Salutary Effects of Overexpression of Rsm22, an Assembly Factor for the Mitochondrial Ribosome, on Frataxin/Yfh1 Depletion Phenotypes in Saccharomyces cerevisiae
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemicals and Reagents
2.2. Yeast Strains and Growth Conditions
2.3. Isolation of Mitochondria and Cytoplasm
2.4. Fe-S Cluster Assembly in Isolated Mitochondria
2.5. Cytoplasmic Fe-S Cluster Assembly
2.6. Miscellaneous
2.7. Data Analysis
3. Results
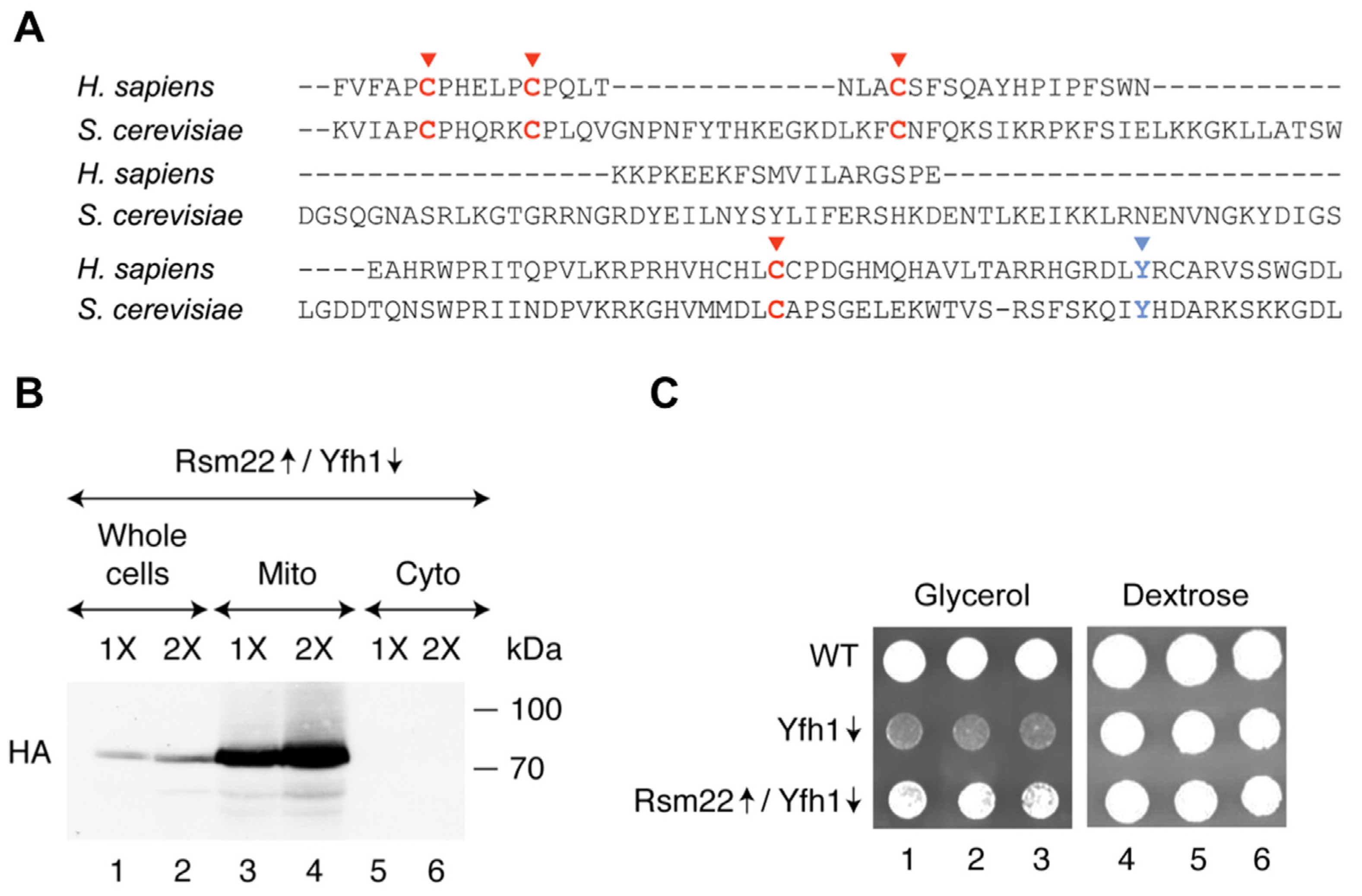
3.1. Rsm22 Overexpression Led to Improved Growth of Yfh1-Depleted Cells on a Non-Fermentable Carbon Source
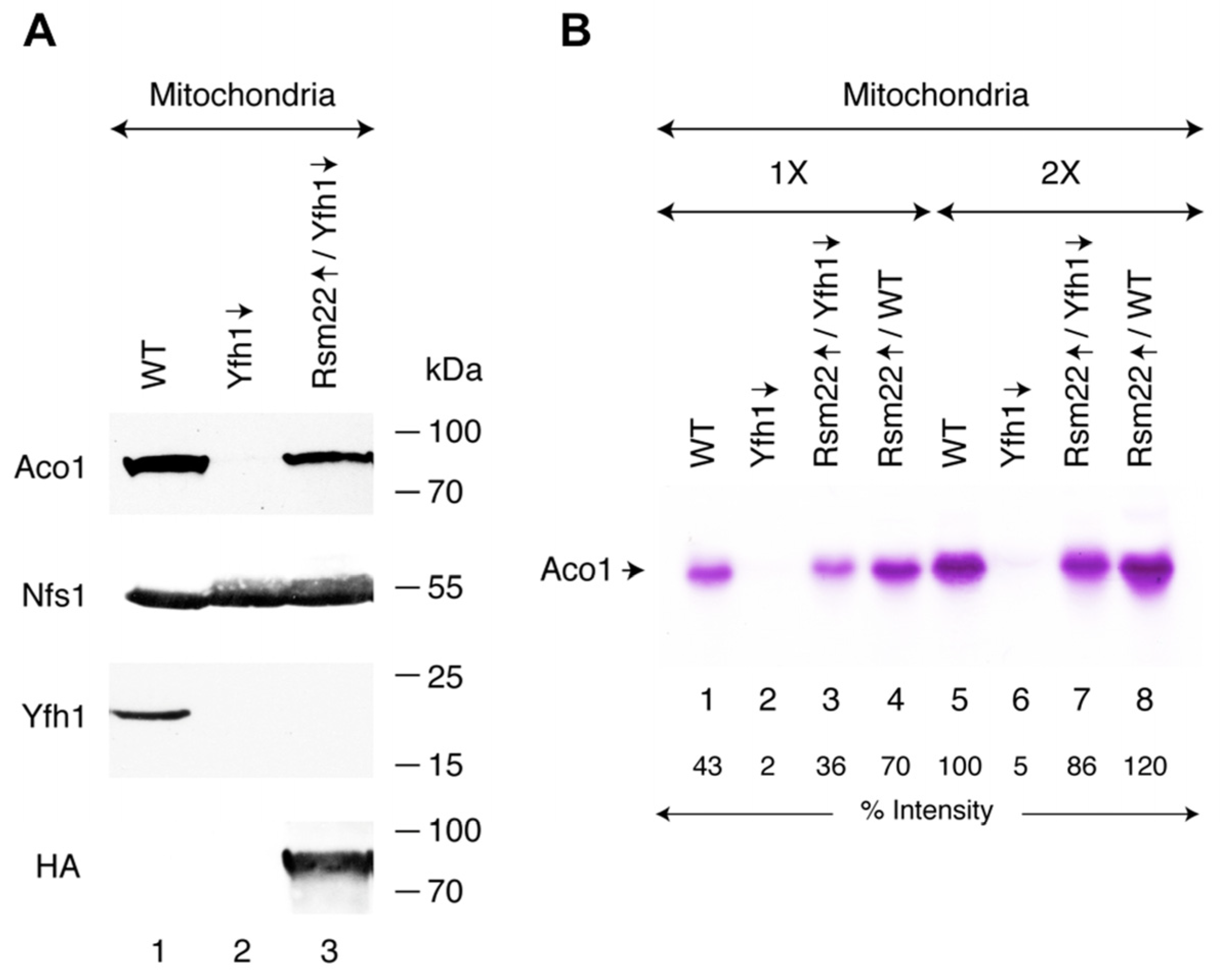
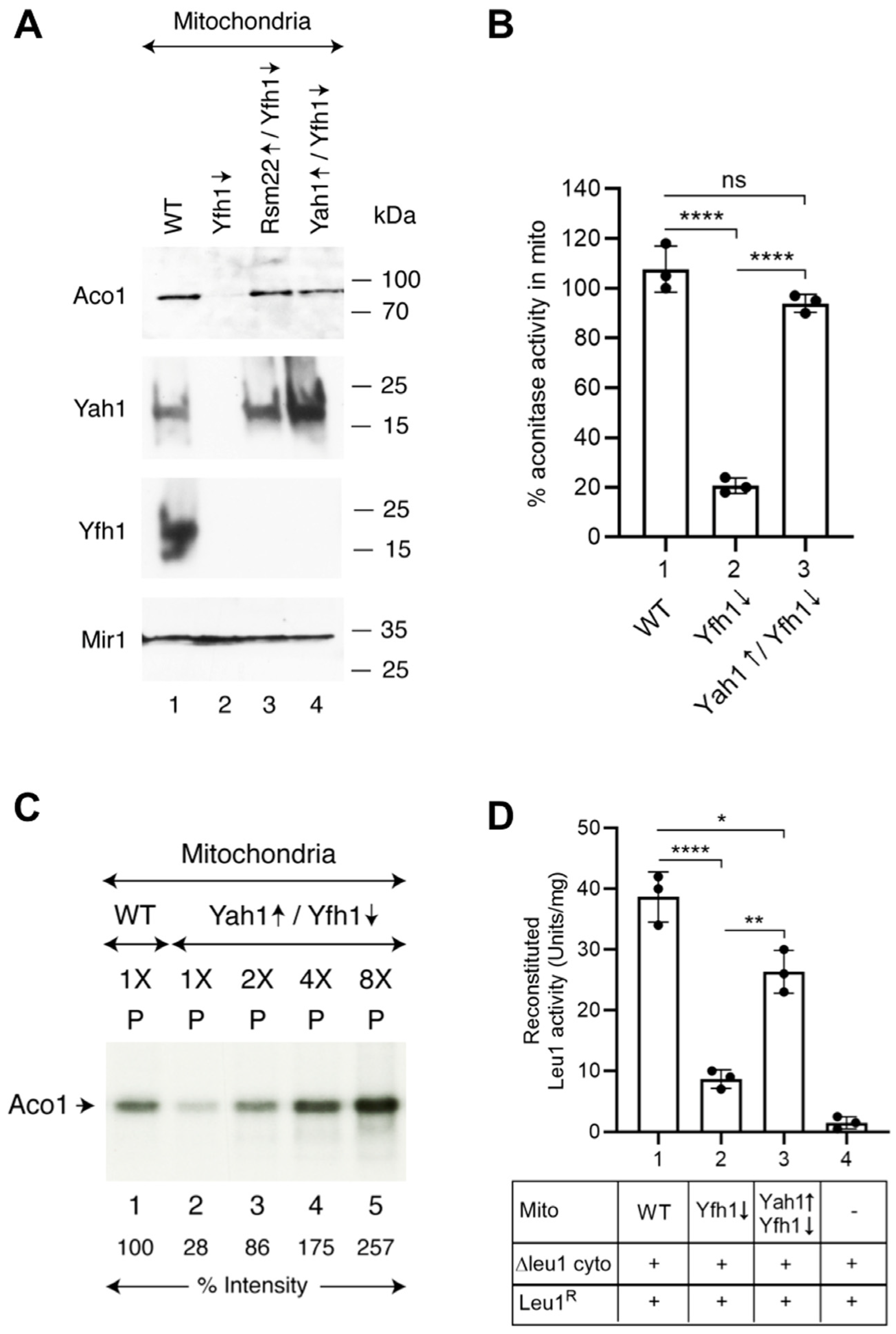
3.2. Rsm22 Overexpression in Yfh1-Depleted Cells Restored Aconitase Protein and Activity
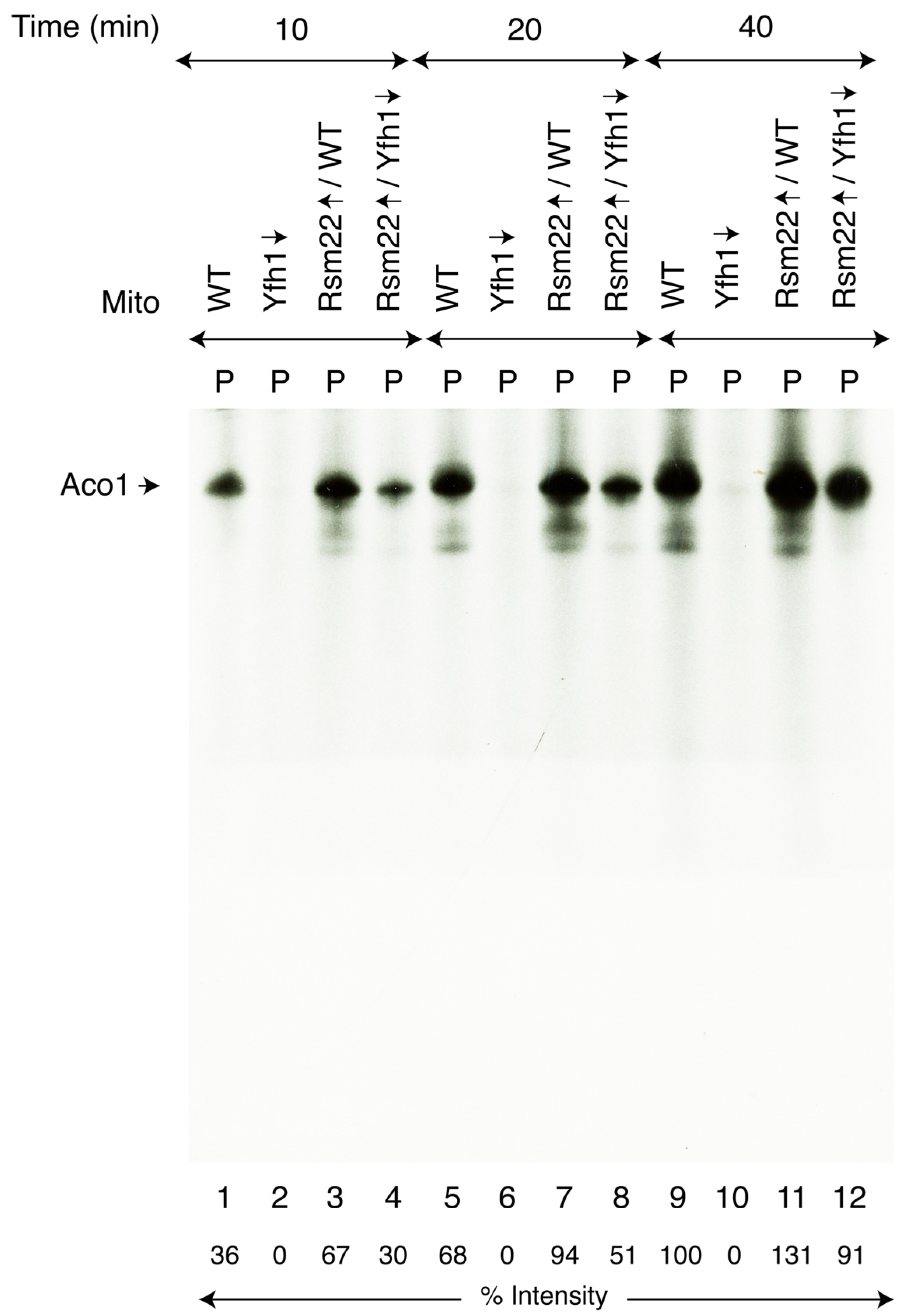
3.3. Rsm22 Overexpression Restored New Fe-S Cluster Synthesis/Assembly in Isolated Mitochondria Lacking Yfh1
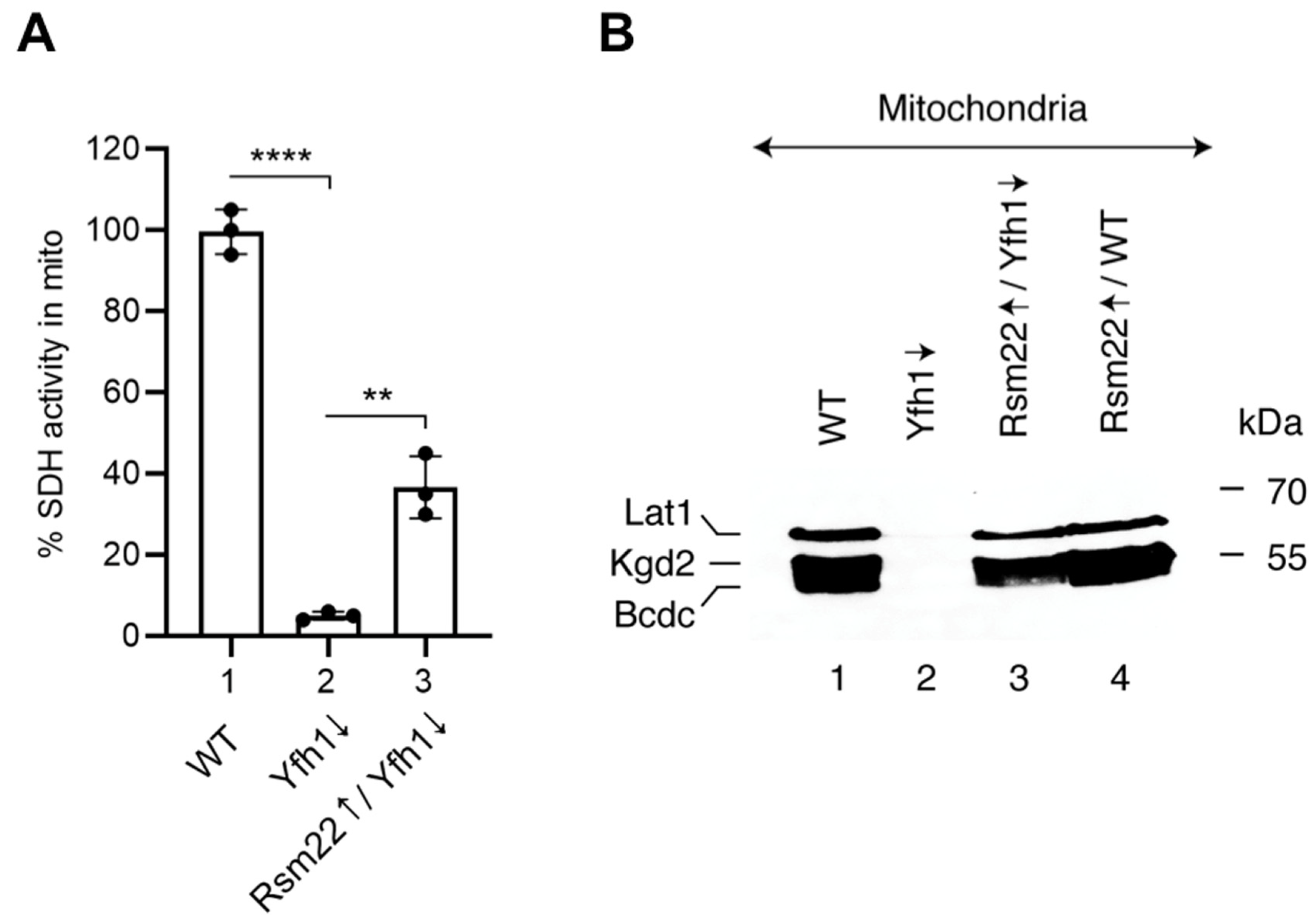
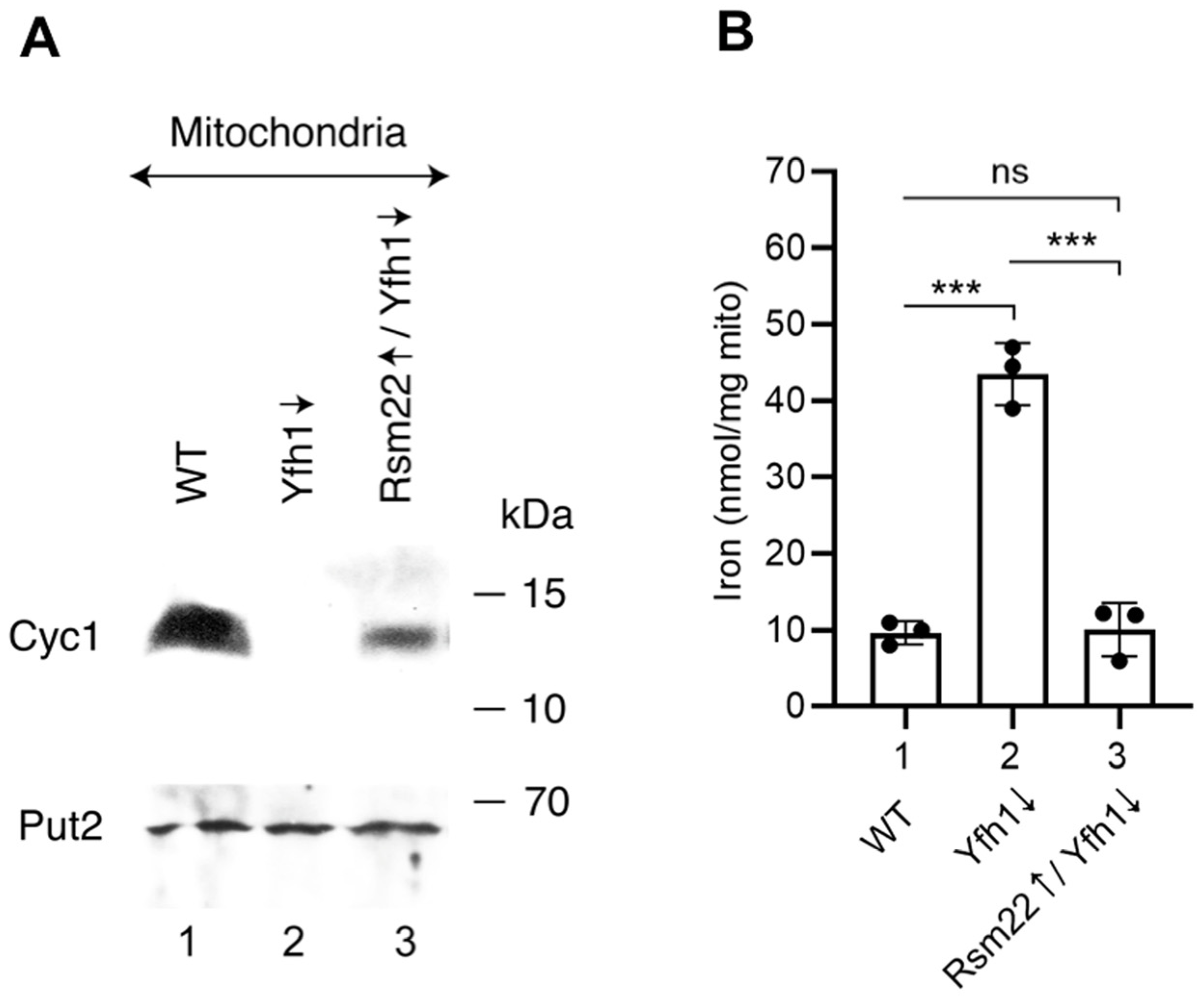
3.4. Overexpression of Rsm22 Rescued Other Iron Proteins, Restored Iron Homeostasis, and Conferred Membrane Potential in Mitochondria Lacking Yfh1
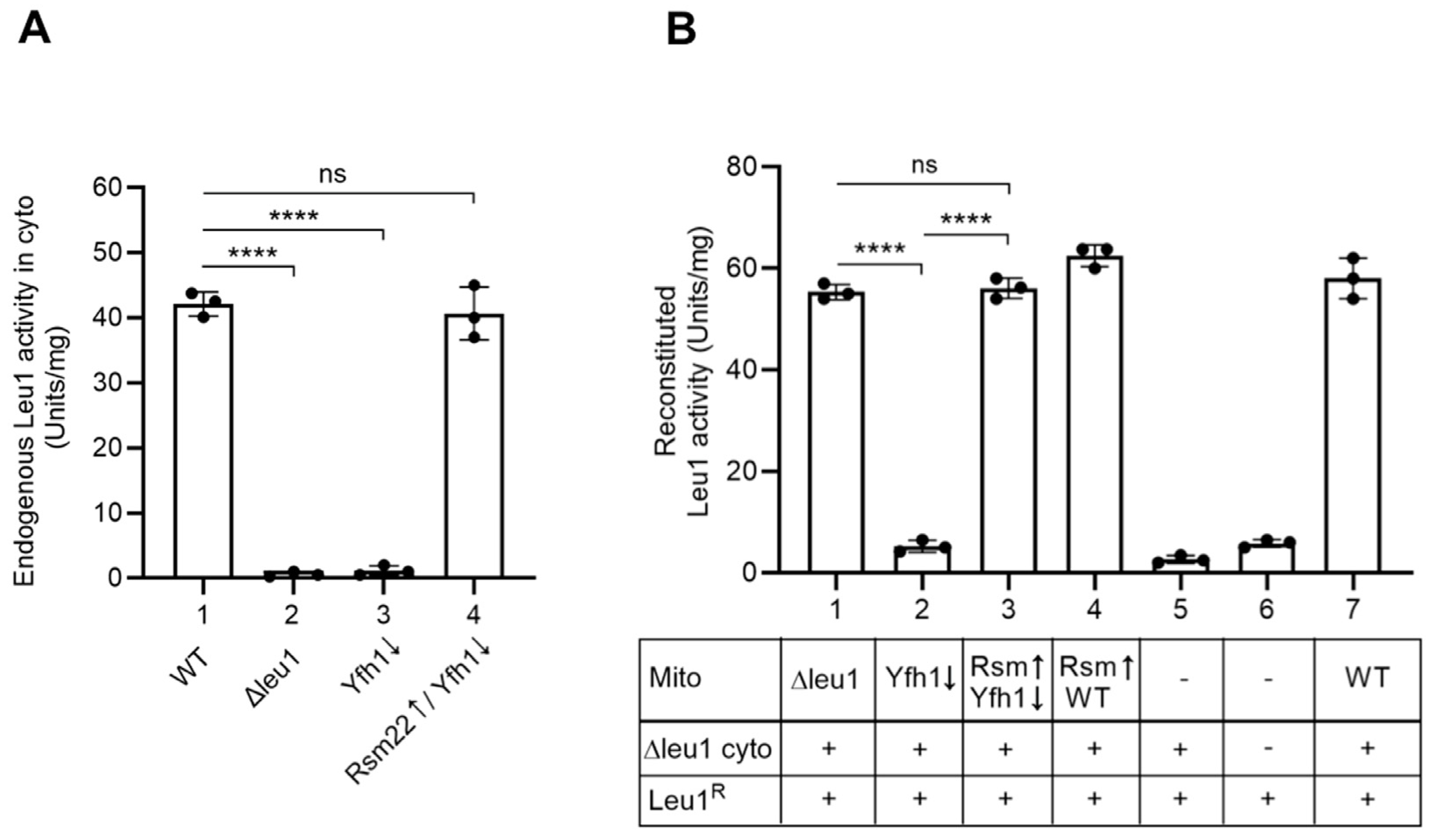
3.5. Yfh1↓ Mitochondria with Overexpressed Rsm22 Regained Capability to Promote Cytoplasmic Fe-S Cluster Assembly
3.6. A Possible Suppression Mechanism
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
| (Fe-S)int | (Fe-S) intermediate |
| ΔNYah1 | N-terminal 60 amino acids, including the mitochondrial targeting signal, removed from the Yah1 precursor protein (pYah1) |
| Leu1R | Recombinant Leu1 |
| ISC | Mitochondrial iron–sulfur cluster machinery |
| CIA | Cytoplasmic iron–sulfur protein assembly machinery |
References
- Keita, M.; McIntyre, K.; Rodden, L.N.; Schadt, K.; Lynch, D.R. Friedreich ataxia: Clinical features and new developments. Neurodegener. Dis. Manag. 2022, 12, 267–283. [Google Scholar] [CrossRef] [PubMed]
- Pandolfo, M. Friedreich Ataxia: An (Almost) 30-Year History After Gene Discovery. Neurol. Genet. 2025, 11, e200236. [Google Scholar] [CrossRef]
- Krasilnikova, M.M.; Humphries, C.L.; Shinsky, E.M. Friedreich’s ataxia: New insights. Emerg. Top. Life Sci. 2023, 7, 313–323. [Google Scholar] [PubMed]
- Campuzano, V.; Montermini, L.; Lutz, Y.; Cova, L.; Hindelang, C.; Jiralerspong, S.; Trottier, Y.; Kish, S.J.; Faucheux, B.; Trouillas, P.; et al. Frataxin is reduced in Friedreich ataxia patients and is associated with mitochondrial membranes. Hum. Mol. Genet. 1997, 6, 1771–1780. [Google Scholar] [CrossRef]
- Lill, R.; Freibert, S.A. Mechanisms of Mitochondrial Iron-Sulfur Protein Biogenesis. Annu. Rev. Biochem. 2020, 89, 471–499. [Google Scholar] [CrossRef]
- Dancis, A.; Pandey, A.K.; Pain, D. Mitochondria function in cytoplasmic FeS protein biogenesis. Biochim. Biophys. Acta Mol. Cell Res. 2024, 1871, 119733. [Google Scholar] [CrossRef]
- Uzarska, M.A.; Grochowina, I.; Soldek, J.; Jelen, M.; Schilke, B.; Marszalek, J.; Craig, E.A.; Dutkiewicz, R. During FeS cluster biogenesis, ferredoxin and frataxin use overlapping binding sites on yeast cysteine desulfurase Nfs1. J. Biol. Chem. 2022, 298, 101570. [Google Scholar] [CrossRef]
- Johnson, D.C.; Dean, D.R.; Smith, A.D.; Johnson, M.K. Structure, function, and formation of biological iron-sulfur clusters. Annu. Rev. Biochem. 2005, 74, 247–281. [Google Scholar] [CrossRef] [PubMed]
- Parent, A.; Elduque, X.; Cornu, D.; Belot, L.; Le Caer, J.P.; Grandas, A.; Toledano, M.B.; D’Autreaux, B. Mammalian frataxin directly enhances sulfur transfer of NFS1 persulfide to both ISCU and free thiols. Nat. Commun. 2015, 6, 5686. [Google Scholar] [CrossRef]
- Schulz, V.; Steinhilper, R.; Oltmanns, J.; Freibert, S.A.; Krapoth, N.; Linne, U.; Welsch, S.; Hoock, M.H.; Schunemann, V.; Murphy, B.J.; et al. Mechanism and structural dynamics of sulfur transfer during de novo [2Fe-2S] cluster assembly on ISCU2. Nat. Commun. 2024, 15, 3269. [Google Scholar] [CrossRef]
- Gervason, S.; Larkem, D.; Mansour, A.B.; Botzanowski, T.; Müller, C.S.; Pecqueur, L.; Le Pavec, G.; Delaunay-Moisan, A.; Brun, O.; Agramunt, J.; et al. Physiologically relevant reconstitution of iron-sulfur cluster biosynthesis uncovers persulfide-processing functions of ferredoxin-2 and frataxin. Nat. Commun. 2019, 10, 3566. [Google Scholar] [CrossRef]
- Steinhilper, R.; Boss, L.; Freibert, S.A.; Schulz, V.; Krapoth, N.; Kaltwasser, S.; Lill, R.; Murphy, B.J. Two-stage binding of mitochondrial ferredoxin-2 to the core iron-sulfur cluster assembly complex. Nat. Commun. 2024, 15, 10559. [Google Scholar] [CrossRef] [PubMed]
- Ast, T.; Itoh, Y.; Sadre, S.; McCoy, J.G.; Namkoong, G.; Wengrod, J.C.; Chicherin, I.; Joshi, P.R.; Kamenski, P.; Suess, D.L.M.; et al. METTL17 is an Fe-S cluster checkpoint for mitochondrial translation. Mol. Cell 2024, 84, 359–374 e358. [Google Scholar] [CrossRef]
- Gerber, J.; Muhlenhoff, U.; Lill, R. An interaction between frataxin and Isu1/Nfs1 that is crucial for Fe/S cluster synthesis on Isu1. EMBO Rep. 2003, 4, 906–911. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Craig, E.A. Binding of yeast frataxin to the scaffold for Fe-S cluster biogenesis, Isu. J. Biol. Chem. 2008, 283, 12674–12679. [Google Scholar] [CrossRef]
- Maio, N.; Jain, A.; Rouault, T.A. Mammalian iron-sulfur cluster biogenesis: Recent insights into the roles of frataxin, acyl carrier protein and ATPase-mediated transfer to recipient proteins. Curr. Opin. Chem. Biol. 2020, 55, 34–44. [Google Scholar] [CrossRef] [PubMed]
- Yoon, H.; Golla, R.; Lesuisse, E.; Pain, J.; Donald, J.E.; Lyver, E.R.; Pain, D.; Dancis, A. Mutation in the Fe-S scaffold protein Isu bypasses frataxin deletion. Biochem. J. 2012, 441, 473–480. [Google Scholar] [CrossRef]
- Das, D.; Patra, S.; Bridwell-Rabb, J.; Barondeau, D.P. Mechanism of frataxin “bypass” in human iron-sulfur cluster biosynthesis with implications for Friedreich’s ataxia. J. Biol. Chem. 2019, 294, 9276–9284. [Google Scholar] [CrossRef]
- Harper, N.J.; Burnside, C.; Klinge, S. Principles of mitoribosomal small subunit assembly in eukaryotes. Nature 2023, 614, 175–181. [Google Scholar] [CrossRef]
- Boß, L.; Stehling, O.; Elsasser, H.P.; Lill, R. Crucial role and conservation of the three [2Fe-2S] clusters in the human mitochondrial ribosome. J. Biol. Chem. 2025, 301, 108087. [Google Scholar] [CrossRef]
- Zhong, H.; Janer, A.; Khalimonchuk, O.; Antonicka, H.; Shoubridge, E.A.; Barrientos, A. BOLA3 and NFU1 link mitoribosome iron-sulfur cluster assembly to multiple mitochondrial dysfunctions syndrome. Nucleic Acids Res. 2023, 51, 11797–11812. [Google Scholar] [CrossRef] [PubMed]
- Longtine, M.S.; McKenzie, A., 3rd; Demarini, D.J.; Shah, N.G.; Wach, A.; Brachat, A.; Philippsen, P.; Pringle, J.R. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 1998, 14, 953–961. [Google Scholar] [CrossRef]
- Sherman, F. Getting started with yeast. Methods Enzymol. 2002, 350, 3–41. [Google Scholar]
- Pandey, A.K.; Pain, J.; Brindha, J.; Dancis, A.; Pain, D. Essential mitochondrial role in iron-sulfur cluster assembly of the cytoplasmic isopropylmalate isomerase Leu1 in Saccharomyces cerevisiae. Mitochondrion 2023, 69, 104–115. [Google Scholar] [CrossRef] [PubMed]
- Pandey, A.K.; Pain, J.; Singh, P.; Dancis, A.; Pain, D. Mitochondrial glutaredoxin Grx5 functions as a central hub for cellular iron-sulfur cluster assembly. J. Biol. Chem. 2025, 301, 108391. [Google Scholar] [CrossRef]
- Pandey, A.K.; Pain, J.; Dancis, A.; Pain, D. Mitochondria export iron-sulfur and sulfur intermediates to the cytoplasm for iron-sulfur cluster assembly and tRNA thiolation in yeast. J. Biol. Chem. 2019, 294, 9489–9502. [Google Scholar] [CrossRef] [PubMed]
- Pierik, A.J.; Netz, D.J.; Lill, R. Analysis of iron-sulfur protein maturation in eukaryotes. Nat. Protoc. 2009, 4, 753–766. [Google Scholar] [CrossRef]
- Sipos, K.; Lange, H.; Fekete, Z.; Ullmann, P.; Lill, R.; Kispal, G. Maturation of cytosolic iron-sulfur proteins requires glutathione. J. Biol. Chem. 2002, 277, 26944–26949. [Google Scholar] [CrossRef]
- Li, J.; Kogan, M.; Knight, S.A.; Pain, D.; Dancis, A. Yeast mitochondrial protein, Nfs1p, coordinately regulates iron-sulfur cluster proteins, cellular iron uptake, and iron distribution. J. Biol. Chem. 1999, 274, 33025–33034. [Google Scholar] [CrossRef]
- Munujos, P.; Coll-Canti, J.; Gonzalez-Sastre, F.; Gella, F.J. Assay of succinate dehydrogenase activity by a colorimetric-continuous method using iodonitrotetrazolium chloride as electron acceptor. Anal. Biochem. 1993, 212, 506–509. [Google Scholar] [CrossRef]
- Geissler, A.; Krimmer, T.; Bomer, U.; Guiard, B.; Rassow, J.; Pfanner, N. Membrane potential-driven protein import into mitochondria. The sorting sequence of cytochrome b2 modulates the Δψ-dependence of translocation of the matrix-targeting sequence. Mol. Biol. Cell 2000, 11, 3977–3991. [Google Scholar] [CrossRef] [PubMed]
- Alam, J.; Rahman, F.T.; Sah-Teli, S.K.; Venkatesan, R.; Koski, M.K.; Autio, K.J.; Hiltunen, J.K.; Kastaniotis, A.J. Expression and analysis of the SAM-dependent RNA methyltransferase Rsm22 from Saccharomyces cerevisiae. Acta Cryst. 2021, D77, 840–853. [Google Scholar] [CrossRef]
- Maio, N.; Singh, A.; Uhrigshardt, H.; Saxena, N.; Tong, W.H.; Rouault, T.A. Cochaperone binding to LYR motifs confers specificity of iron sulfur cluster delivery. Cell Metab. 2014, 19, 445–457. [Google Scholar] [CrossRef] [PubMed]
- Saveanu, C.; Fromont-Racine, M.; Harington, A.; Ricard, F.; Namane, A.; Jacquier, A. Identification of 12 new yeast mitochondrial ribosomal proteins including 6 that have no prokaryotic homologues. J. Biol. Chem. 2001, 276, 15861–15867. [Google Scholar] [CrossRef]
- Onder, O.; Yoon, H.; Naumann, B.; Hippler, M.; Dancis, A.; Daldal, F. Modifications of the lipoamide-containing mitochondrial subproteome in a yeast mutant defective in cysteine desulfurase. Mol. Cell. Proteomics 2006, 5, 1426–1436. [Google Scholar] [CrossRef] [PubMed]
- Dickinson, J.R. Branched-chain keto acid dehydrogenase of yeast. Methods Enzymol. 2000, 324, 389–398. [Google Scholar]
- Sherman, F. The importance of mutation, then and now: Studies with yeast cytochrome c. Mutat. Res. 2005, 589, 1–16. [Google Scholar] [CrossRef]
- Masud, A.J.; Kastaniotis, A.J.; Rahman, M.T.; Autio, K.J.; Hiltunen, J.K. Mitochondrial acyl carrier protein (ACP) at the interface of metabolic state sensing and mitochondrial function. Biochim. Biophys. Acta Mol. Cell Res. 2019, 1866, 118540. [Google Scholar] [CrossRef]
- Hoffman, M.; Gora, M.; Rytka, J. Identification of rate-limiting steps in yeast heme biosynthesis. Biochem. Biophys. Res. Commun. 2003, 310, 1247–1253. [Google Scholar] [CrossRef]
- Outten, C.E.; Albetel, A.N. Iron sensing and regulation in Saccharomyces cerevisiae: Ironing out the mechanistic details. Curr. Opin. Microbiol. 2013, 16, 662–668. [Google Scholar] [CrossRef]
- Miao, R.; Kim, H.; Koppolu, U.M.; Ellis, E.A.; Scott, R.A.; Lindahl, P.A. Biophysical characterization of the iron in mitochondria from Atm1p-depleted Saccharomyces cerevisiae. Biochemistry 2009, 48, 9556–9568. [Google Scholar] [CrossRef] [PubMed]
- Kispal, G.; Csere, P.; Prohl, C.; Lill, R. The mitochondrial proteins Atm1p and Nfs1p are essential for biogenesis of cytosolic Fe/S proteins. EMBO J. 1999, 18, 3981–3989. [Google Scholar] [CrossRef] [PubMed]
- Wilson, R.B.; Roof, D.M. Respiratory deficiency due to loss of mitochondrial DNA in yeast lacking the frataxin homologue. Nat. Genet. 1997, 16, 352–357. [Google Scholar] [CrossRef] [PubMed]
- Babcock, M.; de Silva, D.; Oaks, R.; Davis-Kaplan, S.; Jiralerspong, S.; Montermini, L.; Pandolfo, M.; Kaplan, J. Regulation of mitochondrial iron accumulation by Yfh1p, a putative homolog of frataxin. Science 1997, 276, 1709–1712. [Google Scholar] [CrossRef]
- Yan, R.; Konarev, P.V.; Iannuzzi, C.; Adinolfi, S.; Roche, B.; Kelly, G.; Simon, L.; Martin, S.R.; Py, B.; Barras, F.; et al. Ferredoxin competes with bacterial frataxin in binding to the desulfurase IscS. J. Biol. Chem. 2013, 288, 24777–24787. [Google Scholar] [CrossRef]
- Kim, J.H.; Frederick, R.O.; Reinen, N.M.; Troupis, A.T.; Markley, J.L. [2Fe-2S]-ferredoxin binds directly to cysteine desulfurase and supplies an electron for iron-sulfur cluster assembly but is displaced by the scaffold protein or bacterial frataxin. J. Am. Chem. Soc. 2013, 135, 8117–8120. [Google Scholar] [CrossRef]
- Weiler, B.D.; Brück, M.C.; Kothe, I.; Bill, E.; Lill, R.; Mühlenhoff, U. Mitochondrial [4Fe-4S] protein assembly involves reductive [2Fe-2S] cluster fusion on ISCA1-ISCA2 by electron flow from ferredoxin FDX2. Proc. Natl. Acad. Sci. USA 2020, 117, 20555–20565. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pandey, A.K.; Singh, P.; Pain, J.; Dancis, A.; Pain, D. Salutary Effects of Overexpression of Rsm22, an Assembly Factor for the Mitochondrial Ribosome, on Frataxin/Yfh1 Depletion Phenotypes in Saccharomyces cerevisiae. Biomolecules 2025, 15, 785. https://doi.org/10.3390/biom15060785
Pandey AK, Singh P, Pain J, Dancis A, Pain D. Salutary Effects of Overexpression of Rsm22, an Assembly Factor for the Mitochondrial Ribosome, on Frataxin/Yfh1 Depletion Phenotypes in Saccharomyces cerevisiae. Biomolecules. 2025; 15(6):785. https://doi.org/10.3390/biom15060785
Chicago/Turabian StylePandey, Ashutosh K., Pratibha Singh, Jayashree Pain, Andrew Dancis, and Debkumar Pain. 2025. "Salutary Effects of Overexpression of Rsm22, an Assembly Factor for the Mitochondrial Ribosome, on Frataxin/Yfh1 Depletion Phenotypes in Saccharomyces cerevisiae" Biomolecules 15, no. 6: 785. https://doi.org/10.3390/biom15060785
APA StylePandey, A. K., Singh, P., Pain, J., Dancis, A., & Pain, D. (2025). Salutary Effects of Overexpression of Rsm22, an Assembly Factor for the Mitochondrial Ribosome, on Frataxin/Yfh1 Depletion Phenotypes in Saccharomyces cerevisiae. Biomolecules, 15(6), 785. https://doi.org/10.3390/biom15060785





