J.; Lausberg, F.; Blümmel, A.S.; Heide, A.K.; Müller, M.; Freudl, R. The h-region of twin-arginine signal peptides supports productive binding of bacterial Tat precursor proteins to the TatBC receptor complex. J. Biol. Chem. 2017, 292, 10865–10882. [Google Scholar] [CrossRef] [PubMed]
- Hikita, C.; Mizushima, S. Effects of total hydrophobicity and length of the hydrophobic domain of a signal peptide on in vitro translocation efficiency. J. Biol. Chem. 1992, 267, 4882–4888. [Google Scholar] [CrossRef]
- Haryadi, R.; Ho, S.; Kok, Y.J.; Pu, H.X.; Zheng, L.; Pereira, N.A.; Li, B.; Bi, X.; Goh, L.T.; Yang, Y.; et al. Optimization of heavy chain and light chain signal peptides for high level expression of therapeutic antibodies in CHO cells. PLoS ONE 2015, 10, e0116878. [Google Scholar] [CrossRef] [PubMed]
- Owji, H.; Nezafat, N.; Negahdaripour, M.; Hajiebrahimi, A.; Ghasemi, Y. A comprehensive review of signal peptides: Structure, roles, and applications. Eur. J. Cell Biol. 2018, 97, 422–441. [Google Scholar] [CrossRef] [PubMed]
- Huang, Q.; Palmer, T. Signal Peptide Hydrophobicity Modulates Interaction with the Twin-Arginine Translocase. Mbio 2017, 8, e00909-17. [Google Scholar] [CrossRef]
- Han, S.; Machhi, S.; Berge, M.; Xi, G.; Linke, T.; Schoner, R. Novel signal peptides improve the secretion of recombinant Staphylococcus aureus Alpha toxinH35L in Escherichia coli. AMB Express 2017, 7, 93. [Google Scholar] [CrossRef]
- Palmer, T.; Stansfeld, P.J. Targeting of proteins to the twin-arginine translocation pathway. Mol. Microbiol. 2020, 113, 861–871. [Google Scholar] [CrossRef]
- Nguyen, D.; Stutz, R.; Schorr, S.; Lang, S.; Pfeffer, S.; Freeze, H.H.; Förster, F.; Helms, V.; Dudek, J.; Zimmermann, R. Proteomics reveals signal peptide features determining the client specificity in human TRAP-dependent ER protein import. Nat. Commun. 2018, 9, 3765. [Google Scholar] [CrossRef]
- Mayol, E.; Campillo, M.; Cordomí, A.; Olivella, M. Inter-residue interactions in alpha-helical transmembrane proteins. Bioinformatics 2019, 35, 2578–2584. [Google Scholar] [CrossRef]
- Naderi, M.; Ghaderi, R.; Khezri, J.; Karkhane, A.; Bambai, B. Crucial role of non-hydrophobic residues in H-region signal peptide on secretory production of l-asparaginase II in Escherichia coli. Biochem. Biophys. Res. Commun. 2022, 636 Pt 1, 105–111. [Google Scholar] [CrossRef]
- Snapp, E.L.; McCaul, N.; Quandte, M.; Cabartova, Z.; Bontjer, I.; Källgren, C.; Nilsson, I.; Land, A.; von Heijne, G.; Sanders, R.W.; et al. Structure and topology around the cleavage site regulate post-translational cleavage of the HIV-1 gp160 signal peptide. eLife 2017, 6, e26067. [Google Scholar] [CrossRef] [PubMed]
- Gao, M.; Chen, L.; Yang, J.; Dong, S.; Cao, Q.; Cui, Z.; Dong, Y.; Liu, H.; Shen, Y.; Yang, H.; et al. Multimodal mechanisms of pathogenic variants in the signal peptide of FIX leading to hemophilia B. Blood Adv. 2024, 8, 3893–3905. [Google Scholar] [CrossRef] [PubMed]
- Pottier, C.; Ravenscroft, T.A.; Sanchez-Contreras, M.; Rademakers, R. Genetics of FTLD: Overview and what else we can expect from genetic studies. J. Neurochem. 2016, 138 (Suppl. 1), 32–53. [Google Scholar] [CrossRef]
- Karamysheva, Z.N.; Tikhonova, E.B.; Karamyshev, A.L. Granulin in Frontotemporal Lobar Degeneration: Molecular Mechanisms of the Disease. Front. Neurosci. 2019, 13, 395. [Google Scholar] [CrossRef]
- Pross, E.; Kuhn, A. Two Signal Recognition Particle Sequences Are Present in the Amino-Terminal Domain of the C-Tailed Protein SciP. J. Bacteriol. 2020, 203, e00312-20. [Google Scholar] [CrossRef]
- Ling, W.L.; Su, C.T.; Lua, W.H.; Poh, J.J.; Ng, Y.L.; Wipat, A.; Gan, S.K. Essentially Leading Antibody Production: An Investigation of Amino Acids, Myeloma, and Natural V-Region Signal Peptides in Producing Pertuzumab and Trastuzumab Variants. Front. Immunol. 2020, 11, 604318. [Google Scholar] [CrossRef]
- Jain, R.G.; Rusch, S.L.; Kendall, D.A. Signal peptide cleavage regions. Functional limits on length and topological implications. J. Biol. Chem. 1994, 269, 16305–16310. [Google Scholar] [CrossRef]
- Xue, S.; Liu, X.; Pan, Y.; Xiao, C.; Feng, Y.; Zheng, L.; Zhao, M.; Huang, M. Comprehensive Analysis of Signal Peptides in Saccharomyces cerevisiae Reveals Features for Efficient Secretion. Adv. Sci. 2023, 10, e2203433. [Google Scholar] [CrossRef] [PubMed]
- Zubair, M.; Khan, F.A.; Menghwar, H.; Faisal, M.; Ashraf, M.; Rasheed, M.A.; Marawan, M.A.; Dawood, A.; Chen, Y.; Chen, H.; et al. Progresses on bacterial secretomes enlighten research on Mycoplasma secretome. Microb. Pathog. 2020, 144, 104160. [Google Scholar] [CrossRef]
- Rutz, C.; Klein, W.; Schülein, R. N-Terminal Signal Peptides of G Protein-Coupled Receptors: Significance for Receptor Biosynthesis, Trafficking, and Signal Transduction. Prog. Mol. Biol. Transl. Sci. 2015, 132, 267–287. [Google Scholar] [CrossRef]
- Rozov, S.M.; Deineko, E.V. Increasing the Efficiency of the Accumulation of Recombinant Proteins in Plant Cells: The Role of Transport Signal Peptides. Plants 2022, 11, 2561. [Google Scholar] [CrossRef] [PubMed]
- Ramirez, N.A.; Wu, C.; Chang, C.; Siegel, S.D.; Das, A.; Ton-That, H. A conserved signal-peptidase antagonist modulates membrane homeostasis of actinobacterial sortase critical for surface morphogenesis. Proc. Natl. Acad. Sci. USA 2022, 119, e2203114119. [Google Scholar] [CrossRef] [PubMed]
- Sidorczuk, K.; Mackiewicz, P.; Pietluch, F.; Gagat, P. Characterization of signal and transit peptides based on motif composition and taxon-specific patterns. Sci. Rep. 2023, 13, 15751. [Google Scholar] [CrossRef]
- Payne, S.H.; Bonissone, S.; Wu, S.; Brown, R.N.; Ivankov, D.N.; Frishman, D.; Pasa-Tolić, L.; Smith, R.D.; Pevzner, P.A. Unexpected diversity of signal peptides in prokaryotes. Mbio 2012, 3, e00339-12. [Google Scholar] [CrossRef] [PubMed]
- Aza, P.; Molpeceres, G.; de Salas, F.; Camarero, S. Design of an improved universal signal peptide based on the α-factor mating secretion signal for enzyme production in yeast. Cell. Mol. Life Sci. 2021, 78, 3691–3707. [Google Scholar] [CrossRef]
- Musik, J.E.; Poole, J.; Day, C.J.; Haselhorst, T.; Jen, F.E.; Ve, T.; Masic, V.; Jennings, M.P.; Zalucki, Y.M. New Perspectives on Escherichia coli Signal Peptidase I Substrate Specificity: Investigating Why the TasA Cleavage Site Is Incompatible with LepB Cleavage. Microbiol. Spectr. 2023, 11, e0500522. [Google Scholar] [CrossRef]
- Zhang, Z.; Wan, X.; Li, X.; Wan, C. Effects of a Shift of the Signal Peptide Cleavage Site in Signal Peptide Variant on the Synthesis and Secretion of SARS-CoV-2 Spike Protein. Molecules 2022, 27, 6688. [Google Scholar] [CrossRef]
- Qu, L.; Yi, Z.; Shen, Y.; Lin, L.; Chen, F.; Xu, Y.; Wu, Z.; Tang, H.; Zhang, X.; Tian, F.; et al. Circular RNA vaccines against SARS-CoV-2 and emerging variants. Cell 2022, 185, 1728–1744.e16. [Google Scholar] [CrossRef]
- Fu, Y.; Xu, Y.; Zhang, M.; Lv, F. Removal of signal peptide variants by cation exchange chromatography: A case study. Protein Expr. Purif. 2025, 225, 106581. [Google Scholar] [CrossRef]
- Kikuchi, Y.; Nagaya, S.; Togashi, T.; Imai, Y.; Togashi, M.; Araiso, Y.; Nishiuchi, T.; Morishita, E. Mechanism of antithrombin deficiency due to the novel variant C32W in the C-terminus of the signal peptide. Int. J. Hematol. 2025. Advance online publication. [Google Scholar] [CrossRef]
- Hsieh, H.H.; Shan, S.O. Fidelity of Cotranslational Protein Targeting to the Endoplasmic Reticulum. Int. J. Mol. Sci. 2021, 23, 281. [Google Scholar] [CrossRef]
- Jomaa, A.; Eitzinger, S.; Zhu, Z.; Chandrasekar, S.; Kobayashi, K.; Shan, S.O.; Ban, N. Molecular mechanism of cargo recognition and handover by the mammalian signal recognition particle. Cell Rep. 2021, 36, 109350. [Google Scholar] [CrossRef] [PubMed]
- Kellogg, M.K.; Miller, S.C.; Tikhonova, E.B.; Karamyshev, A.L. SRPassing Co-translational Targeting: The Role of the Signal Recognition Particle in Protein Targeting and mRNA Protection. Int. J. Mol. Sci. 2021, 22, 6284. [Google Scholar] [CrossRef] [PubMed]
- Pauwels, E.; Schülein, R.; Vermeire, K. Inhibitors of the Sec61 Complex and Novel High Throughput Screening Strategies to Target the Protein Translocation Pathway. Int. J. Mol. Sci. 2021, 22, 12007. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Jomaa, A.; Chung, S.; Hwang Fu, Y.H.; Qian, R.; Sun, X.; Hsieh, H.H.; Chandrasekar, S.; Bi, X.; Mattei, S.; et al. Receptor compaction and GTPase rearrangement drive SRP-mediated cotranslational protein translocation into the ER. Sci. Adv. 2021, 7, eabg0942. [Google Scholar] [CrossRef]
- Das, S.; Vera, M.; Gandin, V.; Singer, R.H.; Tutucci, E. Intracellular mRNA transport and localized translation. Nat. Rev. Mol. Cell Biol. 2021, 22, 483–504. [Google Scholar] [CrossRef]
- Juaire, K.D.; Lapouge, K.; Becker, M.M.M.; Kotova, I.; Michelhans, M.; Carapito, R.; Wild, K.; Bahram, S.; Sinning, I. Structural and Functional Impact of SRP54 Mutations Causing Severe Congenital Neutropenia. Structure 2021, 29, 15–28.e7. [Google Scholar] [CrossRef]
- Wild, K.; Becker, M.M.M.; Kempf, G.; Sinning, I. Structure, dynamics and interactions of large SRP variants. Biol. Chem. 2019, 401, 63–80. [Google Scholar] [CrossRef]
- Issa, A.; Schlotter, F.; Flayac, J.; Chen, J.; Wacheul, L.; Philippe, M.; Sardini, L.; Mostefa, L.; Vandermoere, F.; Bertrand, E.; et al. The nucleolar phase of signal recognition particle assembly. Life Sci. Alliance 2024, 7, e202402614. [Google Scholar] [CrossRef]
- Soni, K.; Kempf, G.; Manalastas-Cantos, K.; Hendricks, A.; Flemming, D.; Guizetti, J.; Simon, B.; Frischknecht, F.; Svergun, D.I.; Wild, K.; et al. Structural analysis of the SRP Alu domain from Plasmodium falciparum reveals a non-canonical open conformation. Commun. Biol. 2021, 4, 600. [Google Scholar] [CrossRef]
- Pei, D.; Dalbey, R.E. Membrane translocation of folded proteins. J Biol Chem. 2022, 298, 102107. [Google Scholar] [CrossRef]
- Bariya, P.; Randall, L.L. Coassembly of SecYEG and SecA Fully Restores the Properties of the Native Translocon. J. Bacteriol. 2018, 201, e00493-18. [Google Scholar] [CrossRef] [PubMed]
- Komarudin, A.G.; Driessen, A.J.M. SecA-Mediated Protein Translocation through the SecYEG Channel. Microbiol. Spectr. 2019, 7, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Sanganna Gari, R.R.; Chattrakun, K.; Marsh, B.P.; Mao, C.; Chada, N.; Randall, L.L.; King, G.M. Direct visualization of the E. coli Sec translocase engaging precursor proteins in lipid bilayers. Sci. Adv. 2019, 5, eaav9404. [Google Scholar] [CrossRef] [PubMed]
- Grayczyk, J.P.; Harvey, C.J.; Laczkovich, I.; Alonzo, F., 3rd. A Lipoylated Metabolic Protein Released by Staphylococcus aureus Suppresses Macrophage Activation. Cell Host Microbe 2017, 22, 678–687.e9. [Google Scholar] [CrossRef]
- OOswald, J.; Njenga, R.; Natriashvili, A.; Sarmah, P.; Koch, H.G. The Dynamic SecYEG Translocon. Front. Mol. Biosci. 2021, 8, 664241. [Google Scholar] [CrossRef]
- Krishnamurthy, S.; Sardis, M.F.; Eleftheriadis, N.; Chatzi, K.E.; Smit, J.H.; Karathanou, K.; Gouridis, G.; Portaliou, A.G.; Bondar, A.N.; Karamanou, S.; et al. Preproteins couple the intrinsic dynamics of SecA to its ATPase cycle to translocate via a catch and release mechanism. Cell Rep. 2022, 38, 110346. [Google Scholar] [CrossRef]
- Furukawa, A.; Yoshikaie, K.; Mori, T.; Mori, H.; Morimoto, Y.V.; Sugano, Y.; Iwaki, S.; Minamino, T.; Sugita, Y.; Tanaka, Y.; et al. Tunnel Formation Inferred from the I-Form Structures of the Proton-Driven Protein Secretion Motor SecDF. Cell Rep. 2017, 19, 895–901. [Google Scholar] [CrossRef]
- Hao, B.; Zhou, W.; Theg, S.M. Hydrophobic mismatch is a key factor in protein transport across lipid bilayer membranes via the Tat pathway. J. Biol. Chem. 2022, 298, 101991. [Google Scholar] [CrossRef]
- Severi, E.; Bunoro Batista, M.; Lannoy, A.; Stansfeld, P.J.; Palmer, T. Characterization of a TatA/TatB binding site on the TatC component of the Escherichia coli twin arginine translocase. Microbiology 2023, 169, 001298. [Google Scholar] [CrossRef]
- Schäfer, K.; Künzler, P.; Schneider, K.; Klingl, A.; Eubel, H.; Carrie, C. The Plant Mitochondrial TAT Pathway Is Essential for Complex III Biogenesis. Curr. Biol. CB 2020, 30, 840–853.e5. [Google Scholar] [CrossRef]
- Hao, B.; Zhou, W.; Theg, S.M. The polar amino acid in the TatA transmembrane helix is not strictly necessary for protein function. J. Biol. Chem. 2023, 299, 102998. [Google Scholar] [CrossRef] [PubMed]
- TTaw, M.N.; Boock, J.T.; Sotomayor, B.; Kim, D.; Rocco, M.A.; Waraho-Zhmayev, D.; DeLisa, M.P. Twin-arginine translocase component TatB performs folding quality control via a chaperone-like activity. Sci. Rep. 2022, 12, 14862. [Google Scholar] [CrossRef]
- Massai, F.; Saleeb, M.; Doruk, T.; Elofsson, M.; Forsberg, Å. Development, Optimization, and Validation of a High Throughput Screening Assay for Identification of Tat and Type II Secretion Inhibitors of Pseudomonas aeruginosa. Front. Cell. Infect. Microbiol. 2019, 9, 250. [Google Scholar] [CrossRef] [PubMed]
- Dolata, K.M.; Montero, I.G.; Miller, W.; Sievers, S.; Sura, T.; Wolff, C.; Schlüter, R.; Riedel, K.; Robinson, C. Far-reaching cellular consequences of tat deletion in Escherichia coli revealed by comprehensive proteome analyses. Microbiol. Res. 2019, 218, 97–107. [Google Scholar] [CrossRef]
- Liu, J.; Yin, F.; Liu, T.; Li, S.; Tan, C.; Li, L.; Zhou, R.; Huang, Q. The Tat system and its dependent cell division proteins are critical for virulence of extra-intestinal pathogenic Escherichia coli. Virulence 2020, 11, 1279–1292. [Google Scholar] [CrossRef] [PubMed]
- Meloni, S.; Rey, L.; Sidler, S.; Imperial, J.; Ruiz-Argüeso, T.; Palacios, J.M. The twin-arginine translocation (Tat) system is essential for Rhizobium-legume symbiosis. Mol. Microbiol. 2003, 48, 1195–1207. [Google Scholar] [CrossRef]
- Kleiner-Grote, G.R.M.; Risse, J.M.; Friehs, K. Secretion of recombinant proteins from E. coli. Eng. Life Sci. 2018, 18, 532–550. [Google Scholar] [CrossRef]
- Frain, K.M.; van Dijl, J.M.; Robinson, C. The Twin-Arginine Pathway for Protein Secretion. EcoSal Plus 2019, 8, 10.1128/ecosalplus.ESP-0040-2018. [Google Scholar] [CrossRef]
- Zhang, M.; Liu, L.; Lin, X.; Wang, Y.; Li, Y.; Guo, Q.; Li, S.; Sun, Y.; Tao, X.; Zhang, D.; et al. A Translocation Pathway for Vesicle-Mediated Unconventional Protein Secretion. Cell 2020, 181, 637–652.e15. [Google Scholar] [CrossRef]
- Zou, C.; Lu, L.; Wang, S.; Zhang, C.; Chen, X.; Lin, Y.; Huang, Y. The α-mating factor secretion signals and endogenous signal peptides for recombinant protein secretion in Komagataella phaffii. Biotechnol. Biofuels Bioprod. 2022, 15, 140. [Google Scholar] [CrossRef]
- Mambula, S.S.; Calderwood, S.K. Heat shock protein 70 is secreted from tumor cells by a nonclassical pathway involving lysosomal endosomes. J. Immunol. 2006, 177, 7849–7857. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Tao, X.; Han, Y.; Lin, X.; Tian, R.; Wang, H.; Chang, P.; Sun, Q.; Ge, L.; Zhang, M. A dual role of ERGIC-localized Rabs in TMED10-mediated unconventional protein secretion. Nat. Cell Biol. 2024, 26, 1077–1092. [Google Scholar] [CrossRef]
- Pouresmaeil, M.; Azizi-Dargahlou, S. Factors involved in heterologous expression of proteins in E. coli host. Arch. Microbiol. 2023, 205, 212. [Google Scholar] [CrossRef]
- Klein, G.; Wojtkiewicz, P.; Biernacka, D.; Stupak, A.; Gorzelak, P.; Raina, S. Identification of Substrates of Cytoplasmic Peptidyl-Prolyl Cis/Trans Isomerases and Their Collective Essentiality in Escherichia Coli. Int. J. Mol. Sci. 2020, 21, 4212. [Google Scholar] [CrossRef]
- Mirzadeh, K.; Shilling, P.J.; Elfageih, R.; Cumming, A.J.; Cui, H.L.; Rennig, M.; Nørholm, M.H.H.; Daley, D.O. Increased production of periplasmic proteins in Escherichia coli by directed evolution of the translation initiation region. Microb. Cell Factories 2020, 19, 85. [Google Scholar] [CrossRef] [PubMed]
- Souza, C.C.; Guimarães, J.M.; Pereira, S.D.S.; Mariúba, L.A.M. The multifunctionality of expression systems in Bacillus subtilis: Emerging devices for the production of recombinant proteins. Exp. Biol. Med. 2021, 246, 2443–2453. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Chen, J.; Sun, J.; Zhang, D. Multimer recognition and secretion by the non-classical secretion pathway in Bacillus subtilis. Sci. Rep. 2017, 7, 44023. [Google Scholar] [CrossRef]
- Eom, T.Y.; Gang, Y.; Lee, Y.; Kang, Y.H.; Jo, E.; Marasinghe, S.D.; Park, H.S.; Park, G.H.; Oh, C. Comparative Secretory Efficiency of Two Chitosanase Signal Peptides from Bacillus subtilis in Escherichia coli. J. Microbiol. 2024, 62, 1155–1164. [Google Scholar] [CrossRef]
- Zhang, X.; Qin, Y.; Han, X.; Li, Q.; Wang, Z.; Wang, X.; Zhao, L.; Zhang, H.; Cai, K.; Chu, Y.; et al. Signal peptide replacement of Ag43 enables an efficient bacterial cell surface display of receptor-binding domain of coronavirus. Biochem. Biophys. Res. Commun. 2024, 721, 150146. [Google Scholar] [CrossRef]
- Shen, Q.; Zhou, X.T.; Guo, Q.; Xue, Y.Z.; Xue, Y.P.; Zheng, Y.G. Potential of the Signal Peptide Derived from the PAS_chr3_0030 Gene Product for Secretory Expression of Valuable Enzymes in Pichia pastoris. Appl. Environ. Microbiol. 2022, 88, e0029622. [Google Scholar] [CrossRef]
- Liu, Z.G.; Ding, W.F.; Xie, S.C.; Sun, N.; Zhang, X.; Li, X.; Feng, Y. Establishment and characterization of cell clones from the Papilio cell line RIRI-PaDe-3 by a high-efficiency clonal method. Cytotechnology 2018, 70, 1235–1245. [Google Scholar] [CrossRef] [PubMed]
- Struble, L.R.; Smith, A.L.; Lutz, W.E.; Grubbs, G.; Sagar, S.; Bayles, K.W.; Radhakrishnan, P.; Khurana, S.; El-Gamal, D.; Borgstahl, G.E.O. Insect cell expression and purification of recombinant SARS-COV-2 spike proteins that demonstrate ACE2 binding. Protein Sci. 2022, 31, e4300. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Hernández, A.P.; Martínez-Flores, D.; Cruz-Reséndiz, A.; Padilla-Flores, T.; González-Flores, R.; Estrada, K.; Sampieri, A.; Camacho-Zarco, A.R.; Vaca, L. Baculovirus Display of Peptides and Proteins for Medical Applications. Viruses 2023, 15, 411. [Google Scholar] [CrossRef]
- Yuan, Y.; Zhang, Z.P.; He, Y.N.; Fan, W.S.; Dong, Z.H.; Zhang, L.H.; Sun, X.K.; Song, L.L.; Wei, T.C.; Mo, M.L.; et al. Protection against Virulent Infectious Bronchitis Virus Challenge Conferred by a Recombinant Baculovirus Co-Expressing S1 and N Proteins. Viruses 2018, 10, 347. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, M.S.; He, J.L.; Wu, T.Y.; Juang, R.H. A secretary bi-cistronic baculovirus expression system with improved production of the HA1 protein of H6 influenza virus in insect cells and Spodoptera litura larvae. J. Immunol. Methods 2018, 459, 81–89. [Google Scholar] [CrossRef]
- Poborilova, Z.; Plchova, H.; Cerovska, N.; Gunter, C.J.; Hitzeroth, I.I.; Rybicki, E.P.; Moravec, T. Transient protein expression in tobacco BY-2 plant cell packs using single and multi-cassette replicating vectors. Plant Cell Rep. 2020, 39, 1115–1127. [Google Scholar] [CrossRef]
- Chen, Q. Development of plant-made monoclonal antibodies against viral infections. Curr. Opin. Virol. 2022, 52, 148–160. [Google Scholar] [CrossRef]
- Chang, S.; Xiao, F. Comprehensive review of plant small signaling peptides: From stress adaptation mechanisms to practical solutions for crop resilience. Int. J. Biol. Macromol. 2025, 299, 139971. [Google Scholar] [CrossRef]
- Malm, M.; Kuo, C.C.; Barzadd, M.M.; Mebrahtu, A.; Wistbacka, N.; Razavi, R.; Volk, A.L.; Lundqvist, M.; Kotol, D.; Tegel, H.; et al. Harnessing secretory pathway differences between HEK293 and CHO to rescue production of difficult to express proteins. Metab. Eng. 2022, 72, 171–187. [Google Scholar] [CrossRef]
- van der Woude, R.; Turner, H.L.; Tomris, I.; Bouwman, K.M.; Ward, A.B.; de Vries, R.P. Drivers of recombinant soluble influenza A virus hemagglutinin and neuraminidase expression in mammalian cells. Protein Sci. 2020, 29, 1975–1982. [Google Scholar] [CrossRef]
- Attallah, C.; Etcheverrigaray, M.; Kratje, R.; Oggero, M. A highly efficient modified human serum albumin signal peptide to secrete proteins in cells derived from different mammalian species. Protein Expr. Purif. 2017, 132, 27–33. [Google Scholar] [CrossRef] [PubMed]
- Pulkina, A.; Vasilyev, K.; Muzhikyan, A.; Sergeeva, M.; Romanovskaya-Romanko, E.; Shurygina, A.P.; Shuklina, M.; Vasin, A.; Stukova, M.; Egorov, A. IgGκ Signal Peptide Enhances the Efficacy of an Influenza Vector Vaccine against Respiratory Syncytial Virus Infection in Mice. Int. J. Mol. Sci. 2023, 24, 11445. [Google Scholar] [CrossRef]
- Reuten, R.; Nikodemus, D.; Oliveira, M.B.; Patel, T.R.; Brachvogel, B.; Breloy, I.; Stetefeld, J.; Koch, M. Maltose-Binding Protein (MBP), a Secretion-Enhancing Tag for Mammalian Protein Expression Systems. PLoS ONE 2016, 11, e0152386. [Google Scholar] [CrossRef]
- Chapple, S.D.; Crofts, A.M.; Shadbolt, S.P.; McCafferty, J.; Dyson, M.R. Multiplexed expression and screening for recombinant protein production in mammalian cells. BMC Biotechnol. 2006, 6, 49. [Google Scholar] [CrossRef]
- Kovjazin, R.; Volovitz, I.; Daon, Y.; Vider-Shalit, T.; Azran, R.; Tsaban, L.; Carmon, L.; Louzoun, Y. Signal peptides and trans-membrane regions are broadly immunogenic and have high CD8+ T cell epitope densities: Implications for vaccine development. Mol. Immunol. 2011, 48, 1009–1018. [Google Scholar] [CrossRef]
- Houser, K.V.; Gaudinski, M.R.; Happe, M.; Narpala, S.; Verardi, R.; Sarfo, E.K.; Corrigan, A.R.; Wu, R.; Rothwell, R.S.; Novik, L.; et al. Safety and immunogenicity of an HIV-1 prefusion-stabilized envelope trimer (Trimer 4571) vaccine in healthy adults: A first-in-human open-label, randomized, dose-escalation, phase 1 clinical trial. EClinicalMedicine 2022, 48, 101477. [Google Scholar] [CrossRef]
- Timofeeva, A.; Sedykh, S.; Nevinsky, G. Post-Immune Antibodies in HIV-1 Infection in the Context of Vaccine Development: A Variety of Biological Functions and Catalytic Activities. Vaccines 2022, 10, 384. [Google Scholar] [CrossRef] [PubMed]
- McCaul, N.; Quandte, M.; Bontjer, I.; van Zadelhoff, G.; Land, A.; Crooks, E.T.; Binley, J.M.; Sanders, R.W.; Braakman, I. Intramolecular quality control: HIV-1 envelope gp160 signal-peptide cleavage as a functional folding checkpoint. Cell Rep. 2021, 36, 109646. [Google Scholar] [CrossRef] [PubMed]
- Upadhyay, C.; Rao, P.; Behzadi, M.A.; Feyznezhad, R.; Lambert, G.S.; Kumar, R.; Kumar, M.; Yang, W.; Jiang, X.; Luo, C.C.; et al. Signal peptide exchange alters HIV-1 envelope antigenicity and immunogenicity. Front. Immunol. 2024, 15, 1476924. [Google Scholar] [CrossRef]
- Kang, X.; Huang, T.; Shen, H.; Meng, C.; Jiao, X.; Pan, Z. Salmonella Enteritidis Subunit Vaccine Candidate Based on SseB Protein Co-Delivered with Simvastatin as Adjuvant. Pathogens 2022, 11, 443. [Google Scholar] [CrossRef]
- Rani, D.; Nayak, B.; Srivastava, S. Immunogenicity of gold nanoparticle-based truncated ORF2 vaccine in mice against Hepatitis E virus. 3 Biotech 2021, 11, 49. [Google Scholar] [CrossRef] [PubMed]
- Alves, K.C.S.; Guimarães, J.M.; Almeida, M.E.M.; Mariúba, L.A.M. Plasmodium falciparum merozoite surface protein 3 as a vaccine candidate: A brief review. Rev. Do Inst. De Med. Trop. De Sao Paulo 2022, 64, e23. [Google Scholar] [CrossRef] [PubMed]
- Panatto, D.; Amicizia, D.; Arata, L.; Lai, P.L.; Gasparini, R. A comprehensive analysis of Italian web pages mentioning squalene-based influenza vaccine adjuvants reveals a high prevalence of misinformation. Hum. Vaccines Immunother. 2018, 14, 969–977. [Google Scholar] [CrossRef]
- Umar, M.; Afzal, H.; Murtaza, A.; Cheng, L.T. Lipoprotein Signal Peptide as Adjuvants: Leveraging Lipobox-Driven TLR2 Activation in Modern Vaccine Design. Vaccines 2025, 13, 36. [Google Scholar] [CrossRef]
- Kaminska, P.; Tempes, A.; Scholz, E.; Malik, A.R. Cytokines on the way to secretion. Cytokine Growth Factor Rev. 2024, 79, 52–65. [Google Scholar] [CrossRef]
- Pérez-Trujillo, J.J.; Robles-Rodríguez, O.A.; Garza-Morales, R.; García-García, A.; Rodríguez-Rocha, H.; Villanueva-Olivo, A.; Segoviano-Ramírez, J.C.; Esparza-González, S.C.; Saucedo-Cárdenas, O.; Montes-de-Oca-Luna, R.; et al. Antitumor Response by Endoplasmic Reticulum-Targeting DNA Vaccine Is Improved by Adding a KDEL Retention Signal. Nucleic Acid Ther. 2018, 28, 252–261. [Google Scholar] [CrossRef]
- Han, Y.; RNa XJiang Jiang, X.; Wu, J.; Han, Y.; Zeng, Y.E.G.; Liang, A.; Yang, L.; Zhao, Y.; Huang, Y. Effect of a novel somatostatin-14 DNA vaccine fused to tPA signal peptide and CpG adjuvant on goat lactation and milk composition. Small Rumin. Res. 2020, 187, 106107. [Google Scholar] [CrossRef]
- Momin, T.; Kansagra, K.; Patel, H.; Sharma, S.; Sharma, B.; Patel, J.; Mittal, R.; Sanmukhani, J.; Maithal, K.; Dey, A.; et al. Safety and Immunogenicity of a DNA SARS-CoV-2 vaccine (ZyCoV-D): Results of an open-label, non-randomized phase I part of phase I/II clinical study by intradermal route in healthy subjects in India. EClinicalMedicine 2021, 38, 101020. [Google Scholar] [CrossRef]
- Yadav, P.D.; Kumar, S.; Agarwal, K.; Jain, M.; Patil, D.R.; Maithal, K.; Mathapati, B.; Giri, S.; Mohandas, S.; Shete, A.; et al. Needle-free injection system delivery of ZyCoV-D DNA vaccine demonstrated improved immunogenicity and protective efficacy in rhesus macaques against SARS-CoV-2. J. Med. Virol. 2023, 95, e28484. [Google Scholar] [CrossRef]
- Makhluf, H.; Shresta, S. Development of Zika Virus Vaccines. Vaccines 2018, 6, 7. [Google Scholar] [CrossRef]
- Fonseca, J.A.; McCaffery, J.N.; Caceres, J.; Kashentseva, E.; Singh, B.; Dmitriev, I.P.; Curiel, D.T.; Moreno, A. Inclusion of the murine IgGκ signal peptide increases the cellular immunogenicity of a simian adenoviral vectored Plasmodium vivax multistage vaccine. Vaccine 2018, 36, 2799–2808. [Google Scholar] [CrossRef] [PubMed]
- Jang, S.O.; Lee, J.H.; Chung, Y.J.; Oh, H.S.; Shin, M.; Kim, S.O.; Hong, S.P. Chimeric adenovirus-based herpes zoster vaccine with the tPA signal peptide elicits a robust T-cell immune response. Virology 2024, 600, 110243. [Google Scholar] [CrossRef] [PubMed]
- Sheerin, D.; Dold, C.; Silva-Reyes, L.; Linder, A.; Pollard, A.J.; Rollier, C.S. Inclusion of a dual signal sequence enhances the immunogenicity of a novel viral vectored vaccine against the capsular group B meningococcus. Cell Biosci. 2022, 12, 86. [Google Scholar] [CrossRef] [PubMed]
- Halperin, S.A.; Ye, L.; MacKinnon-Cameron, D.; Smith, B.; Cahn, P.E.; Ruiz-Palacios, G.M.; Ikram, A.; Lanas, F.; Lourdes Guerrero, M.; Muñoz Navarro, S.R.; et al. Final efficacy analysis, interim safety analysis, and immunogenicity of a single dose of recombinant novel coronavirus vaccine (adenovirus type 5 vector) in adults 18 years and older: An international, multicentre, randomised, double-blinded, placebo-controlled phase 3 trial. Lancet 2022, 399, 237–248. [Google Scholar] [CrossRef]
- Mo, C.; Li, X.; Wu, Q.; Fan, Y.; Liu, D.; Zhu, Y.; Yang, Y.; Liao, X.; Zhou, Z.; Zhou, L.; et al. SARS-CoV-2 mRNA vaccine requires signal peptide to induce antibody responses. Vaccine 2023, 41, 6863–6869. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhai, S.; Huang, H.; Qin, S.; Sun, M.; Chen, Y.; Lan, X.; Li, G.; Huang, Z.; Wang, D.; et al. Efficient signal sequence of mRNA vaccines enhances the antigen expression to expand the immune protection against viral infection. J. Nanobiotechnol. 2024, 22, 295. [Google Scholar] [CrossRef]
- Hou, F.; Zhang, Y.; Liu, X.; Murad, Y.M.; Xu, J.; Yu, Z.; Hua, X.; Song, Y.; Ding, J.; Huang, H.; et al. mRNA vaccines encoding fusion proteins of monkeypox virus antigens protect mice from vaccinia virus challenge. Nat. Commun. 2023, 14, 5925. [Google Scholar] [CrossRef]
- Huang, T.; Che, S.; Lv, Z.; Hao, D.; Wang, R.; Yi, Q.; Mei, L.; Yuan, Y.; Zou, H.; Guo, Y.; et al. mRNA-LNP vaccines combined with tPA signal sequence elicit strong protective immunity against Klebsiella pneumoniae. mSphere 2025, 10, e0077524. [Google Scholar] [CrossRef]
- Wei, Z.; Zhang, S.; Wang, X.; Xue, Y.; Dang, S.; Zhai, J. Technological breakthroughs and advancements in the application of mRNA vaccines: A comprehensive exploration and future prospects. Front. Immunol. 2025, 16, 1524317. [Google Scholar] [CrossRef]
- Goswami, J.; Baqui, A.H.; Doreski, P.A.; Perez Marc, G.; Jimenez, G.; Ahmed, S.; Zaman, K.; Duncan, C.J.A.; Ujiie, M.; Rämet, M.; et al. Humoral Immunogenicity of mRNA-1345 RSV Vaccine in Older Adults. J. Infect. Dis. 2024, 230, e996–e1006. [Google Scholar] [CrossRef]
- Chong, C.S.; Kunze, M.; Hochreiter, B.; Krenn, M.; Berger, J.; Maurer-Stroh, S. Rare Human Missense Variants can affect the Function of Disease-Relevant Proteins by Loss and Gain of Peroxisomal Targeting Motifs. Int. J. Mol. Sci. 2019, 20, 4609. [Google Scholar] [CrossRef]
- Brocard, C.; Hartig, A. Peroxisome targeting signal 1: Is it really a simple tripeptide? Biochim. Et Biophys. Acta 2006, 1763, 1565–1573. [Google Scholar] [CrossRef]
- O’Neill, P.; Mistry, R.K.; Brown, A.J.; James, D.C. Protein-Specific Signal Peptides for Mammalian Vector Engineering. ACS Synth. Biol. 2023, 12, 2339–2352. [Google Scholar] [CrossRef] [PubMed]
- Luft, C.; Freeman, J.; Elliott, D.; Al-Tamimi, N.; Kriston-Vizi, J.; Heintze, J.; Lindenschmidt, I.; Seed, B.; Ketteler, R. Application of Gaussia luciferase in bicistronic and non-conventional secretion reporter constructs. BMC Biochem. 2014, 15, 14. [Google Scholar] [CrossRef]
- Xie, J.; Zhuang, Z.; Gou, S.; Zhang, Q.; Wang, X.; Lan, T.; Lian, M.; Li, N.; Liang, Y.; Ouyang, Z.; et al. Precise genome editing of the Kozak sequence enables bidirectional and quantitative modulation of protein translation to anticipated levels without affecting transcription. Nucleic Acids Res. 2023, 51, 10075–10093. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.; Loo, B.Z.L.; Cheng, Y.Y.; Song, P.; Fan, H.; Latypov, O.; Kittelmann, S. Genome-wide high-throughput signal peptide screening via plasmid pUC256E improves protease secretion in Lactiplantibacillus plantarum and Pediococcus acidilactici. BMC Genom. 2022, 23, 48. [Google Scholar] [CrossRef] [PubMed]
- Niu, D.; Wu, Y.; Lian, J. Circular RNA vaccine in disease prevention and treatment. Signal Transduct. Target. Ther. 2023, 8, 341. [Google Scholar] [CrossRef]
- Bu, T.; Yang, Z.; Zhao, J.; Gao, Y.; Li, F.; Yang, R. Expanding the Potential of Circular RNA (CircRNA) Vaccines: A Promising Therapeutic Approach. Int. J. Mol. Sci. 2025, 26, 379. [Google Scholar] [CrossRef]
| Pathway | Srp | Sec | Tat |
|---|---|---|---|
| Scope | All life groups. More common in eukaryotes | Most Gram-negative bacteria, some Gram-positive bacteria | Archaea, Gram-positive bacteria, plant chloroplasts, and some plant mitochondria |
| Transported proteins | Mainly transports proteins that remain unmodified in the inner membrane | Unfolded proteins | Completely folded proteins, especially proteins that require co-factor binding for folding and multi-protein complexes |
| Mechanism | Co-translational translocation | Post-translational translocation | Post-translational translocation |
| Recognition | In eukaryotes, the SP length and amino acids are highly compatible with ER Sec61; in prokaryotes, it binds FtsY to deliver the complex to the SecYEG translocon | Specific SecB SP | S/T-RRXFLK motif |
| Energy | GTP | ATP | PMF |
| Characteristics | Classical Pathway | Atypical Pathway |
|---|---|---|
| SP | Essential | Non-essential |
| Vesicular transport | Strictly dependent on COPII (from ER to Golgi apparatus), COPI (retrograde transport), and secretory vesicles | Types I and II have no vesicles, partially dependent (Type III: autophagosomes/lysosomes; Type IV: Golgi bypass) |
| Energy consumption per transport event | High (SRP needs to bind GTP, Sec61 translocation, and vesicular transport) | Relatively low (passive diffusion, ABC transporters require ATP but with low efficiency) |
| Secretion efficiency | Relatively high efficiency | Relatively low (selective secretion: stress proteins, inflammatory factors) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, S.; He, Z.; Wang, H.; Zhai, J. Signal Peptides: From Molecular Mechanisms to Applications in Protein and Vaccine Engineering. Biomolecules 2025, 15, 897. https://doi.org/10.3390/biom15060897
Zhang S, He Z, Wang H, Zhai J. Signal Peptides: From Molecular Mechanisms to Applications in Protein and Vaccine Engineering. Biomolecules. 2025; 15(6):897. https://doi.org/10.3390/biom15060897
Chicago/Turabian StyleZhang, Shuai, Zhihui He, Hui Wang, and Jingbo Zhai. 2025. "Signal Peptides: From Molecular Mechanisms to Applications in Protein and Vaccine Engineering" Biomolecules 15, no. 6: 897. https://doi.org/10.3390/biom15060897
APA StyleZhang, S., He, Z., Wang, H., & Zhai, J. (2025). Signal Peptides: From Molecular Mechanisms to Applications in Protein and Vaccine Engineering. Biomolecules, 15(6), 897. https://doi.org/10.3390/biom15060897


_Kwok.png)




