Molecular Characterization and Nutritional Regulation of Two Fatty Acid Elongase (elovl8) Genes in Chinese Perch (Siniperca chuatsi)
Abstract
1. Introduction
2. Materials and Methods
2.1. Fish Sampling
2.2. Identification of the Scelovl8a and Scelovl8b Genes in Chinese Perch
2.3. Bioinformatics Analyses of the Scelovl8a and Scelovl8b Genes
2.4. Total RNA Extraction and cDNA Preparation
2.5. Quantitative Real-Time PCR
2.6. Statistical Analysis
3. Results
3.1. Presence of Both elovl8a and elovl8b Genes in Chinese Perch
3.2. Predicted 3D Structures of Both Elovl8a and Elovl8b in Chinese Perch
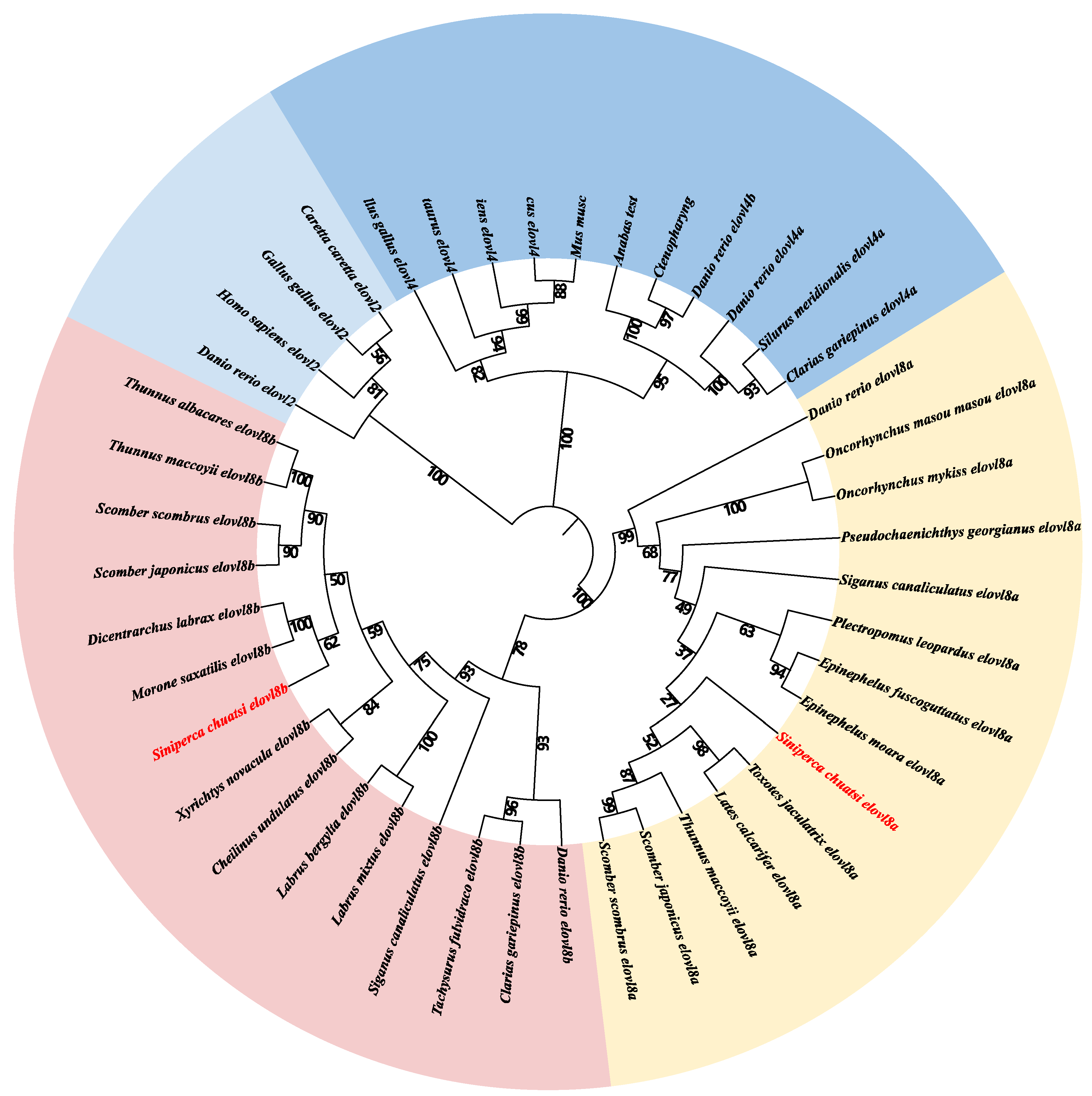
3.3. Phylogenetic Tree of the Examined Elovl Genes in Teleost
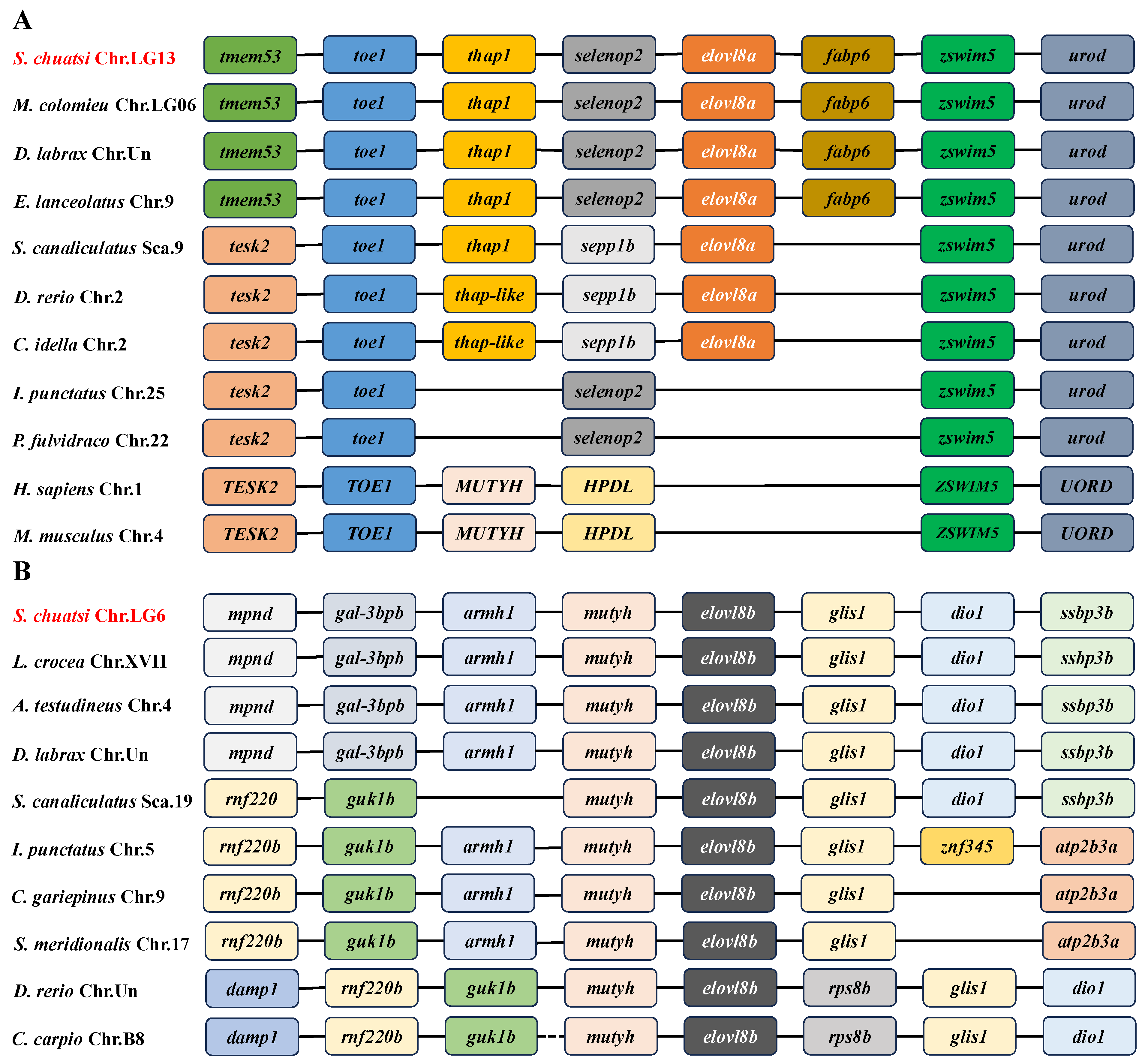
3.4. Gene Structures and Genomic Synteny of Both elovl8 Genes in Teleost
3.5. Tissue Distribution Pattern of Both elovl8a and elovl8b Genes in Chinese Perch
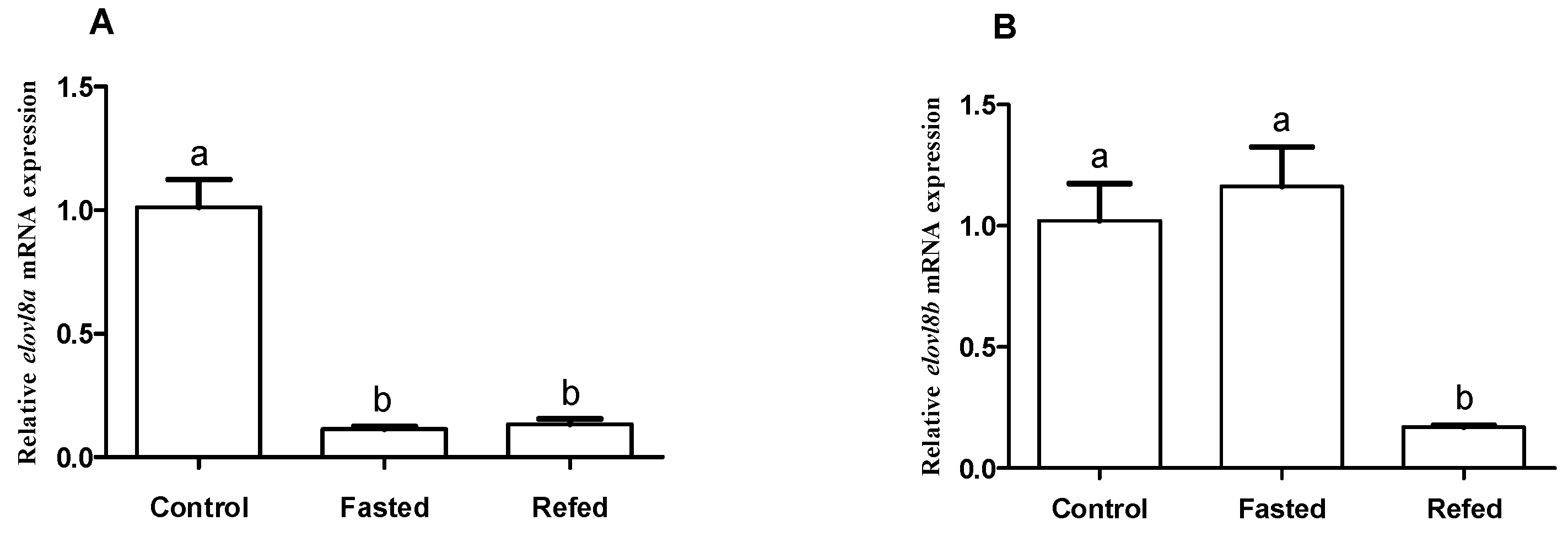
3.6. Effects of Different Nutritional Status on Transcriptional Changes of elovl8a and elovl8b Genes in Chinese Perch
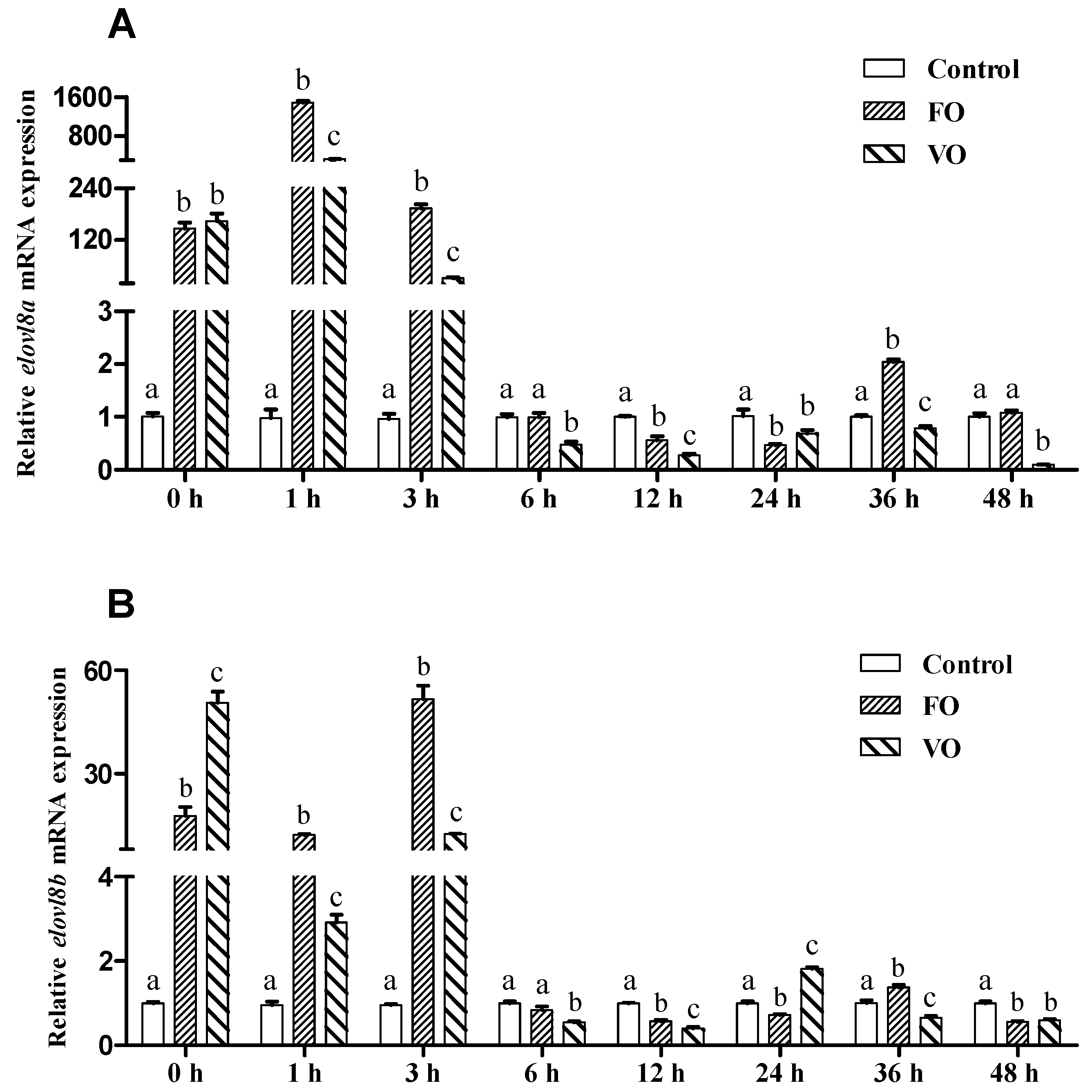
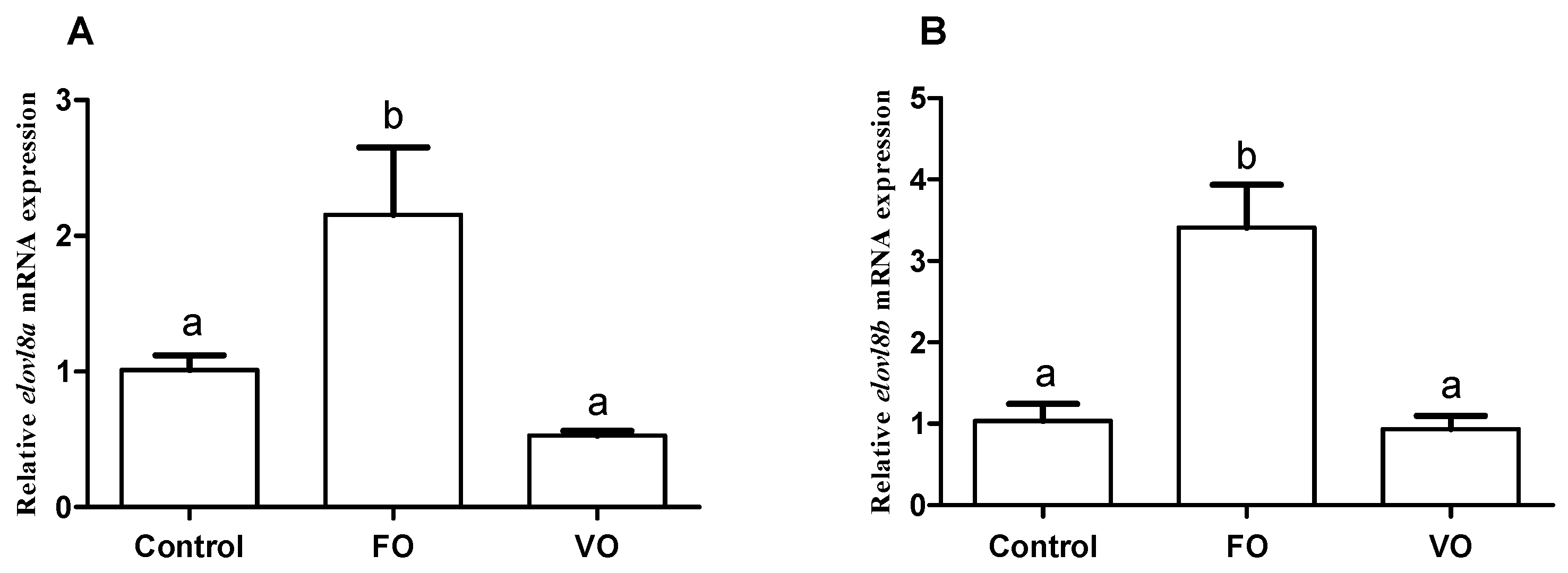
3.7. Effects of Fish Oil and Vegetable Oil Administration on the Expression Levels of Both elovl8a and elovl8b Genes in Chinese Perch
3.8. Effects of Dietary Fish Oil and Vegetable Oil Supplementation on Both elovl8a and elovl8b Transcriptions in Chinese Perch
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Castro, L.F.C.; Tocher, D.R.; Monroig, O. Long-chain polyunsaturated fatty acid biosynthesis in chordates: Insights into the evolution of Fads and Elovl gene repertoire. Prog. Lipid Res. 2016, 62, 25–40. [Google Scholar] [CrossRef] [PubMed]
- Monroig, Ó.; Lopes-Marques, M.; Navarro, J.C.; Hontoria, F.; Ruivo, R.; Santos, M.M.; Venkatesh, B.; Tocher, D.R.; Castro, L.F.C. Evolutionary functional elaboration of the Elovl2/5 gene family in chordates. Sci. Rep. 2016, 6, 20510. [Google Scholar] [CrossRef]
- Oboh, A.; Kabeya, N.; Carmona-Antoñanzas, G.; Castro, L.F.C.; Dick, J.R.; Tocher, D.R.; Monroig, O. Two alternative pathways for docosahexaenoic acid (DHA, 22: 6n-3) biosynthesis are widespread among teleost fish. Sci. Rep. 2017, 7, 3889. [Google Scholar] [CrossRef] [PubMed]
- Oboh, A.; Navarro, J.C.; Tocher, D.R.; Monroig, O. Elongation of very long-chain (> C 24) fatty acids in Clarias gariepinus: Cloning, functional characterization and tissue expression of elovl4 elongases. Lipids 2017, 52, 837–848. [Google Scholar] [CrossRef] [PubMed]
- Keim, S.A.; Branum, A.M. Dietary intake of polyunsaturated fatty acids and fish among US children 12-60 months of age. Matern. Child Nutr. 2015, 11, 987–998. [Google Scholar] [CrossRef]
- Sargent, J.R.; Tocher, D.R.; Bell, J.G. The lipids. In Fish Nutrition; Elsevier: Amsterdam, The Netherlands, 2003; pp. 181–257. [Google Scholar] [CrossRef]
- Monroig, O.; Tocher, D.R.; Castro, L.F.C. Polyunsaturated fatty acid biosynthesis and metabolism in fish. In Polyunsaturated Fatty Acid Metabolism; Elsevier: Amsterdam, The Netherlands, 2018; pp. 31–60. [Google Scholar] [CrossRef]
- Guillou, H.; Zadravec, D.; Martin, P.G.; Jacobsson, A. The key roles of elongases and desaturases in mammalian fatty acid metabolism: Insights from transgenic mice. Prog. Lipid Res. 2010, 49, 186–199. [Google Scholar] [CrossRef]
- Nakamura, M.T.; Nara, T.Y. Structure, function, and dietary regulation of delta6, delta5, and delta9 desaturases. Annu. Rev. Nutr. 2004, 24, 345–376. [Google Scholar] [CrossRef]
- Xie, D.; Chen, C.; Dong, Y.; You, C.; Wang, S.; Monroig, Ó.; Tocher, D.R.; Li, Y. Regulation of long-chain polyunsaturated fatty acid biosynthesis in teleost fish. Prog. Lipid Res. 2021, 82, 101095. [Google Scholar] [CrossRef]
- Li, Y.; Wen, Z.; You, C.; Xie, Z.; Tocher, D.R.; Zhang, Y.; Wang, S.; Li, Y. Genome wide identification and functional characterization of two LC-PUFA biosynthesis elongase (elovl8) genes in rabbitfish (Siganus canaliculatus). Aquaculture 2020, 522, 735127. [Google Scholar] [CrossRef]
- Monroig, Ó.; Rotllant, J.; Cerdá-Reverter, J.M.; Dick, J.R.; Figueras, A.; Tocher, D.R. Expression and role of Elovl4 elongases in biosynthesis of very long-chain fatty acids during zebrafish Danio rerio early embryonic development. Biochim. Biophys. Acta-Mol. Cell Biol. Lipids 2010, 1801, 1145–1154. [Google Scholar] [CrossRef]
- Jakobsson, A.; Westerberg, R.; Jacobsson, A. Fatty acid elongases in mammals: Their regulation and roles in metabolism. Prog. Lipid Res. 2006, 45, 237–249. [Google Scholar] [CrossRef]
- Xu, W.; Wang, S.; You, C.; Zhang, Y.; Monroig, Ó.; Tocher, D.R.; Li, Y. The catadromous teleost Anguilla japonica has a complete enzymatic repertoire for the biosynthesis of docosahexaenoic acid from α-linolenic acid: Cloning and functional characterization of an Elovl2 elongase. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2020, 240, 110373. [Google Scholar] [CrossRef]
- Xue, X.; Feng, C.Y.; Hixson, S.M.; Johnstone, K.; Anderson, D.M.; Parrish, C.C.; Rise, M.L. Characterization of the fatty acyl elongase (elovl) gene family, and hepatic elovl and delta-6 fatty acyl desaturase transcript expression and fatty acid responses to diets containing camelina oil in Atlantic cod (Gadus morhua). Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2014, 175, 9–22. [Google Scholar] [CrossRef] [PubMed]
- Oboh, A. Investigating the Long-Chain Polyunsaturated Fatty Acid Biosynthesis of the African Catfish Clarias gariepinus (Burchell, 1822). Ph.D. Thesis, University of Stirling, Stirling, UK, 2018. [Google Scholar]
- Sun, S.; Wang, Y.; Goh, P.-T.; Lopes-Marques, M.; Castro, L.F.C.; Monroig, Ó.; Kuah, M.-K.; Cao, X.; Shu-Chien, A.C.; Gao, J. Evolution andfunctional characteristics of the novel elovl8 that play pivotal roles in fatty acid biosynthesis. Genes 2021, 12, 1287. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Sun, S.; He, J.; Yang, G.; Gao, J. Effects of elovl8 deletion on survival and lipid metabolism under cold stress of zebrafish. Acta Hydrobiol. Ainica 2022, 46, 303–315. [Google Scholar] [CrossRef]
- Wu, Q.; Zheng, Z.; Wang, C.; Wang, Y.; Sun, Y.; Gao, Y. Molecular characterization, tissue distribution and differential nutritional regulation of three n-3 LC-PUFA biosynthesis-related genes in hybrid grouper (Epinephelus fuscoguttatus ♀ × Epinephelus lanceolatus ♂). Animals 2022, 12, 234. [Google Scholar] [CrossRef]
- Zeng, W.-H.; Wei, X.-Y.; Qin, W.; Qin, C.-J.; Shi, Q.; Guo, S.-T.; Prathomya, P.; Zhang, S.-Y.; Fu, P.; Hu, W.; et al. Molecular characterization, spatiotemporal expression patterns of fatty acid elongase (elovl8) gene, and its transcription changes in response to different diet stimuli in yellow catfish (Pelteobagrus fulvidraco). Front. Mar. Sci. 2023, 10, 1270776. [Google Scholar] [CrossRef]
- Zhao, B.; Peng, Y.; Itakura, Y.; Lizanda, M.; Haga, Y.; Satoh, S.; Navarro, J.C.; Monroig, Ó.; Kabeya, N. A complete biosynthetic pathway of the long-chain polyunsaturated fatty acids in an amphidromous fish, ayu sweetfish Plecoglossus altivelis (Stomiati; Osmeriformes). Biochim. Biophys. Acta-Mol. Cell Biol. Lipids 2024, 1869, 159498. [Google Scholar] [CrossRef]
- Han, C.; Zhu, Q.; Lu, H.; Wang, C.; Zhou, X.; Peng, C.; Tang, L.; Han, L.; Chen, J.; Li, S.; et al. Screening and characterization of sex-specific markers developed by a simple NGS method in mandarin fish (Siniperca chuatsi). Aquaculture 2020, 527, 735495. [Google Scholar] [CrossRef]
- Zhao, J.; Li, C.; Zhao, L.; Wang, W.; Cao, Y. Mitochondrial diversity and phylogeography of the Chinese perch, Siniperca chuatsi (Perciformes: Sinipercidae). Mol. Phylogenet. Evol. 2008, 49, 399–404. [Google Scholar] [CrossRef]
- Sun, C.; Niu, Y.; Ye, X.; Dong, J.; Hu, W.; Zeng, Q.; Chen, Z.; Tian, Y.; Zhang, J.; Lu, M. Construction of a high-density linkage map and mapping of sex determination and growth-related loci in the mandarin fish (Siniperca chuatsi). BMC Genom. 2017, 18, 446. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; He, S.; Liang, X.-F.; Li, L.; Wen, Z.; Zhu, T.A.O.; Shen, D.A.N. Identification of SNPs in NPY and LEP and the association with food habit domestication traits in mandarin fish. J. Genet. 2015, 94, 118–122. [Google Scholar] [CrossRef]
- Ding, W.; Zhang, X.; Zhao, X.; Jing, W.; Cao, Z.; Li, J.; Huang, Y.; You, X.; Wang, M.; Shi, Q.; et al. A chromosome-level genome assembly of the mandarin fish (Siniperca chuatsi). Front Genet. 2021, 12, 671650. [Google Scholar] [CrossRef] [PubMed]
- Wen, Z.Y.; Liu, T.; Qin, C.J.; Zou, Y.C.; Wang, J.; Li, R.; Tao, Y.X. MRAP2 interaction with melanocortin-4 receptor in snakehead (Channa argus). Biomolecules 2021, 11, 481. [Google Scholar] [CrossRef]
- Wen, Z.-Y.; Qin, C.-J.; Wang, J.; He, Y.; Li, H.-T.; Li, R.; Wang, X.-D. Molecular characterization of two leptin genes and their transcriptional changes in response to fasting and refeeding in Northern snakehead (Channa argus). Gene 2020, 736, 144420. [Google Scholar] [CrossRef]
- Shen, Y.; Li, H.; Zhao, J.; Tang, S.; Zhao, Y.; Bi, Y.; Chen, X. The digestive system of mandarin fish (Siniperca chuatsi) can adapt to domestication by feeding with artificial diet. Aquaculture 2021, 538, 736546. [Google Scholar] [CrossRef]
- Wen, Z.-Y.; Wang, J.; Bian, C.; Zhang, X.; Li, J.; Peng, Y.; Zhan, Q.; Shi, Q.; Li, Y.-Y. Molecular cloning of two kcnk3 genes from the Northern snakehead (Channa argus) for quantification of their transcriptions in response to fasting and refeeding. Gen. Comp. Endocrinol. 2019, 281, 49–57. [Google Scholar] [CrossRef]
- Tan, K.; Ma, H.; Li, S.; Zheng, H. Bivalves as future source of sustainable natural omega-3 polyunsaturated fatty acids. Food Chem. 2020, 311, 125907. [Google Scholar] [CrossRef]
- Tan, K.; Zhang, H.; Zheng, H. Climate change and n-3 LC-PUFA availability. Prog. Lipid Res. 2022, 86, 101161. [Google Scholar] [CrossRef]
- Xiao, J.; Wang, W.-X. Genome-wide identification and expression profile of Elovl genes in threadfin fish Eleutheronema. Sci. Rep. 2023, 13, 1080. [Google Scholar] [CrossRef]
- Liu, M.Y.; Wang, Q.C.; Li, J.Q.; Zhang, D.; Mu, J.H.; Shen, X.H. Genome-wide identification and comparative analysis of elongation of very long-chain fatty acid (Elovl) Genes in Echinoderms. Russ. J. Genet. 2024, 60, 450–459. [Google Scholar] [CrossRef]
- Yi, T.L.; Yang, L.K.; Ruan, G.L.; Yang, D.Q.; Tao, Y.X. Melanocortin-4 receptor in swamp eel (Monopterus albus): Cloning, tissue distribution, and pharmacology. Gene 2018, 678, 79–89. [Google Scholar] [CrossRef]
- Parey, E.; Louis, A.; Montfort, J.; Guiguen, Y.; Crollius, H.R.; Berthelot, C. An atlas of fish genome evolution reveals delayed rediploidization following the teleost whole-genome duplication. Genome Res. 2022, 32, 1685–1697. [Google Scholar] [CrossRef] [PubMed]
- Sanchez, R.; Riddle, M.; Woo, J.; Momand, J. Prediction of reversibly oxidized protein cysteine thiols using protein structure properties. Int. J. Dev. Biol. 2009, 53, 765–773. [Google Scholar] [CrossRef]
- Galindo, A.; Garrido, D.; Monroig, Ó.; Pérez, J.A.; Betancor, M.B.; Acosta, N.G.; Kabeya, N.; Marrero, M.A.; Bolaños, A.; Rodríguez, C. Polyunsaturated fatty acid metabolism in three fish species with different trophic level. Aquaculture 2021, 530, 735761. [Google Scholar] [CrossRef]
- Garrido, D.; Monroig, Ó.; Galindo, A.; Betancor, M.B.; Pérez, J.A.; Kabeya, N.; Marrero, M.; Rodríguez, C. Lipid metabolism in Tinca tinca and its n-3 LC-PUFA biosynthesis capacity. Aquaculture 2020, 523, 735147. [Google Scholar] [CrossRef]
- Morais, S.; Mourente, G.; Martínez, A.; Gras, N.; Tocher, D.R. Docosahexaenoic acid biosynthesis via fatty acyl elongase and Δ4-desaturase and its modulation by dietary lipid level and fatty acid composition in a marine vertebrate. Biochim. Biophys. Acta 2015, 1851, 588–597. [Google Scholar] [CrossRef] [PubMed]
- Sprague, M.; Xu, G.; Betancor, M.B.; Olsen, R.E.; Torrissen, O.; Glencross, B.D.; Tocher, D.R. Endogenous production of n-3 long-chain PUFA from first feeding and the influence of dietary linoleic acid and the α-linolenic:linoleic ratio in Atlantic salmon (Salmo salar). Br. J. Nutr. 2019, 122, 1091–1102. [Google Scholar] [CrossRef]
- Torres, M.; Navarro, J.C.; Varó, I.; Monroig, Ó.; Hontoria, F. Nutritional regulation of genes responsible for long-chain (C20-24) and very long-chain (>C24) polyunsaturated fatty acid biosynthesis in post-larvae of gilthead seabream (Sparus aurata) and Senegalese sole (Solea senegalensis). Aquaculture 2020, 525, 735314. [Google Scholar] [CrossRef]
- Trushenski, J.T.; Rombenso, A.N. Trophic levels predict the nutritional essentiality of polyunsaturated fatty acids in fish—Introduction to a special section and a brief synthesis. N. Am. J. Aquac. 2020, 82, 241–250. [Google Scholar] [CrossRef]
- Zhou, B.; Wei, X.-Y.; Wen, Z.-Y.; Wang, B.; Zhao, Y.-Y.; Zeng, W.-H.; He, Y.; Prathomya, P.; Lv, Y.-Y.; Li, Y.-P.; et al. Molecular characterization of fad6 gene and its transcriptional changes in response to different initial diets and nutritional status in yellow catfish (Pelteobagrus fulvidraco). Front. Mar. Sci. 2024, 11, 1453516. [Google Scholar] [CrossRef]
- Xie, D.; Chen, F.; Lin, S.; You, C.; Wang, S.; Zhang, Q.; Monroig, Ó.; Tocher, D.R.; Li, Y. Long-chain polyunsaturated fatty acid biosynthesis in the euryhaline herbivorous teleost Scatophagus argus: Functional characterization, tissue expression and nutritional regulation of two fatty acyl elongases. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2016, 198, 37–45. [Google Scholar] [CrossRef] [PubMed]




| Category | Experimental Diets | ||
|---|---|---|---|
| Control | 2% FO | 2% VO | |
| Proximate composition (%) | |||
| Moisture | 7.30 | 7.70 | 7.30 |
| Crude protein | 50.83 | 49.23 | 49.13 |
| Crude lipid | 12.20 | 13.70 | 13.60 |
| Ash | 13.40 | 13.40 | 13.00 |
| Fatty acids (%Crude lipid) | |||
| C12:0 | 0.046 | 0.044 | 0.040 |
| C13:0 | 0.018 | 0.024 | 0.016 |
| C14:0 | 2.00 | 2.18 | 1.60 |
| C15:0 | 0.26 | 0.21 | 0.15 |
| C16:0 | 16.20 | 16.50 | 15.80 |
| C17:0 | 0.68 | 0.69 | 0.50 |
| C18:0 | 5.05 | 5.18 | 4.70 |
| C20:0 | 0.40 | 0.49 | 0.41 |
| C21:0 | 0.056 | 0.071 | 0.049 |
| C23:0 | 0.076 | 0.068 | 0.0644 |
| C24:0 | 0.33 | 0.31 | 0.31 |
| Total SFA | 25.11 | 25.76 | 23.64 |
| C14:1n-5 | 0.025 | 0.026 | 0.028 |
| C16:1n-7 | 2.21 | 2.80 | 1.91 |
| C17:1n-7 | 0.31 | 0.4 | 0.277 |
| C18:1n-9 | 21.2 | 21.4 | 22.5 |
| C20:1n-11 | 0.71 | 0.71 | 0.59 |
| C22:1n-9 | 0.12 | 0.14 | 0.12 |
| Total MUFA | 24.58 | 25.51 | 25.42 |
| C18:2n-6 | 33.80 | 32.20 | 37.50 |
| C20:3n-6 | 0.072 | 0.10 | 0.067 |
| C20:4n-6 | 0.57 | 0.70 | 0.51 |
| C22:2n-6 | 0.017 | 0.012 | 0.011 |
| Total n-6 PUFA | 34.45 | 33.02 | 38.08 |
| C18:3n-3 | 4.56 | 4.20 | 3.84 |
| C20:3n-3 | 0.055 | 0.056 | 0.053 |
| C20:5n-3 | 4.35 | 4.52 | 3.41 |
| C22:6n-3 | 6.72 | 6.79 | 5.43 |
| Total n-3 PUFA | 15.68 | 15.57 | 12.73 |
| Fatty acids (%Experimental diets) | |||
| C18:2n-6 | 4.12 | 4.52 | 5.10 |
| C18:3n-3 | 0.55 | 0.58 | 0.52 |
| C20:5n-3 | 0.53 | 0.62 | 0.46 |
| C22:6n-3 | 0.82 | 0.93 | 0.74 |
| Primer Name | Primer Sequence (5′-3′) |
|---|---|
| elovl8a-qF | ATTGGCTGCTGGTCTACTC |
| elovl8a-qR | CAGGAAGAGGCTGTGAACT |
| elovl8b-qF | GGCGGGAGTCAAGTATGT |
| elovl8b-qR | TAGGGACGTGAGGTATCG |
| β-actin-qF | GTGCGTGACATCAAGGAGAAG |
| β-actin-qR | GGAAGGAAGGCTGGAAGAGG |
| Number | Species | Protein | Protein ID |
|---|---|---|---|
| 1 | Scomber scombrus | Elovl8a | XP_062285127.1 |
| 2 | Scomber japonicus | Elovl8a | XP_053185701.1 |
| 3 | Thunnus maccoyii | Elovl8a | XP_042283429.1 |
| 4 | Lates calcarifer | Elovl8a | XP_018531677.1 |
| 5 | Toxotes jaculatrix | Elovl8a | XP_040911096.1 |
| 6 | Plectropomus leopardus | Elovl8a | XP_042354264.1 |
| 7 | Epinephelus moara | Elovl8a | XP_049888834.1 |
| 8 | Epinephelus fuscoguttatus | Elovl8a | XP_049453958.1 |
| 9 | Pseudochaenichthys georgianus | Elovl8a | KAA0719250.1 |
| 10 | Danio rerio | Elovl8a | NP_001070061.1 |
| 11 | Oncorhynchus mykiss | Elovl8a | XP_021466941.1 |
| 12 | Oncorhynchus masou masou | Elovl8a | XP_064784052.1 |
| 13 | Siganus canaliculatus | Elovl8a | QMU95576.1 |
| 14 | Clarias gariepinus | Elovl8b | XP_053360794.1 |
| 15 | Cheilinus undulatus | Elovl8b | XP_041647543.1 |
| 16 | Danio rerio | Elovl8b | NP_001191453.1 |
| 17 | Dicentrarchus labrax | Elovl8b | XP_051234026.1 |
| 18 | Morone saxatilis | Elovl8b | XP_035509917.1 |
| 19 | Labrus bergylta | Elovl8b | XP_052466620.1 |
| 20 | Labrus mixtus | Elovl8b | XP_060889255.1 |
| 21 | Scomber japonicus | Elovl8b | XP_053178828.1 |
| 22 | Scomber scombrus | Elovl8b | XP_062279998.1 |
| 23 | Siganus canaliculatus | Elovl8b | QMU95577.1 |
| 24 | Tachysurus fulvidraco | Elovl8b | XP_047655843.1 |
| 25 | Thunnus albacares | Elovl8b | XP_044218547.1 |
| 26 | Thunnus maccoyii | Elovl8b | XP_042273790.1 |
| 27 | Xyrichtys novacula | Elovl8b | CAJ1060369.1 |
| 28 | Danio rerio | Elovl2 | NP_001035452.1 |
| 29 | Homo sapiens | ELOVL2 | NP_060240.3 |
| 30 | Caretta caretta | Elovl2 | XP_048695790.1 |
| 31 | Gallus gallus 2 | ELOVL2 | NP_001184237.1 |
| 32 | Clarias gariepinus | Elovl4a | XP_053351126.1 |
| 33 | Silurus meridionalis | Elovl4a | XP_046702911.1 |
| 34 | Danio rerio | Elovl4a | NP_957090.1 |
| 35 | Ctenopharyngodon idella | Elovl4b | XP_051736330.1 |
| 36 | Anabas testudineus | Elovl4b | XP_026196457.1 |
| 37 | Danio rerio | Elovl4b | NP_956266.1 |
| 38 | Bos taurus | ELOVL4 | NP_001092520.1 |
| 40 | Gallus gallus | ELOVL4 | XP_046794870.1 |
| 41 | Homo sapiens | ELOVL4 | NP_073563.1 |
| 42 | Mus musculus | ELOVL4 | NP_683743.2 |
| 43 | Rattus norvegicus | ELOVL4 | NP_001178725.1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
He, Y.; Wen, Z.; Zhou, L.; Zeng, W.; Prathomya, P.; Yi, T.; Shi, Q. Molecular Characterization and Nutritional Regulation of Two Fatty Acid Elongase (elovl8) Genes in Chinese Perch (Siniperca chuatsi). Biomolecules 2025, 15, 567. https://doi.org/10.3390/biom15040567
He Y, Wen Z, Zhou L, Zeng W, Prathomya P, Yi T, Shi Q. Molecular Characterization and Nutritional Regulation of Two Fatty Acid Elongase (elovl8) Genes in Chinese Perch (Siniperca chuatsi). Biomolecules. 2025; 15(4):567. https://doi.org/10.3390/biom15040567
Chicago/Turabian StyleHe, Yu, Zhengyong Wen, Luo Zhou, Wanhong Zeng, Panita Prathomya, Tilin Yi, and Qiong Shi. 2025. "Molecular Characterization and Nutritional Regulation of Two Fatty Acid Elongase (elovl8) Genes in Chinese Perch (Siniperca chuatsi)" Biomolecules 15, no. 4: 567. https://doi.org/10.3390/biom15040567
APA StyleHe, Y., Wen, Z., Zhou, L., Zeng, W., Prathomya, P., Yi, T., & Shi, Q. (2025). Molecular Characterization and Nutritional Regulation of Two Fatty Acid Elongase (elovl8) Genes in Chinese Perch (Siniperca chuatsi). Biomolecules, 15(4), 567. https://doi.org/10.3390/biom15040567







