Genome-Wide Identification of DNA Methyltransferases (Dnmts) in Fish and Its Potential Roles During Sex Change in Blackhead Seabream
Abstract
1. Introduction
2. Materials and Methods
2.1. Sequence Collection and Characterization of Dnmt Genes
2.2. Phylogenetic Analysis, Motif Identification, and Domain Prediction
2.3. Synteny Analysis
2.4. Evolutionary Homology and Protein Structure of Dnmt
2.5. Experimental Fish
2.6. RNA Extraction, Quantitative Reverse-Transcription PCR (qRT-PCR) Analysis, and Transcriptomic Analysis
3. Results
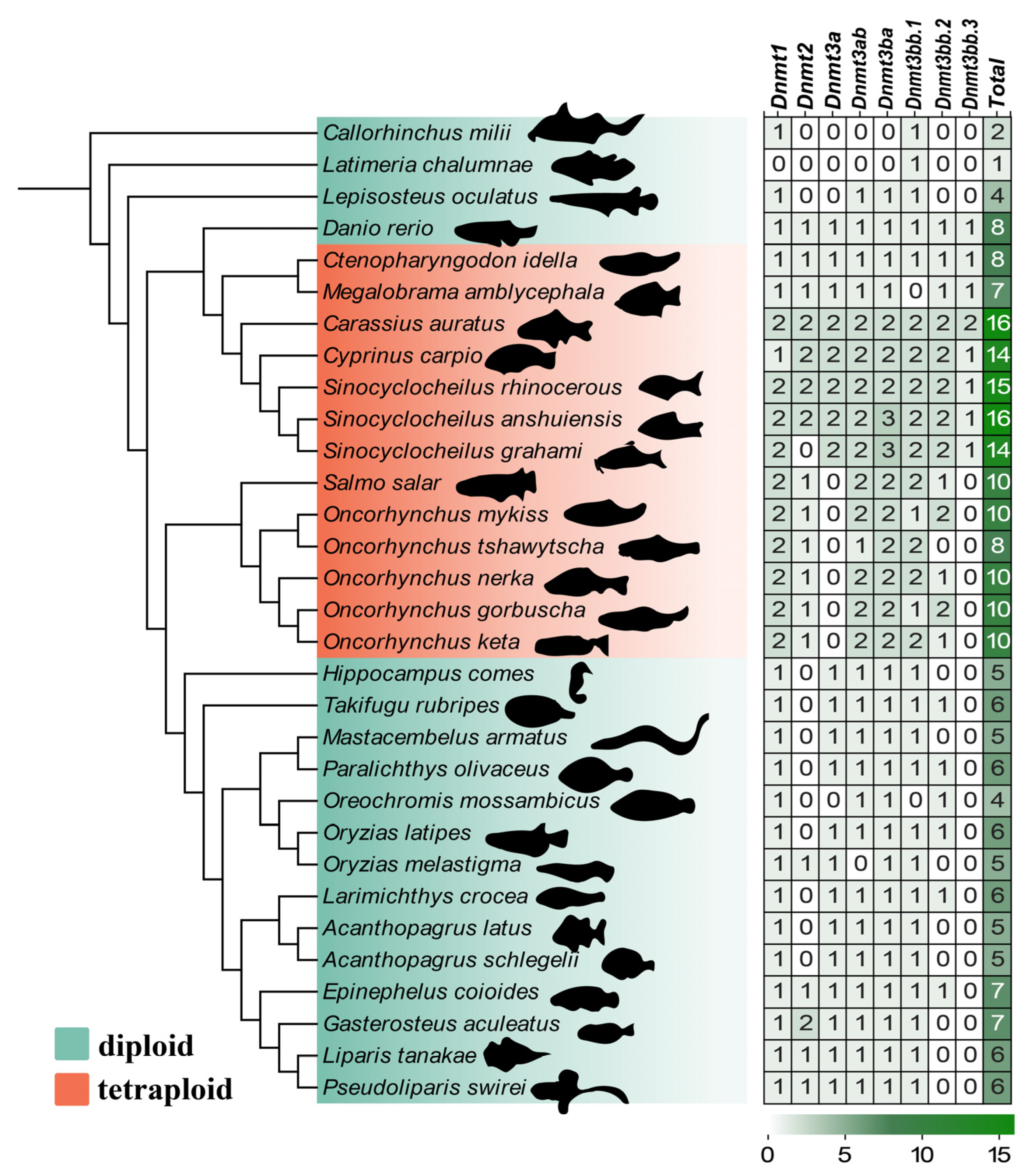
3.1. Genome-Wide Identification of the Dnmt Gene Family
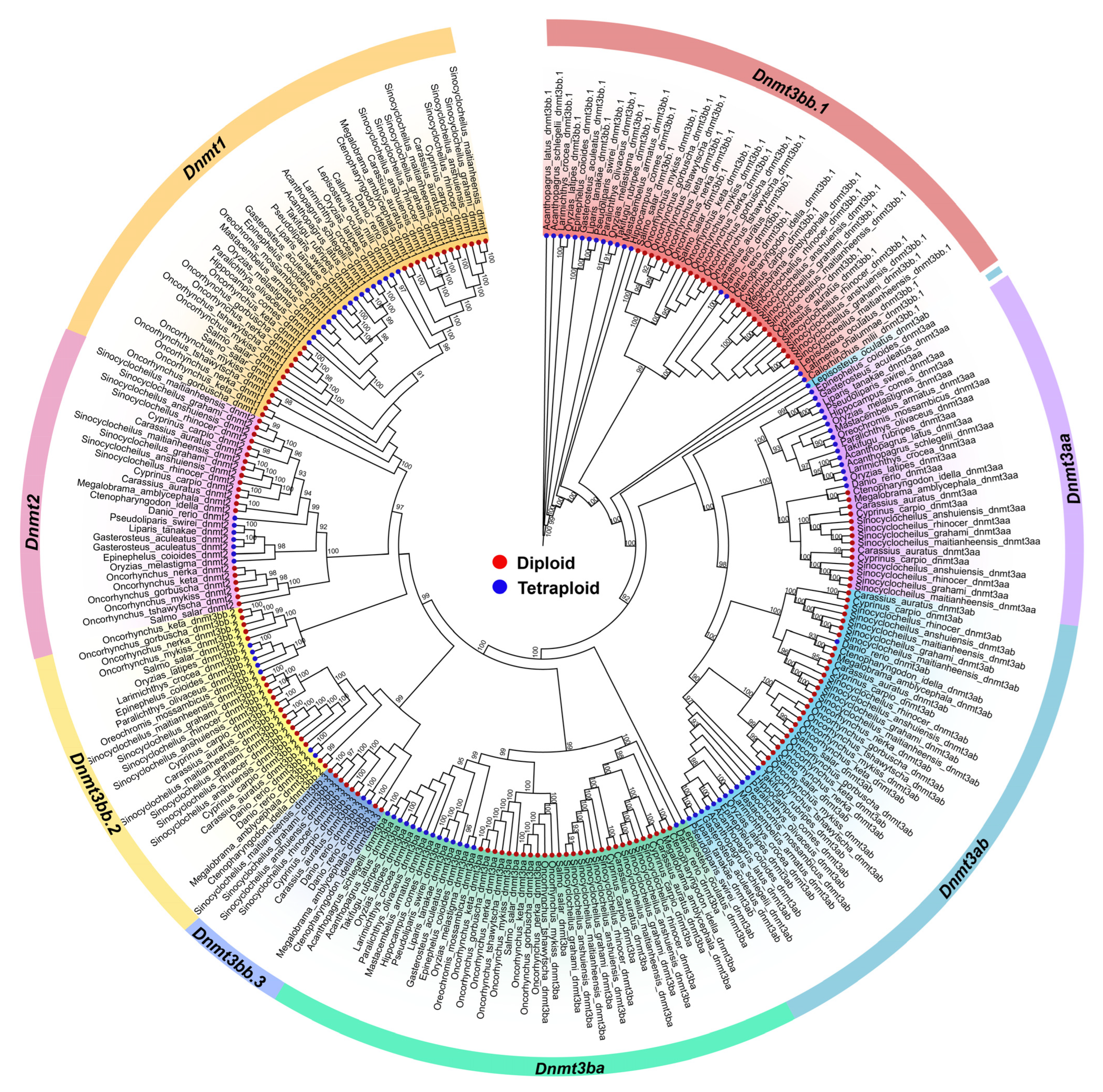
3.2. Phylogenetic Analysis of the Dnmt Family
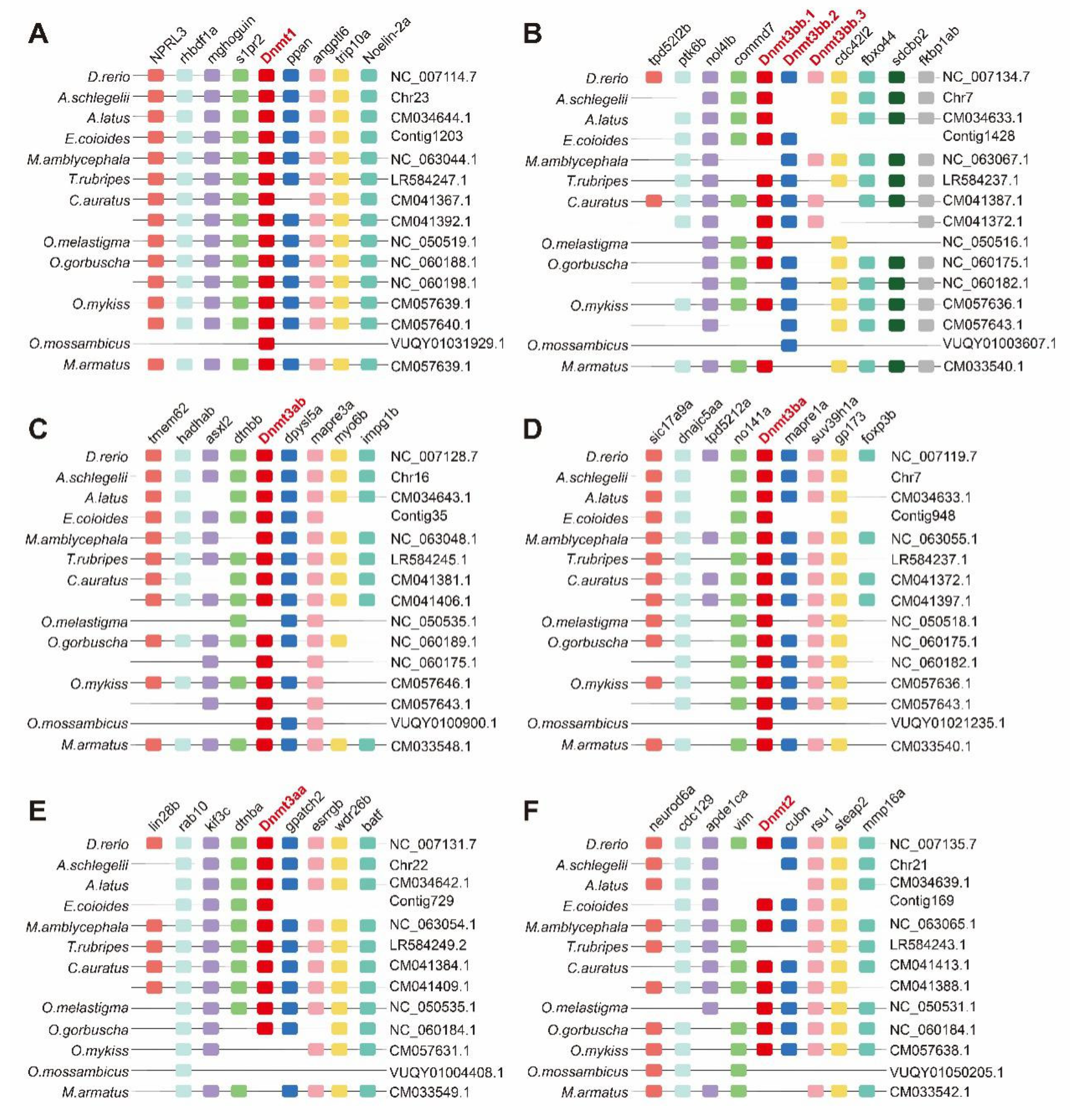
3.3. Synteny Analysis
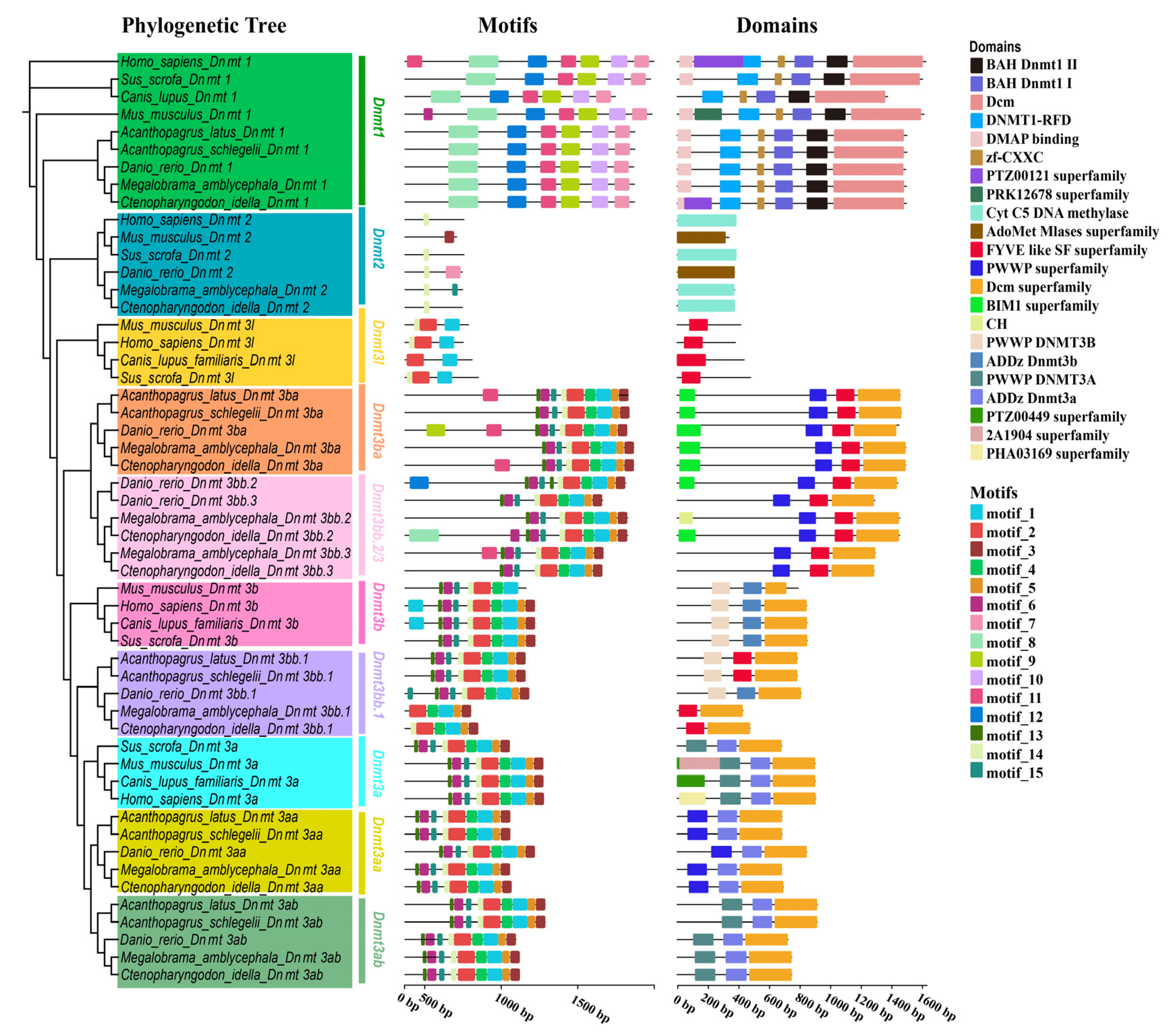
3.4. Structural Analysis of Dnmt Proteins
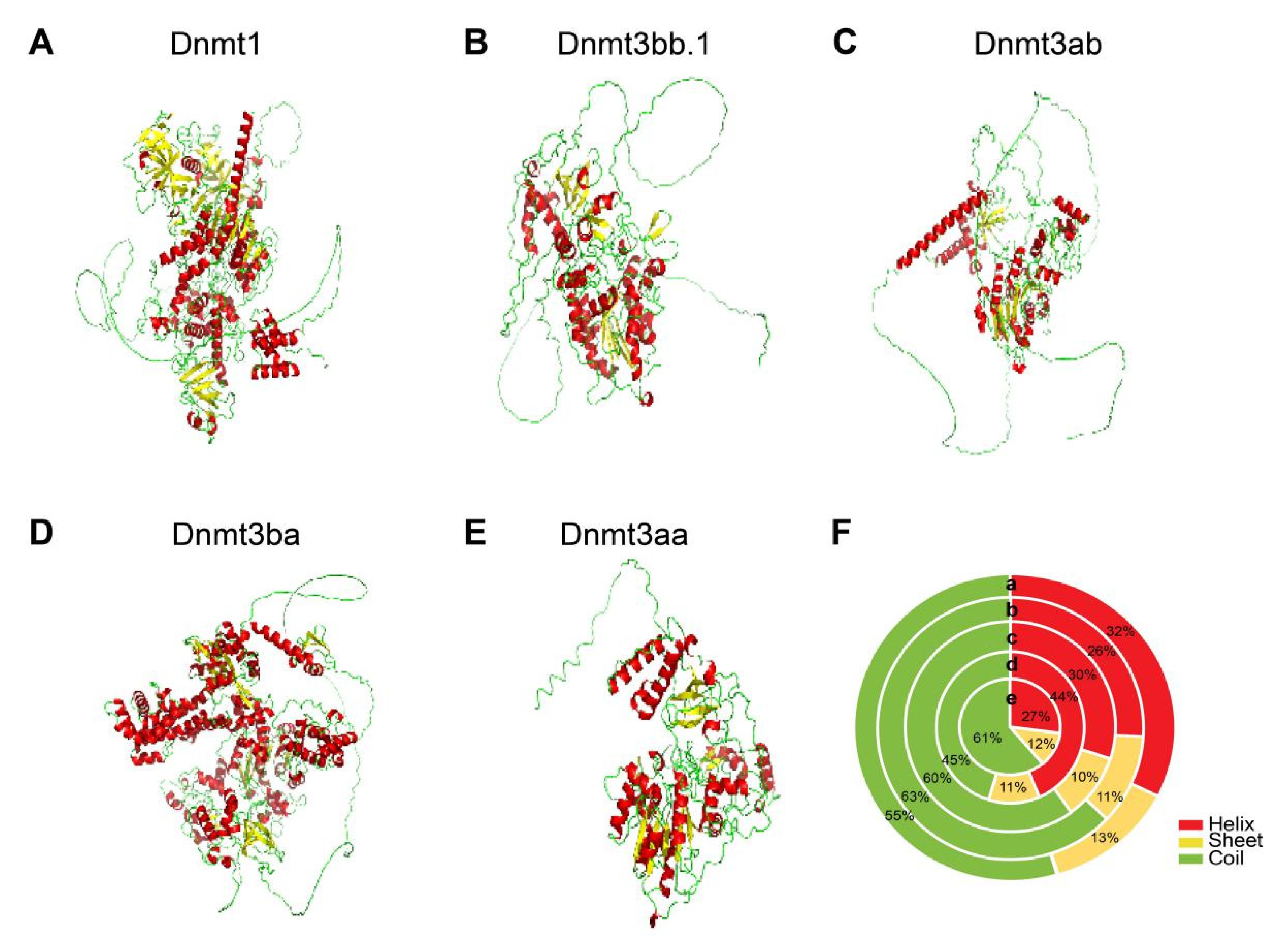
3.5. 3D Structures of Dnmts in Blackhead Seabream
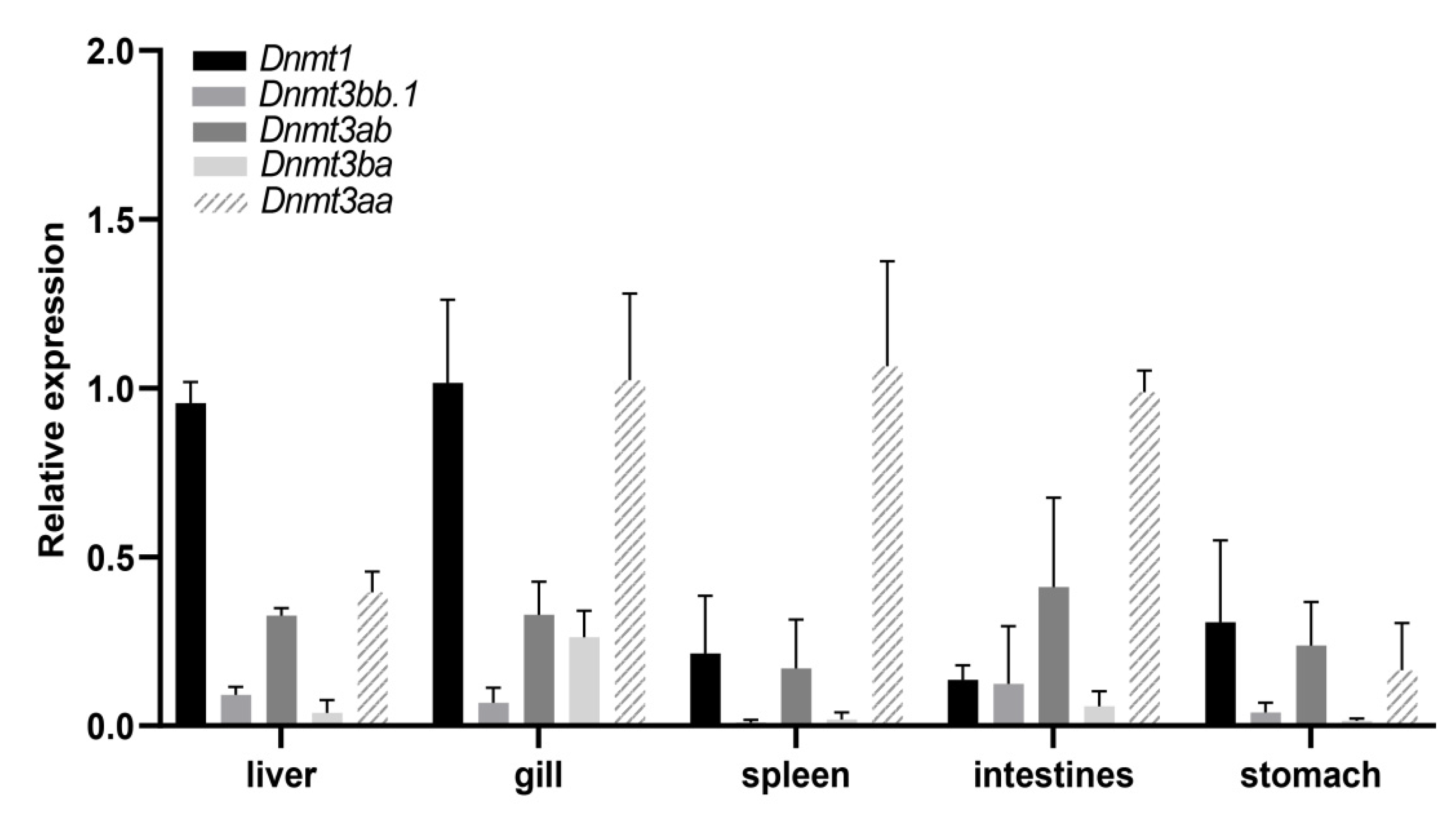
3.6. The Expression Profile of Dnmts in Different Tissues
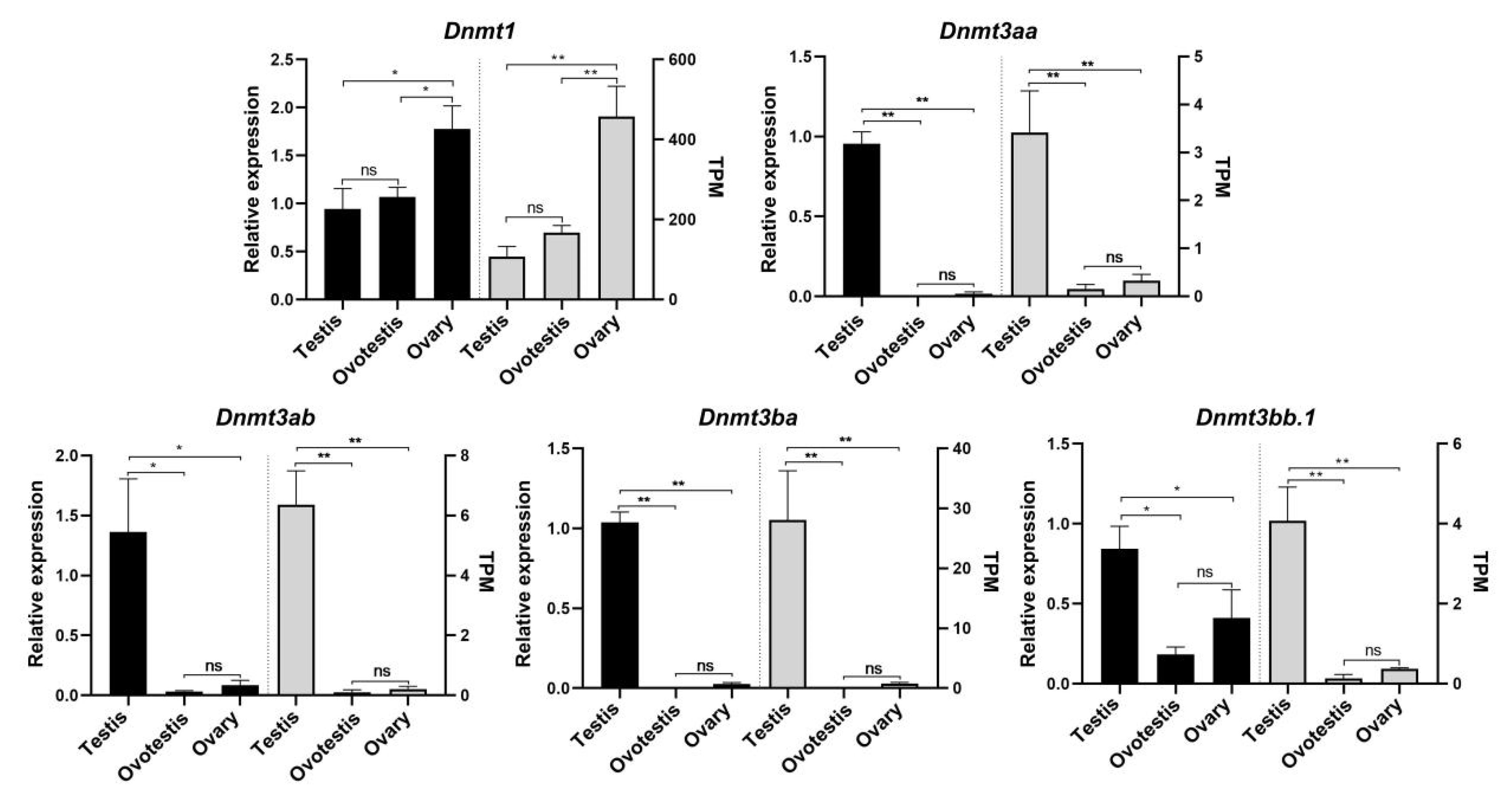
3.7. The Expression Profiles of Dnmts During the Natural Sex Change of Blackhead Seabream
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Schubeler, D. Function and Information Content of DNA Methylation. Nature 2015, 517, 321–326. [Google Scholar] [CrossRef] [PubMed]
- Dura, M.; Teissandier, A.; Armand, M.; Barau, J.; Lapoujade, C.; Fouchet, P.; Bonneville, L.; Schulz, M.; Weber, M.; Baudrin, L.G.; et al. DNMT3A-Dependent DNA Methylation Is Required for Spermatogonial Stem Cells to Commit to Spermatogenesis. Nat. Genet. 2022, 54, 469–480. [Google Scholar] [CrossRef] [PubMed]
- Valdivieso, A.; Anastasiadi, D.; Ribas, L.; Piferrer, F. Development of Epigenetic Biomarkers for the Identification of Sex and Thermal Stress in Fish Using DNA Methylation Analysis and Machine Learning Procedures. Mol. Ecol. Resour. 2023, 23, 453–470. [Google Scholar] [CrossRef]
- Moore, L.D.; Le, T.; Fan, G. DNA Methylation and Its Basic Function. Neuropsychopharmacology 2013, 38, 23–38. [Google Scholar] [CrossRef]
- Dussoix, D.; Arber, W. Host Specificity of DNA Produced by Escherichia coli. II. Control over Acceptance of DNA from Infecting Phage Lambda. J. Mol. Biol. 1962, 5, 37–49. [Google Scholar] [CrossRef]
- Jurkowska, R.Z.; Jurkowski, T.P.; Jeltsch, A. Structure and Function of Mammalian DNA Methyltransferases. ChemBioChem 2011, 12, 206–222. [Google Scholar] [CrossRef] [PubMed]
- Okano, M.; Bell, D.W.; Haber, D.A.; Li, E. DNA Methyltransferases Dnmt3a and Dnmt3b Are Essential for de Novo Methylation and Mammalian Development. Cell 1999, 99, 247–257. [Google Scholar] [CrossRef]
- Kim, B.M.; Mirbahai, L.; Mally, A.; Chipman, J.K.; Rhee, J.S.; Lee, J.S. Correlation between the DNA Methyltransferase (Dnmt) Gene Family and Genome-Wide 5-Methylcytosine (5mC) in Rotifer, Copepod, and Fish. Genes Genom. 2016, 38, 13–23. [Google Scholar] [CrossRef]
- Lyko, F. The DNA Methyltransferase Family: A Versatile Toolkit for Epigenetic Regulation. Nat. Rev. Genet. 2018, 19, 81–92. [Google Scholar] [CrossRef]
- Wolfe, K.H. Yesterday’s Polyploids and the Mystery of Diploidization. Nat. Rev. Genet. 2001, 2, 333–341. [Google Scholar] [CrossRef]
- Ahn, D.; You, K.H.; Kim, C.H. Evolution of the Tbx6/16 Subfamily Genes in Vertebrates: Insights from Zebrafish. Mol. Biol. Evol. 2012, 29, 3959–3983. [Google Scholar] [CrossRef] [PubMed]
- Voldoire, E.; Brunet, F.; Naville, M.; Volff, J.N.; Galiana, D. Expansion by Whole Genome Duplication and Evolution of the Sox Gene Family in Teleost Fish. PLoS ONE 2017, 12, e0180936. [Google Scholar] [CrossRef] [PubMed]
- Ravi, V.; Venkatesh, B. Rapidly Evolving Fish Genomes and Teleost Diversity. Curr. Opin. Genet. Dev. 2008, 18, 544–550. [Google Scholar] [CrossRef] [PubMed]
- Glasauer, S.M.; Neuhauss, S.C. Whole-Genome Duplication in Teleost Fishes and Its Evolutionary Consequences. Mol. Genet. Genom. 2014, 289, 1045–1060. [Google Scholar] [CrossRef]
- Van de Peer, Y.; Maere, S.; Meyer, A. The Evolutionary Significance of Ancient Genome Duplications. Nat. Rev. Genet. 2009, 10, 725–732. [Google Scholar] [CrossRef]
- Ravi, V.; Venkatesh, B. The Divergent Genomes of Teleosts. Annu. Rev. Anim. Biosci. 2018, 6, 47–68. [Google Scholar] [CrossRef]
- Berthelot, C.; Brunet, F.; Chalopin, D.; Juanchich, A.; Bernard, M.; Noel, B.; Bento, P.; Da Silva, C.; Labadie, K.; Alberti, A.; et al. The Rainbow Trout Genome Provides Novel Insights into Evolution after Whole-Genome Duplication in Vertebrates. Nat. Commun. 2014, 5, 3657. [Google Scholar] [CrossRef] [PubMed]
- Xu, P.; Zhang, X.; Wang, X.; Li, J.; Liu, G.; Kuang, Y.; Xu, J.; Zheng, X.; Ren, L.; Wang, G.; et al. Genome Sequence and Genetic Diversity of the Common Carp, Cyprinus carpio. Nat. Genet. 2014, 46, 1212–1219. [Google Scholar] [CrossRef]
- Chen, D.; Zhang, Q.; Tang, W.; Huang, Z.; Wang, G.; Wang, Y.; Shi, J.; Xu, H.; Lin, L.; Li, Z.; et al. The Evolutionary Origin and Domestication History of Goldfish (Carassius auratus). Proc. Natl. Acad. Sci. USA 2020, 117, 29775–29785. [Google Scholar] [CrossRef]
- Guyomard, R.; Boussaha, M.; Krieg, F.; Hervet, C.; Quillet, E. A Synthetic Rainbow Trout Linkage Map Provides New Insights into the Salmonid Whole Genome Duplication and the Conservation of Synteny among Teleosts. BMC Genet. 2012, 13, 15. [Google Scholar] [CrossRef]
- Smith, T.H.; Collins, T.M.; McGowan, R.A. Expression of the Dnmt3 Genes in Zebrafish Development: Similarity to Dnmt3a and Dnmt3b. Dev. Genes Evol. 2011, 220, 347–353. [Google Scholar] [CrossRef] [PubMed]
- Zhou, R.; Shang, R.J.; Gong, D.B.; Xu, X.J.; Tang, Q.R.; Tao, M.; Zhao, R.R.; Liu, S.J. Characterization of De Novo DNA Methyltransferase Dnmt3 Regulating Sterility in Female Allotriploid Fish. Aquaculture 2019, 504, 345–353. [Google Scholar] [CrossRef]
- Sun, L.N.; Jiang, X.L.; Xie, Q.P.; Yuan, J.; Huang, B.F.; Tao, W.J.; Zhou, L.Y.; Nagahama, Y.; Wang, D.S. Transdifferentiation of Differentiated Ovary into Functional Testis by Long-Term Treatment of Aromatase Inhibitor in Nile Tilapia. Endocrinology 2014, 155, 1476–1488. [Google Scholar] [CrossRef] [PubMed]
- Todd, E.V.; Ortega-Recalde, O.; Liu, H.; Lamm, M.S.; Rutherford, K.M.; Cross, H.; Black, M.A.; Kardailsky, O.; Marshall Graves, J.A.; Hore, T.A.; et al. Stress, Novel Sex Genes, and Epigenetic Reprogramming Orchestrate Socially Controlled Sex Change. Sci. Adv. 2019, 5, eaaw7006. [Google Scholar] [CrossRef]
- Warner, R.R.; Swearer, S.E. Social Control of Sex Change in the Bluehead Wrasse, Thalassoma bifasciatum (Pisces: Labridae). Biol. Bull. 1991, 181, 199–204. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Lai, F.; Xiong, J.; Zhu, W.; Yuan, B.; Cheng, H.; Zhou, R. DNA Methylation Modification Is Associated with Gonadal Differentiation in Monopterus albus. Cell Biosci. 2020, 10, 129. [Google Scholar] [CrossRef]
- Zhang, Y.; Sun, X.; Zhang, L.; Zhang, W. Testicular Dnmt3 Expression and Global DNA Methylation Are Down-Regulated by Gonadotropin Releasing Hormones in the Ricefield Eel Monopterus albus. Sci. Rep. 2017, 7, 43158. [Google Scholar] [CrossRef]
- Zhang, K.; Xu, J.; Zhang, Z.; Huang, Y.; Ruan, Z.; Chen, S.; Zhu, F.; You, X.; Jia, C.; Meng, Q.; et al. A Comparative Transcriptomic Study on Developmental Gonads Provides Novel Insights into Sex Change in the Protandrous Black Porgy (Acanthopagrus Schlegelii). Genomics 2019, 111, 277–283. [Google Scholar] [CrossRef]
- Wu, G.C.; Dufour, S.; Chang, C.F. Molecular and Cellular Regulation on Sex Change in Hermaphroditic Fish, with a Special Focus on Protandrous Black Porgy, Acanthopagrus schlegelii. Mol. Cell. Endocrinol. 2021, 520, 111069. [Google Scholar] [CrossRef]
- Zhang, K.; Guo, S.; Yang, S.; Zhou, W.; Wu, J.; Zhang, X.; Shi, Q.; Deng, L. A Telomere-to-Telomere Genome Assembly of the Protandrous Hermaphrodite Blackhead Seabream, Acanthopagrus schlegelii. Sci. Data 2025, 12, 350. [Google Scholar] [CrossRef]
- Birney, E.; Clamp, M.; Durbin, R. GeneWise and Genomewise. Genome Res. 2004, 14, 988–995. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Nguyen, L.T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for Motif Discovery and Searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and Applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef]
- Paysan-Lafosse, T.; Blum, M.; Chuguransky, S.; Grego, T.; Pinto, B.L.; Salazar, G.A.; Bileschi, M.L.; Bork, P.; Bridge, A.; Colwell, L.; et al. InterPro in 2022. Nucleic Acids Res. 2023, 51, D418–D427. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Chen, L.; Li, Q.; Nasif, K.F.A.; Xie, Y.; Deng, B.; Niu, S.; Pouriyeh, S.; Dai, Z.; Chen, J.; Xie, C.Y. AI-Driven Deep Learning Techniques in Protein Structure Prediction. Int. J. Mol. Sci. 2024, 25, 8426. [Google Scholar] [CrossRef]
- Rosignoli, S.; Paiardini, A. Boosting the Full Potential of PyMOL with Structural Biology Plugins. Biomolecules 2022, 12, 1764. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Abera, D.A.; Larbie, C.; Abugri, J.; Ofosu, M.; Mutocheluh, M.; Dongsogo, J. Prevalence and Predictors of Gestational Diabetes Mellitus in Sub-Saharan Africa: A 10-Year Systematic Review. Endocrinol. Diabetes Metab. 2024, 7, e00478. [Google Scholar] [CrossRef]
- Zhang, K.; Chen, M.; He, H.; Kou, H.; Lin, L.; Liang, R. Genome-Wide Identification and Characterization of Toll-like Receptor 5 (TLR5) in Fishes. Front. Genet 2022, 13, 1083578. [Google Scholar] [CrossRef] [PubMed]
- Jurkowska, R.Z.; Jeltsch, A. Enzymology of Mammalian DNA Methyltransferases. Adv. Exp. Med. Biol. 2022, 1389, 69–110. [Google Scholar] [CrossRef] [PubMed]
- Naseeb, S.; Ames, R.M.; Delneri, D.; Lovell, S.C. Rapid Functional and Evolutionary Changes Follow Gene Duplication in Yeast. Proc. Biol. Sci. 2017, 284, 20171393. [Google Scholar] [CrossRef]
- Maere, S.; De Bodt, S.; Raes, J.; Casneuf, T.; Van Montagu, M.; Kuiper, M.; Van de Peer, Y. Modeling Gene and Genome Duplications in Eukaryotes. Proc. Natl. Acad. Sci. USA 2005, 102, 5454–5459. [Google Scholar] [CrossRef]
- Liu, Y.; Li, M.; Zhang, M.; Yang, Z.; Chen, X.; Wu, X. Evolution and Expression Analysis of Carotenoid Cleavage Oxygenase Gene Family in Chinese Mitten Crab Eriocheir sinensis. Int. J. Biol. Macromol. 2024, 257, 128475. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Nielsen, R. Codon-Substitution Models for Detecting Molecular Adaptation at Individual Sites along Specific Lineages. Mol. Biol. Evol. 2002, 19, 908–917. [Google Scholar] [CrossRef]
- Wu, G.C.; Li, H.W.; Huang, C.H.; Lin, H.J.; Lin, C.J.; Chang, C.F. The Testis Is a Primary Factor That Contributes to Epigenetic Modifications in the Ovaries of the Protandrous Black Porgy, Acanthopagrus schlegelii. Biol. Reprod. 2016, 94, 132. [Google Scholar] [CrossRef]
- Wang, F.L.; Yan, L.X.; Shi, H.J.; Liu, X.Y.; Zheng, Q.Y.; Sun, L.N.; Wang, D.S. Genome-Wide Identification, Evolution of DNA Methyltransferases and Their Expression during Gonadal Development in Nile Tilapia. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2018, 226, 73–84. [Google Scholar] [CrossRef]
- Wang, F.; Qin, Z.; Li, Z.; Yang, S.; Gao, T.; Sun, L.; Wang, D. Dnmt3aa but Not Dnmt3ab Is Required for Maintenance of Gametogenesis in Nile Tilapia (Oreochromis niloticus). Int. J. Mol. Sci. 2021, 22, 10170. [Google Scholar] [CrossRef]
- Guo, C.Y.; Tseng, P.W.; Hwang, J.S.; Wu, G.C.; Chang, C.F. Potential Role of DNA Methylation of Cyp19a1a Promoter during Sex Change in Protogynous Orange-Spotted Grouper, Epinephelus coioides. Gen. Comp. Endocrinol. 2021, 311, 113840. [Google Scholar] [CrossRef] [PubMed]
- Hu, Q.; Lian, Z.; Xia, X.; Tian, H.; Li, Z. Integrated Chromatin Accessibility and DNA Methylation Analysis to Reveal the Critical Epigenetic Modification and Regulatory Mechanism in Gonadal Differentiation of the Sequentially Hermaphroditic Fish, Monopterus albus. Biol. Sex Differ. 2022, 13, 73. [Google Scholar] [CrossRef] [PubMed]
- Domingos, J.A.; Budd, A.M.; Banh, Q.Q.; Goldsbury, J.A.; Zenger, K.R.; Jerry, D.R. Sex-Specific Dmrt1 and Cyp19a1 Methylation and Alternative Splicing in Gonads of the Protandrous Hermaphrodite Barramundi. PLoS ONE 2018, 13, e0204182. [Google Scholar] [CrossRef] [PubMed]
| Primer | Forward Primer (5′-3′) | Reverse Primer (5′-3′) |
|---|---|---|
| Dnmt1 | TCCCACAGCACAAGATTACA | AGGAACACCACCATCCAAGC |
| Dnmt3bb.1 | CCGTTCTTCTGGCTGTTCG | TTCTGGGAGGCTGTGATGG |
| Dnmt3ab | ATGTCAGCCTTGAGCACCCG | TCGTCCTCCGCAGCAGATAG |
| Dnmt3ba | CCCTGGCATGAACAGACCC | CCATCTTGCCCTGCCGTAT |
| Dnmt3aa | CTCCTGCGGAAGCCTCAAT | CTGGTAGCCGTCATCGTCAT |
| β-actin | ACAGGGAGAAGATGACCCAGAT | CACCGGAGTCCATGACGATA |
| Protein | Chr | Number of Amino Acids | Molecular Weight | Theoretical pI | Instability Index | Aliphatic Index | GRAVY | Predicted Location |
|---|---|---|---|---|---|---|---|---|
| Dnmt1 | 23 | 1508 | 170,325.94 | 5.81 | 47.95 | 65.79 | −0.701 | Nucleus |
| Dnmt3bb.1 | 7 | 792 | 89,338.44 | 7.49 | 46.73 | 70.29 | −0.509 | Nucleus |
| Dnmt3ab | 16 | 921 | 104,840.84 | 7.24 | 61.31 | 63.6 | −0.72 | Nucleus, cytoplasm |
| Dnmt3ba | 7 | 1471 | 166,048.65 | 6.87 | 44.89 | 76.51 | −0.404 | Nucleus |
| Dnmt3aa | 22 | 691 | 78,163.04 | 6.05 | 51.93 | 69.29 | −0.431 | Nucleus, cytoplasm |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guo, S.; Duan, B.; Chen, J.; Cui, M.; You, C.; Wei, H.; Huang, X.; Deng, L.; Zhang, K. Genome-Wide Identification of DNA Methyltransferases (Dnmts) in Fish and Its Potential Roles During Sex Change in Blackhead Seabream. Biomolecules 2025, 15, 896. https://doi.org/10.3390/biom15060896
Guo S, Duan B, Chen J, Cui M, You C, Wei H, Huang X, Deng L, Zhang K. Genome-Wide Identification of DNA Methyltransferases (Dnmts) in Fish and Its Potential Roles During Sex Change in Blackhead Seabream. Biomolecules. 2025; 15(6):896. https://doi.org/10.3390/biom15060896
Chicago/Turabian StyleGuo, Sixin, Binwei Duan, Jianchao Chen, Mingyang Cui, Canbei You, Hanyin Wei, Xiazi Huang, Li Deng, and Kai Zhang. 2025. "Genome-Wide Identification of DNA Methyltransferases (Dnmts) in Fish and Its Potential Roles During Sex Change in Blackhead Seabream" Biomolecules 15, no. 6: 896. https://doi.org/10.3390/biom15060896
APA StyleGuo, S., Duan, B., Chen, J., Cui, M., You, C., Wei, H., Huang, X., Deng, L., & Zhang, K. (2025). Genome-Wide Identification of DNA Methyltransferases (Dnmts) in Fish and Its Potential Roles During Sex Change in Blackhead Seabream. Biomolecules, 15(6), 896. https://doi.org/10.3390/biom15060896






