LncRNA–Protein Interactions: A Key to Deciphering LncRNA Mechanisms
Abstract
1. Introduction
2. Mechanisms of LncRNA–Protein Interactions
3. Biological Consequences of LPIs
3.1. Subcellular Localization and Functional Compartmentalization
3.2. Regulation of Gene Expression
3.3. Modulation of Cellular Processes
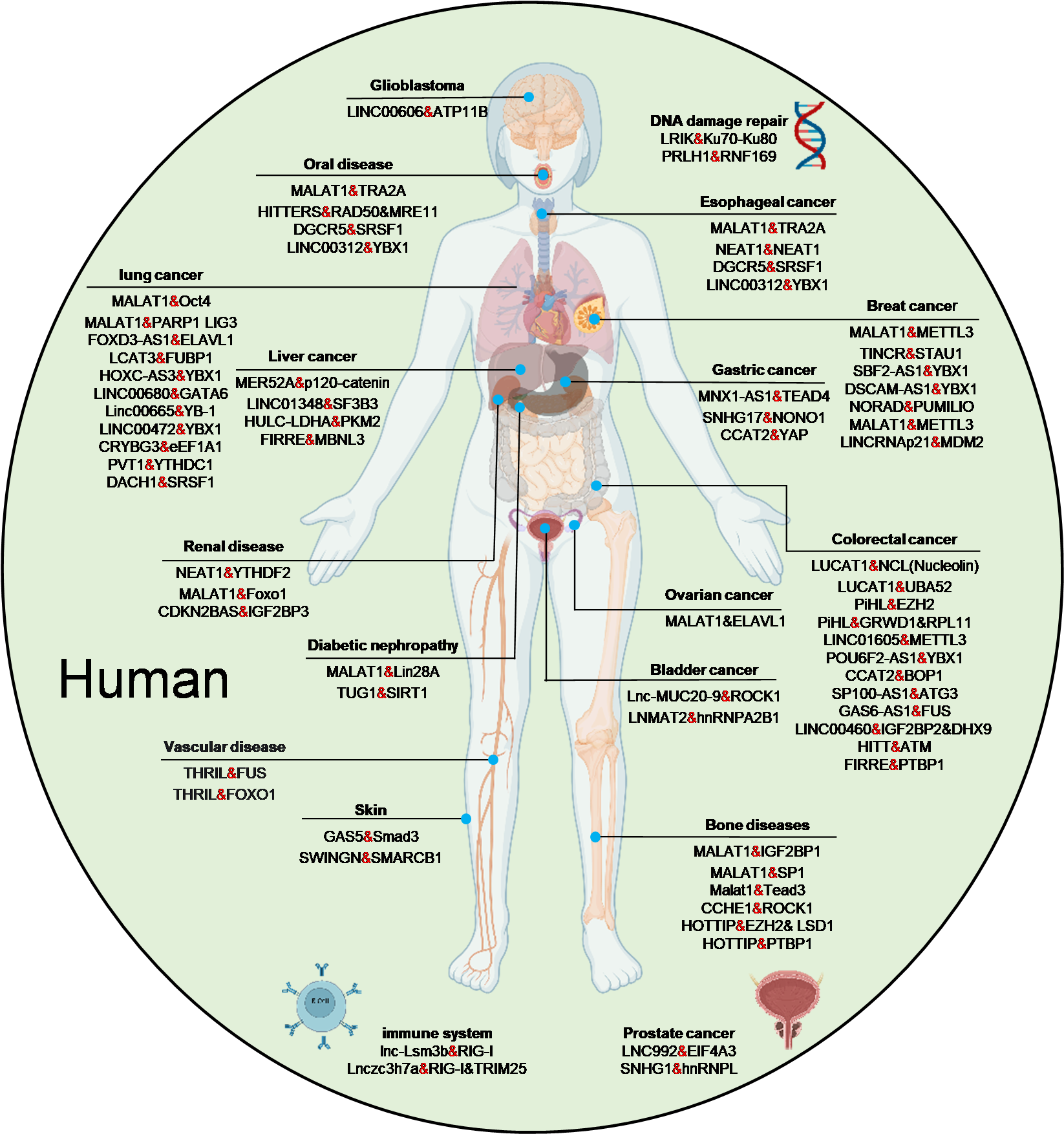
3.4. Implications in Human Diseases
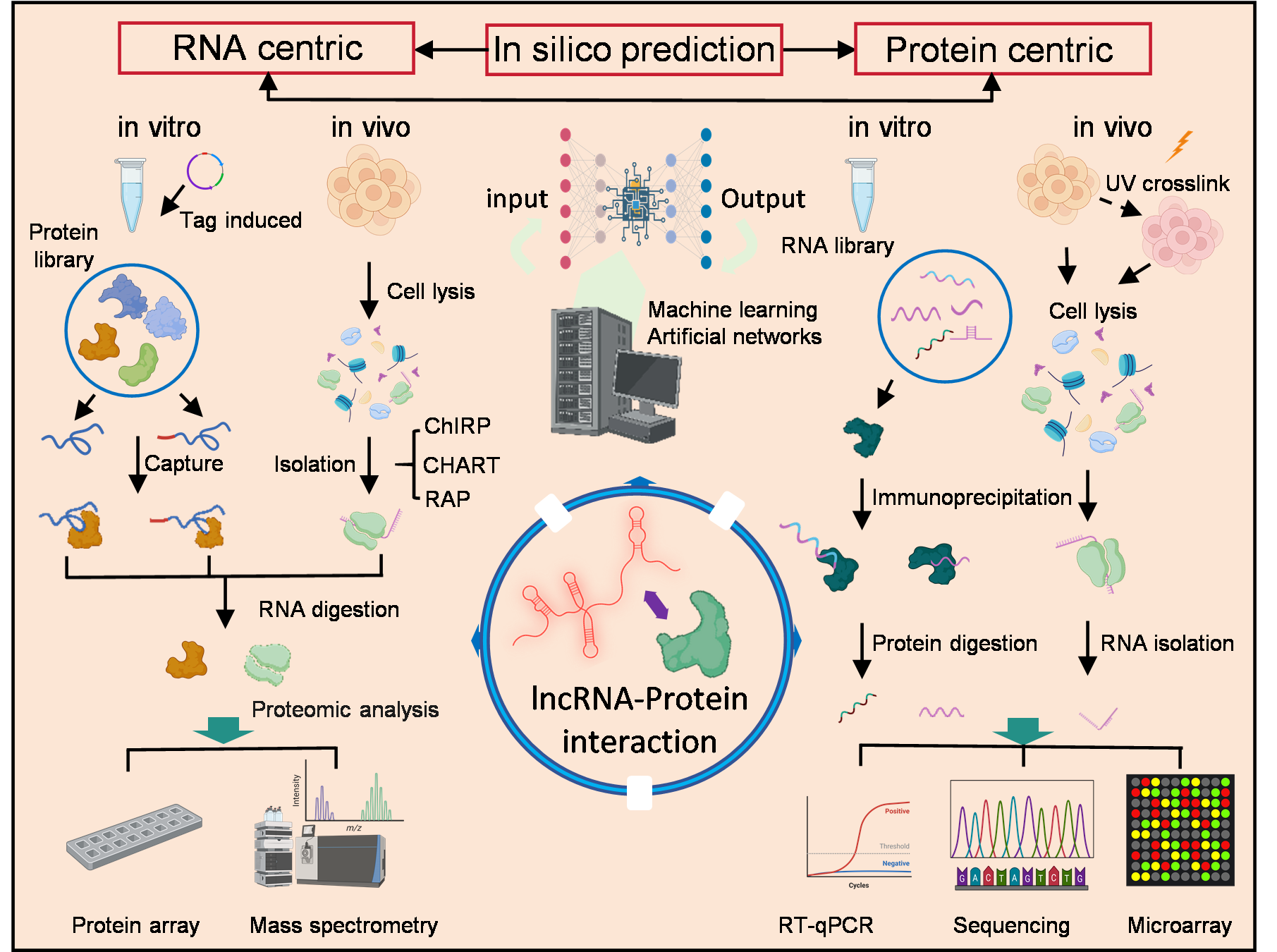
4. Experimental Approaches for Studying LPIs
4.1. Identification of LPIs
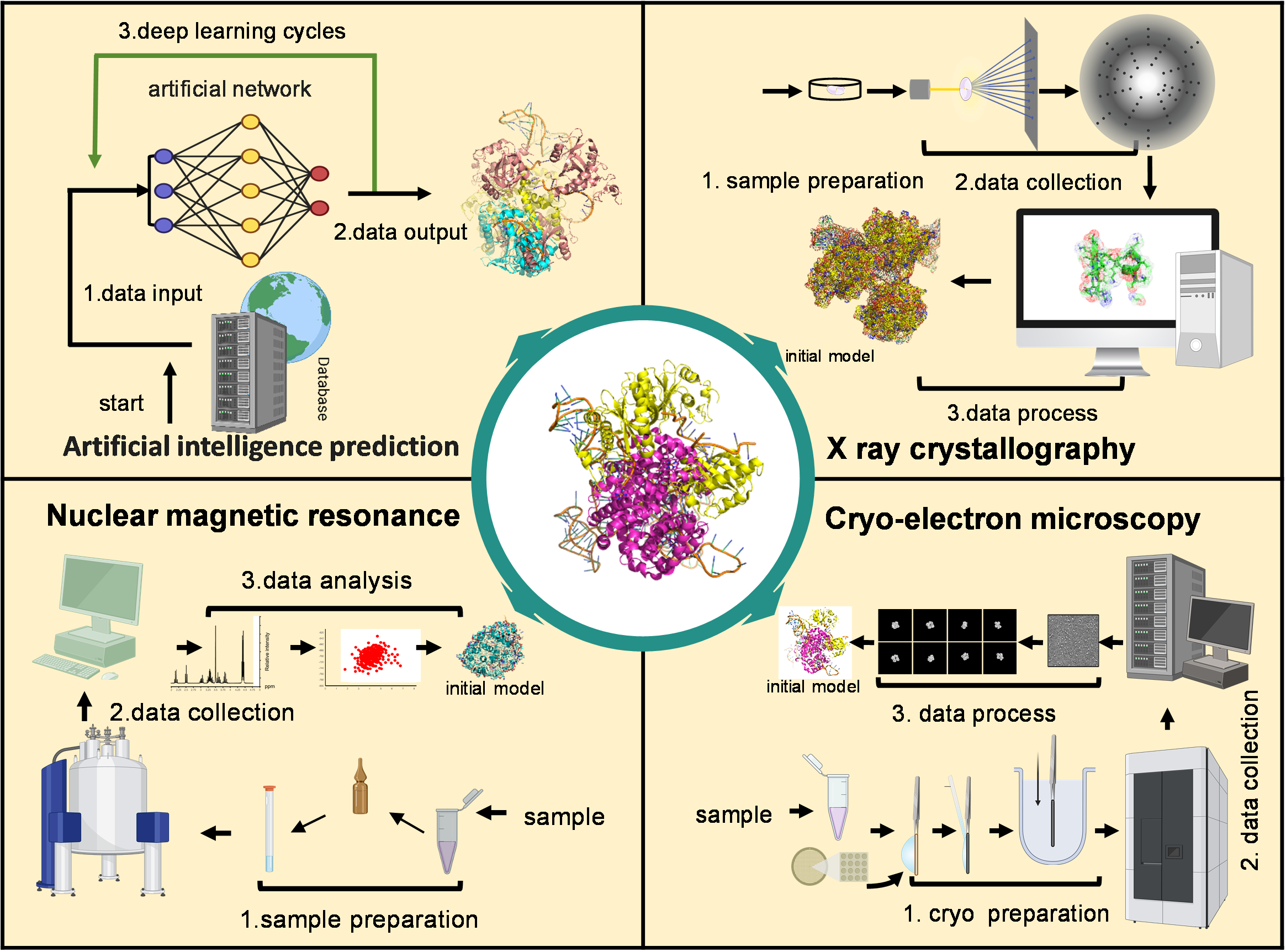
4.2. Structural Insights into LncRNA–Protein Binding
4.2.1. Probing LncRNA Secondary Structure
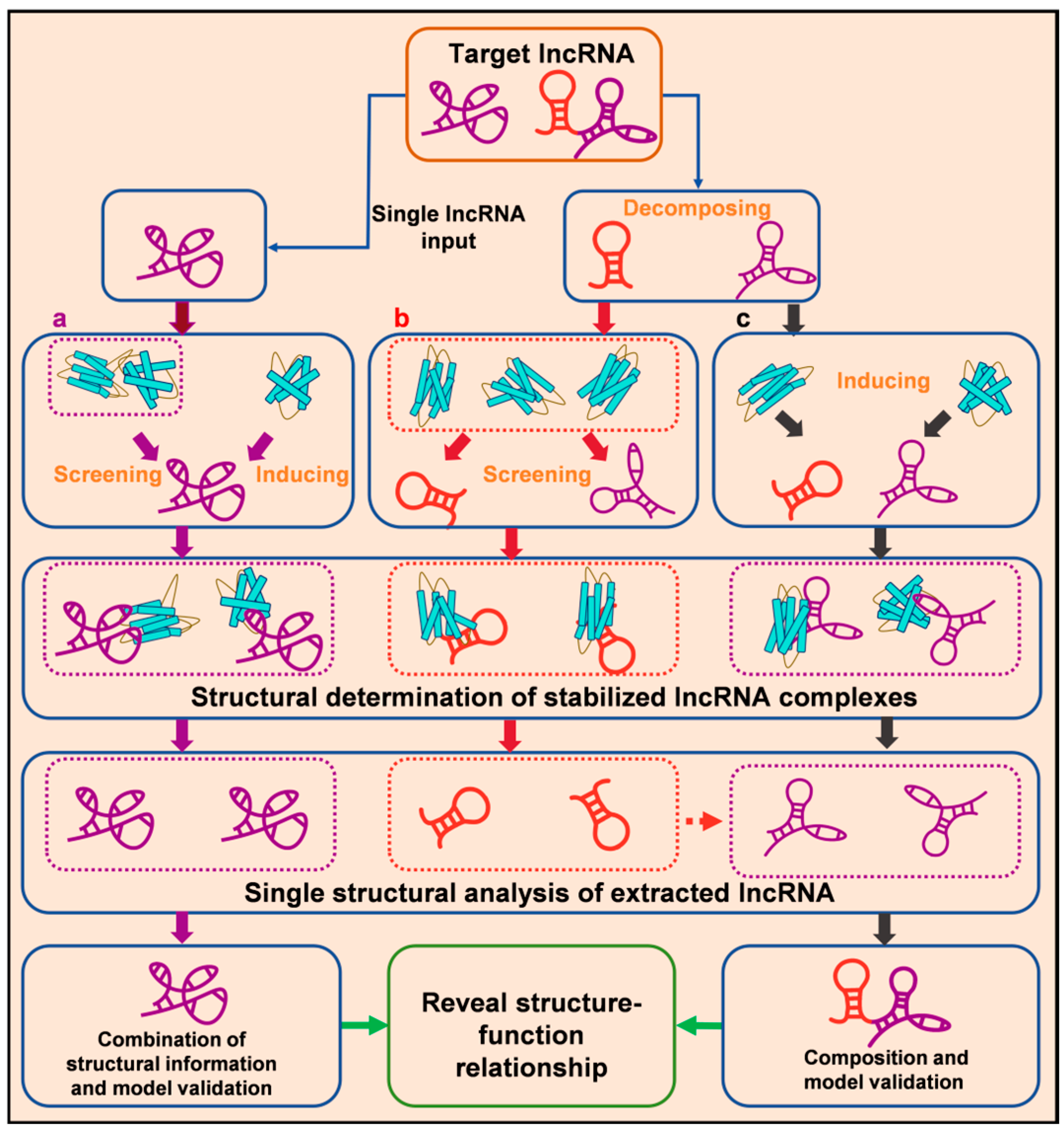
4.2.2. Analysis of LncRNA Tertiary Structure
Single LncRNA–Protein Complex Structural Analysis
| Tool Name | RNA Struct. | lPI Struct. | Key Features | Suitability | Ref. |
|---|---|---|---|---|---|
| RNA-Composer | Yes | No | Fast conversion from 2D to 3D structure | Suitable for short lncRNA fragments with known secondary structure, web-based and fast | [257] |
| 3dRNA v2.0 | Yes | Limited | Assembles 3D structure from motifs and 2D input | Suitable for moderate-length lncRNAs, accuracy depends on quality of 2D input | [258] |
| P-FARFAR2 | Yes | Limited | Parallelized enhancement of FARFAR2, multithreaded greedy sampling for low-energy structure assembly | Suitable for high-precision RNA 3D modeling, better efficiency and accuracy over FARFAR2, especially for complex/larger RNAs | [259] |
| FARNA/FARFAR2 | Yes | Limited | High-precision folding, but computationally intensive | Best for small lncRNA segments, high-quality modeling with heavy compute requirements | [260] |
| RNA-MSM | Indirectly | No | MSA-based RNA language model, captures structural information via attention maps and embeddings | Outperforms BERT-like RNA models in predicting base-pairing and solvent accessibility, suitable for structure-function tasks | [261] |
| MoEFold2D | No (2D) | No | Mixture-of-experts model combining DL and physics-based predictions, auto-detects in/out-of-distribution sequences | Offers high accuracy for in-distribution and robust predictions, useful when training/test domains differ | [262] |
| RNA-LLM Folding | No | No | Evaluates multiple pretrained RNA language models for RNA secondary structure prediction | Provides benchmark datasets and unified experimental framework, shows LLMs can improve prediction in high-homology regions, with limitations in generalization | [263] |
| RPI-SE | No | Yes | Ensemble model using PWM, Legendre moments (proteins), and k-mer features (ncRNAs), high accuracy and robustness | Effective computational predictor for ncRNA-protein interactions, suitable for accelerating interaction studies with sequence data only | [264] |
| SPOT-RNA2 | Yes | No | High-accuracy secondary structure prediction, supports pseudoknots | Great for generating input structures for 3D modeling of lncRNA | [261] |
| MXfold2 | No | No | Combines neural networks and thermodynamics for better generalization | Fast and accurate, ideal for batch secondary structure prediction | [265] |
| RNAstructure | No | No | Supports SHAPE data and pseudoknots, energy minimization | Stable and widely used, excellent for combining with experimental data | [266] |
| IntaRNA | No | Yes | Predicts RNA–protein interaction regions | Useful for predicting lncRNA binding regions with proteins | [267] |
| catRAPID | No | Yes | Sequence-based scoring of RNA–protein interaction strength | Useful for large-scale screening of LPIs | [196] |
| RPI-EDLCN | No | Yes | Ensemble deep learning framework based on CapsuleNet, integrates sequence, secondary structure, motif, and physicochemical features, used for feature extraction | High accuracy in predicting ncRNA-protein interactions across multiple datasets, effective across species | [268] |
| LPI-MFF | No | Yes | Multi-source information fusion (PPI, sequence, structure, physico-chemical), feature selection by random forest | High accuracy and robustness across multiple datasets, good generalization for RPI prediction | [269] |
| DRPScore | Yes | Yes | Deep learning model for identifying native-like RNA-protein complex structures across docking types (bound/unbound) | Suitable for RNA-protein complex modeling, high accuracy in selecting native-like structures even under flexible docking scenarios | [25] |
| AlphaFold3 | Yes | Yes | Accurate protein 3D structure prediction, limited in RNA/RNA-protein complex | Provides protein structure for docking; essential if structure is unknown | [254] |
| RoseTTAFold | Yes | Yes | Deep learning-based 3D structure prediction for RNA-protein complexes, with confidence scores | Suitable for predicting structures of complexes without known homology | [250] |
Integrated Structural Approaches
5. Development of Novel Biomedical Interventions Targeting LPIs
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mattick, J.S.; Amaral, P.P.; Carninci, P.; Carpenter, S.; Chang, H.Y.; Chen, L.-L.; Chen, R.; Dean, C.; Dinger, M.E.; Fitzgerald, K.A.; et al. Long Non-Coding RNAs: Definitions, Functions, Challenges and Recommendations. Nat. Rev. Mol. Cell Biol. 2023, 24, 430–447. [Google Scholar] [CrossRef]
- Wang, Z.; Xie, C.; Chen, X. Diagnostic and Therapeutic Role of Non-Coding RNAs Regulating Programmed Cell Death in Melanoma. Front. Oncol. 2024, 14, 1476684. [Google Scholar] [CrossRef]
- Ponting, C.P.; Haerty, W. Genome-Wide Analysis of Human Long Noncoding RNAs: A Provocative Review. Annu. Rev. Genom. Hum. Genet. 2022, 23, 153–172. [Google Scholar] [CrossRef]
- Chen, L.-L.; Kim, V.N. Small and Long Non-Coding RNAs: Past, Present, and Future. Cell 2024, 187, 6451–6485. [Google Scholar] [CrossRef]
- Rocca, R.; Grillone, K.; Citriniti, E.L.; Gualtieri, G.; Artese, A.; Tagliaferri, P.; Tassone, P.; Alcaro, S. Targeting Non-Coding RNAs: Perspectives and Challenges of in-Silico Approaches. Eur. J. Med. Chem. 2023, 261, 115850. [Google Scholar] [CrossRef]
- Constanty, F.; Shkumatava, A. lncRNAs in Development and Differentiation: From Sequence Motifs to Functional Characterization. Development 2021, 148, dev182741. [Google Scholar] [CrossRef]
- Ma, H.; Jia, X.; Zhang, K.; Su, Z. Cryo-EM Advances in RNA Structure Determination. Sig. Transduct. Target Ther. 2022, 7, 58. [Google Scholar] [CrossRef]
- Deng, J.; Fang, X.; Huang, L.; Li, S.; Xu, L.; Ye, K.; Zhang, J.; Zhang, K.; Zhang, Q.C. RNA Structure Determination: From 2D to 3D. Fundam. Res. 2023, 3, 727–737. [Google Scholar] [CrossRef]
- Alvarez-Dominguez, J.R.; Lodish, H.F. Emerging Mechanisms of Long Noncoding RNA Function during Normal and Malignant Hematopoiesis. Blood 2017, 130, 1965–1975. [Google Scholar] [CrossRef]
- Ribeiro, D.M.; Zanzoni, A.; Cipriano, A.; Delli Ponti, R.; Spinelli, L.; Ballarino, M.; Bozzoni, I.; Tartaglia, G.G.; Brun, C. Protein Complex Scaffolding Predicted as a Prevalent Function of Long Non-Coding RNAs. Nucleic Acids Res. 2018, 46, 917–928. [Google Scholar] [CrossRef]
- Chu, C.; Spitale, R.C.; Chang, H.Y. Technologies to Probe Functions and Mechanisms of Long Noncoding RNAs. Nat. Struct. Mol. Biol. 2015, 22, 29–35. [Google Scholar] [CrossRef]
- Graf, J.; Kretz, M. From Structure to Function: Route to Understanding lncRNA Mechanism. BioEssays 2020, 42, 2000027. [Google Scholar] [CrossRef]
- Statello, L.; Guo, C.-J.; Chen, L.-L.; Huarte, M. Author Correction: Gene Regulation by Long Non-Coding RNAs and Its Biological Functions. Nat. Rev. Mol. Cell Biol. 2021, 22, 159. [Google Scholar] [CrossRef]
- Shaath, H.; Vishnubalaji, R.; Elango, R.; Kardousha, A.; Islam, Z.; Qureshi, R.; Alam, T.; Kolatkar, P.R.; Alajez, N.M. Long Non-Coding RNA and RNA-Binding Protein Interactions in Cancer: Experimental and Machine Learning Approaches. Semin. Cancer Biol. 2022, 86, 325–345. [Google Scholar] [CrossRef]
- Nadhan, R.; Isidoro, C.; Song, Y.S.; Dhanasekaran, D.N. Signaling by LncRNAs: Structure, Cellular Homeostasis, and Disease Pathology. Cells 2022, 11, 2517. [Google Scholar] [CrossRef]
- Yadav, V.K.; Kumar, A.; Tripathi, P.P.; Gupta, J. Long Noncoding RNAs in Intestinal Homeostasis, Regeneration, and Cancer. J. Cell. Physiol. 2021, 236, 7801–7813. [Google Scholar] [CrossRef]
- He, D.; Zheng, J.; Hu, J.; Chen, J.; Wei, X. Long Non-Coding RNAs and Pyroptosis. Clin. Chim. Acta 2020, 504, 201–208. [Google Scholar] [CrossRef]
- Gerber, A.P. RNA-Centric Approaches to Profile the RNA–Protein Interaction Landscape on Selected RNAs. ncRNA 2021, 7, 11. [Google Scholar] [CrossRef]
- Honorato, R.V.; Trellet, M.E.; Jiménez-García, B.; Schaarschmidt, J.J.; Giulini, M.; Reys, V.; Koukos, P.I.; Rodrigues, J.P.G.L.M.; Karaca, E.; van Zundert, G.C.P.; et al. The HADDOCK2.4 Web Server for Integrative Modeling of Biomolecular Complexes. Nat. Protoc. 2024, 19, 3219–3241. [Google Scholar] [CrossRef]
- Zheng, J.; Hong, X.; Xie, J.; Tong, X.; Liu, S. P3DOCK: A Protein–RNA Docking Webserver Based on Template-Based and Template-Free Docking. Bioinformatics 2020, 36, 96–103. [Google Scholar] [CrossRef]
- Caudron-Herger, M.; Jansen, R.E.; Wassmer, E.; Diederichs, S. RBP2GO: A Comprehensive Pan-Species Database on RNA-Binding Proteins, Their Interactions and Functions. Nucleic Acids Res. 2021, 49, D425–D436. [Google Scholar] [CrossRef]
- Chojnowski, G.; Waleń, T.; Bujnicki, J.M. RNA Bricks—A Database of RNA 3D Motifs and Their Interactions. Nucl. Acids Res. 2014, 42, D123–D131. [Google Scholar] [CrossRef]
- Kirsanov, D.D.; Zanegina, O.N.; Aksianov, E.A.; Spirin, S.A.; Karyagina, A.S.; Alexeevski, A.V. NPIDB: Nucleic Acid—Protein Interaction Database. Nucleic Acids Res. 2012, 41, D517–D523. [Google Scholar] [CrossRef]
- Philip, M.; Chen, T.; Tyagi, S. A Survey of Current Resources to Study lncRNA-Protein Interactions. ncRNA 2021, 7, 33. [Google Scholar] [CrossRef]
- Zeng, C.; Jian, Y.; Vosoughi, S.; Zeng, C.; Zhao, Y. Evaluating Native-like Structures of RNA-Protein Complexes through the Deep Learning Method. Nat. Commun. 2023, 14, 1060. [Google Scholar] [CrossRef]
- Yang, Y.; Wen, L.; Zhu, H. Unveiling the Hidden Function of Long Non-Coding RNA by Identifying Its Major Partner-Protein. Cell Biosci. 2015, 5, 59. [Google Scholar] [CrossRef]
- Kaczynski, T.J.; Au, E.D.; Farkas, M.H. Exploring the lncRNA Localization Landscape within the Retinal Pigment Epithelium under Normal and Stress Conditions. BMC Genom. 2022, 23, 539. [Google Scholar] [CrossRef]
- Wang, K.C.; Chang, H.Y. Molecular Mechanisms of Long Noncoding RNAs. Mol. Cell 2011, 43, 904–914. [Google Scholar] [CrossRef]
- Ahmad, P.; Bensaoud, C.; Mekki, I.; Rehman, M.; Kotsyfakis, M. Long Non-Coding RNAs and Their Potential Roles in the Vector–Host–Pathogen Triad. Life 2021, 11, 56. [Google Scholar] [CrossRef]
- Kopp, F.; Mendell, J.T. Functional Classification and Experimental Dissection of Long Noncoding RNAs. Cell 2018, 172, 393–407. [Google Scholar] [CrossRef]
- Palihati, M.; Saitoh, N. RNA in Chromatin Organization and Nuclear Architecture. Curr. Opin. Genet. Dev. 2024, 86, 102176. [Google Scholar] [CrossRef]
- Bridges, M.C.; Daulagala, A.C.; Kourtidis, A. LNCcation: lncRNA Localization and Function. J. Cell Biol. 2021, 220, e202009045. [Google Scholar] [CrossRef]
- Poloni, J.d.F.; Oliveira, F.H.S.d.; Feltes, B.C. Localization Is the Key to Action: Regulatory Peculiarities of lncRNAs. Front. Genet. 2024, 15, 1478352. [Google Scholar] [CrossRef]
- Li, Z.; Tian, J.-M.; Chu, Y.; Zhu, H.-Y.; Wang, J.-J.; Huang, J. Long Non-Coding RNA PVT1 (PVT1) Affects the Expression of CCND1 and Promotes Doxorubicin Resistance in Osteosarcoma Cells. J. Bone Oncol. 2023, 43, 100512. [Google Scholar] [CrossRef]
- Zuckerman, B.; Ulitsky, I. Predictive Models of Subcellular Localization of Long RNAs. RNA 2019, 25, 557–572. [Google Scholar] [CrossRef]
- Leucci, E.; Vendramin, R.; Spinazzi, M.; Laurette, P.; Fiers, M.; Wouters, J.; Radaelli, E.; Eyckerman, S.; Leonelli, C.; Vanderheyden, K.; et al. Melanoma Addiction to the Long Non-Coding RNA SAMMSON. Nature 2016, 531, 518–522. [Google Scholar] [CrossRef]
- Aznaourova, M.; Janga, H.; Sefried, S.; Kaufmann, A.; Dorna, J.; Volkers, S.M.; Georg, P.; Lechner, M.; Hoppe, J.; Dökel, S.; et al. Noncoding RNA MaIL1 Is an Integral Component of the TLR4–TRIF Pathway. Proc. Natl. Acad. Sci. USA 2020, 117, 9042–9053. [Google Scholar] [CrossRef]
- Tran, K.-V.; Brown, E.L.; DeSouza, T.; Jespersen, N.Z.; Nandrup-Bus, C.; Yang, Q.; Yang, Z.; Desai, A.; Min, S.Y.; Rojas-Rodriguez, R.; et al. Human Thermogenic Adipocyte Regulation by the Long Noncoding RNA LINC00473. Nat. Metab. 2020, 2, 397–412. [Google Scholar] [CrossRef]
- Deng, X.; Xiong, W.; Jiang, X.; Zhang, S.; Li, Z.; Zhou, Y.; Xiang, B.; Zhou, M.; Li, X.; Li, G.; et al. LncRNA LINC00472 Regulates Cell Stiffness and Inhibits the Migration and Invasion of Lung Adenocarcinoma by Binding to YBX1. Cell Death Dis. 2020, 11, 945. [Google Scholar] [CrossRef]
- Zhu, Y.; Jin, L.; Shi, R.; Li, J.; Wang, Y.; Zhang, L.; Liang, C.-Z.; Narayana, V.K.; De Souza, D.P.; Thorne, R.F.; et al. The Long Noncoding RNA glycoLINC Assembles a Lower Glycolytic Metabolon to Promote Glycolysis. Mol. Cell 2022, 82, 542–554.e6. [Google Scholar] [CrossRef]
- Wu, M.; Xu, G.; Han, C.; Luan, P.-F.; Xing, Y.-H.; Nan, F.; Yang, L.-Z.; Huang, Y.; Yang, Z.-H.; Shan, L.; et al. lncRNA SLERT Controls Phase Separation of FC/DFCs to Facilitate Pol I Transcription. Science 2021, 373, 547–555. [Google Scholar] [CrossRef]
- Sledziowska, M.; Winczura, K.; Jones, M.; Almaghrabi, R.; Mischo, H.; Hebenstreit, D.; Garcia, P.; Grzechnik, P. Non-Coding RNAs Associated with Prader–Willi Syndrome Regulate Transcription of Neurodevelopmental Genes in Human Induced Pluripotent Stem Cells. Hum. Mol. Genet. 2023, 32, 608–620. [Google Scholar] [CrossRef]
- Li, R.; Fox, A.H. SPArking Interest in the Long Noncoding RNA World: A New Class of 5′ SnoRNA-Stabilized LncRNA That Influences Alternative Splicing. Mol. Cell 2016, 64, 435–437. [Google Scholar] [CrossRef]
- Wu, H.; Yin, Q.-F.; Luo, Z.; Yao, R.-W.; Zheng, C.-C.; Zhang, J.; Xiang, J.-F.; Yang, L.; Chen, L.-L. Unusual Processing Generates SPA LncRNAs That Sequester Multiple RNA Binding Proteins. Mol. Cell 2016, 64, 534–548. [Google Scholar] [CrossRef]
- Crespi, M. Long Non-Coding RNAs Reveal New Regulatory Mechanisms Controlling Gene Expression. Comptes Rendus Biol. 2023, 345, 15–39. [Google Scholar] [CrossRef]
- Zhao, C.; Xie, W.; Zhu, H.; Zhao, M.; Liu, W.; Wu, Z.; Wang, L.; Zhu, B.; Li, S.; Zhou, Y.; et al. LncRNAs and Their RBPs: How to Influence the Fate of Stem Cells? Stem. Cell Res. Ther. 2022, 13, 175. [Google Scholar] [CrossRef]
- Dodel, M.; Guiducci, G.; Dermit, M.; Krishnamurthy, S.; Alard, E.L.; Capraro, F.; Rekad, Z.; Stojic, L.; Mardakheh, F.K. TREX Reveals Proteins That Bind to Specific RNA Regions in Living Cells. Nat. Methods 2024, 21, 423–434. [Google Scholar] [CrossRef]
- Shaw, A.; Gullerova, M. Home and Away: The Role of Non-Coding RNA in Intracellular and Intercellular DNA Damage Response. Genes 2021, 12, 1475. [Google Scholar] [CrossRef]
- Mangoni, D.; Simi, A.; Lau, P.; Armaos, A.; Ansaloni, F.; Codino, A.; Damiani, D.; Floreani, L.; Di Carlo, V.; Vozzi, D.; et al. LINE-1 Regulates Cortical Development by Acting as Long Non-Coding RNAs. Nat. Commun. 2023, 14, 4974. [Google Scholar] [CrossRef]
- Poltronieri, P. Regulatory RNAs: Role as Scaffolds Assembling Protein Complexes and Their Epigenetic Deregulation. Explor. Target. Anti-Tumor Ther. 2024, 5, 841–876. [Google Scholar] [CrossRef]
- Bai, J.-Y.; Jin, B.; Ma, J.-B.; Liu, T.-J.; Yang, C.; Chong, Y.; Wang, X.; He, D.; Guo, P. HOTAIR and Androgen Receptor Synergistically Increase GLI2 Transcription to Promote Tumor Angiogenesis and Cancer Stemness in Renal Cell Carcinoma. Cancer Lett. 2021, 498, 70–79. [Google Scholar] [CrossRef]
- Kadota, S.; Ou, J.; Shi, Y.; Lee, J.T.; Sun, J.; Yildirim, E. Nucleoporin 153 Links Nuclear Pore Complex to Chromatin Architecture by Mediating CTCF and Cohesin Binding. Nat. Commun. 2020, 11, 2606. [Google Scholar] [CrossRef]
- Farhadova, S.; Ghousein, A.; Charon, F.; Surcis, C.; Gomez-Velazques, M.; Roidor, C.; Di Michele, F.; Borensztein, M.; De Sario, A.; Esnault, C.; et al. The Long Non-Coding RNA Meg3 Mediates Imprinted Gene Expression during Stem Cell Differentiation. Nucleic Acids Res. 2024, 52, 6183–6200. [Google Scholar] [CrossRef]
- Fierro, C.; Gatti, V.; La Banca, V.; De Domenico, S.; Scalera, S.; Corleone, G.; Fanciulli, M.; De Nicola, F.; Mauriello, A.; Montanaro, M.; et al. The Long Non-Coding RNA NEAT1 Is a ΔNp63 Target Gene Modulating Epidermal Differentiation. Nat. Commun. 2023, 14, 3795. [Google Scholar] [CrossRef]
- Huo, Y.; Li, Q.; Yang, L.; Li, X.; Sun, C.; Liu, Y.; Liu, H.; Pan, Z.; Li, Q.; Du, X. SDNOR, a Novel Antioxidative lncRNA, Is Essential for Maintaining the Normal State and Function of Porcine Follicular Granulosa Cells. Antioxidants 2023, 12, 799. [Google Scholar] [CrossRef]
- Wu, X.; Fu, M.; Ge, C.; Zhou, H.; Huang, H.; Zhong, M.; Zhang, M.; Xu, H.; Zhu, G.; Hua, W.; et al. m6A-Mediated Upregulation of lncRNA CHASERR Promotes the Progression of Glioma by Modulating the miR-6893-3p/TRIM14 Axis. Mol. Neurobiol. 2024, 61, 5418–5440. [Google Scholar] [CrossRef]
- Lv, W.; Jiang, W.; Luo, H.; Tong, Q.; Niu, X.; Liu, X.; Miao, Y.; Wang, J.; Guo, Y.; Li, J.; et al. Long Noncoding RNA lncMREF Promotes Myogenic Differentiation and Muscle Regeneration by Interacting with the Smarca5/P300 Complex. Nucleic Acids Res. 2022, 50, 10733–10755. [Google Scholar] [CrossRef]
- Chen, W.; Chen, W.; Liu, P.; Qian, S.; Tao, S.; Huang, M.; Xu, W.; Li, C.; Chen, X.; Lin, H.; et al. Role of lncRNA Has2os in Skeletal Muscle Differentiation and Regeneration. Cells 2022, 11, 3497. [Google Scholar] [CrossRef]
- Liu, H.; Liu, H.; Yang, Q.; Fan, Z. LncRNA SNHG1 Enhances Cartilage Regeneration by Modulating Chondrogenic Differentiation and Angiogenesis Potentials of JBMMSCs via Mitochondrial Function Regulation. Stem. Cell Res. Ther. 2024, 15, 177. [Google Scholar] [CrossRef]
- Lu, J.; Ma, H.; Wang, Q.; Song, Z.; Wang, J. Chemotherapy-Mediated lncRNA-Induced Immune Cell Plasticity in Cancer Immunopathogenesis. Int. Immunopharmacol. 2024, 141, 112967. [Google Scholar] [CrossRef]
- Zhang, X.; Pan, B.; Qiu, J.; Ke, X.; Shen, S.; Wang, X.; Tang, N. lncRNA MIAT Targets miR-411-5p/STAT3/PD-L1 Axis Mediating Hepatocellular Carcinoma Immune Response. Int. J. Exp. Pathol. 2022, 103, 102–111. [Google Scholar] [CrossRef]
- Du, J.; Liu, L.; Yang, F.; Leng, S.; Zhang, L.; Huang, D. Identification of Immune-Related lncRNA Regulatory Network in Pulpitis. Dis. Mark. 2022, 2022, 7222092. [Google Scholar] [CrossRef]
- Srinivas, T.; Mathias, C.; Oliveira-Mateos, C.; Guil, S. Roles of lncRNAs in Brain Development and Pathogenesis: Emerging Therapeutic Opportunities. Mol. Ther. 2023, 31, 1550–1561. [Google Scholar] [CrossRef]
- Song, P.; Chen, Y.; Liu, Z.; Liu, H.; Xiao, L.; Sun, L.; Wei, J.; He, L. LncRNA MALAT1 Aggravates Renal Tubular Injury via Activating LIN28A and the Nox4/AMPK/mTOR Signaling Axis in Diabetic Nephropathy. Front. Endocrinol. 2022, 13, 895360. [Google Scholar] [CrossRef]
- Yi, Y.; Zhao, Y.; Li, C.; Zhang, L.; Huang, H.; Li, Y.; Liu, L.; Hou, P.; Cui, T.; Tan, P.; et al. RAID v2.0: An Updated Resource of RNA-Associated Interactions across Organisms. Nucleic Acids Res. 2017, 45, D115–D118. [Google Scholar] [CrossRef]
- Frankish, A.; Diekhans, M.; Jungreis, I.; Lagarde, J.; Loveland, J.E.; Mudge, J.M.; Sisu, C.; Wright, J.C.; Armstrong, J.; Barnes, I.; et al. GENCODE 2021. Nucleic Acids Research 2021, 49, D916–D923. [Google Scholar] [CrossRef]
- Kang, J.; Tang, Q.; He, J.; Li, L.; Yang, N.; Yu, S.; Wang, M.; Zhang, Y.; Lin, J.; Cui, T.; et al. RNAInter v4.0: RNA Interactome Repository with Redefined Confidence Scoring System and Improved Accessibility. Nucleic Acids Res. 2022, 50, D326–D332. [Google Scholar] [CrossRef]
- He, N.; Li, D.; Xu, F.; Jin, J.; Li, L.; Tian, L.; Chen, B.; Li, X.; Ning, S.; Wang, L.; et al. LncPCD: A Manually Curated Database of Experimentally Supported Associations between lncRNA-Mediated Programmed Cell Death and Diseases. Database 2023, 2023, baad087. [Google Scholar] [CrossRef]
- Gao, Y.; Shang, S.; Guo, S.; Li, X.; Zhou, H.; Liu, H.; Sun, Y.; Wang, J.; Wang, P.; Zhi, H.; et al. Lnc2Cancer 3.0: An Updated Resource for Experimentally Supported lncRNA/circRNA Cancer Associations and Web Tools Based on RNA-Seq and scRNA-Seq Data. Nucleic Acids Res. 2021, 49, D1251–D1258. [Google Scholar] [CrossRef]
- Zhao, Y.; Ning, J.; Teng, H.; Deng, Y.; Sheldon, M.; Shi, L.; Martinez, C.; Zhang, J.; Tian, A.; Sun, Y.; et al. Long Noncoding RNA Malat1 Protects against Osteoporosis and Bone Metastasis. Nat. Commun. 2024, 15, 2384. [Google Scholar] [CrossRef]
- Zhao, X.; Chen, Q.; Cai, Y.; Chen, D.; Bei, M.; Dong, H.; Xu, J. TRA2A Binds With LncRNA MALAT1 To Promote Esophageal Cancer Progression By Regulating EZH2/β-Catenin Pathway. J. Cancer 2021, 12, 4883–4890. [Google Scholar] [CrossRef]
- Wu, C.; Chen, W.; Yu, F.; Yuan, Y.; Chen, Y.; Hurst, D.R.; Li, Y.; Li, L.; Liu, Z. Long Noncoding RNA HITTERS Protects Oral Squamous Cell Carcinoma Cells from Endoplasmic Reticulum Stress-Induced Apoptosis via Promoting MRE11-RAD50-NBS1 Complex Formation. Adv. Sci. 2020, 7, 2002747. [Google Scholar] [CrossRef]
- Duan, Y.; Jia, Y.; Wang, J.; Liu, T.; Cheng, Z.; Sang, M.; Lv, W.; Qin, J.; Liu, L. Long Noncoding RNA DGCR5 Involves in Tumorigenesis of Esophageal Squamous Cell Carcinoma via SRSF1-Mediated Alternative Splicing of Mcl-1. Cell Death Dis. 2021, 12, 587. [Google Scholar] [CrossRef]
- Yu, C.-H.; Fang, C.-Y.; Yu, C.-C.; Hsieh, P.-L.; Liao, Y.-W.; Tsai, L.-L.; Chu, P.-M. LINC00312/YBX1 Axis Regulates Myofibroblast Activities in Oral Submucous Fibrosis. Int. J. Mol. Sci. 2020, 21, 2979. [Google Scholar] [CrossRef]
- Wu, Y.; Zhao, Y.; Huan, L.; Zhao, J.; Zhou, Y.; Xu, L.; Hu, Z.; Liu, Y.; Chen, Z.; Wang, L.; et al. An LTR Retrotransposon-Derived Long Noncoding RNA lncMER52A Promotes Hepatocellular Carcinoma Progression by Binding P120-Catenin. Cancer Res. 2020, 80, 976–987. [Google Scholar] [CrossRef]
- Lin, Y.-H.; Wu, M.-H.; Liu, Y.-C.; Lyu, P.-C.; Yeh, C.-T.; Lin, K.-H. LINC01348 Suppresses Hepatocellular Carcinoma Metastasis through Inhibition of SF3B3-Mediated EZH2 Pre-mRNA Splicing. Oncogene 2021, 40, 4675–4685. [Google Scholar] [CrossRef]
- Liao, M.; Yao, D.; Wu, L.; Luo, C.; Wang, Z.; Zhang, J.; Liu, B. Targeting the Warburg Effect: A Revisited Perspective from Molecular Mechanisms to Traditional and Innovative Therapeutic Strategies in Cancer. Acta Pharm. Sin. B 2024, 14, 953–1008. [Google Scholar] [CrossRef]
- Wang, C.; Li, Y.; Yan, S.; Wang, H.; Shao, X.; Xiao, M.; Yang, B.; Qin, G.; Kong, R.; Chen, R.; et al. Interactome Analysis Reveals That lncRNA HULC Promotes Aerobic Glycolysis through LDHA and PKM2. Nat. Commun. 2020, 11, 3162. [Google Scholar] [CrossRef]
- Haga, Y.; Bandyopadhyay, D.; Khatun, M.; Tran, E.; Steele, R.; Banerjee, S.; Ray, R.; Nazzal, M.; Ray, R.B. Increased Expression of Long Non-Coding RNA FIRRE Promotes Hepatocellular Carcinoma by HuR-CyclinD1 Axis Signaling. J. Biol. Chem. 2024, 300, 107247. [Google Scholar] [CrossRef]
- Zhou, J.; Liu, T.; Xu, H.; Wang, Y.; Yang, T.; Liu, L. LncRNA FIRRE Promotes the Proliferation and Metastasis of Hepatocellular Carcinoma by Regulating the Expression of PXN through Interacting with MBNL3. Biochem. Biophys. Res. Commun. 2022, 625, 188–195. [Google Scholar] [CrossRef]
- Malakoti, F.; Targhazeh, N.; Karimzadeh, H.; Mohammadi, E.; Asadi, M.; Asemi, Z.; Alemi, F. Multiple Function of lncRNA MALAT1 in Cancer Occurrence and Progression. Chem. Biol. Drug Des. 2023, 101, 1113–1137. [Google Scholar] [CrossRef]
- Jen, J.; Tang, Y.-A.; Lu, Y.-H.; Lin, C.-C.; Lai, W.-W.; Wang, Y.-C. Oct4 Transcriptionally Regulates the Expression of Long Non-Coding RNAs NEAT1 and MALAT1 to Promote Lung Cancer Progression. Mol. Cancer 2017, 16, 104. [Google Scholar] [CrossRef]
- Huang, J.; Lin, C.; Dong, H.; Piao, Z.; Jin, C.; Han, H.; Jin, D. Targeting MALAT1 Induces DNA Damage and Sensitize Non-Small Cell Lung Cancer Cells to Cisplatin by Repressing BRCA1. Cancer Chemother. Pharmacol. 2020, 86, 663–672. [Google Scholar] [CrossRef]
- Mao, G.; Mu, Z.; Wu, D.A. Exosomal lncRNA FOXD3-AS1 Upregulates ELAVL1 Expression and Activates PI3K/Akt Pathway to Enhance Lung Cancer Cell Proliferation, Invasion, and 5-Fluorouracil Resistance. Acta Biochim. Biophys. Sin. 2021, 53, 1484–1494. [Google Scholar] [CrossRef]
- Qian, X.; Yang, J.; Qiu, Q.; Li, X.; Jiang, C.; Li, J.; Dong, L.; Ying, K.; Lu, B.; Chen, E.; et al. LCAT3, a Novel m6A-Regulated Long Non-Coding RNA, Plays an Oncogenic Role in Lung Cancer via Binding with FUBP1 to Activate c-MYC. J. Hematol. Oncol. 2021, 14, 112. [Google Scholar] [CrossRef]
- Hussain, S.A.; Venkatesh, T. YBX1/lncRNA SBF2-AS1 Interaction Regulates Proliferation and Tamoxifen Sensitivity via PI3K/AKT/MTOR Signaling in Breast Cancer Cells. Mol. Biol. Rep. 2023, 50, 3413–3428. [Google Scholar] [CrossRef]
- Sun, X.; Wang, R.; Tan, M.; Tian, X.; Meng, J. LncRNA LINC00680 Promotes Lung Adenocarcinoma Growth via Binding to GATA6 and Canceling GATA6-Mediated Suppression of SOX12 Expression. Exp. Cell Res. 2021, 405, 112653. [Google Scholar] [CrossRef]
- Cong, Z.; Diao, Y.; Li, X.; Jiang, Z.; Xu, Y.; Zhou, H.; Qiang, Y.; Wu, H.; Shen, Y. Long Non-Coding RNA Linc00665 Interacts with YB-1 and Promotes Angiogenesis in Lung Adenocarcinoma. Biochem. Biophys. Res. Commun. 2020, 527, 545–552. [Google Scholar] [CrossRef]
- Wu, A.; Tang, J.; Guo, Z.; Dai, Y.; Nie, J.; Hu, W.; Liu, N.; Ye, C.; Li, S.; Pei, H.; et al. Long Non-Coding RNA CRYBG3 Promotes Lung Cancer Metastasis via Activating the eEF1A1/MDM2/MTBP Axis. Int. J. Mol. Sci. 2021, 22, 3211. [Google Scholar] [CrossRef]
- Bao, T.; Liu, X.; Hu, J.; Ma, M.; Li, J.; Cao, L.; Yu, B.; Cheng, H.; Zhao, S.; Tian, Z. Recruitment of PVT1 Enhances YTHDC1-Mediated m6A Modification of IL-33 in Hyperoxia-Induced Lung Injury During Bronchopulmonary Dysplasia. Inflammation 2023, 47, 469–482. [Google Scholar] [CrossRef]
- Sun, J.; Jin, T.; Niu, Z.; Guo, J.; Guo, Y.; Yang, R.; Wang, Q.; Gao, H.; Zhang, Y.; Li, T.; et al. LncRNA DACH1 Protects against Pulmonary Fibrosis by Binding to SRSF1 to Suppress CTNNB1 Accumulation. Acta Pharm. Sin. B 2022, 12, 3602–3617. [Google Scholar] [CrossRef]
- Zhou, L.; Xu, D.; Sha, W.; Shen, L.; Lu, G. Long Non-Coding RNA MALAT1 Interacts with Transcription Factor Foxo1 to Regulate SIRT1 Transcription in High Glucose-Induced HK-2 Cells Injury. Biochem. Biophys. Res. Commun. 2018, 503, 849–855. [Google Scholar] [CrossRef]
- Liu, T.; Wang, H.; Fu, Z.; Wang, Z.; Wang, J.; Gan, X.; Wang, A.; Wang, L. Methyltransferase-like 14 Suppresses Growth and Metastasis of Renal Cell Carcinoma by Decreasing Long Noncoding RNA NEAT1. Cancer Sci. 2022, 113, 446–458. [Google Scholar] [CrossRef]
- Xie, X.; Lin, J.; Fan, X.; Zhong, Y.; Chen, Y.; Liu, K.; Ren, Y.; Chen, X.; Lai, D.; Li, X.; et al. LncRNA CDKN2B-AS1 Stabilized by IGF2BP3 Drives the Malignancy of Renal Clear Cell Carcinoma through Epigenetically Activating NUF2 Transcription. Cell Death Dis. 2021, 12, 201. [Google Scholar] [CrossRef]
- Hjazi, A.; Ghaffar, E.; Asghar, W.; Alauldeen Khalaf, H.; Ikram Ullah, M.; Mireya Romero-Parra, R.; Hussien, B.M.; Abdulally Abdulhussien Alazbjee, A.; Singh Bisht, Y.; Fakri Mustafa, Y.; et al. CDKN2B-AS1 as a Novel Therapeutic Target in Cancer: Mechanism and Clinical Perspective. Biochem. Pharmacol. 2023, 213, 115627. [Google Scholar] [CrossRef]
- Ageeli Hakami, M. Diabetes and Diabetic Associative Diseases: An Overview of Epigenetic Regulations of TUG1. Saudi J. Biol. Sci. 2024, 31, 103976. [Google Scholar] [CrossRef]
- Wang, S.; Yi, P.; Wang, N.; Song, M.; Li, W.; Zheng, Y. LncRNA TUG1/miR-29c-3p/SIRT1 Axis Regulates Endoplasmic Reticulum Stress-Mediated Renal Epithelial Cells Injury in Diabetic Nephropathy Model in Vitro. PLoS ONE 2021, 16, e0252761. [Google Scholar] [CrossRef]
- Song, X.; Gao, F.; Li, H.; Qin, W.; Chai, C.; Shi, G.; Yang, H. Long Noncoding RNA THRIL Promotes Foam Cell Formation and Inflammation in Macrophages. Cell Biol. Int. 2023, 47, 156–166. [Google Scholar] [CrossRef]
- Xiao, J.; Lu, Y.; Yang, X. THRIL Mediates Endothelial Progenitor Cells Autophagy via AKT Pathway and FUS. Mol. Med. 2020, 26, 86. [Google Scholar] [CrossRef]
- Zhang, Y.-Y.; Tan, R.-Z.; Yu, Y.; Niu, Y.-Y.; Yu, C. LncRNA GAS5 Protects against TGF-β-Induced Renal Fibrosis via the Smad3/miRNA-142-5p Axis. Am. J. Physiol. Ren. Physiol. 2021, 321, F517–F526. [Google Scholar] [CrossRef]
- Tang, R.; Wang, Y.-C.; Mei, X.; Shi, N.; Sun, C.; Ran, R.; Zhang, G.; Li, W.; Staveley-O’Carroll, K.F.; Li, G.; et al. LncRNA GAS5 Attenuates Fibroblast Activation through Inhibiting Smad3 Signaling. Am. J. Physiol. Cell Physiol. 2020, 319, C105–C115. [Google Scholar] [CrossRef]
- Wang, Y.; Chen, D.; Xie, H.; Zhou, S.; Jia, M.; He, X.; Guo, F.; Lai, Y.; Tang, X.X. LncRNA GAS5 Suppresses TGF-Β1-Induced Transformation of Pulmonary Pericytes into Myofibroblasts by Recruiting KDM5B and Promoting H3K4me2/3 Demethylation of the PDGFRα/β Promoter. Mol. Med. 2023, 29, 32. [Google Scholar] [CrossRef]
- Grossi, E.; Raimondi, I.; Goñi, E.; González, J.; Marchese, F.P.; Chapaprieta, V.; Martín-Subero, J.I.; Guo, S.; Huarte, M. A lncRNA-SWI/SNF Complex Crosstalk Controls Transcriptional Activation at Specific Promoter Regions. Nat. Commun. 2020, 11, 936. [Google Scholar] [CrossRef]
- Jiang, M.; Zhang, S.; Yang, Z.; Lin, H.; Zhu, J.; Liu, L.; Wang, W.; Liu, S.; Liu, W.; Ma, Y.; et al. Self-Recognition of an Inducible Host lncRNA by RIG-I Feedback Restricts Innate Immune Response. Cell 2018, 173, 906–919.e13. [Google Scholar] [CrossRef]
- Lin, H.; Jiang, M.; Liu, L.; Yang, Z.; Ma, Z.; Liu, S.; Ma, Y.; Zhang, L.; Cao, X. The Long Noncoding RNA Lnczc3h7a Promotes a TRIM25-Mediated RIG-I Antiviral Innate Immune Response. Nat. Immunol. 2019, 20, 812–823. [Google Scholar] [CrossRef]
- Zou, S.; Gou, X.; Wen, K. Advances in the Role of Long Non-coding RNAs and RNA-binding Proteins in Regulating DNA Damage Repair in Cancer Cells. Int. J. Mol. Med. 2023, 52, 93. [Google Scholar] [CrossRef]
- Wang, D.; Zhou, Z.; Wu, E.; Ouyang, C.; Wei, G.; Wang, Y.; He, D.; Cui, Y.; Zhang, D.; Chen, X.; et al. LRIK Interacts with the Ku70–Ku80 Heterodimer Enhancing the Efficiency of NHEJ Repair. Cell Death Differ. 2020, 27, 3337–3353. [Google Scholar] [CrossRef]
- Deng, B.; Xu, W.; Wang, Z.; Liu, C.; Lin, P.; Li, B.; Huang, Q.; Yang, J.; Zhou, H.; Qu, L. An LTR Retrotransposon-Derived lncRNA Interacts with RNF169 to Promote Homologous Recombination. EMBO Rep. 2019, 20, e47650. [Google Scholar] [CrossRef]
- Typas, D.; Mailand, N. An Unorthodox Partnership in DNA Repair Pathway Choice. EMBO Rep. 2019, 20, e49105. [Google Scholar] [CrossRef]
- Wang, J.; Chen, H.; Deng, Q.; Chen, Y.; Wang, Z.; Yan, Z.; Wang, Y.; Tang, H.; Liang, H.; Jiang, Y. High Expression of RNF169 Is Associated with Poor Prognosis in Pancreatic Adenocarcinoma by Regulating Tumour Immune Infiltration. Front. Genet. 2023, 13, 1022626. [Google Scholar] [CrossRef]
- Shuai, Y.; Ma, Z.; Liu, W.; Yu, T.; Yan, C.; Jiang, H.; Tian, S.; Xu, T.; Shu, Y. TEAD4 Modulated LncRNA MNX1-AS1 Contributes to Gastric Cancer Progression Partly through Suppressing BTG2 and Activating BCL2. Mol. Cancer 2020, 19, 6. [Google Scholar] [CrossRef]
- Han, T.; Jing, X.; Bao, J.; Zhao, L.; Zhang, A.; Miao, R.; Guo, H.; Zhou, B.; Zhang, S.; Sun, J.; et al. H. Pylori Infection Alters Repair of DNA Double-Strand Breaks via SNHG17. J. Clin. Investig. 2020, 130, 3901–3918. [Google Scholar] [CrossRef]
- Pirlog, R.; Drula, R.; Nutu, A.; Calin, G.A.; Berindan-Neagoe, I. The Roles of the Colon Cancer Associated Transcript 2 (CCAT2) Long Non-Coding RNA in Cancer: A Comprehensive Characterization of the Tumorigenic and Molecular Functions. Int. J. Mol. Sci. 2021, 22, 12491. [Google Scholar] [CrossRef]
- Chen, B.; Dragomir, M.P.; Fabris, L.; Bayraktar, R.; Knutsen, E.; Liu, X.; Tang, C.; Li, Y.; Shimura, T.; Ivkovic, T.C.; et al. The Long Noncoding RNA CCAT2 Induces Chromosomal Instability Through BOP1-AURKB Signaling. Gastroenterology 2020, 159, 2146–2162.e33. [Google Scholar] [CrossRef]
- Zhao, C.; Ling, X.; Xia, Y.; Yan, B.; Guan, Q. The m6A Methyltransferase METTL3 Controls Epithelial-Mesenchymal Transition, Migration and Invasion of Breast Cancer through the MALAT1/miR-26b/HMGA2 Axis. Cancer Cell Int. 2021, 21, 441. [Google Scholar] [CrossRef]
- Lu, D.; Di, S.; Zhuo, S.; Zhou, L.; Bai, R.; Ma, T.; Zou, Z.; Chen, C.; Sun, M.; Tang, J.; et al. The Long Noncoding RNA TINCR Promotes Breast Cancer Cell Proliferation and Migration by Regulating OAS1. Cell Death Discov. 2021, 7, 41. [Google Scholar] [CrossRef]
- Zhang, Y.; Huang, Y.-X.; Wang, D.-L.; Yang, B.; Yan, H.-Y.; Lin, L.-H.; Li, Y.; Chen, J.; Xie, L.-M.; Huang, Y.-S.; et al. LncRNA DSCAM-AS1 Interacts with YBX1 to Promote Cancer Progression by Forming a Positive Feedback Loop That Activates FOXA1 Transcription Network. Theranostics 2020, 10, 10823–10837. [Google Scholar] [CrossRef]
- Abo Elwafa, R.; Abd Elrahman, A.; Ghallab, O. Long Intergenic Non-Coding RNA-P21 Is Associated with Poor Prognosis in Chronic Lymphocytic Leukemia. Clin. Transl. Oncol. 2021, 23, 92–99. [Google Scholar] [CrossRef]
- Amirinejad, R.; Rezaei, M.; Shirvani-Farsani, Z. An Update on Long Intergenic Noncoding RNA P21: A Regulatory Molecule with Various Significant Functions in Cancer. Cell Biosci. 2020, 10, 82. [Google Scholar] [CrossRef]
- Huang, Y.; Yi, Q.; Feng, J.; Xie, W.; Sun, W.; Sun, W. The Role of lincRNA-P21 in Regulating the Biology of Cancer Cells. Hum. Cell 2022, 35, 1640–1649. [Google Scholar] [CrossRef]
- Xiong, J.; Lian, W.; Zhao, R.; Gao, K. METTL3/MALAT1/ELAVL1 Axis Promotes Tumor Growth in Ovarian Cancer. OncoTargets Ther. 2024, 17, 85–97. [Google Scholar] [CrossRef]
- Wang, Z.; Wang, X.; Zhang, T.; Su, L.; Liu, B.; Zhu, Z.; Li, C. LncRNA MALAT1 Promotes Gastric Cancer Progression via Inhibiting Autophagic Flux and Inducing Fibroblast Activation. Cell Death Dis. 2021, 12, 368. [Google Scholar] [CrossRef]
- Farooqi, A.; Zahid, R.; Naureen, H.; Attar, R.; Gazouli, M.; Berardi, R.; Szelachowska, J.; Matkowski, R.; Pawlak, E. Regulation of ROCK1/2 by Long Non-coding RNAs and Circular RNAs in Different Cancer Types (Review). Oncol. Lett. 2022, 23, 159. [Google Scholar] [CrossRef]
- Dai, R.; Zhou, Y.; Chen, Z.; Zou, Z.; Pan, Z.; Liu, P.; Gao, X. Lnc-MUC20-9 Binds to ROCK1 and Functions as a Tumor Suppressor in Bladder Cancer. J. Cell Biochem. 2020, 121, 4214–4225. [Google Scholar] [CrossRef]
- Mehmandar-Oskuie, A.; Jahankhani, K.; Rostamlou, A.; Arabi, S.; Sadat Razavi, Z.; Mardi, A. Molecular Landscape of LncRNAs in Bladder Cancer: From Drug Resistance to Novel LncRNA-Based Therapeutic Strategies. Biomed. Pharmacother. 2023, 165, 115242. [Google Scholar] [CrossRef]
- Chen, C.; Luo, Y.; He, W.; Zhao, Y.; Kong, Y.; Liu, H.; Zhong, G.; Li, Y.; Li, J.; Huang, J.; et al. Exosomal Long Noncoding RNA LNMAT2 Promotes Lymphatic Metastasis in Bladder Cancer. J. Clin. Investig. 2019, 130, 404–421. [Google Scholar] [CrossRef]
- Wu, R.; Li, L.; Bai, Y.; Yu, B.; Xie, C.; Wu, H.; Zhang, Y.; Huang, L.; Yan, Y.; Li, X.; et al. The Long Noncoding RNA LUCAT1 Promotes Colorectal Cancer Cell Proliferation by Antagonizing Nucleolin to Regulate MYC Expression. Cell Death Dis. 2020, 11, 908. [Google Scholar] [CrossRef]
- Zhou, Q.; Hou, Z.; Zuo, S.; Zhou, X.; Feng, Y.; Sun, Y.; Yuan, X. LUCAT1 Promotes Colorectal Cancer Tumorigenesis by Targeting the Ribosomal Protein L40-MDM2-P53 Pathway through Binding with UBA52. Cancer Sci. 2019, 110, 1194–1207. [Google Scholar] [CrossRef]
- Deng, X.; Kong, F.; Li, S.; Jiang, H.; Dong, L.; Xu, X.; Zhang, X.; Yuan, H.; Xu, Y.; Chu, Y.; et al. A KLF4/PiHL/EZH2/HMGA2 Regulatory Axis and Its Function in Promoting Oxaliplatin-Resistance of Colorectal Cancer. Cell Death Dis. 2021, 12, 485. [Google Scholar] [CrossRef]
- Deng, X.; Li, S.; Kong, F.; Ruan, H.; Xu, X.; Zhang, X.; Wu, Z.; Zhang, L.; Xu, Y.; Yuan, H.; et al. Long Noncoding RNA PiHL Regulates P53 Protein Stability through GRWD1/RPL11/MDM2 Axis in Colorectal Cancer. Theranostics 2020, 10, 265–280. [Google Scholar] [CrossRef]
- Yue, M.; Liu, T.; Yan, G.; Luo, X.; Wang, L. LINC01605, Regulated by the EP300-SMYD2 Complex, Potentiates the Binding between METTL3 and SPTBN2 in Colorectal Cancer. Cancer Cell Int. 2021, 21, 504. [Google Scholar] [CrossRef]
- Jiang, T.; Qi, J.; Xue, Z.; Liu, B.; Liu, J.; Hu, Q.; Li, Y.; Ren, J.; Song, H.; Xu, Y.; et al. The m6A Modification Mediated-lncRNA POU6F2-AS1 Reprograms Fatty Acid Metabolism and Facilitates the Growth of Colorectal Cancer via Upregulation of FASN. Mol. Cancer 2024, 23, 55. [Google Scholar] [CrossRef]
- Zhou, Y.; Shao, Y.; Hu, W.; Zhang, J.; Shi, Y.; Kong, X.; Jiang, J. A Novel Long Noncoding RNA SP100-AS1 Induces Radioresistance of Colorectal Cancer via Sponging miR-622 and Stabilizing ATG3. Cell Death Differ. 2023, 30, 111–124. [Google Scholar] [CrossRef]
- Chen, Q.; Zhou, L.; Ma, D.; Hou, J.; Lin, Y.; Wu, J.; Tao, M. LncRNA GAS6-AS1 Facilitates Tumorigenesis and Metastasis of Colorectal Cancer by Regulating TRIM14 through miR-370-3p/miR-1296-5p and FUS. J. Transl. Med. 2022, 20, 356. [Google Scholar] [CrossRef]
- Chen, X.; Song, J.; Wang, X.; Sun, D.; Liu, Y.; Jiang, Y. LncRNA LINC00460: Function and Mechanism in Human Cancer. Thorac. Cancer 2022, 13, 3–14. [Google Scholar] [CrossRef]
- Hou, P.; Meng, S.; Li, M.; Lin, T.; Chu, S.; Li, Z.; Zheng, J.; Gu, Y.; Bai, J. LINC00460/DHX9/IGF2BP2 Complex Promotes Colorectal Cancer Proliferation and Metastasis by Mediating HMGA1 mRNA Stability Depending on m6A Modification. J. Exp. Clin. Cancer Res. 2021, 40, 52. [Google Scholar] [CrossRef]
- Zhao, K.; Wang, X.; Xue, X.; Li, L.; Hu, Y. A Long Noncoding RNA Sensitizes Genotoxic Treatment by Attenuating ATM Activation and Homologous Recombination Repair in Cancers. PLoS Biol. 2020, 18, e3000666. [Google Scholar] [CrossRef]
- Cai, J.; Wang, R.; Chen, Y.; Zhang, C.; Fu, L.; Fan, C. LncRNA FIRRE Regulated Endometrial Cancer Radiotherapy Sensitivity via the miR-199b-5p/SIRT1/BECN1 Axis-Mediated Autophagy. Genomics 2024, 116, 110750. [Google Scholar] [CrossRef]
- Wang, Y.; Li, Z.; Xu, S.; Li, W.; Chen, M.; Jiang, M.; Fan, X. LncRNA FIRRE Functions as a Tumor Promoter by Interaction with PTBP1 to Stabilize BECN1 mRNA and Facilitate Autophagy. Cell Death Dis. 2022, 13, 98. [Google Scholar] [CrossRef]
- Liu, Q.; Dai, G.; Wu, Y.; Wang, X.; Song, M.; Li, X.; Wu, Z.; Xia, R. LncRNA FIRRE Stimulates PTBP1-induced Smurf2 Decay, Stabilizes B-cell Receptor, and Promotes the Development of Diffuse Large B-cell Lymphoma. Hematol. Oncol. 2022, 40, 554–566. [Google Scholar] [CrossRef]
- Gu, N.; Wang, Y.; Li, L.; Sui, X.; Liu, Z. The Mechanism of lncRNA MALAT1 Targeting the miR-124-3p/IGF2BP1 Axis to Regulate Osteogenic Differentiation of Periodontal Ligament Stem Cells. Clin. Oral Invest. 2024, 28, 219. [Google Scholar] [CrossRef]
- Ding, A.; Li, C.-H.; Yu, C.-Y.; Zhou, H.-T.; Zhang, Z.-H. Long Non-Coding RNA MALAT1 Enhances Angiogenesis during Bone Regeneration by Regulating the miR-494/SP1 Axis. Lab. Investig. 2021, 101, 1458–1466. [Google Scholar] [CrossRef]
- Ghafouri-Fard, S.; Azimi, T.; Taheri, M. Cervical Carcinoma High Expressed 1 (CCHE1): An Oncogenic lncRNA in Diverse Neoplasms. Biomed. Pharmacother. 2021, 142, 112003. [Google Scholar] [CrossRef]
- Zhang, Z.; Yu, T.; Geng, W. Long Non-Coding RNA CCHE1 Participates in Postoperative Distant Recurrence but Not Local Recurrence of Osteosarcoma Possibly by Interacting with ROCK1. BMC Musculoskelet. Disord. 2020, 21, 462. [Google Scholar] [CrossRef]
- Yao, X.-Y.; Liu, J.-F.; Luo, Y.; Xu, X.-Z.; Bu, J. LncRNA HOTTIP Facilitates Cell Proliferation, Invasion, and Migration in Osteosarcoma by Interaction with PTBP1 to Promote KHSRP Level. Cell Cycle 2021, 20, 283–297. [Google Scholar] [CrossRef]
- Fu, C.; Xin, J.; Zhang, W.; Lai, J.; Huang, Z. LINC00992 Exerts Oncogenic Activities in Prostate Cancer via Regulation of SOX4. Exp. Cell Res. 2021, 408, 112855. [Google Scholar] [CrossRef]
- Tan, X.; Chen, W.; Lv, D.; Yang, T.; Wu, K.; Zou, L.; Luo, J.; Zhou, X.; Liu, G.; Shu, F.; et al. LncRNA SNHG1 and RNA Binding Protein hnRNPL Form a Complex and Coregulate CDH1 to Boost the Growth and Metastasis of Prostate Cancer. Cell Death Dis. 2021, 12, 138. [Google Scholar] [CrossRef]
- Muller, C.S.M.; Giner, I.S.; Zambalde, É.P.; Carvalho, T.M.; Ribeiro, E.M.D.S.F.; Carvalho De Oliveira, J.; Mathias, C.; Gradia, D.F. The Potential of NORAD–PUMILIO–RALGAPB Regulatory Axis as a Biomarker in Breast Cancer. ncRNA 2022, 8, 76. [Google Scholar] [CrossRef]
- Elguindy, M.M.; Mendell, J.T. NORAD-Induced Pumilio Phase Separation Is Required for Genome Stability. Nature 2021, 595, 303–308. [Google Scholar] [CrossRef]
- Zhao, D.; Wang, C.; Yan, S.; Chen, R. Advances in the Identification of Long Non-Coding RNA Binding Proteins. Anal. Biochem. 2022, 639, 114520. [Google Scholar] [CrossRef]
- Lambert, N.J.; Robertson, A.D.; Burge, C.B. RNA Bind-n-Seq: Measuring the Binding Affinity Landscape of RNA-Binding Proteins. In Methods in Enzymology; Elsevier: Amsterdam, The Netherlands, 2015; Volume 558, pp. 465–493. ISBN 978-0-12-801934-4. [Google Scholar]
- Hanelt, T.N.; Treiber, N.; Treiber, T.; Lehmann, G.; Eichner, N.; Rothmeier, T.; Schmid, G.; Reichelt, R.; Zambelli, F.; Pavesi, G.; et al. Endo-Bind-n-Seq: Identifying RNA Motifs of RNA Binding Proteins Isolated from Endogenous Sources. Life Sci. Alliance 2025, 8, e202402782. [Google Scholar] [CrossRef]
- Ye, X.; Jankowsky, E. High Throughput Approaches to Study RNA-Protein Interactions in Vitro. Methods 2020, 178, 3–10. [Google Scholar] [CrossRef]
- Baldini, L.; Labialle, S. Using Native RIP, UV-CLIP or fCLIP to Address Protein–RNA Interactions In Vivo. In Small Non-Coding RNAs; Rederstorff, M., Ed.; Methods in Molecular Biology; Springer: New York, NY, USA, 2021; Volume 2300, pp. 89–98. ISBN 978-1-07-161385-6. [Google Scholar]
- Martindale, J.; Gorospe, M.; Idda, M. Ribonucleoprotein Immunoprecipitation (RIP) Analysis. BIO-Protocol 2020, 10, e3488. [Google Scholar] [CrossRef]
- Kapral, T.H.; Farnhammer, F.; Zhao, W.; Lu, Z.J.; Zagrovic, B. Widespread Autogenous mRNA–Protein Interactions Detected by CLIP-Seq. Nucleic Acids Res. 2022, 50, 9984–9999. [Google Scholar] [CrossRef]
- Chen, X.; Castro, S.A.; Liu, Q.; Hu, W.; Zhang, S. Practical Considerations on Performing and Analyzing CLIP-Seq Experiments to Identify Transcriptomic-Wide RNA-Protein Interactions. Methods 2019, 155, 49–57. [Google Scholar] [CrossRef]
- Zheng, M.; Lin, Y.; Wang, W.; Zhao, Y.; Bao, X. Application of Nucleoside or Nucleotide Analogues in RNA Dynamics and RNA-Binding Protein Analysis. WIREs RNA 2022, 13, e1722. [Google Scholar] [CrossRef]
- Kiefer, L.; Schofield, J.A.; Simon, M.D. Expanding the Nucleoside Recoding Toolkit: Revealing RNA Population Dynamics with 6-Thioguanosine. J. Am. Chem. Soc. 2018, 140, 14567–14570. [Google Scholar] [CrossRef]
- Van Nostrand, E.L.; Pratt, G.A.; Shishkin, A.A.; Gelboin-Burkhart, C.; Fang, M.Y.; Sundararaman, B.; Blue, S.M.; Nguyen, T.B.; Surka, C.; Elkins, K.; et al. Robust Transcriptome-Wide Discovery of RNA-Binding Protein Binding Sites with Enhanced CLIP (eCLIP). Nat. Methods 2016, 13, 508–514. [Google Scholar] [CrossRef]
- Kim, B.; Kim, V.N. fCLIP-Seq for Transcriptomic Footprinting of dsRNA-Binding Proteins: Lessons from DROSHA. Methods 2019, 152, 3–11. [Google Scholar] [CrossRef]
- Sahadevan, S.; Sekaran, T.; Ashaf, N.; Fritz, M.; Hentze, M.W.; Huber, W.; Schwarzl, T. Htseq-Clip: A Toolset for the Preprocessing of eCLIP/iCLIP Datasets. Bioinformatics 2023, 39, btac747. [Google Scholar] [CrossRef]
- Castello, A.; Fischer, B.; Frese, C.K.; Horos, R.; Alleaume, A.-M.; Foehr, S.; Curk, T.; Krijgsveld, J.; Hentze, M.W. Comprehensive Identification of RNA-Binding Domains in Human Cells. Mol. Cell 2016, 63, 696–710. [Google Scholar] [CrossRef]
- Shan, M.; Gregory, B.D. Using RNA Affinity Purification Followed by Mass Spectrometry to Identify RNA-Binding Proteins (RBPs). In RNA Tagging; Heinlein, M., Ed.; Methods in Molecular Biology; Springer: New York, NY, USA, 2020; Volume 2166, pp. 241–253. ISBN 978-1-07-160711-4. [Google Scholar]
- Fernandez-Chamorro, J.; Francisco-Velilla, R.; Embarc-Buh, A.; Martinez-Salas, E. Identification of RNA-Binding Proteins Associated to RNA Structural Elements. In RNA Scaffolds; Ponchon, L., Ed.; Methods in Molecular Biology; Springer: New York, NY, USA, 2021; Volume 2323, pp. 109–119. ISBN 978-1-07-161498-3. [Google Scholar]
- Azizi, H.; Papadopoulou, B. In Vivo Tethering System to Isolate RNA-Binding Proteins Regulating mRNA Decay in Leishmania. In Trypanosomatids; Michels, P.A.M., Ginger, M.L., Zilberstein, D., Eds.; Methods in Molecular Biology; Springer: New York, NY, USA, 2020; Volume 2116, pp. 325–338. ISBN 978-1-07-160293-5. [Google Scholar]
- Janvier, A.; Hayek, H.; Alghoul, F.; Gross, L.; Allmang, C.; Martin, F.; Eriani, G. Purification of In Vivo or In Vitro-Assembled RNA-Protein Complexes by RNA Centric Methods. In Advanced Technologies for Protein Complex Production and Characterization; Vega, M.C., Fernández, F.J., Eds.; Advances in Experimental Medicine and Biology; Springer Nature Switzerland: Cham, Switzerland, 2024; Volume 3234, pp. 17–29. ISBN 978-3-031-52192-8. [Google Scholar]
- Dvir, S.; Argoetti, A.; Lesnik, C.; Roytblat, M.; Shriki, K.; Amit, M.; Hashimshony, T.; Mandel-Gutfreund, Y. Uncovering the RNA-Binding Protein Landscape in the Pluripotency Network of Human Embryonic Stem Cells. Cell Rep. 2021, 35, 109198. [Google Scholar] [CrossRef]
- Zheng, J.; Dou, R.; Zhang, X.; Zhong, B.; Fang, C.; Xu, Q.; Di, Z.; Huang, S.; Lin, Z.; Song, J.; et al. LINC00543 Promotes Colorectal Cancer Metastasis by Driving EMT and Inducing the M2 Polarization of Tumor Associated Macrophages. J. Transl. Med. 2023, 21, 153. [Google Scholar] [CrossRef]
- Alfeghaly, C.; Behm-Ansmant, I.; Maenner, S. Study of Genome-Wide Occupancy of Long Non-Coding RNAs Using Chromatin Isolation by RNA Purification (ChIRP). In Small Non-Coding RNAs; Rederstorff, M., Ed.; Methods in Molecular Biology; Springer: New York, NY, USA, 2021; Volume 2300, pp. 107–117. ISBN 978-1-07-161385-6. [Google Scholar]
- Kim, J.; Ma, L. Capturing Endogenous Long Noncoding RNAs and Their Binding Proteins Using Chromatin Isolation by RNA Purification. In Long Non-Coding RNAs; Zhang, L., Hu, X., Eds.; Methods in Molecular Biology; Springer: New York, NY, USA, 2021; Volume 2372, pp. 85–92. ISBN 978-1-07-161696-3. [Google Scholar]
- Xing, S.; Wang, J.; Wu, R.; Hefti, M.M.; Crary, J.F.; Lu, Y. Identification of HnRNPC as a Novel Tau Exon 10 Splicing Factor Using RNA Antisense Purification Mass Spectrometry. RNA Biol. 2022, 19, 104–116. [Google Scholar] [CrossRef]
- Pant, P.; Kumarswamy, R. Multiple Oligo Assisted RNA Pulldown via Hybridization Followed by Mass Spectrometry (MORPH-MS) for Exploring the RNA-Protein Interactions. RNA Biol. 2024, 21, 56–64. [Google Scholar] [CrossRef]
- Weissinger, R.; Heinold, L.; Akram, S.; Jansen, R.-P.; Hermesh, O. RNA Proximity Labeling: A New Detection Tool for RNA-Protein Interactions. Molecules 2021, 26, 2270. [Google Scholar] [CrossRef]
- Yi, W.; Li, J.; Zhu, X.; Wang, X.; Fan, L.; Sun, W.; Liao, L.; Zhang, J.; Li, X.; Ye, J.; et al. CRISPR-Assisted Detection of RNA-Protein Interactions in Living Cells. Nat. Methods 2020, 17, 685–688. [Google Scholar] [CrossRef]
- Teng, Z.; Zhang, Y.; Dai, Q.; Wu, C.; Li, D. Constructing Discriminative Feature Space for LncRNA–Protein Interaction Based on Deep Autoencoder and Marginal Fisher Analysis. Comput. Biol. Med. 2023, 157, 106711. [Google Scholar] [CrossRef]
- Shaw, D.; Chen, H.; Xie, M.; Jiang, T. DeepLPI: A Multimodal Deep Learning Method for Predicting the Interactions between lncRNAs and Protein Isoforms. BMC Bioinform. 2021, 22, 24. [Google Scholar] [CrossRef]
- Chen, Y.; Fu, X.; Li, Z.; Peng, L.; Zhuo, L. Prediction of lncRNA–Protein Interactions via the Multiple Information Integration. Front. Bioeng. Biotechnol. 2021, 9, 647113. [Google Scholar] [CrossRef]
- Zhao, J.; Sun, J.; Shuai, S.C.; Zhao, Q.; Shuai, J. Predicting Potential Interactions between lncRNAs and Proteins via Combined Graph Auto-Encoder Methods. Brief. Bioinform. 2023, 24, bbac527. [Google Scholar] [CrossRef]
- Zhao, X.; Zhang, Y.; Du, X. DFpin: Deep Learning–Based Protein-Binding Site Prediction with Feature-Based Non-Redundancy from RNA Level. Comput. Biol. Med. 2022, 142, 105216. [Google Scholar] [CrossRef]
- Wang, Y.; Pan, Z.; Mou, M.; Xia, W.; Zhang, H.; Zhang, H.; Liu, J.; Zheng, L.; Luo, Y.; Zheng, H.; et al. A Task-Specific Encoding Algorithm for RNAs and RNA-Associated Interactions Based on Convolutional Autoencoder. Nucleic Acids Res. 2023, 51, e110. [Google Scholar] [CrossRef]
- Wei, J.; Chen, S.; Zong, L.; Gao, X.; Li, Y. Protein–RNA Interaction Prediction with Deep Learning: Structure Matters. Brief. Bioinform. 2022, 23, bbab540. [Google Scholar] [CrossRef]
- Agarwal, A.; Kant, S.; Bahadur, R.P. Efficient Mapping of RNA-binding Residues in RNA-binding Proteins Using Local Sequence Features of Binding Site Residues in protein-RNA Complexes. Proteins 2023, 91, 1361–1379. [Google Scholar] [CrossRef]
- Li, P.; Liu, Z.-P. PST-PRNA: Prediction of RNA-Binding Sites Using Protein Surface Topography and Deep Learning. Bioinformatics 2022, 38, 2162–2168. [Google Scholar] [CrossRef]
- Wang, Y.; Chen, Z.; Pan, Z.; Huang, S.; Liu, J.; Xia, W.; Zhang, H.; Zheng, M.; Li, H.; Hou, T.; et al. RNAincoder: A Deep Learning-Based Encoder for RNA and RNA-Associated Interaction. Nucleic Acids Res. 2023, 51, W509–W519. [Google Scholar] [CrossRef]
- Zhang, H.; Lu, M.; Lin, G.; Zheng, L.; Zhang, W.; Xu, Z.; Zhu, F. SoCube: An Innovative End-to-End Doublet Detection Algorithm for Analyzing scRNA-Seq Data. Brief. Bioinform. 2023, 24, bbad104. [Google Scholar] [CrossRef]
- Agarwal, A.; Singh, K.; Kant, S.; Bahadur, R.P. A Comparative Analysis of Machine Learning Classifiers for Predicting Protein-Binding Nucleotides in RNA Sequences. Comput. Struct. Biotechnol. J. 2022, 20, 3195–3207. [Google Scholar] [CrossRef]
- Du, X.; Zhao, X.; Zhang, Y. DeepBtoD: Improved RNA-Binding Proteins Prediction via Integrated Deep Learning. J. Bioinform. Comput. Biol. 2022, 20, 2250006. [Google Scholar] [CrossRef]
- Du, Z.; Xiao, X.; Uversky, V.N. DeepA-RBPBS: A Hybrid Convolution and Recurrent Neural Network Combined with Attention Mechanism for Predicting RBP Binding Site. J. Biomol. Struct. Dyn. 2022, 40, 4250–4258. [Google Scholar] [CrossRef]
- Zhao, W.; Zhang, S.; Zhu, Y.; Xi, X.; Bao, P.; Ma, Z.; Kapral, T.H.; Chen, S.; Zagrovic, B.; Yang, Y.T.; et al. POSTAR3: An Updated Platform for Exploring Post-Transcriptional Regulation Coordinated by RNA-Binding Proteins. Nucleic Acids Res. 2022, 50, D287–D294. [Google Scholar] [CrossRef]
- Hafner, M.; Katsantoni, M.; Köster, T.; Marks, J.; Mukherjee, J.; Staiger, D.; Ule, J.; Zavolan, M. CLIP and Complementary Methods. Nat. Rev. Methods Primers 2021, 1, 20. [Google Scholar] [CrossRef]
- Blue, S.M.; Yee, B.A.; Pratt, G.A.; Mueller, J.R.; Park, S.S.; Shishkin, A.A.; Starner, A.C.; Van Nostrand, E.L.; Yeo, G.W. Transcriptome-Wide Identification of RNA-Binding Protein Binding Sites Using seCLIP-Seq. Nat. Protoc. 2022, 17, 1223–1265. [Google Scholar] [CrossRef]
- Ramanathan, M.; Porter, D.F.; Khavari, P.A. Methods to Study RNA-Protein Interactions. Nat. Methods 2019, 16, 225–234. [Google Scholar] [CrossRef]
- Wei, B.; Zhang, Y.; Gong, X. DeepLPI: A Novel Deep Learning-Based Model for Protein–Ligand Interaction Prediction for Drug Repurposing. Sci. Rep. 2022, 12, 18200. [Google Scholar] [CrossRef]
- Ding, Y.; Sun, Y.; Wang, H.; Zhao, H.; Yin, R.; Zhang, M.; Pan, X.; Zhu, X. Atherosis-Associated Lnc_000048 Activates PKR to Enhance STAT1-Mediated Polarization of THP-1 Macrophages to M1 Phenotype. Neural Regen. Res. 2024, 19, 2488–2498. [Google Scholar] [CrossRef]
- Armaos, A.; Colantoni, A.; Proietti, G.; Rupert, J.; Tartaglia, G.G. catRAPID Omics v2.0: Going Deeper and Wider in the Prediction of Protein-RNA Interactions. Nucleic Acids Res. 2021, 49, W72–W79. [Google Scholar] [CrossRef]
- Laverty, K.U.; Jolma, A.; Pour, S.E.; Zheng, H.; Ray, D.; Morris, Q.; Hughes, T.R. PRIESSTESS: Interpretable, High-Performing Models of the Sequence and Structure Preferences of RNA-Binding Proteins. Nucleic Acids Res. 2022, 50, e111. [Google Scholar] [CrossRef]
- Horlacher, M.; Wagner, N.; Moyon, L.; Kuret, K.; Goedert, N.; Salvatore, M.; Ule, J.; Gagneur, J.; Winther, O.; Marsico, A. Towards in Silico CLIP-Seq: Predicting Protein-RNA Interaction via Sequence-to-Signal Learning. Genome Biol. 2023, 24, 180. [Google Scholar] [CrossRef]
- Qiao, Y.; Yang, R.; Liu, Y.; Chen, J.; Zhao, L.; Huo, P.; Wang, Z.; Bu, D.; Wu, Y.; Zhao, Y. DeepFusion: A Deep Bimodal Information Fusion Network for Unraveling Protein-RNA Interactions Using in Vivo RNA Structures. Comput. Struct. Biotechnol. J. 2024, 23, 617–625. [Google Scholar] [CrossRef]
- Zhu, H.; Yang, Y.; Wang, Y.; Wang, F.; Huang, Y.; Chang, Y.; Wong, K.-C.; Li, X. Dynamic Characterization and Interpretation for Protein-RNA Interactions across Diverse Cellular Conditions Using HDRNet. Nat. Commun. 2023, 14, 6824. [Google Scholar] [CrossRef]
- Wu, Z.; Basu, S.; Wu, X.; Kurgan, L. qNABpredict: Quick, Accurate, and Taxonomy-Aware Sequence-Based Prediction of Content of Nucleic Acid Binding Amino Acids. Protein Sci. 2023, 32, e4544. [Google Scholar] [CrossRef]
- Zhang, F.; Li, M.; Zhang, J.; Kurgan, L. HybridRNAbind: Prediction of RNA Interacting Residues across Structure-Annotated and Disorder-Annotated Proteins. Nucleic Acids Res. 2023, 51, e25. [Google Scholar] [CrossRef]
- Chu, L.-C.; Christopoulou, N.; McCaughan, H.; Winterbourne, S.; Cazzola, D.; Wang, S.; Litvin, U.; Brunon, S.; Harker, P.J.; McNae, I.; et al. pyRBDome: A Comprehensive Computational Platform for Enhancing RNA-Binding Proteome Data. Life Sci. Alliance 2024, 7, e202402787. [Google Scholar] [CrossRef]
- Battistelli, C.; Garbo, S.; Riccioni, V.; Montaldo, C.; Santangelo, L.; Vandelli, A.; Strippoli, R.; Tartaglia, G.G.; Tripodi, M.; Cicchini, C. Design and Functional Validation of a Mutant Variant of the LncRNA HOTAIR to Counteract Snail Function in Epithelial-to-Mesenchymal Transition. Cancer Res. 2021, 81, 103–113. [Google Scholar] [CrossRef]
- Owens, M.C.; Clark, S.C.; Yankey, A.; Somarowthu, S. Identifying Structural Domains and Conserved Regions in the Long Non-Coding RNA lncTCF7. Int. J. Mol. Sci. 2019, 20, 4770. [Google Scholar] [CrossRef]
- Fu, J.; Yu, L.; Yan, H.; Tang, S.; Wang, Z.; Dai, T.; Chen, H.; Zhang, S.; Hu, H.; Liu, T.; et al. LncRNAs in Non-Small Cell Lung Cancer: Novel Diagnostic and Prognostic Biomarkers. Front. Mol. Biosci. 2023, 10, 1297198. [Google Scholar] [CrossRef]
- Delli Ponti, R.; Armaos, A.; Marti, S.; Tartaglia, G.G. A Method for RNA Structure Prediction Shows Evidence for Structure in lncRNAs. Front. Mol. Biosci. 2018, 5, 111. [Google Scholar] [CrossRef]
- Spetale, F.E.; Murillo, J.; Villanova, G.V.; Bulacio, P.; Tapia, E. FGGA-Lnc: Automatic Gene Ontology Annotation of lncRNA Sequences Based on Secondary Structures. Interface Focus. 2021, 11, 20200064. [Google Scholar] [CrossRef]
- Chorostecki, U.; Saus, E.; Gabaldón, T. Structural Characterization of NORAD Reveals a Stabilizing Role of Spacers and Two New Repeat Units. Comput. Struct. Biotechnol. J. 2021, 19, 3245–3254. [Google Scholar] [CrossRef]
- Lee, S.; Kopp, F.; Chang, T.-C.; Sataluri, A.; Chen, B.; Sivakumar, S.; Yu, H.; Xie, Y.; Mendell, J.T. Noncoding RNA NORAD Regulates Genomic Stability by Sequestering PUMILIO Proteins. Cell 2016, 164, 69–80. [Google Scholar] [CrossRef]
- Solayman, M.; Litfin, T.; Singh, J.; Paliwal, K.; Zhou, Y.; Zhan, J. Probing RNA Structures and Functions by Solvent Accessibility: An Overview from Experimental and Computational Perspectives. Brief. Bioinform. 2022, 23, bbac112. [Google Scholar] [CrossRef]
- Sanbonmatsu, K. Getting to the Bottom of lncRNA Mechanism: Structure–Function Relationships. Mamm. Genome 2022, 33, 343–353. [Google Scholar] [CrossRef]
- Qi, Y.; Zhang, Y.; Mu, Q.; Zheng, G.; Zhang, M.; Chen, B.; Huang, J.; Ma, C.; Wang, X. RNA Secondary Structurome Revealed Distinct Thermoregulation in Plasmodium Falciparum. Front. Cell Dev. Biol. 2022, 9, 766532. [Google Scholar] [CrossRef]
- Chan, D.; Feng, C.; England, W.E.; Wyman, D.; Flynn, R.A.; Wang, X.; Shi, Y.; Mortazavi, A.; Spitale, R.C. Diverse Functional Elements in RNA Predicted Transcriptome-Wide by Orthogonal RNA Structure Probing. Nucleic Acids Res. 2021, 49, 11868–11882. [Google Scholar] [CrossRef]
- Spitale, R.C.; Incarnato, D. Probing the Dynamic RNA Structurome and Its Functions. Nat. Rev. Genet. 2023, 24, 178–196. [Google Scholar] [CrossRef]
- De Vries, T.; Novakovic, M.; Ni, Y.; Smok, I.; Inghelram, C.; Bikaki, M.; Sarnowski, C.P.; Han, Y.; Emmanouilidis, L.; Padroni, G.; et al. Specific Protein-RNA Interactions Are Mostly Preserved in Biomolecular Condensates. Sci. Adv. 2024, 10, eadm7435. [Google Scholar] [CrossRef]
- Yankey, A.; Clark, S.C.; Owens, M.C.; Somarowthu, S. Purification and Structural Characterization of the Long Noncoding RNAs. Methods Mol. Biol. 2021, 2372, 93–110. [Google Scholar] [CrossRef]
- Saha, K.; Ghosh, G. Chemical Probing of RNA Structure In Vivo Using SHAPE-MaP and DMS-MaP. Methods Mol. Biol. 2023, 2666, 81–93. [Google Scholar] [CrossRef]
- Mitchell, D.; Cotter, J.; Saleem, I.; Mustoe, A.M. Mutation Signature Filtering Enables High-Fidelity RNA Structure Probing at All Four Nucleobases with DMS. Nucleic Acids Res. 2023, 51, 8744–8757. [Google Scholar] [CrossRef]
- Feltes, B.C.; Pinto, É.S.M.; Mangini, A.T.; Dorn, M. NIAS-Server 2.0: A Versatile Complementary Tool for Structural Biology Studies. J. Comput. Chem. 2023, 44, 1610–1623. [Google Scholar] [CrossRef]
- Bugnon, L.A.; Edera, A.A.; Prochetto, S.; Gerard, M.; Raad, J.; Fenoy, E.; Rubiolo, M.; Chorostecki, U.; Gabaldón, T.; Ariel, F.; et al. Secondary Structure Prediction of Long Noncoding RNA: Review and Experimental Comparison of Existing Approaches. Brief. Bioinform. 2022, 23, bbac205. [Google Scholar] [CrossRef]
- Singh, J.; Paliwal, K.; Zhang, T.; Singh, J.; Litfin, T.; Zhou, Y. Improved RNA Secondary Structure and Tertiary Base-Pairing Prediction Using Evolutionary Profile, Mutational Coupling and Two-Dimensional Transfer Learning. Bioinformatics 2021, 37, 2589–2600. [Google Scholar] [CrossRef]
- Matarrese, M.A.G.; Loppini, A.; Nicoletti, M.; Filippi, S.; Chiodo, L. Assessment of Tools for RNA Secondary Structure Prediction and Extraction: A Final-User Perspective. J. Biomol. Struct. Dyn. 2023, 41, 6917–6936. [Google Scholar] [CrossRef]
- Yu, B.; Li, P.; Zhang, Q.C.; Hou, L. Differential Analysis of RNA Structure Probing Experiments at Nucleotide Resolution: Uncovering Regulatory Functions of RNA Structure. Nat. Commun. 2022, 13, 4227. [Google Scholar] [CrossRef]
- Lasher, B.; Hendrix, D.A. bpRNA-Align: Improved RNA Secondary Structure Global Alignment for Comparing and Clustering RNA Structures. RNA 2023, 29, 584–595. [Google Scholar] [CrossRef]
- Ali, S.E.; Mittal, A.; Mathews, D.H. RNA Secondary Structure Analysis Using RNAstructure. Curr. Protoc. 2023, 3, e846. [Google Scholar] [CrossRef]
- Cheng, Y.; Zhang, S.; Xu, X.; Chen, S.-J. Vfold2D-MC: A Physics-Based Hybrid Model for Predicting RNA Secondary Structure Folding. J. Phys. Chem. B 2021, 125, 10108–10118. [Google Scholar] [CrossRef]
- Zhang, H.; Zhang, L.; Liu, K.; Li, S.; Mathews, D.H.; Huang, L. Linear-Time Algorithms for RNA Structure Prediction. In RNA Structure Prediction; Kawaguchi, R.K., Iwakiri, J., Eds.; Methods in Molecular Biology; Springer: New York, NY, USA, 2023; Volume 2586, pp. 15–34. ISBN 978-1-07-162767-9. [Google Scholar]
- Chen, C.-C.; Chan, Y.-M. REDfold: Accurate RNA Secondary Structure Prediction Using Residual Encoder-Decoder Network. BMC Bioinform. 2023, 24, 122. [Google Scholar] [CrossRef]
- Bose, E.; Xiong, S.; Jones, A.N. Probing RNA Structure and Dynamics Using Nanopore and next Generation Sequencing. J. Biol. Chem. 2024, 300, 107317. [Google Scholar] [CrossRef]
- Yang, M.; Zhu, P.; Cheema, J.; Bloomer, R.; Mikulski, P.; Liu, Q.; Zhang, Y.; Dean, C.; Ding, Y. In Vivo Single-Molecule Analysis Reveals COOLAIR RNA Structural Diversity. Nature 2022, 609, 394–399. [Google Scholar] [CrossRef]
- Oleynikov, M.; Jaffrey, S.R. RNA Tertiary Structure and Conformational Dynamics Revealed by BASH MaP. Elife 2024, 13, RP98540. [Google Scholar]
- Olson, S.W.; Turner, A.-M.W.; Arney, J.W.; Saleem, I.; Weidmann, C.A.; Margolis, D.M.; Weeks, K.M.; Mustoe, A.M. Discovery of a Large-Scale, Cell-State-Responsive Allosteric Switch in the 7SK RNA Using DANCE-MaP. Mol. Cell 2022, 82, 1708–1723.e10. [Google Scholar] [CrossRef]
- Luo, L.; Chiu, L.-Y.; Sugarman, A.; Gupta, P.; Rouskin, S.; Tolbert, B.S. HnRNP A1/A2 Proteins Assemble onto 7SK snRNA via Context Dependent Interactions. J. Mol. Biol. 2021, 433, 166885. [Google Scholar] [CrossRef]
- Callaway, E. Revolutionary Cryo-EM Is Taking over Structural Biology. Nature 2020, 578, 201. [Google Scholar] [CrossRef]
- Shepherd, D.C.; Dalvi, S.; Ghosal, D. From Cells to Atoms: Cryo-EM as an Essential Tool to Investigate Pathogen Biology, Host–Pathogen Interaction, and Drug Discovery. Mol. Microbiol. 2022, 117, 610–617. [Google Scholar] [CrossRef]
- Townshend, R.J.L.; Eismann, S.; Watkins, A.M.; Rangan, R.; Karelina, M.; Das, R.; Dror, R.O. Geometric Deep Learning of RNA Structure. Science 2021, 373, 1047–1051. [Google Scholar] [CrossRef]
- El Omari, K.; Duman, R.; Mykhaylyk, V.; Orr, C.M.; Latimer-Smith, M.; Winter, G.; Grama, V.; Qu, F.; Bountra, K.; Kwong, H.S.; et al. Experimental Phasing Opportunities for Macromolecular Crystallography at Very Long Wavelengths. Commun. Chem. 2023, 6, 219. [Google Scholar] [CrossRef]
- Skeparnias, I.; Zhang, J. Structural Basis of NEAT1 lncRNA Maturation and menRNA Instability. Nat. Struct. Mol. Biol. 2024, 31, 1650–1654. [Google Scholar] [CrossRef]
- Kim, D.N.; Thiel, B.C.; Mrozowich, T.; Hennelly, S.P.; Hofacker, I.L.; Patel, T.R.; Sanbonmatsu, K.Y. Zinc-Finger Protein CNBP Alters the 3-D Structure of lncRNA Braveheart in Solution. Nat. Commun. 2020, 11, 148. [Google Scholar] [CrossRef]
- Uroda, T.; Anastasakou, E.; Rossi, A.; Teulon, J.M.; Pellequer, J.L.; Annibale, P.; Pessey, O.; Inga, A.; Chillón, I.; Marcia, M. Conserved Pseudoknots in lncRNA MEG3 Are Essential for Stimulation of the p53 Pathway. Mol. Cell. 2019, 75, 982–995.e9. [Google Scholar] [CrossRef]
- Borodavka, A.; Singaram, S.W.; Stockley, P.G.; Gelbart, W.M.; Ben-Shaul, A.; Tuma, R. Sizes of Long RNA Molecules Are Determined by the Branching Patterns of Their Secondary Structures. Biophys. J. 2016, 111, 2077–2085. [Google Scholar] [CrossRef]
- Jones, A.N.; Sattler, M. Challenges and Perspectives for Structural Biology of lncRNAs—The Example of the Xist lncRNA A-Repeats. J. Mol. Cell Biol. 2019, 11, 845–859. [Google Scholar] [CrossRef]
- Qiu, X.; Li, H.; Ver Steeg, G.; Godzik, A. Advances in AI for Protein Structure Prediction: Implications for Cancer Drug Discovery and Development. Biomolecules 2024, 14, 339. [Google Scholar] [CrossRef]
- Qin, Y.; Chen, Z.; Peng, Y.; Xiao, Y.; Zhong, T.; Yu, X. Deep Learning Methods for Protein Structure Prediction. MedComm–Future Med. 2024, 3, e96. [Google Scholar] [CrossRef]
- Chen, L.; Li, Q.; Nasif, K.F.A.; Xie, Y.; Deng, B.; Niu, S.; Pouriyeh, S.; Dai, Z.; Chen, J.; Xie, C.Y. AI-Driven Deep Learning Techniques in Protein Structure Prediction. Int. J. Mol. Sci. 2024, 25, 8426. [Google Scholar] [CrossRef]
- Brandes, N.; Ofer, D.; Peleg, Y.; Rappoport, N.; Linial, M. ProteinBERT: A Universal Deep-Learning Model of Protein Sequence and Function. Bioinformatics 2022, 38, 2102–2110. [Google Scholar] [CrossRef]
- Krapp, L.F.; Abriata, L.A.; Cortés Rodriguez, F.; Dal Peraro, M. PeSTo: Parameter-Free Geometric Deep Learning for Accurate Prediction of Protein Binding Interfaces. Nat. Commun. 2023, 14, 2175. [Google Scholar] [CrossRef]
- Bryant, P. Deep Learning for Protein Complex Structure Prediction. Curr. Opin. Struct. Biol. 2023, 79, 102529. [Google Scholar] [CrossRef]
- Baek, M.; McHugh, R.; Anishchenko, I.; Jiang, H.; Baker, D.; DiMaio, F. Accurate Prediction of Protein–Nucleic Acid Complexes Using RoseTTAFoldNA. Nat. Methods 2024, 21, 117–121. [Google Scholar] [CrossRef]
- Green, A.G.; Elhabashy, H.; Brock, K.P.; Maddamsetti, R.; Kohlbacher, O.; Marks, D.S. Large-Scale Discovery of Protein Interactions at Residue Resolution Using Co-Evolution Calculated from Genomic Sequences. Nat. Commun. 2021, 12, 1396. [Google Scholar] [CrossRef]
- Liu, H.; Zhuo, C.; Gao, J.; Zeng, C.; Zhao, Y. AI-Integrated Network for RNA Complex Structure and Dynamic Prediction. Biophys. Rev. (Melville) 2024, 5, 041304. [Google Scholar] [CrossRef]
- Zhang, F.; Zhao, B.; Shi, W.; Li, M.; Kurgan, L. DeepDISOBind: Accurate Prediction of RNA-, DNA- and Protein-Binding Intrinsically Disordered Residues with Deep Multi-Task Learning. Brief. Bioinform. 2022, 23, bbab521. [Google Scholar] [CrossRef]
- Hennig, J. Structural Biology of RNA and Protein-RNA Complexes after AlphaFold3. Chembiochem 2025, 26, e202401047. [Google Scholar] [CrossRef]
- Chen, K.; Litfin, T.; Singh, J.; Zhan, J.; Zhou, Y. MARS and RNAcmap3: The Master Database of All Possible RNA Sequences Integrated with RNAcmap for RNA Homology Search. Genom. Proteom. Bioinform. 2024, 22, qzae018. [Google Scholar] [CrossRef]
- Cheng, J.; Lin, Y.; Xu, L.; Chen, K.; Li, Q.; Xu, K.; Ning, L.; Kang, J.; Cui, T.; Huang, Y.; et al. ViRBase v3.0: A Virus and Host ncRNA-Associated Interaction Repository with Increased Coverage and Annotation. Nucleic Acids Res. 2022, 50, D928–D933. [Google Scholar] [CrossRef]
- Sarzynska, J.; Popenda, M.; Antczak, M.; Szachniuk, M. RNA Tertiary Structure Prediction Using RNAComposer in CASP15. Proteins 2023, 91, 1790–1799. [Google Scholar] [CrossRef]
- Zhang, Y.; Xiong, Y.; Yang, C.; Xiao, Y. 3dRNA/DNA: 3D Structure Prediction from RNA to DNA. J. Mol. Biol. 2024, 436, 168742. [Google Scholar] [CrossRef]
- Kamga Youmbi, F.I.; Kengne Tchendji, V.; Tayou Djamegni, C. P-FARFAR2: A Multithreaded Greedy Approach to Sampling Low-Energy RNA Structures in Rosetta FARFAR2. Comput. Biol. Chem. 2023, 104, 107878. [Google Scholar] [CrossRef]
- Watkins, A.M.; Rangan, R.; Das, R. FARFAR2: Improved De Novo Rosetta Prediction of Complex Global RNA Folds. Structure 2020, 28, 963–976.e6. [Google Scholar] [CrossRef]
- Zhang, Y.; Lang, M.; Jiang, J.; Gao, Z.; Xu, F.; Litfin, T.; Chen, K.; Singh, J.; Huang, X.; Song, G.; et al. Multiple Sequence Alignment-Based RNA Language Model and Its Application to Structural Inference. Nucleic Acids Res. 2024, 52, e3. [Google Scholar] [CrossRef]
- Qiu, X. Robust RNA Secondary Structure Prediction with a Mixture of Deep Learning and Physics-Based Experts. Biol. Methods Protoc. 2025, 10, bpae097. [Google Scholar] [CrossRef]
- Zablocki, L.I.; Bugnon, L.A.; Gerard, M.; Di Persia, L.; Stegmayer, G.; Milone, D.H. Comprehensive Benchmarking of Large Language Models for RNA Secondary Structure Prediction. Brief Bioinform. 2025, 26, bbaf137. [Google Scholar] [CrossRef]
- Yi, H.-C.; You, Z.-H.; Wang, M.-N.; Guo, Z.-H.; Wang, Y.-B.; Zhou, J.-R. RPI-SE: A Stacking Ensemble Learning Framework for ncRNA-Protein Interactions Prediction Using Sequence Information. BMC Bioinform. 2020, 21, 60. [Google Scholar] [CrossRef]
- Sato, K.; Akiyama, M.; Sakakibara, Y. RNA Secondary Structure Prediction Using Deep Learning with Thermodynamic Integration. Nat. Commun. 2021, 12, 941. [Google Scholar] [CrossRef]
- Zhu, M.; Mathews, D.H. Sequence Design Using RNAstructure. Methods Mol. Biol. 2025, 2847, 17–31. [Google Scholar] [CrossRef]
- Raden, M.; Miladi, M. How to Do RNA-RNA Interaction Prediction? A Use-Case Driven Handbook Using IntaRNA. Methods Mol. Biol. 2024, 2726, 209–234. [Google Scholar] [CrossRef]
- Li, X.; Qu, W.; Yan, J.; Tan, J. RPI-EDLCN: An Ensemble Deep Learning Framework Based on Capsule Network for ncRNA-Protein Interaction Prediction. J. Chem. Inf. Model. 2024, 64, 2221–2235. [Google Scholar] [CrossRef]
- Liang, Y.; Yin, X.; Zhang, Y.; Guo, Y.; Wang, Y. Predicting lncRNA-Protein Interactions through Deep Learning Framework Employing Multiple Features and Random Forest Algorithm. BMC Bioinform. 2024, 25, 108. [Google Scholar] [CrossRef]
- Shi, F.; Zhang, K.; Cheng, Q.; Che, S.; Zhi, S.; Yu, Z.; Liu, F.; Duan, F.; Wang, Y.; Yang, N. Molecular Mechanism Governing RNA-Binding Property of Mammalian TRIM71 Protein. Sci. Bull. 2024, 69, 72–81. [Google Scholar] [CrossRef]
- Liu, B.; Shi, H.; Al-Hashimi, H.M. Developments in Solution-State NMR Yield Broader and Deeper Views of the Dynamic Ensembles of Nucleic Acids. Curr. Opin. Struct. Biol. 2021, 70, 16–25. [Google Scholar] [CrossRef]
- Zhu, X.; Xie, X.; Das, H.; Tan, B.G.; Shi, Y.; Al-Behadili, A.; Peter, B.; Motori, E.; Valenzuela, S.; Posse, V.; et al. Non-Coding 7S RNA Inhibits Transcription via Mitochondrial RNA Polymerase Dimerization. Cell 2022, 185, 2309–2323.e24. [Google Scholar] [CrossRef]
- Kerr, A.G.; Wang, Z.; Wang, N.; Kwok, K.H.M.; Jalkanen, J.; Ludzki, A.; Lecoutre, S.; Langin, D.; Bergo, M.O.; Dahlman, I.; et al. The Long Noncoding RNA ADIPINT Regulates Human Adipocyte Metabolism via Pyruvate Carboxylase. Nat. Commun. 2022, 13, 2958. [Google Scholar] [CrossRef]
- Schwalbe, H.; Audergon, P.; Haley, N.; Amaro, C.A.; Agirre, J.; Baldus, M.; Banci, L.; Baumeister, W.; Blackledge, M.; Carazo, J.M.; et al. The Future of Integrated Structural Biology. Structure 2024, 32, 1563–1580. [Google Scholar] [CrossRef]
- Ziegler, S.J.; Mallinson, S.J.B.; St. John, P.C.; Bomble, Y.J. Advances in Integrative Structural Biology: Towards Understanding Protein Complexes in Their Cellular Context. Comput. Struct. Biotechnol. J. 2021, 19, 214–225. [Google Scholar] [CrossRef]
- Huang, S.; Hao, X.-Y.; Li, Y.-J.; Wu, J.; Xiang, D.-X.; Luo, S. Nonviral Delivery Systems for Antisense Oligonucleotide Therapeutics. Biomater. Res. 2022, 26, 49. [Google Scholar] [CrossRef]
- Hammond, S.M.; Aartsma-Rus, A.; Alves, S.; Borgos, S.E.; Buijsen, R.A.M.; Collin, R.W.J.; Covello, G.; Denti, M.A.; Desviat, L.R.; Echevarría, L.; et al. Delivery of Oligonucleotide-based Therapeutics: Challenges and Opportunities. EMBO Mol. Med. 2021, 13, e13243. [Google Scholar] [CrossRef]
- Collotta, D.; Bertocchi, I.; Chiapello, E.; Collino, M. Antisense Oligonucleotides: A Novel Frontier in Pharmacological Strategy. Front. Pharmacol. 2023, 14, 1304342. [Google Scholar] [CrossRef]
- Esposito, R.; Bosch, N.; Lanzós, A.; Polidori, T.; Pulido-Quetglas, C.; Johnson, R. Hacking the Cancer Genome: Profiling Therapeutically Actionable Long Non-Coding RNAs Using CRISPR-Cas9 Screening. Cancer Cell 2019, 35, 545–557. [Google Scholar] [CrossRef]
- Jia, Z.; Wang, Y.; Sun, X.; Zhao, X.; Zhang, Y.; Xu, S.; Wang, Y.; Li, Y. Effect of lncRNA XLOC_005950 Knockout by CRISPR/Cas9 Gene Editing on Energy Metabolism and Proliferation in Osteosarcoma MG63 Cells Mediated by hsa-miR-542-3p. Oncol. Lett. 2021, 22, 669. [Google Scholar] [CrossRef]
- Aplin, C.; Milano, S.K.; Zielinski, K.A.; Pollack, L.; Cerione, R.A. Evolving Experimental Techniques for Structure-Based Drug Design. J. Phys. Chem. B 2022, 126, 6599–6607. [Google Scholar] [CrossRef]
- Saur, M.; Hartshorn, M.J.; Dong, J.; Reeks, J.; Bunkoczi, G.; Jhoti, H.; Williams, P.A. Fragment-Based Drug Discovery Using Cryo-EM. Drug Discov. Today 2020, 25, 485–490. [Google Scholar] [CrossRef]
- Stanzione, F.; Giangreco, I.; Cole, J.C. Use of Molecular Docking Computational Tools in Drug Discovery. In Progress in Medicinal Chemistry; Elsevier: Amsterdam, The Netherlands, 2021; Volume 60, pp. 273–343. ISBN 978-0-323-85056-8. [Google Scholar]
- Velmurugan, D.; Pachaiappan, R.; Ramakrishnan, C. Recent Trends in Drug Design and Discovery. Curr. Top. Med. Chem. 2020, 20, 1761–1770. [Google Scholar] [CrossRef]
- Radaeva, M.; Ho, C.-H.; Xie, N.; Zhang, S.; Lee, J.; Liu, L.; Lallous, N.; Cherkasov, A.; Dong, X. Discovery of Novel Lin28 Inhibitors to Suppress Cancer Cell Stemness. Cancers 2022, 14, 5687. [Google Scholar] [CrossRef]
- Byun, W.G.; Lim, D.; Park, S.B. Discovery of Small-Molecule Modulators of Protein–RNA Interactions by Fluorescence Intensity-Based Binding Assay. ChemBioChem 2020, 21, 818–824. [Google Scholar] [CrossRef]
- Borgelt, L.; Li, F.; Hommen, P.; Lampe, P.; Hwang, J.; Goebel, G.L.; Sievers, S.; Wu, P. Trisubstituted Pyrrolinones as Small-Molecule Inhibitors Disrupting the Protein–RNA Interaction of LIN28 and Let-7. ACS Med. Chem. Lett. 2021, 12, 893–898. [Google Scholar] [CrossRef]
- Pedram Fatemi, R.; Salah-Uddin, S.; Modarresi, F.; Khoury, N.; Wahlestedt, C.; Faghihi, M.A. Screening for Small-Molecule Modulators of Long Noncoding RNA-Protein Interactions Using AlphaScreen. SLAS Discov. 2015, 20, 1132–1141. [Google Scholar] [CrossRef]
- Li, A.; Bouhss, A.; Clément, M.-J.; Bauvais, C.; Taylor, J.P.; Bollot, G.; Pastré, D. Using the Structural Diversity of RNA: Protein Interfaces to Selectively Target RNA with Small Molecules in Cells: Methods and Perspectives. Front. Mol. Biosci. 2023, 10, 1298441. [Google Scholar] [CrossRef]
- Mwangi, M.N.; Yonkunas, M.J.; Ageeli, A.A.; McGovern-Gooch, K.R.; Yilmaz, S.; Baird, N.J. A Newly Identified Peripheral Duplex Anchors and Stabilizes the MALAT1 Triplex. Biochemistry 2024, 63, 2280–2292. [Google Scholar] [CrossRef]
- Swain, M.; Ageeli, A.A.; Kasprzak, W.K.; Li, M.; Miller, J.T.; Sztuba-Solinska, J.; Schneekloth, J.S.; Koirala, D.; Piccirili, J.; Fraboni, A.J.; et al. Dynamic Bulge Nucleotides in the KSHV PAN ENE Triple Helix Provide a Unique Binding Platform for Small Molecule Ligands. Nucleic Acids Res. 2021, 49, 13179–13193. [Google Scholar] [CrossRef]

| Method | Principle | Advantages | Limitations | Ref. | |
|---|---|---|---|---|---|
| Protein-Centric | RIP-Seq/RIP-chip | Immunoprecipitation of RBP, followed by RNA identification | Simple, applicable in vivo | Cannot map precise binding sites | [155] |
| CLIP-Seq (HITS-CLIP, iCLIP, PAR-CLIP) | UV crosslinking, IP of RBP, and sequencing | Single-nucleotide resolution; transcriptome-wide | Labor-intensive,RNA loss during purification | [156,168,191] | |
| eCLIP | Enhanced CLIP with size-matched input control | Improved specificity and reproducibility | Requires large sample amounts | [160] | |
| fCLIP | Formaldehyde-based crosslinking CLIP variant | Captures transient or weak interactions | Lower resolution compared to UV-based CLIP | [161] | |
| RBNS | In vitro pull-down of lncRNAs with immobilized RBP | Quantifies binding affinity, statistical modeling | In vitro only, prone to false positives | [152] | |
| seCLIP | Size-matched input and simplified protocol to purify and sequence LPIs | High resolution, efficiency, and scalability | Still relies on UV crosslinking and may require optimization | [192] | |
| RNA-Centric | IVT RNA | Biotin-labeled RNA baits used to isolate RBPs | Straightforward, customizable | In vitro only, RNA misfolding risks | [165] [193] |
| CHIRP/CHART/RAP | Hybridization of biotinylated probes to native RNA, IP of LPIs | In vivo relevance, genome-scale coverage | Requires high expression levels and probe optimization | [170] | |
| MORPH-MS | Multiplex probe hybridization followed by MS | Efficient RNA retrieval with fewer oligos | Still dependent on transcript abundance | [173] | |
| RAP-MS | Biotinylated antisense probes capture target RNA and associated proteins, proteins identified by MS | Specific, enables in vivo RBP identification | Requires effective probe design, low resolution for binding sites | [172] | |
| RPL (RNA Proximity Labeling) | Labeling enzymes are directed to specific RNAs to biotinylate nearby proteins | In vivo compatible, identifies weak/transient interactions | Requires expression optimization | [174] | |
| CARPID | CRISPR assisted RNA–protein interaction detection | Specific to endogenous RNAs, works without crosslinking | Limited by gRNA design and accessibility | [175] | |
| In Silico | LPICGAE, DFRPI, DeepLPI | Machine learning-based prediction from sequence and structural data | Fast screening, captures nonlinear patterns | Dependent on training data quality | [176,179,194] |
| RNAInter, RPISeq, catRAPID | Rule-based or probabilistic sequence feature models | Web-accessible, user-friendly | Limited performance on unseen RNA–protein pairs | [67,195,196] | |
| RNAincoder, SoCube, DeepBtoD | Advanced neural nets trained on interaction databases | Improved performance for large-scale predictions | Limited for low-abundance RNAs | [185,186,188] | |
| DeepA-RBPBS | Deep learning model using CNN, BiGRU, and attention to predict RBP binding sites from RNA sequence and structure | Captures both sequence and structural features, effective across multiple RBPs | Relies on well-annotated training data, may be sensitive to feature encoding quality | [189] | |
| PRIESSTESS | Using regression on binding data, integrating both sequence and secondary structure features | Captures both sequence and structure specificity, applicable to diverse RBPs | May oversimplify complex binding rules, depends on quality of input data | [197] | |
| RBPNet | Predicts CLIP-seq crosslink count distribution from RNA sequence at single-nucleotide resolution, using bias correction and attribution methods | High-resolution predictions, supports eCLIP, iCLIP, and miCLIP, captures known and novel motifs, enables variant impact analysis via in silico mutagenesis | Requires large-scale training data, computationally intensive, performance depends on data quality and assay consistency | [198] | |
| DeepFusion | Integrating RNA sequence and in vivo structural features from DMS-seq to predict RBP binding | Combines local motifs and global context, improved accuracy over sequence-only models, applicable to RNA degradation studies | Depends on high-quality DMS-seq data, computationally intensive | [199] | |
| HDRNet | Deep learning-based framework to predict dynamic RBP binding events under diverse cellular conditions | High accuracy in cross cell type prediction, interpretable motifs, applicable to disease association studies | Requires large, diverse annotated datasets, black-box model may limit mechanistic insight | [200] | |
| qNABpredict | Predicts nucleic acid-binding residue content using support vector regression and computed features from protein sequence | Much faster than residue-level methods, provides informative summary, suitable for large scale analysis | Less detailed than residue-level tool, focuses on content rather than precise binding residues | [201] | |
| HybridRNAbind | Meta-model integrating structure- and disorder-trained predictors for RNA-binding residues | Reduces cross-predictions, works well across structured and disordered regions | Performance depends on quality/diversity of input models, modestly effective on non-matching annotations | [202] | |
| pyRBDome | Pipeline integrating ML-based RBS predictions with structural data to refine RBPs datasets | Enhances confidence in RNA-binding proteome data, improves RBS detection via ensemble learning | Relies on existing structure/ML data quality, structural benchmarks still have known limitations | [203] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Z.; Aftab, M.; Dong, Z.; Jiang, Y.; Liu, K. LncRNA–Protein Interactions: A Key to Deciphering LncRNA Mechanisms. Biomolecules 2025, 15, 881. https://doi.org/10.3390/biom15060881
Wang Z, Aftab M, Dong Z, Jiang Y, Liu K. LncRNA–Protein Interactions: A Key to Deciphering LncRNA Mechanisms. Biomolecules. 2025; 15(6):881. https://doi.org/10.3390/biom15060881
Chicago/Turabian StyleWang, Zuoneng, Muhammad Aftab, Zigang Dong, Yanan Jiang, and Kangdong Liu. 2025. "LncRNA–Protein Interactions: A Key to Deciphering LncRNA Mechanisms" Biomolecules 15, no. 6: 881. https://doi.org/10.3390/biom15060881
APA StyleWang, Z., Aftab, M., Dong, Z., Jiang, Y., & Liu, K. (2025). LncRNA–Protein Interactions: A Key to Deciphering LncRNA Mechanisms. Biomolecules, 15(6), 881. https://doi.org/10.3390/biom15060881






