Effects of 5-Methyl-2′-Deoxycytidine in G-Quadruplex Forming Aptamers d(G3C)4 and d[GCG2(CG3)3C]: Investigating the Key Role of the Loops
Abstract
1. Introduction
2. Materials and Methods
2.1. Oligonucleotide Synthesis and Purification
2.2. CD Spectroscopy
2.3. NMR Spectroscopy
2.4. Gel Electrophoresis
2.5. Isothermal Association Kinetics
2.6. Nuclease Stability Assay
2.7. MTT Assay
3. Results
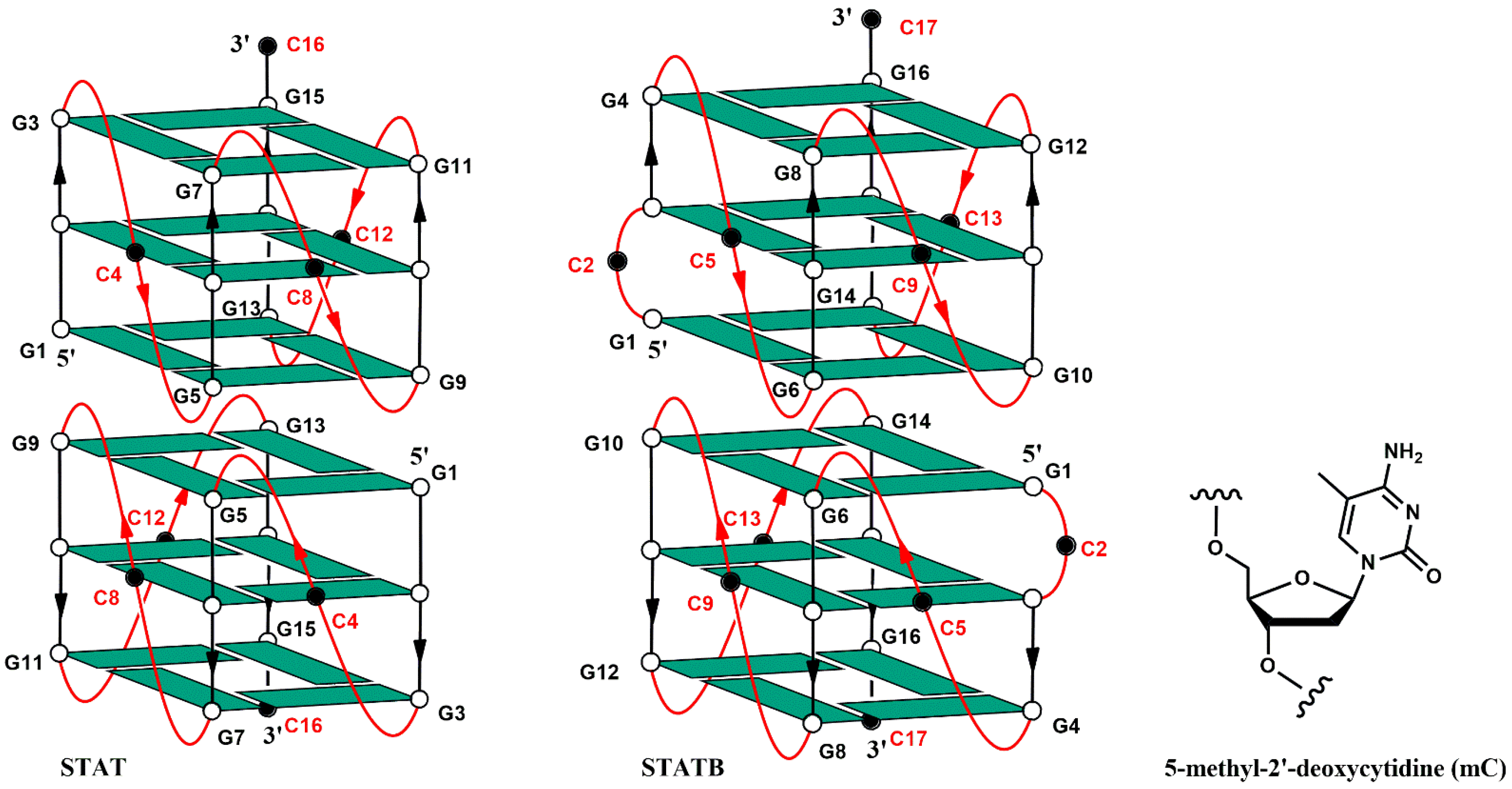
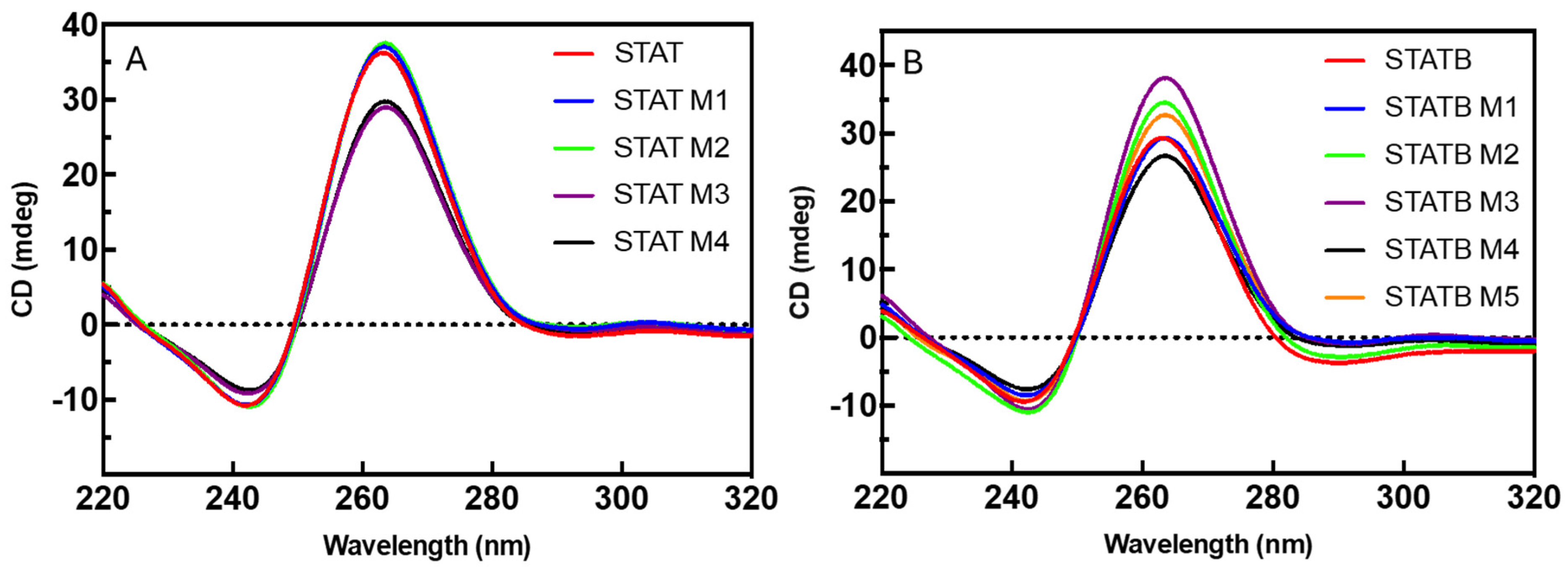
3.1. CD Spectroscopy
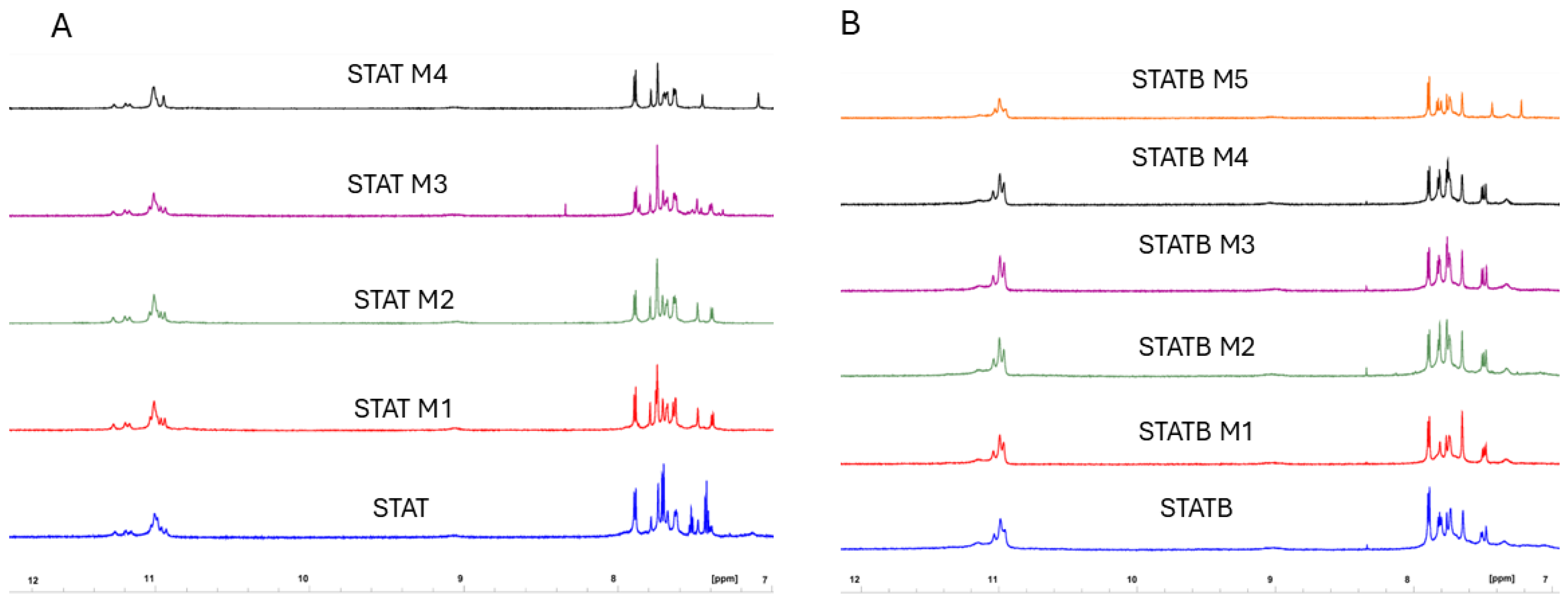
3.2. Nuclear Magnetic Resonance
3.3. Polyacrylamide Gel Electrophoresis (PAGE)
3.4. Association Kinetics
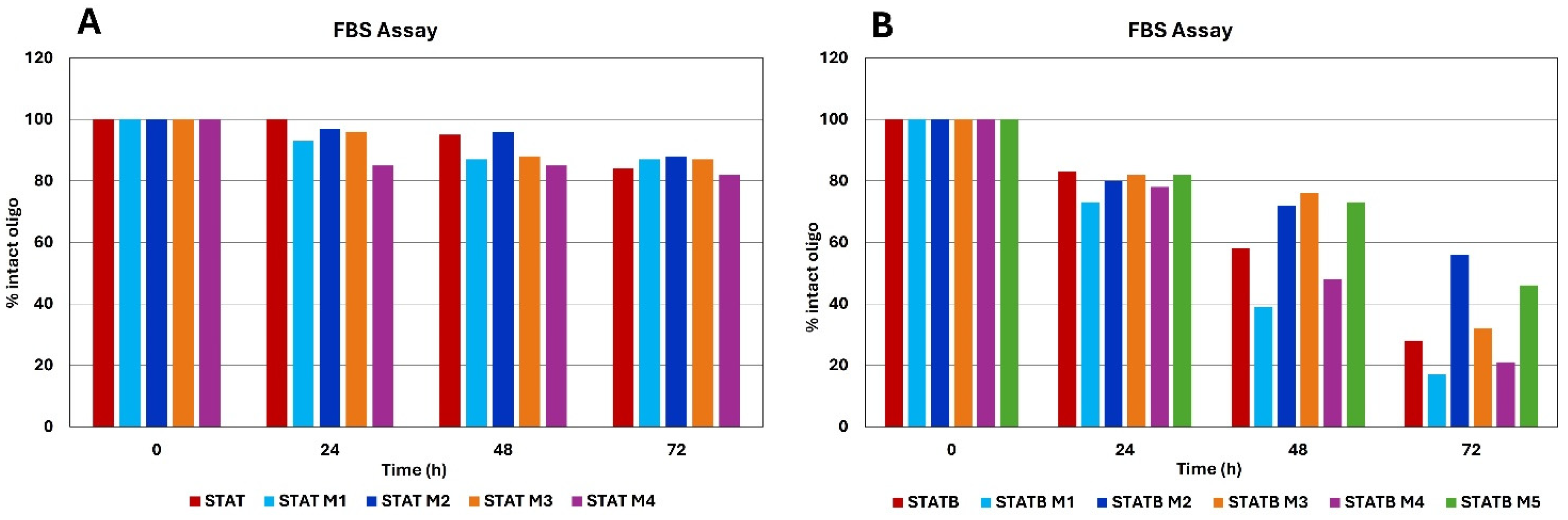
3.5. Nuclease Stability Assay
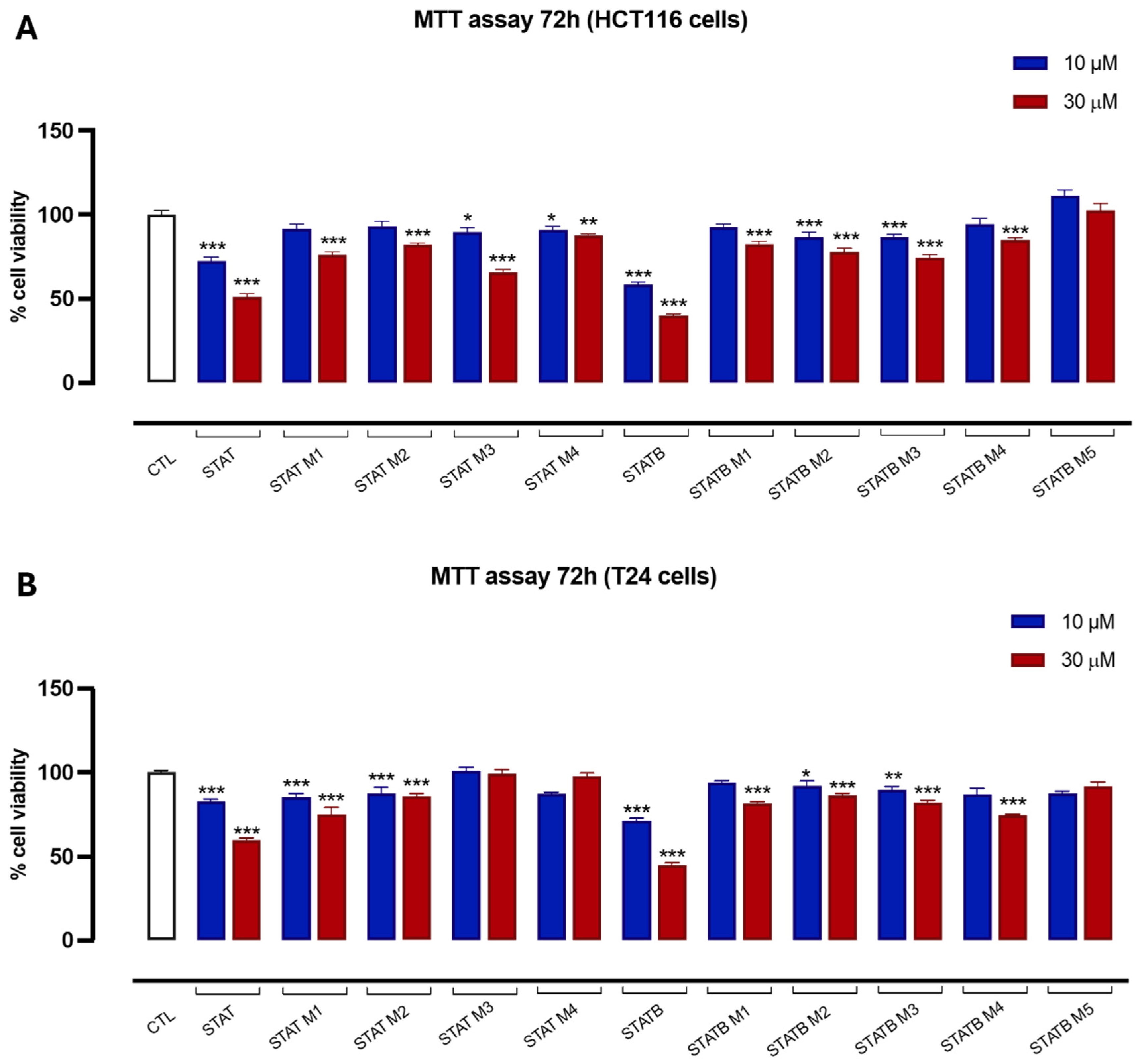
3.6. Antiproliferative Activity
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sujith, S.; Naresh, R.; Srivisanth, B.U.; Sajeevan, A.; Rajaramon, S.; David, H.; Solomon, A.P. Aptamers: Precision Tools for Diagnosing and Treating Infectious Diseases. Front. Cell. Infect. Microbiol. 2024, 14, 1402932. [Google Scholar] [CrossRef] [PubMed]
- Platella, C.; Riccardi, C.; Montesarchio, D.; Roviello, G.N.; Musumeci, D. G-Quadruplex-Based Aptamers against Protein Targets in Therapy and Diagnostics. Biochim. Biophys. Acta Gen. Subj. 2017, 1861, 1429–1447. [Google Scholar] [CrossRef] [PubMed]
- Roxo, C.; Kotkowiak, W.; Pasternak, A. G-Quadruplex-Forming Aptamers—Characteristics, Applications, and Perspectives. Molecules 2019, 24, 3781. [Google Scholar] [CrossRef] [PubMed]
- Kuo, M.H.J.; Wang, Z.F.; Tseng, T.Y.; Li, M.H.; Hsu, S.T.D.; Lin, J.J.; Chang, T.C. Conformational Transition of a Hairpin Structure to G-Quadruplex within the WNT1 Gene Promoter. J. Am. Chem. Soc. 2015, 137, 210–218. [Google Scholar] [CrossRef]
- Reina, C.; Cavalieri, V. Epigenetic Modulation of Chromatin States and Gene Expression by G-Quadruplex Structures. Int. J. Mol. Sci. 2020, 21, 4172. [Google Scholar] [CrossRef]
- Stevens, A.J.; de Jong, L.; Kennedy, M.A. The Dynamic Regulation of G-Quadruplex DNA Structures by Cytosine Methylation. Int. J. Mol. Sci. 2022, 23, 2407. [Google Scholar] [CrossRef]
- Fry, M.; Loebt, L.A. The Fragile X Syndrome d(CGG). Nucleotide Repeats Form a Stable Tetrahelical Structure. Proc. Natl. Acad. Sci. USA 1994, 91, 4950–4954. [Google Scholar] [CrossRef]
- Kettani, A.; Kumar, R.A.; Patel, D.J. Solution Structure of a DNA Quadruplex Containing the Fragile X Syndrome Triplet Repeat. J. Mol. Biol. 1995, 254, 638–656. [Google Scholar] [CrossRef]
- Zamiri, B.; Mirceta, M.; Bomsztyk, K.; Macgregor, R.B.; Pearson, C.E. Quadruplex Formation by Both G-Rich and C-Rich DNA Strands of the C9orf72 (GGGGCC)8•(GGCCCC)8 Repeat: Effect of CpG Methylation. Nucleic Acids Res. 2015, 43, 10055–10064. [Google Scholar] [CrossRef][Green Version]
- Lin, J.; Hou, J.; Xiang, H.; Yan, Y.; Gu, Y.; Tan, J.; Li, D.; Gu, L.; Ou, T.; Huang, Z. Stabilization of G-Quadruplex DNA by C-5-Methyl-Cytosine in Bcl-2 Promoter: Implications for Epigenetic Regulation. Biochem. Biophys. Res. Commun. 2013, 433, 368–373. [Google Scholar] [CrossRef]
- Li, P.T.; Wang, Z.F.; Chu, I.T.; Kuan, Y.M.; Li, M.H.; Huang, M.C.; Chiang, P.C.; Chang, T.C.; Chen, C.T. Expression of the Human Telomerase Reverse Transcriptase Gene Is Modulated by Quadruplex Formation in Its First Exon Due to DNA Methylation. J. Biol. Chem. 2017, 292, 20859–20870. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.F.; Li, M.H.; Chu, I.T.; Winnerdy, F.R.; Phan, A.T.; Chang, T.C. Cytosine Epigenetic Modification Modulates the Formation of an Unprecedented G4 Structure in the WNT1 Promoter. Nucleic Acids Res. 2021, 48, 1120–1130. [Google Scholar] [CrossRef] [PubMed]
- D’Amico, F.; Graziano, R.; D’Aria, F.; Russomanno, P.; Di Fonzo, S.; Amato, J.; Pagano, B. Cytosine Epigenetic Modifications and Conformational Changes in G-quadruplex DNA: An Ultraviolet Resonance Raman Spectroscopy Study. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2023, 300, 122901. [Google Scholar] [CrossRef]
- Nain, N.; Singh, A.; Khan, S.; Kukreti, S. G-Quadruplex Formation at Human DAT1 Gene Promoter: Effect of Cytosine Methylation. Biochem. Biophys. Rep. 2023, 34, 101464. [Google Scholar] [CrossRef]
- Tsukakoshi, K.; Saito, S.; Yoshida, W.; Goto, S.; Ikebukuro, K. CpG Methylation Changes G-Quadruplex Structures Derived from Gene Promoters and Interaction with VEGF and SP1. Molecules 2018, 23, 944. [Google Scholar] [CrossRef]
- Weerasinghe, P.; Garcia, G.E.; Zhu, Q.; Yuan, P.; Feng, L.; Mao, L.; Jing, N. Inhibition of Stat3 Activation and Tumor Growth Suppression of Non-Small Cell Lung Cancer by G-Quartet Oligonucleotides. Int. J. Oncol. 2007, 31, 129–136. [Google Scholar] [CrossRef]
- Weerasinghe, P.; Li, Y.; Guan, Y.; Zhang, R.; Tweardy, D.J.; Jing, N. T40214/PEI Complex: A Potent Therapeutics for Prostate Cancer That Targets STAT3 Signaling. Prostate 2008, 68, 1430–1442. [Google Scholar] [CrossRef]
- Esposito, V.; Benigno, D.; Bello, I.; Panza, E.; Bucci, M.; Virgilio, A.; Galeone, A. Structural and Biological Features of G-Quadruplex Aptamers as Promising Inhibitors of the STAT3 Signaling Pathway. Int. J. Mol. Sci. 2023, 24, 9524. [Google Scholar] [CrossRef] [PubMed]
- Chai, E.Z.P.; Shanmugam, M.K.; Arfuso, F.; Dharmarajan, A.; Wang, C.; Kumar, A.P.; Samy, R.P.; Lim, L.H.K.; Wang, L.; Goh, B.C.; et al. Targeting Transcription Factor STAT3 for Cancer Prevention and Therapy. Pharmacol. Ther. 2016, 162, 86–97. [Google Scholar] [CrossRef]
- Do, N.Q.; Lim, K.W.; Teo, M.H.; Heddi, B.; Phan, A.T. Stacking of G-Quadruplexes: NMR Structure of a G-Rich Oligonucleotide with Potential Anti-HIV and Anticancer Activity. Nucleic Acids Res. 2011, 39, 9448–9457. [Google Scholar] [CrossRef]
- Hwang, T.L.; Shaka, A.J. Water Suppression That Works. Excitation Sculpting Using Arbitrary Wave-Forms and Pulsed-Field Gradients. J. Magn. Reson. A 1995, 112, 275–279. [Google Scholar] [CrossRef]
- Esposito, V.; Galeone, A.; Mayol, L.; Oliviero, G.; Virgilio, A.; Randazzo, L. A Topological Classification of G-Quadruplex Structures. Nucleosides Nucleotides Nucleic Acids 2007, 26, 1155–1159. [Google Scholar] [CrossRef]
- Kypr, J.; Kejnovská, I.; Renčiuk, D.; Vorlíčková, M. Circular Dichroism and Conformational Polymorphism of DNA. Nucleic Acids Res. 2009, 37, 1713–1725. [Google Scholar] [CrossRef] [PubMed]
- Vorlíčková, M.; Kejnovská, I.; Bednářová, K.; Renčiuk, D.; Kypr, J. Circular Dichroism Spectroscopy of DNA: From Duplexes to Quadruplexes. Chirality 2012, 24, 691–698. [Google Scholar] [CrossRef] [PubMed]
- Masiero, S.; Trotta, R.; Pieraccini, S.; De Tito, S.; Perone, R.; Randazzo, A.; Spada, G.P. A Non-Empirical Chromophoric Interpretation of CD Spectra of DNA G-Quadruplex Structures. Org. Biomol. Chem. 2010, 8, 2683–2692. [Google Scholar] [CrossRef]
- Vorlíčková, M.; Kejnovská, I.; Sagi, J.; Renčiuk, D.; Bednářová, K.; Motlová, J.; Kypr, J. Circular Dichroism and Guanine Quadruplexes. Methods 2012, 57, 64–75. [Google Scholar] [CrossRef] [PubMed]
- Olsen, C.M.; Marky, L.A. Monitoring the Temperature Unfolding of G-Quadruplexes by UV and Circular Dichroism Spectroscopies and Calorimetry Techniques. Methods Mol. Biol. 2010, 608, 147–158. [Google Scholar] [CrossRef]
- Do, N.Q.; Phan, A.T. Monomer-Dimer Equilibrium for the 5′-5′ Stacking of Propeller-Type Parallel-Stranded g-Quadruplexes: NMR Structural Study. Chemistry 2012, 18, 14752–14759. [Google Scholar] [CrossRef]
- Zhang, Y.; Lai, B.; Juhas, M. Recent Advances in Aptamer Discovery and Applications. Molecules 2019, 24, 941. [Google Scholar] [CrossRef]
- Varada, M.; Aher, M.; Erande, N.; Kumar, V.A.; Fernandes, M. Methoxymethyl Threofuranosyl Thymidine (4′-MOM-TNA-T) at the T7 Position of the Thrombin-Binding Aptamer Boosts Anticoagulation Activity, Thermal Stability, and Nuclease Resistance. ACS Omega 2020, 5, 498–506. [Google Scholar] [CrossRef]
- Virgilio, A.; Esposito, V.; Pecoraro, A.; Russo, A.; Vellecco, V.; Pepe, A.; Bucci, M.; Russo, G.; Galeone, A. Structural Properties and Anticoagulant/Cytotoxic Activities of Heterochiral Enantiomeric Thrombin Binding Aptamer (TBA) Derivatives. Nucleic Acids Res. 2020, 48, 12556–12565. [Google Scholar] [CrossRef] [PubMed]
- Jana, J.; Weisz, K. Thermodynamic Stability of G-Quadruplexes: Impact of Sequence and Environment. ChemBioChem 2021, 22, 2848–2856. [Google Scholar] [CrossRef] [PubMed]
- Ghimire, C.; Park, S.; Iida, K.; Yangyuoru, P.; Otomo, H.; Yu, Z.; Nagasawa, K.; Sugiyama, H.; Mao, H. Direct Quantification of Loop Interaction and π-π Stacking for G-Quadruplex Stability at the Submolecular Level. J. Am. Chem. Soc. 2014, 136, 15537–15544. [Google Scholar] [CrossRef] [PubMed]
- Esposito, V.; Russo, A.; Amato, T.; Vellecco, V.; Bucci, M.; Mayol, L.; Russo, G.; Virgilio, A.; Galeone, A. The “Janus Face” of the Thrombin Binding Aptamer: Investigating the Anticoagulant and Antiproliferative Properties through Straightforward Chemical Modifications. Bioorg. Chem. 2018, 76, 202–209. [Google Scholar] [CrossRef] [PubMed]
- Benigno, D.; Virgilio, A.; Bello, I.; La Manna, S.; Vellecco, V.; Bucci, M.; Marasco, D.; Panza, E.; Esposito, V.; Galeone, A. Properties and Potential Antiproliferative Activity of Thrombin-Binding Aptamer (TBA) Derivatives with One or Two Additional G-Tetrads. Int. J. Mol. Sci. 2022, 23, 14921. [Google Scholar] [CrossRef]
- Fan, X.; Sun, L.; Wu, Y.; Zhang, L.; Yang, Z. Bioactivity of 2′-Deoxyinosine-Incorporated Aptamer AS1411. Sci. Rep. 2016, 6, 25799. [Google Scholar] [CrossRef]
- Fan, X.; Sun, L.; Li, K.; Yang, X.; Cai, B.; Zhang, Y.; Zhu, Y.; Ma, Y.; Guan, Z.; Wu, Y.; et al. The Bioactivity of D/L-Isonucleoside- and 2′-Deoxyinosine-Incorporated Aptamer AS1411s Including DNA Replication/MicroRNA Expression. Mol. Ther. Nucleic Acids 2017, 9, 218–229. [Google Scholar] [CrossRef]
| NAME | SEQUENCE (5′-3′) | Tm (±1) | ∆Tm |
|---|---|---|---|
| STAT | GGGCGGGCGGGCGGGC | 82 | - |
| STAT M1 | GGGmCGGGCGGGCGGGC | 90 | +8 |
| STAT M2 | GGGCGGGmCGGGCGGGC | 90 | +8 |
| STAT M3 | GGGCGGGCGGGmCGGGC | 87 | +5 |
| STAT M4 | GGGCGGGCGGGCGGGmC | 89 | +7 |
| STATB | GCGGCGGGCGGGCGGGC | 63 | - |
| STATB M1 | GmCGGCGGGCGGGCGGGC | 68 | +5 |
| STATB M2 | GCGGmCGGGCGGGCGGGC | 68 | +5 |
| STATB M3 | GCGGCGGGmCGGGCGGGC | 68 | +5 |
| STATB M4 | GCGGCGGGCGGGmCGGGC | 67 | +4 |
| STATB M5 | GCGGCGGGCGGGCGGGmC | 66 | +3 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Esposito, V.; Benigno, D.; Aliberti, C.; Esposito, C.; Panza, E.; Virgilio, A.; Galeone, A. Effects of 5-Methyl-2′-Deoxycytidine in G-Quadruplex Forming Aptamers d(G3C)4 and d[GCG2(CG3)3C]: Investigating the Key Role of the Loops. Biomolecules 2025, 15, 753. https://doi.org/10.3390/biom15060753
Esposito V, Benigno D, Aliberti C, Esposito C, Panza E, Virgilio A, Galeone A. Effects of 5-Methyl-2′-Deoxycytidine in G-Quadruplex Forming Aptamers d(G3C)4 and d[GCG2(CG3)3C]: Investigating the Key Role of the Loops. Biomolecules. 2025; 15(6):753. https://doi.org/10.3390/biom15060753
Chicago/Turabian StyleEsposito, Veronica, Daniela Benigno, Carla Aliberti, Camilla Esposito, Elisabetta Panza, Antonella Virgilio, and Aldo Galeone. 2025. "Effects of 5-Methyl-2′-Deoxycytidine in G-Quadruplex Forming Aptamers d(G3C)4 and d[GCG2(CG3)3C]: Investigating the Key Role of the Loops" Biomolecules 15, no. 6: 753. https://doi.org/10.3390/biom15060753
APA StyleEsposito, V., Benigno, D., Aliberti, C., Esposito, C., Panza, E., Virgilio, A., & Galeone, A. (2025). Effects of 5-Methyl-2′-Deoxycytidine in G-Quadruplex Forming Aptamers d(G3C)4 and d[GCG2(CG3)3C]: Investigating the Key Role of the Loops. Biomolecules, 15(6), 753. https://doi.org/10.3390/biom15060753










