Advancements in SELEX Technology for Aptamers and Emerging Applications in Therapeutics and Drug Delivery
Abstract
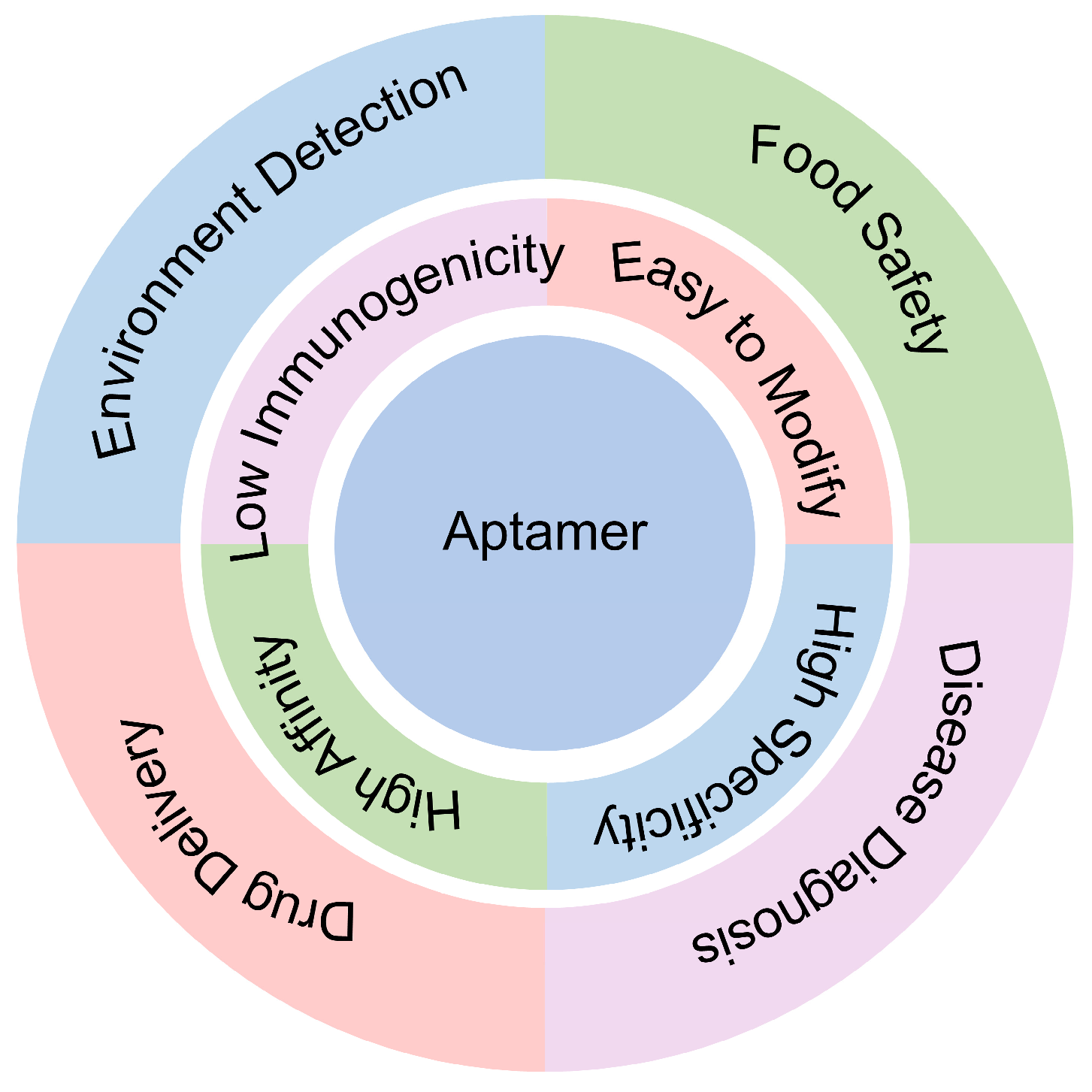
1. Introduction
2. Enhancing SELEX Methodology: Innovations and Refinements
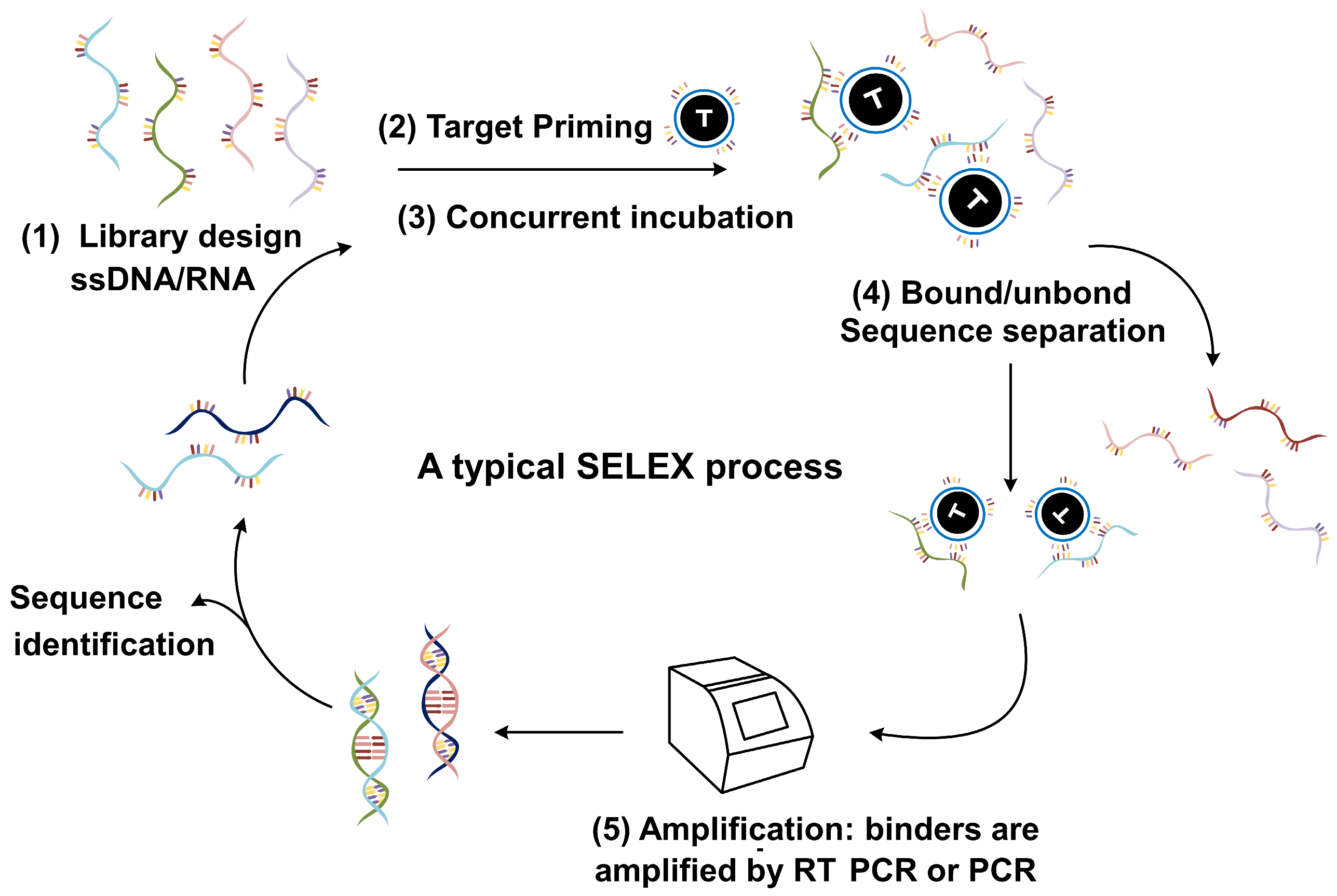
2.1. SELEX Technology
2.2. Advances in SELEX Techniques
2.2.1. Capillary Electrophoresis SELEX
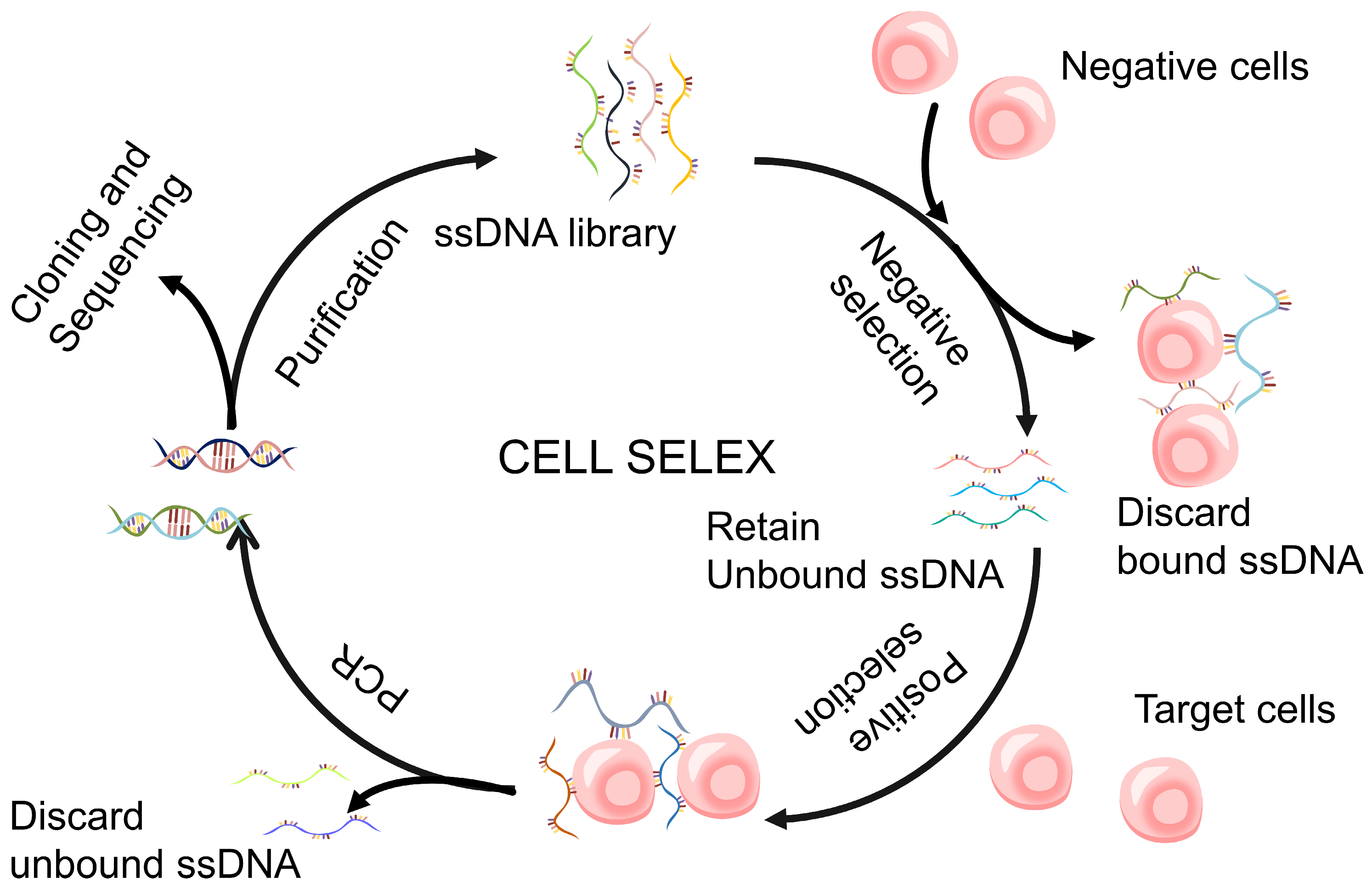
2.2.2. Cell-SELEX
2.2.3. Microfluidic and Microarray SELEX
2.2.4. Integration of NGS into SELEX Protocols
2.3. Challenges and Improvement Strategies for SELEX Technology
3. Application of Aptamers in Therapeutics
3.1. Eye Disorders
3.1.1. Vascular Endothelial Growth Factor
3.1.2. Infections of the Eye
3.2. Thrombosis Disease
3.2.1. Thrombin
3.2.2. von Willebrand Factor
3.2.3. Coagulation Factor Ixa
3.3. Viral and Neurological Diseases
3.4. Cancer
3.5. Challenges in Aptamer-Based Therapeutics
3.5.1. Aptamer Stability
3.5.2. Renal Filtration
3.5.3. Toxicity
4. Application of Aptamers in Drug Delivery
4.1. Targeted Drug Delivery
4.1.1. Prostate-Specific Membrane Antigen (PSMA)
4.1.2. Nucleolin
4.1.3. HIV
4.2. Aptamer–Drug Conjugates
4.2.1. Covalent Conjugation
4.2.2. Noncovalent Conjugation
4.3. Nanomaterial Drug Delivery
5. Conclusions and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Jonczyk, A.; Gottschalk, M.; Mangan, M.S.J.; Majlesain, Y.; Thiem, M.W.; Burbaum, L.-C.; Weighardt, H.; Latz, E.; Mayer, G.; Förster, I. Topical Application of a CCL22-Binding Aptamer Suppresses Contact Allergy. Mol. Ther. Nucleic Acids 2024, 35, 102254. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Chen, C.; Larcher, L.M.; Barrero, R.A.; Veedu, R.N. Three Decades of Nucleic Acid Aptamer Technologies: Lessons Learned, Progress and Opportunities on Aptamer Development. Biotechnol. Adv. Int. Rev. J. 2019, 37, 28–50. [Google Scholar] [CrossRef] [PubMed]
- Xiang, Y.; Lu, Y. DNA as Sensors and Imaging Agents for Metal Ions. Inorg. Chem. 2014, 53, 1925–1942. [Google Scholar] [CrossRef] [PubMed]
- Ruscito, A.; DeRosa, M.C. Small-Molecule Binding Aptamers: Selection Strategies, Characterization, and Applications. Front. Chem. 2016, 4, 14. [Google Scholar] [CrossRef]
- Yan, J.; Xiong, H.; Cai, S.; Wen, N.; Liu, Z. Advances in Aptamer Screening Technologies. Talanta 2019, 200, 124–144. [Google Scholar] [CrossRef]
- Tan, W.; Donovan, M.J.; Jiang, J. Aptamers from Cell-Based Selection for Bioanalytical Applications. Chem. Rev. 2013, 113, 2842–2862. [Google Scholar] [CrossRef]
- Gupta, R.; Rahi Alhachami, F.; Khalid, I.; Majdi, H.S.; Nisar, N.; Mohamed Hasan, Y.; Sivaraman, R.; Romero Parra, R.M.; Al Mashhadani, Z.I.; Fakri Mustafa, Y. Recent Progress in Aptamer-Functionalized Metal-Organic Frameworks-Based Optical and Electrochemical Sensors for Detection of Mycotoxins. Crit. Rev. Anal. Chem. 2024, 54, 1707–1728. [Google Scholar] [CrossRef]
- Tuerk, C.; Gold, L. Systematic Evolution of Ligands by Exponential Enrichment: RNA Ligands to Bacteriophage T4 DNA Polymerase. Science 1990, 249, 505–510. [Google Scholar] [CrossRef]
- Mendonsa, S.D.; Bowser, M.T. In Vitro Selection of Aptamers with Affinity for Neuropeptide Y Using Capillary Electrophoresis. J. Am. Chem. Soc. 2005, 127, 9382–9383. [Google Scholar] [CrossRef]
- Rosch, J.C.; Balikov, D.A.; Gong, F.; Lippmann, E.S. A Systematic Evolution of Ligands by Exponential Enrichment Workflow with Consolidated Counterselection to Efficiently Isolate High-affinity Aptamers. Eng. Rep. 2020, 2, e12089. [Google Scholar] [CrossRef]
- Yang, G.; Zhu, C.; Zhao, L.; Li, L.; Huang, Y.; Zhang, Y.; Qu, F. Pressure Controllable Aptamers Picking Strategy by Targets Competition. Chin. Chem. Lett. 2021, 32, 218–220. [Google Scholar] [CrossRef]
- Dong, H.; Wang, H.; Guo, Z.; Huang, J.; Zhang, P.; Guo, Y.; Sun, X. Combination of Capture-SELEX and Post-SELEX for Procymidone-Specific Aptamer Selection and Broad-Specificity Aptamer Discovery, and Development of Aptamer-Based Lateral Flow Assay. Anal. Chim. Acta 2024, 1318, 342922. [Google Scholar] [CrossRef] [PubMed]
- Torkamanian-Afshar, M.; Nematzadeh, S.; Tabarzad, M.; Najafi, A.; Lanjanian, H.; Masoudi-Nejad, A. In Silico Design of Novel Aptamers Utilizing a Hybrid Method of Machine Learning and Genetic Algorithm. Mol. Divers. 2021, 25, 1395–1407. [Google Scholar] [CrossRef] [PubMed]
- Komarova, N.; Barkova, D.; Kuznetsov, A. Implementation of High-Throughput Sequencing (HTS) in Aptamer Selection Technology. Int. J. Mol. Sci. 2020, 21, 8774. [Google Scholar] [CrossRef]
- Nalewaj, M.; Szabat, M. Examples of Structural Motifs in Viral Genomes and Approaches for RNA Structure Characterization. Int. J. Mol. Sci. 2022, 23, 15917. [Google Scholar] [CrossRef]
- Morita, Y.; Leslie, M.; Kameyama, H.; Volk, D.E.; Tanaka, T. Aptamer Therapeutics in Cancer: Current and Future. Cancers 2018, 10, 80. [Google Scholar] [CrossRef]
- Ma, H.; Liu, J.; Ali, M.M.; Mahmood, M.A.I.; Labanieh, L.; Lu, M.; Iqbal, S.M.; Zhang, Q.; Zhao, W.; Wan, Y. Nucleic Acid Aptamers in Cancer Research, Diagnosis and Therapy. Chem. Soc. Rev. 2015, 44, 1240–1256. [Google Scholar] [CrossRef]
- Chen, K.; Zhang, Y.; Zhu, L.; Chu, H.; Shao, X.; Asakiya, C.; Huang, K.; Xu, W. Insights into Nucleic Acid-Based Self-Assembling Nanocarriers for Targeted Drug Delivery and Controlled Drug Release. J. Control. Release 2022, 341, 869–891. [Google Scholar] [CrossRef]
- Yan, C.; Miao, L.; Zhang, Y.; Zhou, X.; Wang, G.; Li, Y.; Qiao, Q.; Xu, Z. Dye Disaggregation Light-up Aptamer for Super-Resolution RNA Imaging. Sens. Actuators B Chem. 2023, 386, 133731. [Google Scholar] [CrossRef]
- Bush, K. The Interrogation of Cas9 Aptamers and sgRNA Structures Through SELEX. Ph.D. Thesis, Duke University, Durham, NC, USA, 2022. [Google Scholar]
- Mendonsa, S.D.; Bowser, M.T. In Vitro Evolution of Functional DNA Using Capillary Electrophoresis. J. Am. Chem. Soc. 2004, 126, 20–21. [Google Scholar] [CrossRef]
- Ellington, A.D.; Szostak, J.W. In Vitro Selection of RNA Molecules That Bind Specific Ligands. Nature 1990, 346, 818–822. [Google Scholar] [CrossRef] [PubMed]
- Zhu, C.; Yang, G.; Ghulam, M.; Li, L.; Qu, F. Evolution of Multi-Functional Capillary Electrophoresis for High-Efficiency Selection of Aptamers. Biotechnol. Adv. 2019, 37, 107432. [Google Scholar] [CrossRef] [PubMed]
- Kanoatov, M.; Galievsky, V.A.; Krylova, S.M.; Cherney, L.T.; Jankowski, H.K.; Krylov, S.N. Using Nonequilibrium Capillary Electrophoresis of Equilibrium Mixtures (NECEEM) for Simultaneous Determination of Concentration and Equilibrium Constant. Anal. Chem. 2015, 87, 3099–3106. [Google Scholar] [CrossRef] [PubMed]
- Musheev, M.U.; Krylov, S.N. Selection of Aptamers by Systematic Evolution of Ligands by Exponential Enrichment: Addressing the Polymerase Chain Reaction Issue. Anal. Chim. Acta 2006, 564, 91–96. [Google Scholar] [CrossRef]
- Berezovski, M.; Musheev, M.; Drabovich, A.; Krylov, S.N. Non-SELEX Selection of Aptamers. J. Am. Chem. Soc. 2006, 128, 1410–1411. [Google Scholar] [CrossRef]
- Li, Q.; Zhao, X.; Liu, H.; Qu, F. Low pH Capillary Electrophoresis Application to Improve Capillary Electrophoresis-Systematic Evolution of Ligands by Exponential Enrichment. J. Chromatogr. A 2014, 1364, 289–294. [Google Scholar] [CrossRef]
- Zhu, C.; Li, L.; Yang, G.; Fang, S.; Liu, M.; Ghulam, M.; Hao, C.; Chen, Y.; Qu, F. Online Reaction Based Single-Step Capillary Electrophoresis-Systematic Evolution of Ligands by Exponential Enrichment for ssDNA Aptamers Selection. Anal. Chim. Acta 2019, 1070, 112–122. [Google Scholar] [CrossRef]
- Riley, K.R.; Saito, S.; Gagliano, J.; Colyer, C.L. Facilitating Aptamer Selection and Collection by Capillary Transient Isotachophoresis with Laser-Induced Fluorescence Detection. J. Chromatogr. A 2014, 1368, 183–189. [Google Scholar] [CrossRef]
- Luo, Z.; Zhou, H.; Jiang, H.; Ou, H.; Li, X.; Zhang, L. Development of a Fraction Collection Approach in Capillary Electrophoresis SELEX for Aptamer Selection. Analyst 2015, 140, 2664–2670. [Google Scholar] [CrossRef]
- Wakui, K.; Yoshitomi, T.; Yamaguchi, A.; Tsuchida, M.; Saito, S.; Shibukawa, M.; Furusho, H.; Yoshimoto, K. Rapidly Neutralizable and Highly Anticoagulant Thrombin-Binding DNA Aptamer Discovered by MACE SELEX. Mol. Ther. Nucleic Acids 2019, 16, 348–359. [Google Scholar] [CrossRef]
- Takao, J.; Nagai, R.; Endo, T.; Hisamoto, H.; Sueyoshi, K. Aptamer Selection Based on Microscale Electrophoretic Filtration Using a Hydrogel-Plugged Capillary Device. Molecules 2022, 27, 5818. [Google Scholar] [CrossRef]
- Yang, G.; Zhu, C.; Liu, X.-H.; Wang, Y.; Qu, F. Screening of Clenbuterol Hydrochloride Aptamers Based on Capillary Electrophoresis. Chin. J. Anal. Chem. 2018, 46, 1595–1603. [Google Scholar] [CrossRef]
- Lou, B.; Chen, E.; Zhao, X.; Qu, F.; Yan, J. The Application of Capillary Electrophoresis for Assisting Whole-Cell Aptamers Selection by Characterizing Complete ssDNA Distribution. J. Chromatogr. A 2016, 1437, 203–209. [Google Scholar] [CrossRef] [PubMed]
- Yang, G.; Tian, W.; Hu, Y.; Wen, Y.; Zhao, Y.; Jiang, G.; Liu, H.; Zhao, L.; Zhu, C.; Qu, F. Real-Time Visualization of “Small Molecules-ssDNA” Complexes for Aptamer Screening Based on Online Competition CE-SELEX. Talanta 2025, 284, 127199. [Google Scholar] [CrossRef] [PubMed]
- Shaukat, A.; Chrouda, A.; Sadaf, S.; Alhamlan, F.; Eissa, S.; Zourob, M. Cell-SELEX for Aptamer Discovery and Its Utilization in Constructing Electrochemical Biosensor for Rapid and Highly Sensitive Detection of Legionella Pneumophila Serogroup 1. Sci. Rep. 2024, 14, 14132. [Google Scholar] [CrossRef]
- Homann, M.; Göringer, H.U. Combinatorial Selection of High Affinity RNA Ligands to Live African Trypanosomes. Nucleic Acids Res. 1999, 27, 2006–2014. [Google Scholar] [CrossRef]
- Kaur, H. Recent Developments in Cell-SELEX Technology for Aptamer Selection. Biochim. Et Biophys. Acta (BBA) Gen. Subj. 2018, 1862, 2323–2329. [Google Scholar] [CrossRef]
- Hicke, B.J.; Marion, C.; Chang, Y.-F.; Gould, T.; Lynott, C.K.; Parma, D.; Schmidt, P.G.; Warren, S. Tenascin-C Aptamers Are Generated Using Tumor Cells and Purified Protein. J. Biol. Chem. 2001, 276, 48644–48654. [Google Scholar] [CrossRef]
- Razlansari, M.; Jafarinejad, S.; Rahdar, A.; Shirvaliloo, M.; Arshad, R.; Fathi-Karkan, S.; Mirinejad, S.; Sargazi, S.; Sheervalilou, R.; Ajalli, N. Development and Classification of RNA Aptamers for Therapeutic Purposes: An Updated Review with Emphasis on Cancer. Mol. Cell. Biochem. 2023, 478, 1573–1598. [Google Scholar] [CrossRef]
- Zumrut, H.E.; Batool, S.; Argyropoulos, K.V.; Williams, N.; Azad, R.; Mallikaratchy, P.R. Integrating Ligand-Receptor Interactions and in Vitro Evolution for Streamlined Discovery of Artificial Nucleic Acid Ligands. Mol. Ther. Nucleic Acids 2019, 17, 150–163. [Google Scholar] [CrossRef]
- Moccia, F.; Platella, C.; Musumeci, D.; Batool, S.; Zumrut, H.; Bradshaw, J.; Mallikaratchy, P.; Montesarchio, D. The Role of G-Quadruplex Structures of LIGS-Generated Aptamers R1. 2 and R1. 3 in IgM Specific Recognition. Int. J. Biol. Macromol. 2019, 133, 839–849. [Google Scholar] [CrossRef]
- Batool, S.; Argyropoulos, K.V.; Azad, R.; Okeoma, P.; Zumrut, H.; Bhandari, S.; Dekhang, R.; Mallikaratchy, P.R. Dimerization of an Aptamer Generated from Ligand-Guided Selection (LIGS) Yields a High Affinity Scaffold against B-Cells. Biochim. Et Biophys. Acta (BBA) Gen. Subj. 2019, 1863, 232–240. [Google Scholar] [CrossRef] [PubMed]
- Kumar, V.; Raj, S.B.; Kanakaraj, L.; Paul, A.D.; Kavitha, K.; Ravi, M.; Sucharitha, P. Aptamer: A Review on It’s In Vitro Selection and Drug Delivery System. Int. J. Appl. Pharm 2022, 14, 35–42. [Google Scholar] [CrossRef]
- Souza, G.R.; Molina, J.R.; Raphael, R.M.; Ozawa, M.G.; Stark, D.J.; Levin, C.S.; Bronk, L.F.; Ananta, J.S.; Mandelin, J.; Georgescu, M.-M. Three-Dimensional Tissue Culture Based on Magnetic Cell Levitation. Nat. Nanotechnol. 2010, 5, 291–296. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Li, H.; Jia, W.; Chen, Z.; Xu, D. Selection of Aptamers Based on a Protein Microarray Integrated with a Microfluidic Chip. Lab Chip 2017, 17, 178–185. [Google Scholar] [CrossRef]
- Jia, W.; Li, H.; Wilkop, T.; Liu, X.; Yu, X.; Cheng, Q.; Xu, D.; Chen, H.-Y. Silver Decahedral Nanoparticles Empowered SPR Imaging-SELEX for High Throughput Screening of Aptamers with Real-Time Assessment. Biosens. Bioelectron. 2018, 109, 206–213. [Google Scholar] [CrossRef]
- Yu, F.; Li, H.; Sun, W.; Xu, D.; He, F. Rapid Selection of Aptamers Based on Protein Microarray. RSC Adv. 2019, 9, 9762–9768. [Google Scholar] [CrossRef]
- Hybarger, G.; Bynum, J.; Williams, R.F.; Valdes, J.J.; Chambers, J.P. A Microfluidic SELEX Prototype. Anal. Bioanal. Chem. 2006, 384, 191–198. [Google Scholar] [CrossRef]
- Kim, J.; Olsen, T.R.; Zhu, J.; Hilton, J.P.; Yang, K.-A.; Pei, R.; Stojanovic, M.N.; Lin, Q. Integrated Microfluidic Isolation of Aptamers Using Electrophoretic Oligonucleotide Manipulation. Sci. Rep. 2016, 6, 26139. [Google Scholar] [CrossRef]
- McNamara, S.L.; Brudno, Y.; Miller, A.B.; Ham, H.O.; Aizenberg, M.; Chaikof, E.L.; Mooney, D.J. Regenerating Antithrombotic Surfaces through Nucleic Acid Displacement. ACS Biomater. Sci. Eng. 2020, 6, 2159–2166. [Google Scholar] [CrossRef]
- Lorenz, C.; Gesell, T.; Zimmermann, B.; Schoeberl, U.; Bilusic, I.; Rajkowitsch, L.; Waldsich, C.; Von Haeseler, A.; Schroeder, R. Genomic SELEX for Hfq-Binding RNAs Identifies Genomic Aptamers Predominantly in Antisense Transcripts. Nucleic Acids Res. 2010, 38, 3794–3808. [Google Scholar] [CrossRef] [PubMed]
- Didarian, R.; Ozbek, H.K.; Ozalp, V.C.; Erel, O.; Yildirim-Tirgil, N. Enhanced SELEX Platforms for Aptamer Selection with Improved Characteristics: A Review. Mol. Biotechnol. 2024, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Zhou, Z.; Zheng, L.; Gong, Z.; Li, Y.; Jin, Y.; Huang, Y.; Chi, M. Urinary Tract Infections Caused by Uropathogenic Escherichia Coli: Mechanisms of Infection and Treatment Options. Int. J. Mol. Sci. 2023, 24, 10537. [Google Scholar] [CrossRef] [PubMed]
- Reiss, D.J.; Howard, F.M.; Mobley, H.L. A Novel Approach for Transcription Factor Analysis Using SELEX with High-Throughput Sequencing (TFAST). PLoS ONE 2012, 7, e42761. [Google Scholar] [CrossRef]
- Lakhin, A.V.; Tarantul, V.Z.; Gening, L. Aptamers: Problems, Solutions and Prospects. Acta Naturae 2013, 5, 34–43. [Google Scholar] [CrossRef]
- Kohlberger, M.; Gadermaier, G. SELEX: Critical Factors and Optimization Strategies for Successful Aptamer Selection. Biotechnol. Appl. Biochem. 2022, 69, 1771–1792. [Google Scholar] [CrossRef]
- Douaki, A.; Garoli, D.; Inam, A.S.; Angeli, M.A.C.; Cantarella, G.; Rocchia, W.; Wang, J.; Petti, L.; Lugli, P. Smart Approach for the Design of Highly Selective Aptamer-Based Biosensors. Biosensors 2022, 12, 574. [Google Scholar] [CrossRef]
- Deng, J.; Qin, Y. Advancements and Emerging Trends in Ophthalmic Anti-VEGF Therapy: A Bibliometric Analysis. Int. Ophthalmol. 2024, 44, 368. [Google Scholar] [CrossRef]
- Zhang, M.; Lu, X.; Li, G.; Zhang, S.; Zhang, J.; Fu, X.; Sun, F. Advancements in Neovascular Ophthalmopathy: Therapeutic Strategies and Future Prospects. Authorea Prepr. 2023. [Google Scholar] [CrossRef]
- Tucker, C.E.; Chen, L.-S.; Judkins, M.B.; Farmer, J.A.; Gill, S.C.; Drolet, D.W. Detection and Plasma Pharmacokinetics of an Anti-Vascular Endothelial Growth Factor Oligonucleotide-Aptamer (NX1838) in Rhesus Monkeys. J. Chromatogr. B Biomed. Sci. Appl. 1999, 732, 203–212. [Google Scholar] [CrossRef]
- Novack, G.D.; Robin, A.L. Ocular Pharmacology. J. Clin. Pharmacol. 2024, 64, 1068–1082. [Google Scholar] [CrossRef] [PubMed]
- Torkashvand, A.; Izadian, A.; Hajrasouliha, A. Advances in Ophthalmic Therapeutic Delivery: A Comprehensive Overview of Present and Future Directions. Surv. Ophthalmol. 2024, 69, 967–983. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Wasmuth, S.; Bauer, D.; Baehler, H.; Hennig, M.; Heiligenhaus, A. Subconjunctival Antisense Oligonucleotides Targeting TNF-α Influence Immunopathology and Viral Replication in Murine HSV-1 Retinitis. Graefes Arch. Clin. Exp. Ophthalmol. 2008, 246, 1265–1273. [Google Scholar] [CrossRef] [PubMed]
- Moerdyk-Schauwecker, M.; Stein, D.A.; Eide, K.; Blouch, R.E.; Bildfell, R.; Iversen, P.; Jin, L. Inhibition of HSV-1 Ocular Infection with Morpholino Oligomers Targeting ICP0 and ICP27. Antivir. Res. 2009, 84, 131–141. [Google Scholar] [CrossRef]
- Wasmuth, S.; Bauer, D.; Steuhl, K.-P.; Heiligenhaus, A. Topical Antisense-Oligonucleotides Targeting IFN-Gamma mRNA Improve Incidence and Severity of Herpetic Stromal Keratitis by Cytokine Specific and Sequence Unspecific Effects. Graefes Arch. Clin. Exp. Ophthalmol. 2008, 246, 443–451. [Google Scholar] [CrossRef]
- Zyzak, J.; Mitkiewicz, M.; Leszczyńska, E.; Reniewicz, P.; Moynagh, P.N.; Siednienko, J. HSV-1/TLR9-Mediated IFNβ and TNFα Induction Is Mal-Dependent in Macrophages. J. Innate Immun. 2020, 12, 387–398. [Google Scholar] [CrossRef]
- Elg, M.; Gustafsson, D.; Carlsson, S. Antithrombotic Effects and Bleeding Time of Thrombin Inhibitors and Warfarin in the Rat. Thromb. Res. 1999, 94, 187–197. [Google Scholar] [CrossRef]
- Mittal, P.; Sayar, Z.; Cohen, H. Warfarin and Heparin Monitoring in Antiphospholipid Syndrome. Hematology 2024, 2024, 192–199. [Google Scholar] [CrossRef]
- Bock, L.C.; Griffin, L.C.; Latham, J.A.; Vermaas, E.H.; Toole, J.J. Selection of Single-Stranded DNA Molecules That Bind and Inhibit Human Thrombin. Nature 1992, 355, 564–566. [Google Scholar] [CrossRef]
- Liu, M.; Zaman, K.; Fortenberry, Y.M. Overview of the Therapeutic Potential of Aptamers Targeting Coagulation Factors. Int. J. Mol. Sci. 2021, 22, 3897. [Google Scholar] [CrossRef]
- Kretz, C.A.; Stafford, A.R.; Fredenburgh, J.C.; Weitz, J.I. HD1, a Thrombin-Directed Aptamer, Binds Exosite 1 on Prothrombin with High Affinity and Inhibits Its Activation by Prothrombinase. J. Biol. Chem. 2015, 290, 4813. [Google Scholar] [CrossRef] [PubMed]
- Macaya, R.F.; Schultze, P.; Smith, F.W.; Roe, J.A.; Feigon, J. Thrombin-Binding DNA Aptamer Forms a Unimolecular Quadruplex Structure in Solution. Proc. Natl. Acad. Sci. USA 1993, 90, 3745–3749. [Google Scholar] [CrossRef] [PubMed]
- De Fenza, M.; Eremeeva, E.; Troisi, R.; Yang, H.; Esposito, A.; Sica, F.; Herdewijn, P.; D’Alonzo, D.; Guaragna, A. Structure-Activity Relationship Study of a Potent α-Thrombin Binding Aptamer Incorporating Hexitol Nucleotides. Chemistry 2020, 26, 9589–9597. [Google Scholar] [CrossRef] [PubMed]
- De Caterina, R.; Moliterno, D.; Kristensen, S. The ESC Textbook of Thrombosis; Oxford University Press: Oxford, UK, 2024; ISBN 0-19-286922-1. [Google Scholar]
- Induruwa, I.P. Platelet Receptor Glycoprotein VI in Ischaemic Stroke. Ph.D. Thesis, University of Cambridge, Cambridge, UK, 2020. [Google Scholar]
- Tscharre, M.; Michelson, A.D.; Gremmel, T. Novel Antiplatelet Agents in Cardiovascular Disease. J. Cardiovasc. Pharmacol. Ther. 2020, 25, 191–200. [Google Scholar] [CrossRef]
- Kovacevic, K.D.; Greisenegger, S.; Langer, A.; Gelbenegger, G.; Buchtele, N.; Pabinger, I.; Petroczi, K.; Zhu, S.; Gilbert, J.C.; Jilma, B. The Aptamer BT200 Blocks von Willebrand Factor and Platelet Function in Blood of Stroke Patients. Sci. Rep. 2021, 11, 3092. [Google Scholar] [CrossRef]
- Chion, A.; Byrne, C.; Atiq, F.; Doherty, D.; Aguila, S.; Fazavana, J.; Lopes, P.; Karampini, E.; Amin, A.; Preston, R.J. The Aptamer BT200 Blocks Interaction of K1405-K1408 in the VWF-A1 Domain with Macrophage LRP1. Blood 2024, 144, 1445–1456. [Google Scholar] [CrossRef]
- Nimjee, S.M.; Dornbos, D.; Pitoc, G.A.; Wheeler, D.G.; Layzer, J.M.; Venetos, N.; Huttinger, A.; Talentino, S.E.; Musgrave, N.J.; Moody, H. Preclinical Development of a vWF Aptamer to Limit Thrombosis and Engender Arterial Recanalization of Occluded Vessels. Mol. Ther. 2019, 27, 1228–1241. [Google Scholar] [CrossRef]
- Soloveva, P.A.; Podoplelova, N.A.; Panteleev, M.A. Binding of Coagulation Factor IXa to Procoagulant Platelets Revisited: Low Affinity and Interactions with Other Factors. Biochem. Biophys. Res. Commun. 2024, 720, 150099. [Google Scholar] [CrossRef]
- Bos, M.H.; van Diest, R.E.; Monroe, D.M. Blood Coagulation Factor IX: Structural Insights Impacting Hemophilia B Therapy. Blood 2024, 144, 2198–2210. [Google Scholar] [CrossRef]
- Chan, M.Y.; Rusconi, C.P.; Alexander, J.H.; Tonkens, R.M.; Harrington, R.A.; Becker, R.C. A Randomized, Repeat-dose, Pharmacodynamic and Safety Study of an Antidote-controlled Factor IXa Inhibitor. J. Thromb. Haemost. 2008, 6, 789–796. [Google Scholar] [CrossRef]
- Vavalle, J.P.; Cohen, M.G. The REG1 Anticoagulation System: A Novel Actively Controlled Factor IX Inhibitor Using RNA Aptamer Technology for Treatment of Acute Coronary Syndrome. Future Cardiol. 2012, 8, 371–382. [Google Scholar] [CrossRef] [PubMed]
- Gailani, D.; Gruber, A. Targeting Factor XI and Factor XIa to Prevent Thrombosis. Blood 2024, 143, 1465–1475. [Google Scholar] [CrossRef] [PubMed]
- Povsic, T.J.; Vavalle, J.P.; Aberle, L.H.; Kasprzak, J.D.; Cohen, M.G.; Mehran, R.; Bode, C.; Buller, C.E.; Montalescot, G.; Cornel, J.H. A Phase 2, Randomized, Partially Blinded, Active-Controlled Study Assessing the Efficacy and Safety of Variable Anticoagulation Reversal Using the REG1 System in Patients with Acute Coronary Syndromes: Results of the RADAR Trial. Eur. Heart J. 2013, 34, 2481–2489. [Google Scholar] [CrossRef] [PubMed]
- Kharvani, H.R.; Aghanajafi, C. Investigation into the Two-Way Interaction of Coronary Flow and Heart Function in Coronary Artery Disease Predicted by a Computational Model of Autoregulation of Coronary Flow. J. Biomech. 2024, 164, 111970. [Google Scholar] [CrossRef]
- Song, Y.; Song, J.; Wei, X.; Huang, M.; Sun, M.; Zhu, L.; Lin, B.; Shen, H.; Zhu, Z.; Yang, C. Discovery of Aptamers Targeting the Receptor-Binding Domain of the SARS-CoV-2 Spike Glycoprotein. Anal. Chem. 2020, 92, 9895–9900. [Google Scholar] [CrossRef]
- Liu, X.; Wang, Y.; Wu, J.; Qi, J.; Zeng, Z.; Wan, Q.; Chen, Z.; Manandhar, P.; Cavener, V.S.; Boyle, N.R.; et al. Neutralizing Aptamers Block S/RBD-ACE2 Interactions and Prevent Host Cell Infection. Angew. Chem. Int. Ed. 2021, 60, 10273–10278. [Google Scholar] [CrossRef]
- Negahdary, M.; Angnes, L. Electrochemical Aptamer-Based Nanobiosensors for Diagnosing Alzheimer’s Disease: A Review. Biomater. Adv. 2022, 135, 112689. [Google Scholar] [CrossRef]
- Zheng, Y.; Zhang, L.; Zhao, J.; Li, L.; Wang, M.; Gao, P.; Wang, Q.; Zhang, X.; Wang, W. Advances in Aptamers against Aβ and Applications in Aβ Detection and Regulation for Alzheimer’s Disease. Theranostics 2022, 12, 2095. [Google Scholar] [CrossRef]
- You, X.; Gopinath, S.C.B.; Lakshmipriya, T.; Li, D. High-Affinity Detection of Alpha-Synuclein by Aptamer-Gold Conjugates on an Amine-Modified Dielectric Surface. J. Anal. Methods Chem. 2019, 2019, 6526850. [Google Scholar] [CrossRef]
- Yang, X.; Zhao, X.; Liu, F.; Li, H.; Zhang, C.X.; Yang, Z. Simple, Rapid and Sensitive Detection of Parkinson’s Disease Related Alpha-Synuclein Using a DNA Aptamer Assisted Liquid Crystal Biosensor. Soft Matter 2021, 17, 4842–4847. [Google Scholar] [CrossRef]
- Zheng, Y.; Qu, J.; Xue, F.; Zheng, Y.; Yang, B.; Chang, Y.; Yang, H.; Zhang, J. Novel DNA Aptamers for Parkinson’s Disease Treatment Inhibit α-Synuclein Aggregation and Facilitate Its Degradation. Mol. Ther. Nucleic Acids 2018, 11, 228–242. [Google Scholar] [CrossRef] [PubMed]
- Lopes-Nunes, J.; Oliveira, P.A.; Cruz, C. Enhanced Targeted Liposomal Delivery of Imiquimod via Aptamer Functionalization for Head and Neck Cancer Therapy. Colloids Surf. B Biointerfaces 2024, 243, 114121. [Google Scholar] [CrossRef] [PubMed]
- Hazazi, A.; Khan, F.R.; Albloui, F.; Arif, S.; Abdulaziz, O.; Alhomrani, M.; Sindi, A.A.; Abu-Alghayth, M.H.; Abalkhail, A.; Nassar, S.A. Signaling Pathways in HPV-Induced Cervical Cancer: Exploring the Therapeutic Promise of RNA Modulation. Pathol. Res. Pract. 2024, 263, 155612. [Google Scholar] [CrossRef] [PubMed]
- Bamrungsap, S.; Chen, T.; Shukoor, M.I.; Chen, Z.; Sefah, K.; Chen, Y.; Tan, W. Pattern Recognition of Cancer Cells Using Aptamer-Conjugated Magnetic Nanoparticles. ACS Nano 2012, 6, 3974–3981. [Google Scholar] [CrossRef]
- Zhang, N.; Li, J.; Liu, B.; Zhang, D.; Zhang, C.; Guo, Y.; Chu, X.; Wang, W.; Wang, H.; Yan, X. Signal Enhancing Strategies in Aptasensors for the Detection of Small Molecular Contaminants by Nanomaterials and Nucleic Acid Amplification. Talanta 2022, 236, 122866. [Google Scholar] [CrossRef]
- Sun, D.; Ma, Y.; Wu, M.; Chen, Z.; Zhang, L.; Lu, J. Recent Progress in Aptamer-Based Microfluidics for the Detection of Circulating Tumor Cells and Extracellular Vesicles. J. Pharm. Anal. 2023, 13, 340–354. [Google Scholar] [CrossRef]
- Ye, Q.; Zhang, Z.; Liu, J.; Wang, X. Screen-Printed Electrode-Based Biosensors Modified with Functional Nucleic Acid Probes and Their Applications in This Pandemic Age: A Review. Anal. Methods 2022, 14, 2961–2975. [Google Scholar] [CrossRef]
- Qu, L.; Xu, J.; Tan, X.; Liu, Z.; Xu, L.; Peng, R. Dual-Aptamer Modification Generates a Unique Interface for Highly Sensitive and Specific Electrochemical Detection of Tumor Cells. ACS Appl. Mater. Interfaces 2014, 6, 7309–7315. [Google Scholar] [CrossRef]
- Nam, E.J.; Kim, E.J.; Wark, A.W.; Rho, S.; Kim, H.; Lee, H.J. Highly Sensitive Electrochemical Detection of Proteins Using Aptamer-Coated Gold Nanoparticles and Surface Enzyme Reactions. Analyst 2012, 137, 2011–2016. [Google Scholar] [CrossRef]
- Odeh, F.; Nsairat, H.; Alshaer, W.; Ismail, M.A.; Esawi, E.; Qaqish, B.; Bawab, A.A.; Ismail, S.I. Aptamers Chemistry: Chemical Modifications and Conjugation Strategies. Molecules 2019, 25, 3. [Google Scholar] [CrossRef]
- Miranda-Castro, R.; de-los-Santos-Álvarez, N.; Lobo-Castañón, M.J. Characterization of Aptamer-Ligand Complexes. In Aptamers for Analytical Applications: Affinity Acquisition and Method Design; John Wiley & Sons: Hoboken, NJ, USA, 2018; pp. 127–172. [Google Scholar]
- Hussain, R.M.; Ciulla, T.A. Emerging Vascular Endothelial Growth Factor Antagonists to Treat Neovascular Age-Related Macular Degeneration. Expert Opin. Emerg. Drugs 2017, 22, 235–246. [Google Scholar] [CrossRef] [PubMed]
- Kang, C. Avacincaptad Pegol: First Approval. Drugs 2023, 83, 1447–1453. [Google Scholar] [CrossRef] [PubMed]
- Sczepanski, J.T.; Joyce, G.F. Specific Inhibition of microRNA Processing Using L-RNA Aptamers. J. Am. Chem. Soc. 2015, 137, 16032–16037. [Google Scholar] [CrossRef] [PubMed]
- Kumar Kulabhusan, P.; Hussain, B.; Yüce, M. Current Perspectives on Aptamers as Diagnostic Tools and Therapeutic Agents. Pharmaceutics 2020, 12, 646. [Google Scholar] [CrossRef]
- He, F.; Wen, N.; Xiao, D.; Yan, J.; Xiong, H.; Cai, S.; Liu, Z.; Liu, Y. Aptamer-Based Targeted Drug Delivery Systems: Current Potential and Challenges. Curr. Med. Chem. 2020, 27, 2189–2219. [Google Scholar] [CrossRef]
- Ko, S.; Jo, M.; Jung, S.T. Recent Achievements and Challenges in Prolonging the Serum Half-Lives of Therapeutic IgG Antibodies through Fc Engineering. BioDrugs 2021, 35, 147–157. [Google Scholar] [CrossRef]
- Rodríguez-Alfonso, A.; Heck, A.; Ruiz-Blanco, Y.B.; Gilg, A.; Ständker, L.; Kuan, S.L.; Weil, T.; Sanchez-Garcia, E.; Wiese, S.; Münch, J. Advanced EPI-X4 Derivatives Covalently Bind Human Serum Albumin Resulting in Prolonged Plasma Stability. Int. J. Mol. Sci. 2022, 23, 15029. [Google Scholar] [CrossRef]
- Zhang, H.; Vandesompele, J.; Braeckmans, K.; De Smedt, S.C.; Remaut, K. Nucleic Acid Degradation as Barrier to Gene Delivery: A Guide to Understand and Overcome Nuclease Activity. Chem. Soc. Rev. 2024, 53, 317–360. [Google Scholar] [CrossRef]
- Kim, H.; Kim, S.; Lee, D.; Lee, D.; Yoon, J.; Lee, H. Oligonucleotide Therapeutics and Their Chemical Modification Strategies for Clinical Applications. J. Pharm. Investig. 2024, 54, 415–433. [Google Scholar] [CrossRef]
- Ruckman, J.; Green, L.S.; Beeson, J.; Waugh, S.; Gillette, W.L.; Henninger, D.D.; Claesson-Welsh, L.; Janjic, N. 2′-Fluoropyrimidine RNA-Based Aptamers to the 165-Amino Acid Form of Vascular Endothelial Growth Factor (VEGF165): Inhibition of Receptor Binding and VEGF-Induced Vascular Permeability through Interactions Requiring the Exon 7-Encoded Domain. J. Biol. Chem. 1998, 273, 20556–20567. [Google Scholar] [CrossRef]
- Freund, N.; Taylor, A.I.; Arangundy-Franklin, S.; Subramanian, N.; Peak-Chew, S.-Y.; Whitaker, A.M.; Freudenthal, B.D.; Abramov, M.; Herdewijn, P.; Holliger, P. A Two-Residue Nascent-Strand Steric Gate Controls Synthesis of 2′-O-Methyl-and 2′-O-(2-Methoxyethyl)-RNA. Nat. Chem. 2023, 15, 91–100. [Google Scholar] [CrossRef] [PubMed]
- Arese, M.; Mahmoudian, M.; Bussolino, F. RNA Aptamer-Mediated Gene Therapy of Prostate Cancer: Lessons from the Past and Future Directions. Expert Opin. Drug Deliv. 2023, 20, 1609–1621. [Google Scholar] [CrossRef] [PubMed]
- Becette, O.B.; Tran, A.; Jones, J.W.; Marino, J.P.; Brinson, R.G. Structural Fingerprinting of siRNA Therapeutics by Solution NMR Spectroscopy. Nucleic Acid Ther. 2022, 32, 267–279. [Google Scholar] [CrossRef] [PubMed]
- Yin, W.; Pham, C.V.; Wang, T.; Al Shamaileh, H.; Chowdhury, R.; Patel, S.; Li, Y.; Kong, L.; Hou, Y.; Zhu, Y. Inhibition of Autophagy Promotes the Elimination of Liver Cancer Stem Cells by CD133 Aptamer-Targeted Delivery of Doxorubicin. Biomolecules 2022, 12, 1623. [Google Scholar] [CrossRef]
- Schneider, D.J.; Lynch, S.A.; Gelinas, A.D.; Ostroff, R.M.; Rohloff, J.C.; Williams, P.; Janjic, N.; Drolet, D.W. SOMAmer Reagents and the SomaScan Platform: Chemically Modified Aptamers and Their Applications in Therapeutics, Diagnostics, and Proteomics. In RNA Therapeutics; Elsevier: Amsterdam, The Netherlands, 2022; pp. 171–260. [Google Scholar]
- Kundu, N. Directed Evolution of a Novel Heterochiral Ribonuclease Ribozyme and Kinetic Characterization of Heterochiral DNA Strand Displacement Reactions. Doctoral Dissertation, Oxford Academic, Oxford, UK, 2022. [Google Scholar]
- Thevendran, R.; Citartan, M. Assays to Estimate the Binding Affinity of Aptamers. Talanta 2022, 238, 122971. [Google Scholar] [CrossRef]
- Zhang, A.; Meng, K.; Liu, Y.; Pan, Y.; Qu, W.; Chen, D.; Xie, S. Absorption, Distribution, Metabolism, and Excretion of Nanocarriers in Vivo and Their Influences. Adv. Colloid Interface Sci. 2020, 284, 102261. [Google Scholar] [CrossRef]
- Lee, C.H.; Lee, S.-H.; Kim, J.H.; Noh, Y.-H.; Noh, G.-J.; Lee, S.-W. Pharmacokinetics of a Cholesterol-Conjugated Aptamer against the Hepatitis C Virus (HCV) NS5B Protein. Mol. Ther. Nucleic Acids 2015, 4, e254. [Google Scholar] [CrossRef]
- Rusconi, C.P.; Roberts, J.D.; Pitoc, G.A.; Nimjee, S.M.; White, R.R.; Quick Jr, G.; Scardino, E.; Fay, W.P.; Sullenger, B.A. Antidote-Mediated Control of an Anticoagulant Aptamer In Vivo. Nat. Biotechnol. 2004, 22, 1423–1428. [Google Scholar] [CrossRef]
- Heo, K.; Min, S.-W.; Sung, H.J.; Kim, H.G.; Kim, H.J.; Kim, Y.H.; Choi, B.K.; Han, S.; Chung, S.; Lee, E.S. An Aptamer-Antibody Complex (Oligobody) as a Novel Delivery Platform for Targeted Cancer Therapies. J. Control. Release 2016, 229, 1–9. [Google Scholar] [CrossRef]
- Soule, E.E.; Bompiani, K.M.; Woodruff, R.S.; Sullenger, B.A. Targeting Two Coagulation Cascade Proteases with a Bivalent Aptamer Yields a Potent and Antidote-Controllable Anticoagulant. Nucleic Acid Ther. 2016, 26, 1–9. [Google Scholar] [CrossRef]
- Musumeci, D.; Montesarchio, D. Polyvalent Nucleic Acid Aptamers and Modulation of Their Activity: A Focus on the Thrombin Binding Aptamer. Pharmacol. Ther. 2012, 136, 202–215. [Google Scholar] [CrossRef] [PubMed]
- Willis, M.C.; Collins, B.; Zhang, T.; Green, L.S.; Sebesta, D.P.; Bell, C.; Kellogg, E.; Gill, S.C.; Magallanez, A.; Knauer, S. Liposome-Anchored Vascular Endothelial Growth Factor Aptamers. Bioconjug. Chem. 1998, 9, 573–582. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Rashid, F.; Shah, A.; Awan, H.M.; Wu, M.; Liu, A.; Wang, J.; Zhu, T.; Luo, Z.; Shan, G. The Isolation of an RNA Aptamer Targeting to P53 Protein with Single Amino Acid Mutation. Proc. Natl. Acad. Sci. USA 2015, 112, 10002–10007. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, M.; Ramadan, E.; Elsadek, N.E.; Emam, S.E.; Shimizu, T.; Ando, H.; Ishima, Y.; Elgarhy, O.H.; Sarhan, H.A.; Hussein, A.K. Polyethylene Glycol (PEG): The Nature, Immunogenicity, and Role in the Hypersensitivity of PEGylated Products. J. Control. Release 2022, 351, 215–230. [Google Scholar] [CrossRef]
- Turecek, P.L.; Bossard, M.J.; Schoetens, F.; Ivens, I.A. PEGylation of Biopharmaceuticals: A Review of Chemistry and Nonclinical Safety Information of Approved Drugs. J. Pharm. Sci. 2016, 105, 460–475. [Google Scholar] [CrossRef]
- Hinglajia, H.; Prajapati, B.; Patel, G. Targeting Brain Disorders Using Lipid-Based Formulations. In Lipid-Based Drug Delivery Systems; Jenny Stanford Publishing: Singapore, 2024; pp. 549–590. [Google Scholar]
- Drolet, D.W.; Nelson, J.; Tucker, C.E.; Zack, P.M.; Nixon, K.; Bolin, R.; Judkins, M.B.; Farmer, J.A.; Wolf, J.L.; Gill, S.C. Pharmacokinetics and Safety of an Anti-Vascular Endothelial Growth Factor Aptamer (NX1838) Following Injection into the Vitreous Humor of Rhesus Monkeys. Pharm. Res. 2000, 17, 1503–1510. [Google Scholar] [CrossRef]
- Ganson, N.J.; Povsic, T.J.; Sullenger, B.A.; Alexander, J.H.; Zelenkofske, S.L.; Sailstad, J.M.; Rusconi, C.P.; Hershfield, M.S. Pre-Existing Anti–Polyethylene Glycol Antibody Linked to First-Exposure Allergic Reactions to Pegnivacogin, a PEGylated RNA Aptamer. J. Allergy Clin. Immunol. 2016, 137, 1610–1613.e7. [Google Scholar] [CrossRef]
- Sajid, M.I.; Moazzam, M.; Kato, S.; Yeseom Cho, K.; Tiwari, R.K. Overcoming Barriers for siRNA Therapeutics: From Bench to Bedside. Pharmaceuticals 2020, 13, 294. [Google Scholar] [CrossRef]
- Geary, R.S.; Rosie, Z.Y.; Watanabe, T.; Henry, S.P.; Hardee, G.E.; Chappell, A.; Matson, J.; Sasmor, H.; Cummins, L.; Levin, A.A. Pharmacokinetics of a Tumor Necrosis Factor-α Phosphorothioate 2′-O-(2-Methoxyethyl) Modified Antisense Oligonucleotide: Comparison across Species. Drug Metab. Dispos. 2003, 31, 1419–1428. [Google Scholar] [CrossRef]
- Henry, S.P.; Zuckerman, J.E.; Rojko, J.; Hall, W.C.; Harman, R.J.; Kitchen, D.; Crooke, S.T. Toxicological Properties of Several Novel Oligonucleotide Analogs in Mice. Anti-Cancer Drug Des. 1997, 12, 1–14. [Google Scholar]
- Sarmiento, U.M.; Perez, J.R.; Becker, J.M.; Narayanan, R. In Vivo Toxicological Effects of Rel A Antisense Phosphorothioates in CD-1 Mice. Antisense Res. Dev. 1994, 4, 99–107. [Google Scholar] [CrossRef] [PubMed]
- Henry, S.P.; Taylor, J.; Midgley, L.; Levin, A.A.; Kornbrust, D.J. Evaluation of the Toxicity of ISIS 2302, a Phosphorothioate Oligonucleotide, in a 4-Week Study in CD-1 Mice. Antisense Nucleic Acid Drug Dev. 1997, 7, 473–481. [Google Scholar] [CrossRef] [PubMed]
- Bouchard, P.R.; Hutabarat, R.M.; Thompson, K.M. Discovery and Development of Therapeutic Aptamers. Annu. Rev. Pharmacol. Toxicol. 2010, 50, 237–257. [Google Scholar] [CrossRef] [PubMed]
- Singh, M.; Thakur, M.; Mishra, M.; Yadav, M.; Vibhuti, R.; Menon, A.M.; Nagda, G.; Dwivedi, V.P.; Dakal, T.C.; Yadav, V. Gene Regulation of Intracellular Adhesion Molecule-1 (ICAM-1): A Molecule with Multiple Functions. Immunol. Lett. 2021, 240, 123–136. [Google Scholar] [CrossRef]
- Yacyshyn, B.R.; Bowen-Yacyshyn, M.B.; Jewell, L.; Tami, J.A.; Bennett, C.F.; Kisner, D.L.; Shanahan Jr, W.R. A Placebo-Controlled Trial of ICAM-1 Antisense Oligonucleotide in the Treatment of Crohn’s Disease. Gastroenterology 1998, 114, 1133–1142. [Google Scholar] [CrossRef]
- Gao, J.; Nutan, B.; Gargouri, D.; Pisal, N.D.; Do, V.; Zubair, M.; Alanzi, H.; Wang, H.; Lee, D.; Joshi, N. Unlocking the Potential of Chemically Modified Nucleic Acid Therapeutics. Adv. Ther. 2024, 7, 2400231. [Google Scholar] [CrossRef]
- Sioud, M. How the Initial Discovery of Modified RNA Enabled Evasion of Innate Immune Responses and Facilitated the Development of RNA Therapeutics. Scand. J. Immunol. 2023, 98, e13282. [Google Scholar] [CrossRef]
- Tammaro, A.; Kers, J.; Scantlebery, A.M.; Florquin, S. Metabolic Flexibility and Innate Immunity in Renal Ischemia Reperfusion Injury: The Fine Balance between Adaptive Repair and Tissue Degeneration. Front. Immunol. 2020, 11, 1346. [Google Scholar] [CrossRef]
- Foy, J.W.-D.; Rittenhouse, K.; Modi, M.; Patel, M. Local Tolerance and Systemic Safety of Pegaptanib Sodium in the Dog and Rabbit. J. Ocul. Pharmacol. Ther. 2007, 23, 452–466. [Google Scholar] [CrossRef]
- Catani, M.; De Luca, C.; Medeiros Garcia Alcântara, J.; Manfredini, N.; Perrone, D.; Marchesi, E.; Weldon, R.; Müller-Späth, T.; Cavazzini, A.; Morbidelli, M. Oligonucleotides: Current Trends and Innovative Applications in the Synthesis, Characterization, and Purification. Biotechnol. J. 2020, 15, 1900226. [Google Scholar] [CrossRef]
- Jayasinghe, M.K.; Tan, M.; Peng, B.; Yang, Y.; Sethi, G.; Pirisinu, M.; Le, M.T. New Approaches in Extracellular Vesicle Engineering for Improving the Efficacy of Anti-Cancer Therapies. Semin. Cancer Biol. 2021, 74, 62–78. [Google Scholar] [CrossRef] [PubMed]
- Holmes, E.H. PSMA Specific Antibodies and Their Diagnostic and Therapeutic Use. Expert Opin. Investig. Drugs 2001, 10, 511–519. [Google Scholar] [CrossRef] [PubMed]
- Teodori, L.; Omer, M.; Kjems, J. RNA Nanostructures for Targeted Drug Delivery and Imaging. RNA Biol. 2024, 21, 391–409. [Google Scholar] [CrossRef] [PubMed]
- Pieken, W.A.; Olsen, D.B.; Benseler, F.; Aurup, H.; Eckstein, F. Kinetic Characterization of Ribonuclease-Resistant 2′-Modified Hammerhead Ribozymes. Science 1991, 253, 314–317. [Google Scholar] [CrossRef]
- Zhou, J.; Rossi, J. Aptamers as Targeted Therapeutics: Current Potential and Challenges. Nat. Rev. Drug Discov. 2017, 16, 181–202. [Google Scholar] [CrossRef]
- Cruz-Hernández, C.D.; Rodríguez-Martínez, G.; Cortés-Ramírez, S.A.; Morales-Pacheco, M.; Cruz-Burgos, M.; Losada-García, A.; Reyes-Grajeda, J.P.; González-Ramírez, I.; González-Covarrubias, V.; Camacho-Arroyo, I. Aptamers as Theragnostic Tools in Prostate Cancer. Biomolecules 2022, 12, 1056. [Google Scholar] [CrossRef]
- Firlej, V.; Soyeux, P.; Nourieh, M.; Huet, E.; Semprez, F.; Allory, Y.; Londono-Vallejo, A.; de La Taille, A.; Vacherot, F.; Destouches, D. Overexpression of Nucleolin and Associated Genes in Prostate Cancer. Int. J. Mol. Sci. 2022, 23, 4491. [Google Scholar] [CrossRef]
- Yazdian-Robati, R.; Bayat, P.; Oroojalian, F.; Zargari, M.; Ramezani, M.; Taghdisi, S.M.; Abnous, K. Therapeutic Applications of AS1411 Aptamer, an Update Review. Int. J. Biol. Macromol. 2020, 155, 1420–1431. [Google Scholar] [CrossRef]
- Carvalho, J.; Lopes-Nunes, J.; Vialet, B.; Rosado, T.; Gallardo, E.; Vale, J.; Eloy, C.; Ferreira, S.; Palmeira-de-Oliveira, R.; Campello, M.P.C. Nanoaggregate-Forming Lipid-Conjugated AS1411 Aptamer as a Promising Tumor-Targeted Delivery System of Anticancer Agents in Vitro. Nanomed. Nanotechnol. Biol. Med. 2021, 36, 102429. [Google Scholar] [CrossRef]
- Van den Avont, A.; Sharma-Walia, N. Anti-Nucleolin Aptamer AS1411: An Advancing Therapeutic. Front. Mol. Biosci. 2023, 10, 1217769. [Google Scholar] [CrossRef]
- Elalouf, A.; Maoz, H.; Rosenfeld, A.Y. Comprehensive Insights into the Molecular Basis of HIV Glycoproteins. Appl. Sci. 2024, 14, 8271. [Google Scholar] [CrossRef]
- Brelot, A.; Chakrabarti, L.A. CCR5 Revisited: How Mechanisms of HIV Entry Govern AIDS Pathogenesis. J. Mol. Biol. 2018, 430, 2557–2589. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Li, H.; Li, S.; Zaia, J.; Rossi, J.J. Novel Dual Inhibitory Function Aptamer–siRNA Delivery System for HIV-1 Therapy. Mol. Ther. 2008, 16, 1481–1489. [Google Scholar] [CrossRef] [PubMed]
- Bala, J.; Chinnapaiyan, S.; Dutta, R.K.; Unwalla, H. Aptamers in HIV Research Diagnosis and Therapy. RNA Biol. 2018, 15, 327–337. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.-F.; Shangguan, D.; Liu, H.; Phillips, J.A.; Zhang, X.; Chen, Y.; Tan, W. Molecular Assembly of an Aptamer–Drug Conjugate for Targeted Drug Delivery to Tumor Cells. ChemBioChem 2009, 10, 862–868. [Google Scholar] [CrossRef]
- Boyacioglu, O.; Stuart, C.H.; Kulik, G.; Gmeiner, W.H. Dimeric DNA Aptamer Complexes for High-Capacity–Targeted Drug Delivery Using pH-Sensitive Covalent Linkages. Mol. Ther. Nucleic Acids 2013, 2, e107. [Google Scholar] [CrossRef]
- Zhu, G.; Niu, G.; Chen, X. Aptamer–Drug Conjugates. Bioconjug. Chem. 2015, 26, 2186–2197. [Google Scholar] [CrossRef]
- He, J.; Peng, T.; Peng, Y.; Ai, L.; Deng, Z.; Wang, X.-Q.; Tan, W. Molecularly Engineering Triptolide with Aptamers for High Specificity and Cytotoxicity for Triple-Negative Breast Cancer. J. Am. Chem. Soc. 2020, 142, 2699–2703. [Google Scholar] [CrossRef]
- Poon, K.A.; Flagella, K.; Beyer, J.; Tibbitts, J.; Kaur, S.; Saad, O.; Yi, J.-H.; Girish, S.; Dybdal, N.; Reynolds, T. Preclinical Safety Profile of Trastuzumab Emtansine (T-DM1): Mechanism of Action of Its Cytotoxic Component Retained with Improved Tolerability. Toxicol. Appl. Pharmacol. 2013, 273, 298–313. [Google Scholar] [CrossRef]
- Zhu, G.; Zheng, J.; Song, E.; Donovan, M.; Zhang, K.; Liu, C.; Tan, W. Self-Assembled, Aptamer-Tethered DNA Nanotrains for Targeted Transport of Molecular Drugs in Cancer Theranostics. Proc. Natl. Acad. Sci. USA 2013, 110, 7998–8003. [Google Scholar] [CrossRef]
- Mirza, Z.; Karim, S. Nanoparticles-Based Drug Delivery and Gene Therapy for Breast Cancer: Recent Advancements and Future Challenges. Semin. Cancer Biol. 2021, 69, 226–237. [Google Scholar] [CrossRef] [PubMed]
- Mahmoudpour, M.; Ding, S.; Lyu, Z.; Ebrahimi, G.; Du, D.; Dolatabadi, J.E.N.; Torbati, M.; Lin, Y. Aptamer Functionalized Nanomaterials for Biomedical Applications: Recent Advances and New Horizons. Nano Today 2021, 39, 101177. [Google Scholar] [CrossRef]
- Li, N.; Larson, T.; Nguyen, H.H.; Sokolov, K.V.; Ellington, A.D. Directed Evolution of Gold Nanoparticle Delivery to Cells. Chem. Commun. 2010, 46, 392–394. [Google Scholar] [CrossRef] [PubMed]
- Jia, H.-R.; Zhang, Z.; Fang, X.; Jiang, M.; Chen, M.; Chen, S.; Gu, K.; Luo, Z.; Wu, F.-G.; Tan, W. Recent Advances of Cell Surface Modification Based on Aptamers. Mater. Today Nano 2022, 18, 100188. [Google Scholar] [CrossRef]
- Elmowafy, E.M.; Tiboni, M.; Soliman, M.E. Biocompatibility, Biodegradation and Biomedical Applications of Poly (Lactic Acid)/Poly (Lactic-Co-Glycolic Acid) Micro and Nanoparticles. J. Pharm. Investig. 2019, 49, 347–380. [Google Scholar] [CrossRef]
- Zeb, A.; Gul, M.; Nguyen, T.-T.-L.; Maeng, H.-J. Controlled Release and Targeted Drug Delivery with Poly (Lactic-Co-Glycolic Acid) Nanoparticles: Reviewing Two Decades of Research. J. Pharm. Investig. 2022, 52, 683–724. [Google Scholar] [CrossRef]
- Xiong, H.; Liu, L.; Wang, Y.; Jiang, H.; Wang, X. Engineered Aptamer-Organic Amphiphile Self-Assemblies for Biomedical Applications: Progress and Challenges. Small 2022, 18, 2104341. [Google Scholar] [CrossRef]
- Liu, M.; Wang, L.; Lo, Y.; Shiu, S.C.-C.; Kinghorn, A.B.; Tanner, J.A. Aptamer-Enabled Nanomaterials for Therapeutics, Drug Targeting and Imaging. Cells 2022, 11, 159. [Google Scholar] [CrossRef]
- Xu, Z.; Wang, Z.; Deng, Z.; Zhu, G. Recent Advances in the Synthesis, Stability, and Activation of Platinum (IV) Anticancer Prodrugs. Coord. Chem. Rev. 2021, 442, 213991. [Google Scholar] [CrossRef]
- Cao, Z.; Tong, R.; Mishra, A.; Xu, W.; Wong, G.C.; Cheng, J.; Lu, Y. Reversible Cell-specific Drug Delivery with Aptamer-functionalized Liposomes. Angew. Chem. Int. Ed. 2009, 48, 6494–6498. [Google Scholar] [CrossRef]
- Alshaer, W.; Hillaireau, H.; Fattal, E. Aptamer-Guided Nanomedicines for Anticancer Drug Delivery. Adv. Drug Deliv. Rev. 2018, 134, 122–137. [Google Scholar] [CrossRef]
- Wong, K.-Y.; Wong, M.-S.; Liu, J. Aptamer-Functionalized Liposomes for Drug Delivery. Biomed. J. 2023, 47, 100685. [Google Scholar] [CrossRef]




| Criteria | Aptamers | Antibody |
|---|---|---|
| Composition | Short single-stranded DNA or RNA oligonucleotides | Protein molecules composed of two heavy and two light chains |
| Molecular weight | ~5–25 kDa | ~150–180 kDa |
| Target range | Broad target recognition spectrum (e.g., peptides, proteins, small molecules, organic compounds, metal ions, viruses, bacteria, yeast, and mammalian cells) | Narrow target recognition spectrum (e.g., proteins, peptides, and carbohydrates) |
| Generation method | In vitro screening techniques | In vivo immunization or hybridoma technology |
| Affinity | High binding affinity | High binding affinity |
| Specificity | High specificity via sequence and structural optimization | High specificity for antigen epitopes |
| Chemical modification | Easily amenable to chemical modifications | Chemically less modifiable |
| Chemical conjugation | High chemical conjugation flexibility | Limited chemical conjugation capability |
| Degradability | Susceptible to nuclease degradation; modifiable for enhanced stability | Resistant to enzymatic degradation |
| Immunogenicity | Low or none immunogenicity | High immunogenicity |
| Aptamers | Type | Target | Stage | Application | Modifications | References |
|---|---|---|---|---|---|---|
| Pegaptanib | RNA | VEGF165 | Approved | AMD | 2′-Fluoro, PEGylated | [61] |
| Pegpleranib | DNA | VEGF | Phase III | Combination therapy for AMD | PEGylated | [105] |
| Avacincaptad pegol | RNA | Complement C5 | Approved | Geographic atrophy (dry AMD) | 2′-Fluoro, 2′-O-methyl, PEGylated | [106] |
| HD1 | DNA | Thrombin | Phase I | Anticoagulant (HIT therapy) | PDA | [72,73] |
| NU172 | DNA | Thrombin | Phase II | Anticoagulant | None | [74] |
| ARC1779 | RNA | vWF | Phase II | Thrombotic thrombocytopenic purpura | 2′-Fluoro, 2′-O-methyl, PEGylated | [75,76,77] |
| BT200 | DNA | vWF | Phase II | Thrombotic disorders (prevents arterial thrombosis) | PEGylated | [78] |
| DTRI-031 | RNA | vWF | Phase I | Thrombotic disorders | 2′-Fluoro, PEGylated | [79] |
| REG1 system | RNA | FIXa | Phase III | Reversible anticoagulation | dOxa | [83] |
| NOX-A12 | RNA | CXCL12 | Phase II | Cancer immunotherapy | PEGylated | [107] |
| TLS11a | DNA | Human hepatocellular carcinoma cells | Preclinical | Targeted therapy for hepatocellular carcinoma | FC protein | [100] |
| TLS1c | RNA | TLS/FUS protein | Preclinical | Amyotrophic lateral sclerosis and certain cancers | T15 | [101] |
| Aspect | Challenge | Solution Strategies |
|---|---|---|
| Aptamer stability | Susceptibility to nuclease-mediated degradation | 1. Chemical modifications 2. Incorporation of unnatural nucleotide analogs 3. Encapsulation in nanocarriers or liposomes |
| Renal filtration | Rapid renal clearance due to low molecular weight | 1. PEGylation or protein conjugation 2. Multivalent aptamer complex formation 3. Tissue-targeting ligand integration |
| Toxicity | Non-specific immune activation or off-target effects | 1. Rational sequence design to minimize off-target binding 2. Comprehensive in vitro/in vivo toxicity assessment 3. Targeted delivery system development |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Feng, L.; Sun, Y.; Jia, W.; Yu, Y.; Liu, C.; Yang, J.; Luan, Y.; Chen, J.; Wang, F. Advancements in SELEX Technology for Aptamers and Emerging Applications in Therapeutics and Drug Delivery. Biomolecules 2025, 15, 818. https://doi.org/10.3390/biom15060818
Feng L, Sun Y, Jia W, Yu Y, Liu C, Yang J, Luan Y, Chen J, Wang F. Advancements in SELEX Technology for Aptamers and Emerging Applications in Therapeutics and Drug Delivery. Biomolecules. 2025; 15(6):818. https://doi.org/10.3390/biom15060818
Chicago/Turabian StyleFeng, Liangjie, Yu Sun, Wenshen Jia, Yang Yu, Chang Liu, Jing Yang, Yunxia Luan, Jin Chen, and Fengchao Wang. 2025. "Advancements in SELEX Technology for Aptamers and Emerging Applications in Therapeutics and Drug Delivery" Biomolecules 15, no. 6: 818. https://doi.org/10.3390/biom15060818
APA StyleFeng, L., Sun, Y., Jia, W., Yu, Y., Liu, C., Yang, J., Luan, Y., Chen, J., & Wang, F. (2025). Advancements in SELEX Technology for Aptamers and Emerging Applications in Therapeutics and Drug Delivery. Biomolecules, 15(6), 818. https://doi.org/10.3390/biom15060818






