The Role of N6-Methyladenosine (m6A) RNA Modification in the Pathogenesis of Parkinson’s Disease
Abstract
1. Introduction
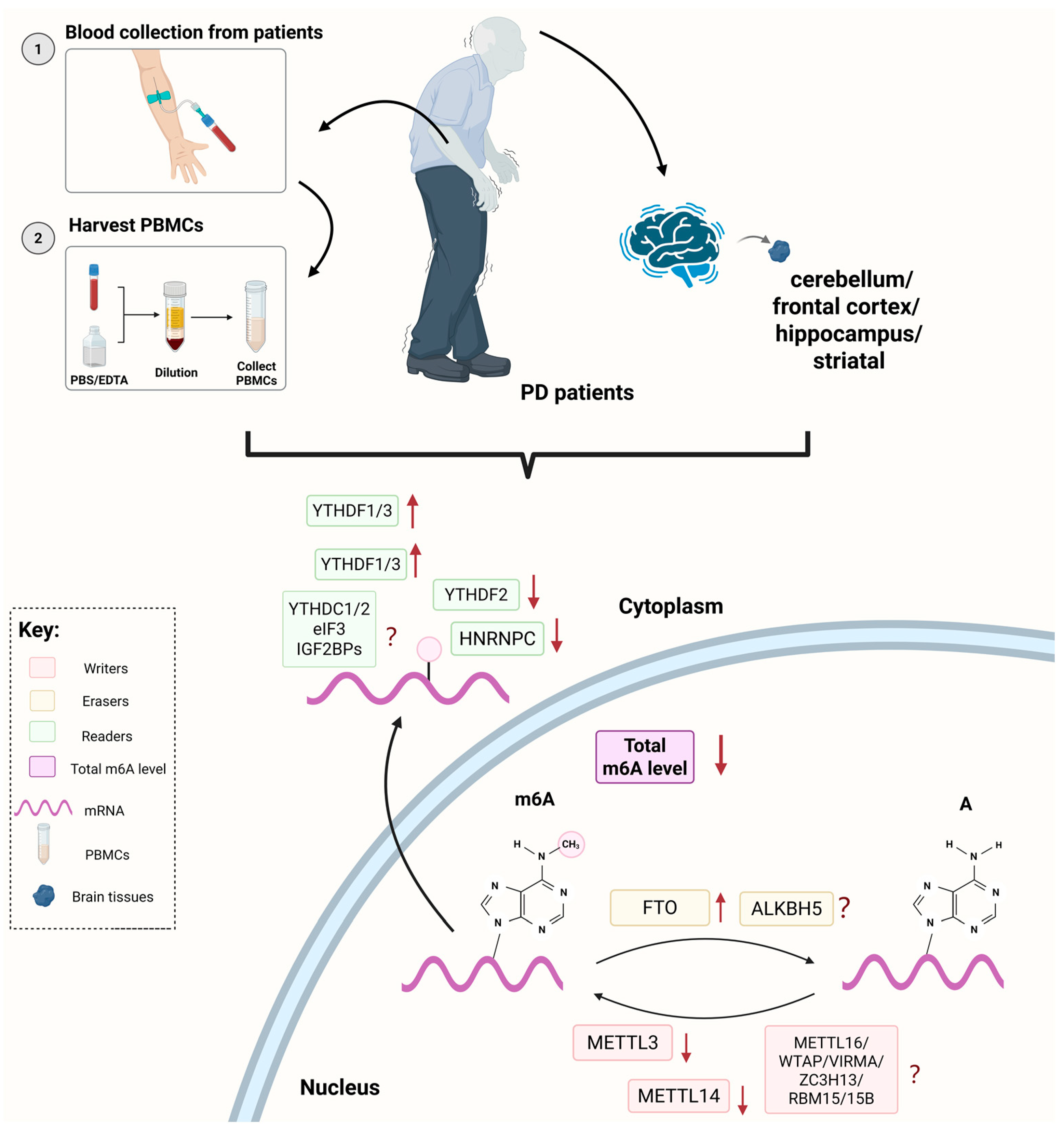
2. Clinical Studies of m6A RNA Modification in PD
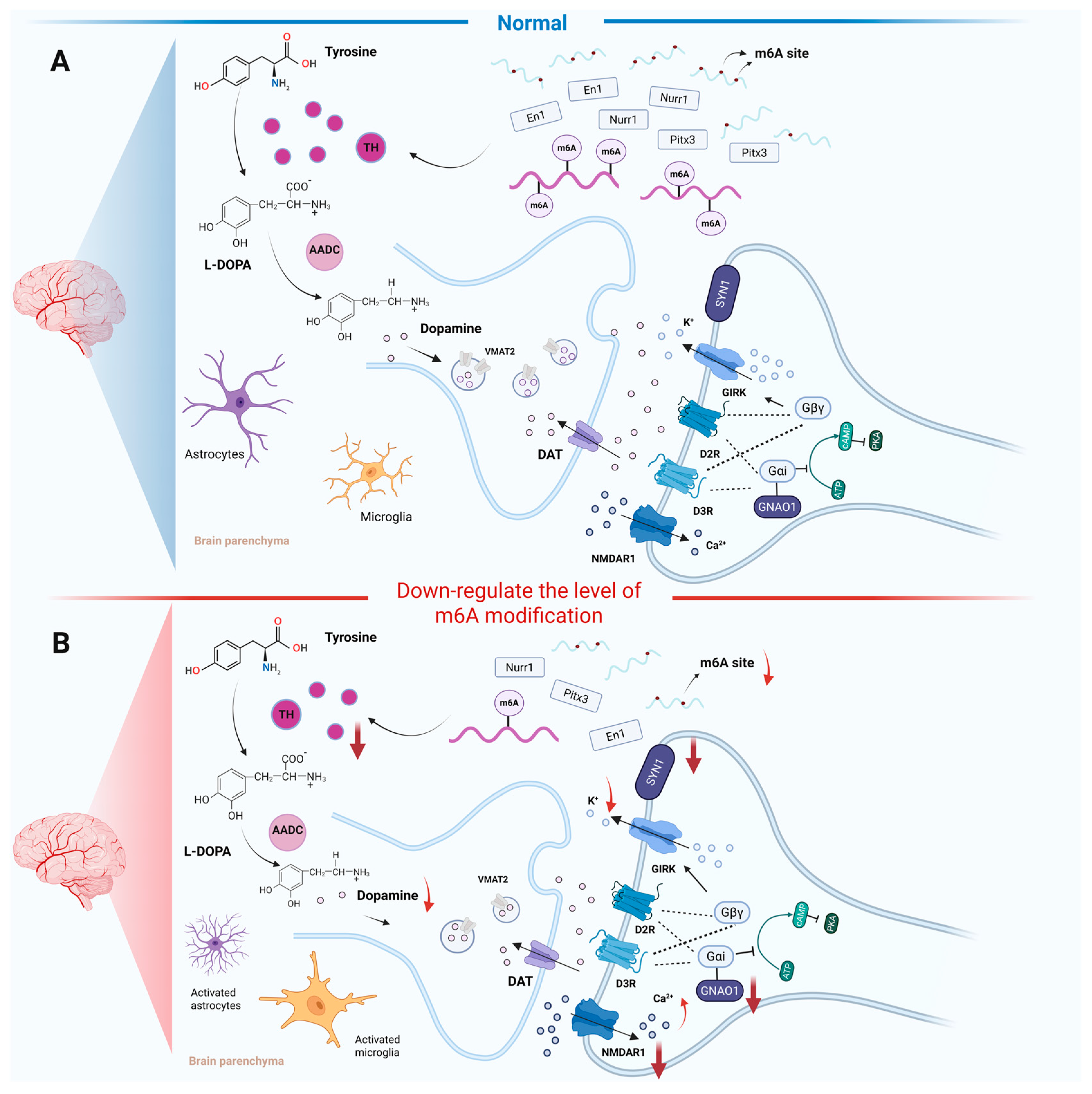
3. Regulation of m6A RNA Modification in Dopaminergic Neurons
3.1. m6A RNA Modification Effect on Dopamine Metabolism
3.2. m6A RNA Modification Effect on the Survival of Dopaminergic Neurons
4. m6A RNA Modification of PD Pathogenic Genes
5. Discussion
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| ALKBH5 | AlkB homologue 5 protein |
| ATM | Ataxia telangiectasia mutated |
| En1 | Engrailed-1 |
| FTO | Fat mass and obesity associated proteins |
| GNAO1 | Guanine nucleotide-binding protein O subunit α |
| HNRNPC | Heterogeneous nuclear ribonucleoprotein C |
| IFN-β | Interferon-β |
| IL-6 | Interleukin-6 |
| LC-MS | Liquid chromatography-tandem mass spectrometry |
| MeRIP-Seq | Methylated RNA immunoprecipitation sequencing |
| Mn | Manganese |
| MPTP | 1-methy1-4-phenyl-1,2,3,6-tetrahydropyridine |
| MPP+ | 1-methyl-4-phenylpyridinium |
| NMDAR1 | N-methyl-D-aspartate receptor 1 |
| Nurr1 | Nuclear receptor-related factor 1 |
| PBMCs | Peripheral blood mononuclear cells |
| PD | Parkinson’s disease |
| Pitx3 | Paired-like homeodomain transcription factor 3 |
| ROS | Reactive oxygen species |
| SN | Substantia nigra |
| SYN1 | Synaptic vesicle protein 1 |
| SLC7A11 | solute carrier family 7 member 11 |
| TH | Tyrosine hydroxylase |
| TRAF6 | Tumor necrosis factor receptor-associated factor 6 |
| TNF-α | Tumor necrosis factor-alpha |
| α-Syn | α-synuclein |
| m6A | N6-methyladenosine |
| m6A-SNPs | m6A-related single nucleotide polymorphisms |
| RBPs | RNA-binding proteins |
References
- Bokar, J.A.; Shambaugh, M.E.; Polayes, D.; Matera, A.G.; Rottman, F.M. Purification and cDNA cloning of the AdoMet-binding subunit of the human mRNA (N6-adenosine)-methyltransferase. Rna 1997, 3, 1233–1247. [Google Scholar] [PubMed]
- Meyer, K.D.; Jaffrey, S.R. The dynamic epitranscriptome: N6-methyladenosine and gene expression control. Nat. Rev. Mol. Cell Biol. 2014, 15, 313–326. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Yue, Y.; Han, D.; Wang, X.; Fu, Y.; Zhang, L.; Jia, G.; Yu, M.; Lu, Z.; Deng, X.; et al. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat. Chem. Biol. 2014, 10, 93–95. [Google Scholar] [CrossRef]
- Pendleton, K.E.; Chen, B.; Liu, K.; Hunter, O.V.; Xie, Y.; Tu, B.P.; Conrad, N.K. The U6 snRNA m(6)A Methyltransferase METTL16 Regulates SAM Synthetase Intron Retention. Cell 2017, 169, 824–835.e814. [Google Scholar] [CrossRef] [PubMed]
- van Tran, N.; Ernst, F.G.M.; Hawley, B.R.; Zorbas, C.; Ulryck, N.; Hackert, P.; Bohnsack, K.E.; Bohnsack, M.T.; Jaffrey, S.R.; Graille, M.; et al. The human 18S rRNA m6A methyltransferase METTL5 is stabilized by TRMT112. Nucleic Acids Res. 2019, 47, 7719–7733. [Google Scholar] [CrossRef]
- Mathoux, J.; Henshall, D.C.; Brennan, G.P. Regulatory Mechanisms of the RNA Modification m(6)A and Significance in Brain Function in Health and Disease. Front. Cell Neurosci. 2021, 15, 671932. [Google Scholar] [CrossRef]
- Jiang, X.; Liu, B.; Nie, Z.; Duan, L.; Xiong, Q.; Jin, Z.; Yang, C.; Chen, Y. The role of m6A modification in the biological functions and diseases. Signal Transduct. Target. Ther. 2021, 6, 74. [Google Scholar] [CrossRef]
- Jia, G.; Fu, Y.; Zhao, X.; Dai, Q.; Zheng, G.; Yang, Y.; Yi, C.; Lindahl, T.; Pan, T.; Yang, Y.G.; et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 2011, 7, 885–887. [Google Scholar] [CrossRef]
- Zheng, G.; Dahl, J.A.; Niu, Y.; Fedorcsak, P.; Huang, C.M.; Li, C.J.; Vågbø, C.B.; Shi, Y.; Wang, W.L.; Song, S.H.; et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol. Cell 2013, 49, 18–29. [Google Scholar] [CrossRef]
- Li, Z.; Weng, H.; Su, R.; Weng, X.; Zuo, Z.; Li, C.; Huang, H.; Nachtergaele, S.; Dong, L.; Hu, C.; et al. FTO Plays an Oncogenic Role in Acute Myeloid Leukemia as a N(6)-Methyladenosine RNA Demethylase. Cancer Cell 2017, 31, 127–141. [Google Scholar] [CrossRef]
- Tang, C.; Klukovich, R.; Peng, H.; Wang, Z.; Yu, T.; Zhang, Y.; Zheng, H.; Klungland, A.; Yan, W. ALKBH5-dependent m6A demethylation controls splicing and stability of long 3′-UTR mRNAs in male germ cells. Proc. Natl. Acad. Sci. USA 2018, 115, E325–E333. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.; Wang, X.; Lu, Z.; Zhao, B.S.; Ma, H.; Hsu, P.J.; Liu, C.; He, C. YTHDF3 facilitates translation and decay of N(6)-methyladenosine-modified RNA. Cell Res. 2017, 27, 315–328. [Google Scholar] [CrossRef]
- Xiao, W.; Adhikari, S.; Dahal, U.; Chen, Y.S.; Hao, Y.J.; Sun, B.F.; Sun, H.Y.; Li, A.; Ping, X.L.; Lai, W.Y.; et al. Nuclear m(6)A Reader YTHDC1 Regulates mRNA Splicing. Mol. Cell 2016, 61, 507–519. [Google Scholar] [CrossRef]
- Hsu, P.J.; Zhu, Y.; Ma, H.; Guo, Y.; Shi, X.; Liu, Y.; Qi, M.; Lu, Z.; Shi, H.; Wang, J.; et al. Ythdc2 is an N(6)-methyladenosine binding protein that regulates mammalian spermatogenesis. Cell Res. 2017, 27, 1115–1127. [Google Scholar] [CrossRef] [PubMed]
- Arguello, A.E.; DeLiberto, A.N.; Kleiner, R.E. RNA Chemical Proteomics Reveals the N(6)-Methyladenosine (m(6)A)-Regulated Protein-RNA Interactome. J. Am. Chem. Soc. 2017, 139, 17249–17252. [Google Scholar] [CrossRef]
- Yen, Y.P.; Chen, J.A. The m(6)A epitranscriptome on neural development and degeneration. J. Biomed. Sci. 2021, 28, 40. [Google Scholar] [CrossRef] [PubMed]
- Walters, B.J.; Mercaldo, V.; Gillon, C.J.; Yip, M.; Neve, R.L.; Boyce, F.M.; Frankland, P.W.; Josselyn, S.A. The Role of The RNA Demethylase FTO (Fat Mass and Obesity-Associated) and mRNA Methylation in Hippocampal Memory Formation. Neuropsychopharmacology 2017, 42, 1502–1510. [Google Scholar] [CrossRef]
- Weng, Y.L.; Wang, X.; An, R.; Cassin, J.; Vissers, C.; Liu, Y.; Liu, Y.; Xu, T.; Wang, X.; Wong, S.Z.H.; et al. Epitranscriptomic m(6)A Regulation of Axon Regeneration in the Adult Mammalian Nervous System. Neuron 2018, 97, 313–325.e316. [Google Scholar] [CrossRef]
- Yoon, K.J.; Ringeling, F.R.; Vissers, C.; Jacob, F.; Pokrass, M.; Jimenez-Cyrus, D.; Su, Y.; Kim, N.S.; Zhu, Y.; Zheng, L.; et al. Temporal Control of Mammalian Cortical Neurogenesis by m(6)A Methylation. Cell 2017, 171, 877–889.e817. [Google Scholar] [CrossRef]
- Zhang, F.; Kang, Y.; Wang, M.; Li, Y.; Xu, T.; Yang, W.; Song, H.; Wu, H.; Shu, Q.; Jin, P. Fragile X mental retardation protein modulates the stability of its m6A-marked messenger RNA targets. Hum. Mol. Genet. 2018, 27, 3936–3950. [Google Scholar] [CrossRef]
- Xia, L.; Zhang, F.; Li, Y.; Mo, Y.; Zhang, L.; Li, Q.; Luo, M.; Hou, X.; Du, Z.; Deng, J.; et al. A new perspective on Alzheimer’s disease: m6A modification. Front. Genet. 2023, 14, 1166831. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Yu, C.; Guo, M.; Zheng, X.; Ali, S.; Huang, H.; Zhang, L.; Wang, S.; Huang, Y.; Qie, S.; et al. Down-Regulation of m6A mRNA Methylation Is Involved in Dopaminergic Neuronal Death. ACS Chem. Neurosci. 2019, 10, 2355–2363. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.; Yang, L.; Wei, W. Role of Fto on CaMKII/CREB signaling pathway of hippocampus in depressive-like behaviors induced by chronic restraint stress mice. Behav. Brain Res. 2021, 406, 113227. [Google Scholar] [CrossRef] [PubMed]
- Yao, Y.; Wen, Y.; Du, T.; Sun, N.; Deng, H.; Ryan, J.; Rao, S. Meta-analysis indicates that SNP rs9939609 within FTO is not associated with major depressive disorder (MDD) in Asian population. J. Affect. Disord. 2016, 193, 27–30. [Google Scholar] [CrossRef]
- Zaman, T.; Helbig, K.L.; Clatot, J.; Thompson, C.H.; Kang, S.K.; Stouffs, K.; Jansen, A.E.; Verstraete, L.; Jacquinet, A.; Parrini, E.; et al. SCN3A-Related Neurodevelopmental Disorder: A Spectrum of Epilepsy and Brain Malformation. Ann. Neurol. 2020, 88, 348–362. [Google Scholar] [CrossRef]
- Hayes, M.T. Parkinson’s Disease and Parkinsonism. Am. J. Med. 2019, 132, 802–807. [Google Scholar] [CrossRef]
- Damier, P.; Hirsch, E.C.; Agid, Y.; Graybiel, A.M. The substantia nigra of the human brain. II. Patterns of loss of dopamine-containing neurons in Parkinson’s disease. Brain 1999, 122 Pt 8, 1437–1448. [Google Scholar] [CrossRef]
- Bloem, B.R.; Okun, M.S.; Klein, C. Parkinson’s disease. Lancet 2021, 397, 2284–2303. [Google Scholar] [CrossRef]
- Polymeropoulos, M.H.; Lavedan, C.; Leroy, E.; Ide, S.E.; Dehejia, A.; Dutra, A.; Pike, B.; Root, H.; Rubenstein, J.; Boyer, R.; et al. Mutation in the alpha-synuclein gene identified in families with Parkinson’s disease. Science 1997, 276, 2045–2047. [Google Scholar] [CrossRef]
- Anglade, P.; Vyas, S.; Javoy-Agid, F.; Herrero, M.T.; Michel, P.P.; Marquez, J.; Mouatt-Prigent, A.; Ruberg, M.; Hirsch, E.C.; Agid, Y. Apoptosis and autophagy in nigral neurons of patients with Parkinson’s disease. Histol. Histopathol. 1997, 12, 25–31. [Google Scholar]
- Michel, P.P.; Hirsch, E.C.; Hunot, S. Understanding Dopaminergic Cell Death Pathways in Parkinson Disease. Neuron 2016, 90, 675–691. [Google Scholar] [CrossRef] [PubMed]
- Munhoz, R.P.; Tumas, V.; Pedroso, J.L.; Silveira-Moriyama, L. The clinical diagnosis of Parkinson’s disease. Arq. Neuropsiquiatr. 2024, 82, 1–10. [Google Scholar] [CrossRef]
- Tolosa, E.; Wenning, G.; Poewe, W. The diagnosis of Parkinson’s disease. Lancet Neurol. 2006, 5, 75–86. [Google Scholar] [CrossRef] [PubMed]
- Emamzadeh, F.N.; Surguchov, A. Parkinson’s Disease: Biomarkers, Treatment, and Risk Factors. Front. Neurosci. 2018, 12, 612. [Google Scholar] [CrossRef]
- Lotankar, S.; Prabhavalkar, K.S.; Bhatt, L.K. Biomarkers for Parkinson’s Disease: Recent Advancement. Neurosci. Bull. 2017, 33, 585–597. [Google Scholar] [CrossRef] [PubMed]
- He, H.; Zhang, Q.; Liao, J.; Lei, J.; Luo, M.; Huang, J.; Chen, M.; Shen, Y.; Wang, J.; Xu, P.; et al. METTL14 is decreased and regulates m(6) A modification of α-synuclein in Parkinson’s disease. J. Neurochem. 2023, 166, 609–622. [Google Scholar] [CrossRef]
- Martinez De La Cruz, B.; Gell, C.; Markus, R.; Macdonald, I.; Fray, R.; Knight, H.M. m(6) A mRNA methylation in human brain is disrupted in Lewy body disorders. Neuropathol. Appl. Neurobiol. 2023, 49, e12885. [Google Scholar] [CrossRef]
- Geng, Y.; Long, X.; Zhang, Y.; Wang, Y.; You, G.; Guo, W.; Zhuang, G.; Zhang, Y.; Cheng, X.; Yuan, Z.; et al. FTO-targeted siRNA delivery by MSC-derived exosomes synergistically alleviates dopaminergic neuronal death in Parkinson’s disease via m6A-dependent regulation of ATM mRNA. J. Transl. Med. 2023, 21, 652. [Google Scholar] [CrossRef]
- Qiu, X.; He, H.; Huang, Y.; Wang, J.; Xiao, Y. Genome-wide identification of m(6)A-associated single-nucleotide polymorphisms in Parkinson’s disease. Neurosci. Lett. 2020, 737, 135315. [Google Scholar] [CrossRef]
- Jiang, S.; Xie, Y.; He, Z.; Zhang, Y.; Zhao, Y.; Chen, L.; Zheng, Y.; Miao, Y.; Zuo, Z.; Ren, J. m6ASNP: A tool for annotating genetic variants by m6A function. Gigascience 2018, 7, giy035. [Google Scholar] [CrossRef]
- Dumitriu, A.; Pacheco, C.D.; Wilk, J.B.; Strathearn, K.E.; Latourelle, J.C.; Goldwurm, S.; Pezzoli, G.; Rochet, J.C.; Lindquist, S.; Myers, R.H. Cyclin-G-associated kinase modifies α-synuclein expression levels and toxicity in Parkinson’s disease: Results from the GenePD Study. Hum. Mol. Genet. 2011, 20, 1478–1487. [Google Scholar] [CrossRef] [PubMed]
- Ma, Z.G.; He, F.; Xu, J. Quantitative assessment of the association between GAK rs1564282 C/T polymorphism and the risk of Parkinson’s disease. J. Clin. Neurosci. 2015, 22, 1077–1080. [Google Scholar] [CrossRef] [PubMed]
- Du, T.; Li, G.; Yang, J.; Ma, K. RNA demethylase Alkbh5 is widely expressed in neurons and decreased during brain development. Brain Res. Bull. 2020, 163, 150–159. [Google Scholar] [CrossRef]
- Quan, W.; Li, J.; Liu, L.; Zhang, Q.; Qin, Y.; Pei, X.; Chen, J. Influence of N6-Methyladenosine Modification Gene HNRNPC on Cell Phenotype in Parkinson’s Disease. Parkinsons Dis. 2021, 2021, 9919129. [Google Scholar] [CrossRef] [PubMed]
- Qin, L.; Min, S.; Shu, L.; Pan, H.; Zhong, J.; Guo, J.; Sun, Q.; Yan, X.; Chen, C.; Tang, B.; et al. Genetic analysis of N6-methyladenosine modification genes in Parkinson’s disease. Neurobiol. Aging 2020, 93, 143.e9–143.e13. [Google Scholar] [CrossRef]
- Nagatsu, T.; Levitt, M.; Udenfriend, S. Tyrosine Hydroxylase. The Initial Step in Norepinephrine Biosynthesis. J. Biol. Chem. 1964, 239, 2910–2917. [Google Scholar] [CrossRef]
- Zhou, Z.D.; Yi, L.X.; Wang, D.Q.; Lim, T.M.; Tan, E.K. Role of dopamine in the pathophysiology of Parkinson’s disease. Transl. Neurodegener. 2023, 12, 44. [Google Scholar] [CrossRef]
- Christenson, J.G.; Dairman, W.; Udenfriend, S. Preparation and properties of a homogeneous aromatic L-amino acid decarboxylase from hog kidney. Arch. Biochem. Biophys. 1970, 141, 356–367. [Google Scholar] [CrossRef]
- Robbins, T.W. Dopamine and cognition. Curr. Opin. Neurol. 2003, 16 (Suppl. S2), S1–S2. [Google Scholar] [CrossRef]
- Blakely, R.D. Neurobiology. Dopamine’s reversal of fortune. Science 2001, 293, 2407–2409. [Google Scholar] [CrossRef]
- Teng, Y.; Liu, Z.; Chen, X.; Liu, Y.; Geng, F.; Le, W.; Jiang, H.; Yang, L. Conditional deficiency of m6A methyltransferase Mettl14 in substantia nigra alters dopaminergic neuron function. J. Cell Mol. Med. 2021, 25, 8567–8572. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.; Huang, L.; Xia, Y.; Cheng, S.; Yang, C.; Chen, C.; Zou, Z.; Wang, X.; Tian, X.; Jiang, X.; et al. Analysis of m6A modification regulators in the substantia nigra and striatum of MPTP-induced Parkinson’s disease mice. Neurosci. Lett. 2022, 791, 136907. [Google Scholar] [CrossRef]
- Beaulieu, J.M.; Gainetdinov, R.R. The physiology, signaling, and pharmacology of dopamine receptors. Pharmacol. Rev. 2011, 63, 182–217. [Google Scholar] [CrossRef]
- Matsumoto, M. Dopamine signals and physiological origin of cognitive dysfunction in Parkinson’s disease. Mov. Disord. 2015, 30, 472–483. [Google Scholar] [CrossRef]
- Hess, M.E.; Hess, S.; Meyer, K.D.; Verhagen, L.A.; Koch, L.; Brönneke, H.S.; Dietrich, M.O.; Jordan, S.D.; Saletore, Y.; Elemento, O.; et al. The fat mass and obesity associated gene (Fto) regulates activity of the dopaminergic midbrain circuitry. Nat. Neurosci. 2013, 16, 1042–1048. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, H.; Glaaser, I.W.; Slesinger, P.A. Direct modulation of G protein-gated inwardly rectifying potassium (GIRK) channels. Front. Physiol. 2024, 15, 1386645. [Google Scholar] [CrossRef] [PubMed]
- Selberg, S.; Yu, L.Y.; Bondarenko, O.; Kankuri, E.; Seli, N.; Kovaleva, V.; Herodes, K.; Saarma, M.; Karelson, M. Small-Molecule Inhibitors of the RNA M6A Demethylases FTO Potently Support the Survival of Dopamine Neurons. Int. J. Mol. Sci. 2021, 22, 4537. [Google Scholar] [CrossRef]
- Lowe, E.K.; Racioppi, C.; Peyriéras, N.; Ristoratore, F.; Christiaen, L.; Swalla, B.J.; Stolfi, A. A cis-regulatory change underlying the motor neuron-specific loss of Ebf expression in immotile tunicate larvae. Evol. Dev. 2021, 23, 72–85. [Google Scholar] [CrossRef]
- Hu, W.; Wang, M.; Sun, G.; Zhang, L.; Lu, H. Early B Cell Factor 3 (EBF3) attenuates Parkinson’s disease through directly regulating contactin-associated protein-like 4 (CNTNAP4) transcription: An experimental study. Cell Signal 2024, 118, 111139. [Google Scholar] [CrossRef]
- He, F.; Ru, X.; Wen, T. NRF2, a Transcription Factor for Stress Response and Beyond. Int. J. Mol. Sci. 2020, 21, 4777. [Google Scholar] [CrossRef]
- Pang, P.; Zhang, S.; Fan, X.; Zhang, S. Knockdown of fat mass and obesity alleviates the ferroptosis in Parkinson’s disease through m6A-NRF2-dependent manner. Cell Biol. Int. 2024, 48, 431–439. [Google Scholar] [CrossRef]
- Li, Z.; Chen, X.; Xiang, W.; Tang, T.; Gan, L. m6A Demethylase FTO-Mediated Upregulation of BAP1 Induces Neuronal Ferroptosis via the p53/SLC7A11 Axis in the MPP(+)/MPTP-Induced Parkinson’s Disease Model. ACS Chem. Neurosci. 2025, 16, 405–416. [Google Scholar] [CrossRef] [PubMed]
- Bai, L.; Tang, Q.; Zou, Z.; Meng, P.; Tu, B.; Xia, Y.; Cheng, S.; Zhang, L.; Yang, K.; Mu, S.; et al. m6A Demethylase FTO Regulates Dopaminergic Neurotransmission Deficits Caused by Arsenite. Toxicol. Sci. 2018, 165, 431–446. [Google Scholar] [CrossRef] [PubMed]
- Qi, Z.; Wang, S.; Li, J.; Wen, Y.; Cui, R.; Zhang, K.; Liu, Y.; Yang, X.; Zhang, L.; Xu, B.; et al. Protective role of mRNA demethylase FTO on axon guidance molecules of nigro-striatal projection system in manganese-induced parkinsonism. J. Hazard. Mater. 2022, 426, 128099. [Google Scholar] [CrossRef]
- Abeliovich, A.; Schmitz, Y.; Fariñas, I.; Choi-Lundberg, D.; Ho, W.H.; Castillo, P.E.; Shinsky, N.; Verdugo, J.M.; Armanini, M.; Ryan, A.; et al. Mice lacking alpha-synuclein display functional deficits in the nigrostriatal dopamine system. Neuron 2000, 25, 239–252. [Google Scholar] [CrossRef] [PubMed]
- Shao, L.; Hu, F.; Xu, R.; Nie, H.; Zhang, H.; Zhang, P. METTL14 Regulates the m6A Modification of TRAF6 to Suppress Mitochondrial Dysfunction and Ferroptosis in Dopaminergic Neurons via the cGAS-STING Pathway. Curr. Mol. Med. 2024, 24, 1518–1528. [Google Scholar] [CrossRef]
- Qin, X.Y.; Zhang, S.P.; Cao, C.; Loh, Y.P.; Cheng, Y. Aberrations in Peripheral Inflammatory Cytokine Levels in Parkinson Disease: A Systematic Review and Meta-analysis. JAMA Neurol. 2016, 73, 1316–1324. [Google Scholar] [CrossRef]
- Gorelenkova Miller, O.; Mieyal, J.J. Critical Roles of Glutaredoxin in Brain Cells-Implications for Parkinson’s Disease. Antioxid. Redox Signal 2019, 30, 1352–1368. [Google Scholar] [CrossRef]
- Gong, X.; Huang, M.; Chen, L. NRF1 mitigates motor dysfunction and dopamine neuron degeneration in mice with Parkinson’s disease by promoting GLRX m(6) A methylation through upregulation of METTL3 transcription. CNS Neurosci. Ther. 2024, 30, e14441. [Google Scholar] [CrossRef]
- Saijo, K.; Winner, B.; Carson, C.T.; Collier, J.G.; Boyer, L.; Rosenfeld, M.G.; Gage, F.H.; Glass, C.K. A Nurr1/CoREST pathway in microglia and astrocytes protects dopaminergic neurons from inflammation-induced death. Cell 2009, 137, 47–59. [Google Scholar] [CrossRef]
- Livneh, I.; Moshitch-Moshkovitz, S.; Amariglio, N.; Rechavi, G.; Dominissini, D. The m(6)A epitranscriptome: Transcriptome plasticity in brain development and function. Nat. Rev. Neurosci. 2020, 21, 36–51. [Google Scholar] [CrossRef]
- Wang, Y.; Jia, G. Detection methods of epitranscriptomic mark N6-methyladenosine. Essays Biochem. 2020, 64, 967–979. [Google Scholar]
- Nagarajan, A.; Janostiak, R.; Wajapeyee, N. Dot Blot Analysis for Measuring Global N(6)-Methyladenosine Modification of RNA. Methods Mol. Biol. 2019, 1870, 263–271. [Google Scholar] [PubMed]
- Dominissini, D.; Nachtergaele, S.; Moshitch-Moshkovitz, S.; Peer, E.; Kol, N.; Ben-Haim, M.S.; Dai, Q.; Di Segni, A.; Salmon-Divon, M.; Clark, W.C.; et al. The dynamic N(1)-methyladenosine methylome in eukaryotic messenger RNA. Nature 2016, 530, 441–446. [Google Scholar] [CrossRef] [PubMed]
- Dominissini, D.; Moshitch-Moshkovitz, S.; Schwartz, S.; Salmon-Divon, M.; Ungar, L.; Osenberg, S.; Cesarkas, K.; Jacob-Hirsch, J.; Amariglio, N.; Kupiec, M.; et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 2012, 485, 201–206. [Google Scholar] [CrossRef] [PubMed]
- Linder, B.; Grozhik, A.V.; Olarerin-George, A.O.; Meydan, C.; Mason, C.E.; Jaffrey, S.R. Single-nucleotide-resolution mapping of m6A and m6Am throughout the transcriptome. Nat. Methods 2015, 12, 767–772. [Google Scholar] [CrossRef]
- Imanishi, M.; Tsuji, S.; Suda, A.; Futaki, S. Detection of N(6)-methyladenosine based on the methyl-sensitivity of MazF RNA endonuclease. Chem. Commun. 2017, 53, 12930–12933. [Google Scholar] [CrossRef]
- Deng, S.; Zhang, H.; Zhu, K.; Li, X.; Ye, Y.; Li, R.; Liu, X.; Lin, D.; Zuo, Z.; Zheng, J. M6A2Target: A comprehensive database for targets of m6A writers, erasers and readers. Brief. Bioinform. 2021, 22, bbaa055. [Google Scholar] [CrossRef]
- Luo, X.; Li, H.; Liang, J.; Zhao, Q.; Xie, Y.; Ren, J.; Zuo, Z. RMVar: An updated database of functional variants involved in RNA modifications. Nucleic Acids Res. 2021, 49, D1405–D1412. [Google Scholar] [CrossRef]
- Xuan, J.; Chen, L.; Chen, Z.; Pang, J.; Huang, J.; Lin, J.; Zheng, L.; Li, B.; Qu, L.; Yang, J. RMBase v3.0: Decode the landscape, mechanisms and functions of RNA modifications. Nucleic Acids Res. 2024, 52, D273–D284. [Google Scholar] [CrossRef]
- Liang, Z.; Ye, H.; Ma, J.; Wei, Z.; Wang, Y.; Zhang, Y.; Huang, D.; Song, B.; Meng, J.; Rigden, D.J.; et al. m6A-Atlas v2.0: Updated resources for unraveling the N6-methyladenosine (m6A) epitranscriptome among multiple species. Nucleic Acids Res. 2024, 52, D194–D202. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Zeng, P.; Li, Y.H.; Zhang, Z.; Cui, Q. SRAMP: Prediction of mammalian N6-methyladenosine (m6A) sites based on sequence-derived features. Nucleic Acids Res. 2016, 44, e91. [Google Scholar] [CrossRef] [PubMed]
- Jia, F.; Fellner, A.; Kumar, K.R. Monogenic Parkinson’s Disease: Genotype, Phenotype, Pathophysiology, and Genetic Testing. Genes 2022, 13, 471. [Google Scholar] [CrossRef]
- Coukos, R.; Krainc, D. Key genes and convergent pathogenic mechanisms in Parkinson disease. Nat. Rev. Neurosci. 2024, 25, 393–413. [Google Scholar] [CrossRef] [PubMed]
- Song, H.; Feng, X.; Zhang, H.; Luo, Y.; Huang, J.; Lin, M.; Jin, J.; Ding, X.; Wu, S.; Huang, H.; et al. METTL3 and ALKBH5 oppositely regulate m(6)A modification of TFEB mRNA, which dictates the fate of hypoxia/reoxygenation-treated cardiomyocytes. Autophagy 2019, 15, 1419–1437. [Google Scholar] [CrossRef]
- Wang, F.; Liao, Y.; Zhang, M.; Zhu, Y.; Wang, W.; Cai, H.; Liang, J.; Song, F.; Hou, C.; Huang, S.; et al. N6-methyladenosine demethyltransferase FTO-mediated autophagy in malignant development of oral squamous cell carcinoma. Oncogene 2021, 40, 3885–3898. [Google Scholar] [CrossRef]
- Suga, N.; Ikeda, Y.; Yoshikawa, S.; Taniguchi, K.; Sawamura, H.; Matsuda, S. In Search of a Function for the N6-Methyladenosine in Epitranscriptome, Autophagy and Neurodegenerative Diseases. Neurol. Int. 2023, 15, 967–979. [Google Scholar] [CrossRef]
- Gao, Z.; Zha, X.; Li, M.; Xia, X.; Wang, S. Insights into the m(6)A demethylases FTO and ALKBH5: Structural, biological function, and inhibitor development. Cell Biosci. 2024, 14, 108. [Google Scholar] [CrossRef]

| Gene | Mod ID | Predictive Reliability | Technique | Region | Data Source |
|---|---|---|---|---|---|
| SNCA Homo | m6A_site_644495 | High confidence | MeRIP-seq | CDS | GSE48037GSE76414 |
| SNCA Homo | m6A_site_644502 | High confidence | MeRIP-seq | CDS | GSE107954GSE93911 |
| SNCA Homo | m6A_site_644494 | Moderate confidence | MAZTER-Seq | CDS/Intron | GSE129842 |
| SNCA Mus | m6A_site_505923 | High confidence | MeRIP-seq | CDS | GSE47215 |
| SNCA Mus | m6A_site_505932 | Moderate confidence | MeRIP-seq | Exon | GSE100528GSE29714 |
| PRKN Homo | m6A_site_742128 | Very High confidence | MeRIP-seq | UTR | GSE93911GSE29714 |
| PRKN Homo | m6A_site_742132 | Very High confidence | MeRIP-seq | UTR | GSE120229GSE29714 |
| PRKN Homo | m6A_site_742134 | Very High confidence | m6A-seq | Exon/Intron | GSE93911 |
| PRKN Homo | m6A_site_742126 | High confidence | m6A-seq | UTR | GSE93911GSE87515 |
| PRKN Homo | m6A_site_742129 | High confidence | MeRIP-seq/m6A-seq | UTR | GSM3396437GSM928399 |
| PRKN Homo | m6A_site_742131 | High confidence | MeRIP-seq/m6A-seq | UTR | GSE120229GSE29714 |
| PRKN Homo | m6A_site_742135 | High confidence | m6A-seq | Exon/Intron | GSE93911 |
| PRKN Homo | m6A_site_742136 | High confidence | m6A-seq | Exon/Intron | GSE93911 |
| PRKN Homo | m6A_site_742137 | High confidence | m6A-seq | Exon/Intron | GSE93911 |
| PRKN Homo | m6A_site_742133 | Moderate confidence | MeRIP-seq | UTR | GSE120229GSE29714 |
| PRKN Mus | m6A_site_250551 | Very High confidence | MeRIP-seq | Exon/Intron | GSE47215GSE100528 |
| PRKN Mus | m6A_site_250552 | Very High confidence | MeRIP-seq | Exon/Intron | GSE47215GSE100528 |
| PRKN Mus | m6A_site_250547 | High confidence | MeRIP-seq | Exon/Intron | GSE47215GSE100528 |
| PRKN Mus | m6A_site_250550 | High confidence | MeRIP-seq | Exon/Intron | GSE47215GSE100528 |
| PRKN Mus | m6A_site_250553 | High confidence | MeRIP-seq | Exon/Intron | GSE47215GSE100528 |
| PRKN Mus | m6A_site_250556 | High confidence | MeRIP-seq | Exon/Intron | GSE47216 |
| PRKN Mus | m6A_site_250546 | Moderate confidence | MeRIP-seq | Exon/Intron | GSE100528 |
| PINK1 Homo | m6A_site_12074 | Very High confidence | MeRIP-seq/m6A-seq | Exon | GSE102493 GSE110320 |
| PINK1 Homo | m6A_site_12076 | Very High confidence | MeRIP-seq/m6A-seq | Exon | GSE102493 GSE110320 |
| PINK1 Homo | m6A_site_12028 | High confidence | m6A-seq | Exon | GSE102493 GSE46705 |
| PINK1 Homo | m6A_site_12013 | Moderate confidence | m6A-seq | CDS | GSE102493GSE125780 |
| PINK1 Homo | m6A_site_12019 | Moderate confidence | m6A-seq | Exon | GSE87190GSE93911 |
| PINK1 Homo | m6A_site_12032 | Moderate confidence | MeRIP-seq/m6A-seq | Exon | GSE102493GSE110320 |
| PINK1 Homo | m6A_site_12055 | Moderate confidence | m6A-seq | Exon | GSE110320GSE87190 |
| PINK1 Mus | m6A_site_440098 | Very High confidence | MeRIP-seq/m6A-seq | CDS | GSE47215GSE53244 |
| PINK1 Mus | m6A_site_440091 | High confidence | MeRIP-seq/m6A-seq | CDS | GSE47215 GSE98085 |
| PINK1 Mus | m6A_site_440082 | Moderate confidence | MeRIP-seq/m6A-seq | UTR | GSE100528GSE47215 |
| PINK1 Mus | m6A_site_440095 | Moderate confidence | m6A-seq | Exon | GSE98085 |
| LRRK2 Homo | m6A_site_182822 | Very High confidence | m6A-CLIP | CDS | GSE71154 |
| LRRK2 Mus | m6A_site_220644 | Very High confidence | m6A-seq | CDS | GSE98085 |
| LRRK2 Mus | m6A_site_220649 | Very High confidence | MeRIP-seq | Exon | GSE47215 |
| LRRK2 Mus | m6A_site_220651 | Very High confidence | MeRIP-seq | Exon | GSE100528 |
| LRRK2 Mus | m6A_site_220652 | Very High confidence | MeRIP-seq | Exon | GSE100528 |
| LRRK2 Mus | m6A_site_220648 | Very High confidence | MeRIP-seq | Exon | GSE47215 |
| LRRK2 Mus | m6A_site_220642 | Moderate confidence | MeRIP-seq/m6A-seq | CDS | GSE115106GSE61995 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Y.; Zhao, T.; Yuan, C.; Chen, X. The Role of N6-Methyladenosine (m6A) RNA Modification in the Pathogenesis of Parkinson’s Disease. Biomolecules 2025, 15, 617. https://doi.org/10.3390/biom15050617
Wang Y, Zhao T, Yuan C, Chen X. The Role of N6-Methyladenosine (m6A) RNA Modification in the Pathogenesis of Parkinson’s Disease. Biomolecules. 2025; 15(5):617. https://doi.org/10.3390/biom15050617
Chicago/Turabian StyleWang, Yulu, Tianyuan Zhao, Chunsen Yuan, and Xuechai Chen. 2025. "The Role of N6-Methyladenosine (m6A) RNA Modification in the Pathogenesis of Parkinson’s Disease" Biomolecules 15, no. 5: 617. https://doi.org/10.3390/biom15050617
APA StyleWang, Y., Zhao, T., Yuan, C., & Chen, X. (2025). The Role of N6-Methyladenosine (m6A) RNA Modification in the Pathogenesis of Parkinson’s Disease. Biomolecules, 15(5), 617. https://doi.org/10.3390/biom15050617






