Unveiling Pharmacogenomics Insights into Circular RNAs: Toward Precision Medicine in Cancer Therapy
Abstract
1. Introduction
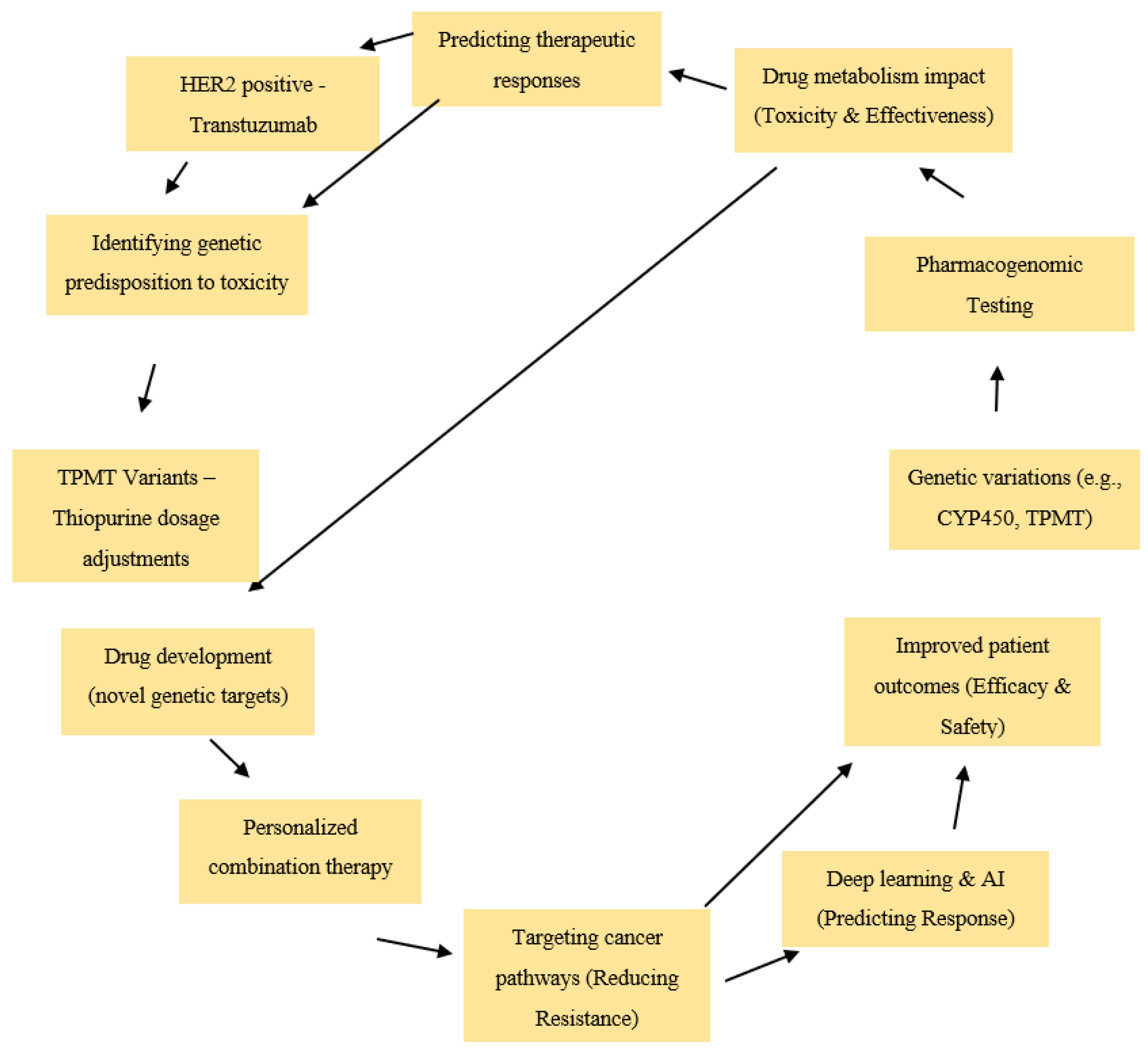
1.1. Background on Precision Medicine and Pharmacogenomics
1.2. Advances in AI-Driven Pharmacogenomics
1.3. The Role of Pharmacogenomics in Personalized Cancer Therapy
1.4. Circular RNAs: Emerging Players in Cancer Biology
1.5. Bridging Pharmacogenomics and circRNAs
2. Pharmacogenomic Potential of Circular RNAs
2.1. circRNAs as Biomarkers for Drug Response
2.2. circRNA Expression Variability and Pharmacogenomic Profiling
2.3. circRNAs in Modulating Drug Metabolism and Transport
3. CircRNA-Mediated Drug Resistance in Cancer
3.1. Mechanistic Insights into circRNA-Driven Drug Resistance
3.2. circRNAs as miRNA Sponges in Drug Resistance
3.3. circRNAs and Epigenetic Regulation in Drug Resistance
4. circRNAs as Therapeutic Targets in Cancer
4.1. CircRNA-Based Therapies for Overcoming Drug Resistance
4.2. Synthetic circRNAs for Drug Delivery
4.3. CircRNA Modulation in Combination Therapies
5. Emerging Tools and Techniques for circRNA-Based Pharmacogenomics
5.1. High-Throughput Technologies for circRNA Profiling
5.2. circRNA-Pharmacogenomic Databases and Resources
5.2.1. Comparative Analysis of circRNA Databases
5.2.2. Reliability and Validation Considerations
5.3. Experimental Models for circRNA Functional Validation
6. Clinical Translation of circRNAs in Pharmacogenomics
6.1. circRNAs as Predictive Biomarkers in Cancer Therapy
| circRNA (ID, Size) | Cancer Type | Proposed Clinical Role | Reference * |
|---|---|---|---|
| circHIPK3 (hsa_circ_0000284; 1099 bp) | Gastric Cancer | Diagnostic biomarker for cisplatin resistance; therapeutic target to enhance ferroptosis | [144] |
| circATIC (hsa_circ_0058063; 1640 bp) | Bladder Cancer | Diagnostic biomarker for cisplatin resistance; therapeutic target to modulate miR-335-5p/B2M axis | [145] |
| circMORC3 (hsa_circ_0001189; 420 bp) | Bladder Cancer | Therapeutic target to affect m6A modification on DNA damage response genes | [146] |
| circPVT1 (hsa_circ_0001821; 410 bp) | Osteosarcoma | Diagnostic biomarker and therapeutic target for doxorubicin and cisplatin resistance | [147] |
| circ-ABCB10 (hsa_circ_0008717; 724 bp) | Lung Cancer | Therapeutic target to enhance cisplatin sensitivity | [148] |
| circSMARCA5 (hsa_circ_0001445; 269 bp) | Prostate Cancer | Inhibits tumor proliferation, migration, and invasion by regulating the miR-181b-5p/miR-17-3p-TIMP3 axis | [149] |
| circELP3 (hsa_circ_0001785; 467 bp) | Breast Cancer | Potential therapeutic target; involved in tumor progression through miRNA sponging | [150] |
| circCNIH4 (hsa_circ_0000190; 254 bp) | Colorectal Cancer | Diagnostic biomarker; upregulated in tissues and plasma | [151] |
6.2. circRNAs in Personalized Therapeutic Regimens
6.3. Challenges and Opportunities for Clinical Applications
7. Future Perspectives in circRNA Pharmacogenomics
7.1. Toward Personalized Medicine: The Role of circRNAs
7.2. Unexplored Frontiers in circRNA Pharmacogenomics
7.3. Bridging Basic Research and Clinical Practice
8. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Al Meslamanis, A.Z. The future of precision medicine in oncology. Expert. Rev. Precis. Med. Drug Dev. 2023, 8, 43–47. [Google Scholar] [CrossRef]
- Schwartzberg, L.; Kim, E.S.; Liu, D.; Schrag, D. Precision oncology: Who, how, what, when, and when not? In American Society of Clinical Oncology Educational Book, Proceedings of the Annual Meeting 2017, Chicago, IL, USA, 2–6 June 2017; American Society of Clinical Oncology: Orlando, FL, USA, 2017; Volume 37, pp. 160–169. [Google Scholar]
- Schmidt, K.T.; Chau, C.H.; Price, D.K.; Figg, W.D. Precision oncology medicine: The clinical relevance of patient-specific biomarkers used to optimize cancer treatment. J. Clin. Pharmacol. 2016, 56, 1484–1499. [Google Scholar] [PubMed]
- Fountzilas, E.; Tsimberidou, A.M. Overview of precision oncology trials: Challenges and opportunities. Expert Rev. Clin. Pharmacol. 2018, 11, 797–804. [Google Scholar] [CrossRef] [PubMed]
- Malone, E.R.; Oliva, M.; Sabatini, P.J.; Stockley, T.L.; Siu, L.L. Molecular profiling for precision cancer therapies. Genome Med. 2020, 12, 8. [Google Scholar] [PubMed]
- Verma, A.K.; Singh, K.; Gupta, J.K.; Kumar, S.; Jain, D. Pharmacological Approaches and Innovative Strategies for Individualized Patient Care. Recent Pat. Biotechnol. 2025; in press. [Google Scholar]
- Lajmi, N.; Alves-Vasconcelos, S.; Tsiachristas, A.; Haworth, A.; Woods, K.; Crichton, C.; Noble, T.; Salih, H.; Várnai, K.A.; Branford-White, H.; et al. Challenges and solutions to system-wide use of precision oncology as the standard of care paradigm. Camb. Prism. Precis. Med. 2024, 2, e4. [Google Scholar]
- Chua, I.S.; Gaziel-Yablowitz, M.; Korach, Z.T.; Kehl, K.L.; Levitan, N.A.; Arriaga, Y.E.; Jackson, G.P.; Bates, D.W.; Hassett, M. Artificial intelligence in oncology: Path to implementation. Cancer Med. 2021, 10, 4138–4149. [Google Scholar] [PubMed]
- Obermeyer, Z.; Emanuel, E.J. Predicting the future—Big data, machine learning, and clinical medicine. N. Engl. J. Med. 2016, 375, 1216–1219. [Google Scholar] [CrossRef]
- Alsharif, F. Artificial Intelligence in Oncology: Applications, Challenges and Future Frontiers. Int. J. Pharm. Investig. 2024, 14, 647–656. [Google Scholar]
- Li, L.; Sun, M.; Wang, J.; Wan, S. Multi-omics based artificial intelligence for cancer research. Adv. Cancer Res. 2024, 163, 303–356. [Google Scholar]
- Cai, Z.; Poulos, R.C.; Aref, A.; Robinson, P.J.; Reddel, R.R.; Zhong, Q. DeePathNet: A Transformer-Based Deep Learning Model Integrating Multiomic Data with Cancer Pathways. Cancer Res. Commun. 2024, 4, 3151–3164. [Google Scholar] [CrossRef] [PubMed]
- Gervits, A.; Sharan, R. Predicting genetic interactions, cell line dependencies and drug sensitivities with variational graph auto-encoder. Front. Bioinform. 2022, 2, 1025783. [Google Scholar] [CrossRef] [PubMed]
- Topol, E.J. High-performance medicine: The convergence of human and artificial intelligence. Nat. Med. 2019, 25, 44–56. [Google Scholar] [CrossRef] [PubMed]
- Wan, X.; Xiao, J.; Tam, S.S.; Cai, M.; Sugimura, R.; Wang, Y.; Wan, X.; Lin, Z.; Wu, A.R.; Yang, C. Integrating spatial and single-cell transcriptomics data using deep generative models with SpatialScope. Nat. Commun. 2023, 14, 7848. [Google Scholar] [CrossRef] [PubMed]
- Shin, J.; Piao, Y.; Bang, D.; Kim, S.; Jo, K. DRPreter: Interpretable anticancer drug response prediction using knowledge-guided graph neural networks and transformer. Int. J. Mol. Sci. 2022, 23, 13919. [Google Scholar] [CrossRef] [PubMed]
- Friedman, A.A.; Letai, A.; Fisher, D.E.; Flaherty, K.T. Precision medicine for cancer with next-generation functional diagnostics. Nat. Rev. Cancer 2015, 15, 747–756. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.C.; Wang, Z. Precision medicine: Disease subtyping and tailored treatment. Cancers 2023, 15, 3837. [Google Scholar] [CrossRef]
- Pinker, K.; Chin, J.; Melsaether, A.N.; Morris, E.A.; Moy, L. Precision medicine and radiogenomics in breast cancer: New approaches toward diagnosis and treatment. Radiology 2018, 287, 732–747. [Google Scholar] [CrossRef]
- Francis, A.P.; Khan, S.A.; Fuloria, S.; Fuloria, N.K.; Meenakshi, D.U. Big Data and Precision Oncology in Healthcare. In Big Data in Oncology: Impact, Challenges, and Risk Assessment; River Publishers: Aalborg, Denmark, 2023; pp. 47–74. [Google Scholar]
- Char, D.S.; Shah, N.H.; Magnus, D. Implementing machine learning in health care—Addressing ethical challenges. N. Engl. J. Med. 2018, 378, 981–983. [Google Scholar] [CrossRef]
- Mittelstadt, B.D.; Floridi, L. The ethics of big data: Current and foreseeable issues in biomedical contexts. The Ethics of Biomedical Big Data; Springer: Cham, Switzerland, 2016; pp. 445–480. [Google Scholar]
- Franczyk, B.; Rysz, J.; Gluba-Brzózka, A. Pharmacogenetics of drugs used in the treatment of cancers. Genes 2022, 13, 311. [Google Scholar] [CrossRef] [PubMed]
- Miteva-Marcheva, N.N.; Ivanov, H.Y.; Dimitrov, D.K.; Stoyanova, V.K. Application of pharmacogenetics in oncology. Biomark. Res. 2020, 8, 32. [Google Scholar] [CrossRef] [PubMed]
- Novelli, G.; Ciccacci, C.; Borgiani, P.; Papaluca Amati, M.; Abadie, E. Genetic tests and genomic biomarkers: Regulation, qualification and validation. Clin. Cases Min. Bone Metab. 2008, 5, 149–154. [Google Scholar] [PubMed] [PubMed Central]
- Burstein, H.J. The distinctive nature of HER2-positive breast cancers. N. Engl. J. Med. 2005, 353, 1652–1654. [Google Scholar] [CrossRef] [PubMed]
- Phillips, K.A.; Veenstra, D.L.; Oren, E.; Lee, J.K.; Sadee, W.; Haddow, J.E. Potential role of pharmacogenomics in reducing adverse drug reactions: A systematic review. JAMA 2014, 281, 927–934. [Google Scholar] [CrossRef] [PubMed]
- Relling, M.V.; Klein, T.E. CPIC: Clinical Pharmacogenetics Implementation Consortium of the Pharmacogenomics Research Network. Clin. Pharmacol. Ther. 2011, 89, 464–467. [Google Scholar] [CrossRef]
- Winterfeld, S. Pharmacogenomics in Cancer Treatment Tailoring Therapy for Better Outcomes. Health Sci. J. 2024, 18, 1–3. [Google Scholar]
- Liao, C.Y.; Chen, Y.M.; Wu, Y.T.; Chao, H.S.; Chiu, H.Y.; Wang, T.W.; Chen, J.R.; Shiao, T.H.; Lu, C.F. Personalized prediction of immunotherapy response in lung cancer patients using advanced radiomics and deep learning. Cancer Imaging 2024, 24, 129. [Google Scholar] [CrossRef] [PubMed]
- Su, M.; Xiao, Y.; Ma, J.; Tang, Y.; Tian, B.; Zhang, Y.; Li, X.; Wu, Z.; Yang, D.; Zhou, Y.; et al. Circular RNAs in Cancer: Emerging functions in hallmarks, stemness, resistance and roles as potential biomarkers. Mol. Cancer 2019, 18, 90. [Google Scholar] [CrossRef]
- Hansen, T.B.; Jensen, T.I.; Clausen, B.H.; Bramsen, J.B.; Finsen, B.; Damgaard, C.K.; Kjems, J. Natural RNA circles function as efficient microRNA sponges. Nature 2013, 495, 384–388. [Google Scholar] [CrossRef]
- Memczak, S.; Jens, M.; Elefsinioti, A.; Torti, F.; Krueger, J.; Rybak, A.; Maier, L.; Mackowiak, S.D.; Gregersen, L.H.; Munschauer, M.; et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013, 495, 333. [Google Scholar] [CrossRef] [PubMed]
- Haga, S.B. Artificial intelligence, medications, pharmacogenomics, and ethics. Pharmacogenomics 2024, 25, 611–622. [Google Scholar] [CrossRef] [PubMed]
- Pietrzak, J.; Świechowski, R.; Wosiak, A.; Wcisło, S.; Balcerczak, E. ADAMTS gene-derived circRNA molecules in non-small-cell lung cancer: Expression profiling, clinical correlations and survival analysis. Int. J. Mol. Sci. 2024, 25, 1897. [Google Scholar] [CrossRef] [PubMed]
- Geng, Y.; Zheng, X.; Zhang, D.; Wei, S.; Feng, J.; Wang, W.; Zhang, L.; Wu, C.; Hu, W. CircHIF1A induces cetuximab resistance in colorectal cancer by promoting HIF1α-mediated glycometabolism alteration. Biol. Direct 2024, 19, 36. [Google Scholar] [CrossRef]
- Zhang, M.; Wang, S. Roles of circular RNAs in colorectal cancer. Oncol. Lett. 2021, 22, 602. [Google Scholar] [CrossRef] [PubMed]
- Yu, N.; Gong, H.; Chen, W.; Peng, W. CircRNA ZKSCAN1 promotes lung adenocarcinoma progression by miR-185-5p/TAGLN2 axis. Thorac. Cancer 2023, 14, 1467–1476. [Google Scholar] [CrossRef] [PubMed]
- Cai, N.; Cheng, K.; Ma, Y.; Liu, S.; Tao, R.; Li, Y.; Li, D.; Guo, B.; Jia, W.; Liang, H.; et al. Targeting MMP9 in CTNNB1 mutant hepatocellular carcinoma restores CD8+ T cell-mediated antitumour immunity and improves anti-PD-1 efficacy. Gut 2024, 73, 985–999. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Zhang, C.; Zhang, B.; Yu, H.; Yu, Q. circRNA of AR-suppressed PABPC1 91 bp enhances the cytotoxicity of natural killer cells against hepatocellular carcinoma via upregulating UL16 binding protein 1. Oncol. Lett. 2019, 17, 388–397. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.L.; Xiao, Q.; Yu, B.; Lv, Q.L.; Liu, Z.Q.; Yin, J.Y. CircRNAs in tumor immunity and immunotherapy: Perspectives from innate and adaptive immunity. Cancer Lett. 2023, 564, 216219. [Google Scholar] [CrossRef]
- Yan, T.; Tian, X.; Liu, F.; Liu, Q.; Sheng, Q.; Wu, J.; Jiang, S. The emerging role of circular RNAs in drug resistance of non-small cell lung cancer. Front. Oncol. 2022, 12, 1003230. [Google Scholar] [CrossRef]
- Mirzaei, S.; Gholami, M.H.; Hushmandi, K.; Hashemi, F.; Zabolian, A.; Canadas, I.; Zarrabi, A.; Nabavi, N.; Aref, A.R.; Crea, F.; et al. The long and short non-coding RNAs modulating EZH2 signaling in cancer. J. Hematol. Oncol. 2022, 15, 18. [Google Scholar] [CrossRef] [PubMed]
- Gao, D.; Zhang, X.; Liu, B.; Meng, D.; Fang, K.; Guo, Z.; Li, L. Screening circular RNA related to chemotherapeutic resistance in breast cancer. Epigenomics 2017, 9, 1175–1188. [Google Scholar] [CrossRef] [PubMed]
- Christowitz, C.; Davis, T.; Isaacs, A.; van Niekerk, G.; Hattingh, S.; Engelbrecht, A.M. Mechanisms of doxorubicin-induced drug resistance and drug resistanttumour growth in a murine breast tumour model. BMC Cancer 2019, 19, 757. [Google Scholar] [CrossRef] [PubMed]
- Cui, C.; Yang, J.; Li, X.; Liu, D.; Fu, L.; Wang, X. Functions and mechanisms of circular RNAs in cancer radiotherapy and chemotherapy resistance. Mol. Cancer 2020, 19, 58. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.M.; Bai, J.Y.; Yang, K.X. Effect of resveratrol on doxorubicin resistance in breast neoplasm cells by modulating PI3K/Akt signaling pathway. IUBMB Life 2018, 70, 491–500. [Google Scholar]
- Qiao, X.; Zhu, L.; Song, R.; Shang, C.; Guo, Y. CD44 occurring alternative splicing promotes cisplatin resistance and evokes tumor immune response in oral squamous cell carcinoma cells. Transl. Oncol. 2023, 31, 101644. [Google Scholar] [CrossRef]
- Huang, P.; Li, F.; Mo, Z.; Geng, C.; Wen, F.; Zhang, C.; Guo, J.; Wu, S.; Li, L.; Brünner, N.; et al. A comprehensive RNA study to identify circRNA and miRNA biomarkers for docetaxel resistance in breast cancer. Front. Oncol. 2021, 11, 669270. [Google Scholar] [CrossRef]
- Kulcheski, F.R.; Christoff, A.P.; Margis, R. Circular RNAs are miRNA Sponges and can be Used as a New Class of Biomarker. J. Biotechnol. 2016, 238, 42–51. [Google Scholar] [CrossRef] [PubMed]
- Meng, S.; Zhou, H.; Feng, Z.; Xu, Z.; Tang, Y.; Li, P.; Wu, M. CircRNA: Functions and properties of a novel potential biomarker for cancer. Mol. Cancer 2017, 16, 94. [Google Scholar] [CrossRef]
- Zhang, Q.; Zhang, B.; Sun, L.; Yan, Q.; Zhang, Y.; Zhang, Z.; Su, Y.; Wang, C. MicroRNA-130b targets PTEN to induce resistance to cisplatin in lung cancer cells by activating Wnt/β-catenin pathway. Cell Biochem. Funct. 2018, 36, 194–202. [Google Scholar] [CrossRef]
- Li, X.; Yang, L.; Chen, L.L. The biogenesis, functions, and challenges of circular RNAs. Mol. Cell 2018, 71, 428–442. [Google Scholar] [CrossRef] [PubMed]
- Lei, K.; Bai, H.; Wei, Z.; Xie, C.; Wang, J.; Li, J.; Chen, Q. The mechanism and function of circular RNAs in human diseases. Exp. Cell Res. 2018, 368, 147–158. [Google Scholar] [CrossRef] [PubMed]
- Sasagawa, S.; Honma, Y.; Peng, X.; Maejima, K.; Nagaoka, K.; Kobayashi, Y.; Oosawa, A.; Johnson, T.A.; Okawa, Y.; Liang, H.; et al. Predicting chemotherapy responsiveness in gastric cancer through machine learning analysis of genome, immune, and neutrophil signatures. Gastric Cancer 2024, 28, 228–244. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Ran, Y.; Tao, C.; Li, S.; Chen, J.; Yang, E. Detection of circular RNA expression and related quantitative trait loci in the human dorsolateral prefrontal cortex. Genome Biol. 2019, 20, 99. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Tao, L.; Qiu, J.; Xu, J.; Yang, X.; Zhang, Y.; Tian, X.; Guan, X.; Cen, X.; Zhao, Y. Tumor biomarkers for diagnosis, prognosis and targeted therapy. Signal Transduct. Target. Ther. 2024, 9, 132. [Google Scholar] [PubMed]
- Li, L.; Zheng, Y.C.; Kayani, M.U.; Xu, W.; Wang, G.Q.; Sun, P.; Ao, N.; Zhang, L.N.; Gu, Z.Q.; Wu, L.C.; et al. Comprehensive analysis of circRNA expression profiles in humans by RAISE. Int. J. Oncol. 2017, 51, 1625–1638. [Google Scholar] [CrossRef] [PubMed]
- Kedlian, V.R.; Donertas, H.M.; Thornton, J.M. The widespread increase in inter-individual variability of gene expression in the human brain with age. Aging 2019, 11, 2253. [Google Scholar] [CrossRef] [PubMed]
- Digby, B.; Finn, S.; Broin, P.Ó. Computational approaches and challenges in the analysis of circRNA data. BMC Genom. 2024, 25, 527. [Google Scholar] [CrossRef] [PubMed]
- Vromman, M.; Vandesompele, J. Circular RNA detection pipelines yield divergent sets of circular RNAs. Nat. Methods 2023, 20, 1135–1136. [Google Scholar]
- Dodbele, S.; Mutlu, N.; Wilusz, J.E. Best practices to ensure robust investigation of circular RNAs: Pitfalls and tips. EMBO Rep. 2021, 22, e52072. [Google Scholar] [CrossRef] [PubMed]
- Hutchins, E.; Reiman, R.; Winarta, J.; Beecroft, T.; Richholt, R.; De Both, M.; Shahbander, K.; Carlson, E.; Janss, A.; Siniard, A.; et al. Extracellular circular RNA profiles in plasma and urine of healthy, male college athletes. Sci. Data 2021, 8, 276. [Google Scholar] [CrossRef]
- Li, S.; Han, L. Circular RNAs as promising biomarkers in cancer: Detection, function, and beyond. Genome Med. 2019, 11, 15. [Google Scholar] [CrossRef] [PubMed]
- Thompson, J.A.; Tan, J.; Greene, C.S. Cross-platform normalization of microarray and RNA-seq data for machine learning applications. PeerJ 2016, 4, e1621. [Google Scholar] [CrossRef]
- Zanger, U.M.; Schwab, M. Cytochrome P450 enzymes in drug metabolism: Regulation of gene expression, enzyme activities, and impact of genetic variation. Pharmacol. Ther. 2013, 138, 103–141. [Google Scholar] [PubMed]
- Lin, H.; Wang, Y.; Wang, P.; Long, F.; Wang, T. Mutual regulation between N6-methyladenosine (m6A) modification and circular RNAs in cancer: Impacts on therapeutic resistance. Mol. Cancer 2022, 21, 148. [Google Scholar] [CrossRef] [PubMed]
- Heming, C.P.; Muriithi, W.; Macharia, L.W.; Niemeyer Filho, P.; Moura-Neto, V.; Aran, V. P-glycoprotein and cancer: What do we currently know? Heliyon 2022, 8, e11171. [Google Scholar]
- Dong, Y.; Gao, Q.; Chen, Y.; Zhang, Z.; Du, Y.; Liu, Y.; Zhang, G.; Li, S.; Wang, G.; Chen, X.; et al. Identification of CircRNA signature associated with tumor immune infiltration to predict therapeutic efficacy of immunotherapy. Nat. Commun. 2023, 14, 2540. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Zhang, M.; Liu, J.; Li, Z.; Ni, B. Potential therapies for HCC involving targeting the ferroptosis pathway. Am. J. Cancer Res. 2024, 14, 1446. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Huang, Z.; He, M.; Liao, J.; Zhang, Q.; Wang, S.; Xie, L.; Ouyang, L.; Koeffler, H.P.; Yin, D.; et al. Circular RNA MAPK4 (circ-MAPK4) inhibits cell apoptosis via MAPK signaling pathway by sponging miR-125a-3p in gliomas. Mol. Cancer 2020, 19, 17. [Google Scholar] [CrossRef] [PubMed]
- Xu, Q.; Deng, B.; Li, M.; Chen, Y.; Zhuan, L. circRNA-UBAP2 promotes the proliferation and inhibits apoptosis of ovarian cancer though miR-382-5p/PRPF8 axis. J. Ovarian Res. 2020, 13, 81. [Google Scholar] [CrossRef]
- Kocaturk, N.M.; Akkoc, Y.; Kig, C.; Bayraktar, O.; Gozuacik, D.; Kutlu, O. Autophagy as a molecular target for cancer treatment. Eur. J. Pharm. Sci. 2019, 134, 116–137. [Google Scholar]
- Levine, B.; Kroemer, G. Biological functions of autophagy genes: A disease perspective. Cell 2019, 176, 11–42. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.J.; Lei, Y.H.; Yao, N.; Wang, C.R.; Hu, N.; Ye, W.C.; Zhang, D.M.; Chen, Z.S. Autophagy and multidrug resistance in cancer. Chin. J. Cancer 2017, 36, 52. [Google Scholar] [CrossRef] [PubMed]
- Rocha, C.R.; Silva, M.M.; Quinet, A.; Cabral-Neto, J.B.; Menck, C.F. DNA repair pathways and cisplatin resistance: An intimate relationship. Clinics 2018, 73 (Suppl. S1), e478s. [Google Scholar] [CrossRef] [PubMed]
- Du, W.W.; Yang, W.; Li, X.; Awan, F.M.; Yang, Z.; Fang, L.; Lyu, J.; Li, F.; Peng, C.; Krylov, S.N.; et al. A circular RNA circ-DNMT1 enhances breast cancer progression by activating autophagy. Oncogene 2018, 37, 5829–5842. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Lu, B. The roles of ceRNAs-mediated autophagy in cancer chemoresistance and metastasis. Cancers 2020, 12, 2926. [Google Scholar] [CrossRef]
- Lagunas-Rangel, F.A. Role of circular RNAs in DNA repair. RNA Biol. 2024, 21, 1226–1238. [Google Scholar] [CrossRef] [PubMed]
- Brognard, J.; Clark, A.S.; Ni, Y.; Dennis, P.A. Akt/protein kinase B is constitutively active in non-small cell lung cancer cells and promotes cellular survival and resistance to chemotherapy and radiation. Cancer Res. 2001, 61, 3986–3997. [Google Scholar]
- Ma, S.; Kong, S.; Wang, F.; Ju, S. CircRNAs: Biogenesis, functions, and role in drug-resistant Tumours. Mol. Cancer 2020, 19, 119. [Google Scholar] [CrossRef]
- Xu, T.; Wang, M.; Jiang, L.; Ma, L.; Wan, L.; Chen, Q.; Wei, C.; Wang, Z. CircRNAs in anticancer drug resistance: Recent advances and future potential. Mol. Cancer 2020, 19, 127. [Google Scholar] [CrossRef]
- Liu, L.Z.; Zhou, X.D.; Qian, G.; Shi, X.; Fang, J.; Jiang, B.H. AKT1 amplification regulates cisplatin resistance in human lung cancer cells through the mammalian target of rapamycin/p70S6K1 pathway. Cancer Res. 2007, 67, 6325–6332. [Google Scholar] [CrossRef]
- Stoica, G.E.; Franke, T.F.; Moroni, M.; Mueller, S.; Morgan, E.; Iann, M.C.; Winder, A.D.; Reiter, R.; Wellstein, A.; Martin, M.B.; et al. Effect of estradiol on estrogen receptor-α gene expression and activity can be modulated by the ErbB2/PI 3-K/Akt pathway. Oncogene 2003, 22, 7998–8011. [Google Scholar] [CrossRef] [PubMed]
- Lastwika, K.J.; Wilson, I.I.I.W.; Li, Q.K.; Norris, J.; Xu, H.; Ghazarian, S.R.; Kitagawa, H.; Kawabata, S.; Taube, J.M.; Yao, S.; et al. Control of PD-L1 expression by oncogenic activation of the AKT–mTOR pathway in non–small cell lung cancer. Cancer Res. 2016, 76, 227–238. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Xiao, Q.; Xiao, J.; Niu, C.; Li, Y.; Zhang, X.; Zhou, Z.; Shu, G.; Yin, G. Wnt/β-catenin signalling: Function, biological mechanisms, and therapeutic opportunities. Signal Transduct. Target. Ther. 2022, 7, 3. [Google Scholar] [CrossRef] [PubMed]
- Vromman, M.; Anckaert, J.; Bortoluzzi, S.; Buratin, A.; Chen, C.Y.; Chu, Q.; Chuang, T.J.; Dehghannasiri, R.; Dieterich, C.; Dong, X.; et al. Large-scale benchmarking of circRNA detection tools reveals large differences in sensitivity but not in precision. Nat. Methods 2023, 20, 1159–1169. [Google Scholar] [CrossRef]
- Zhang, M.; Xu, K.; Fu, L.; Wang, Q.; Chang, Z.; Zou, H.; Zhang, Y.; Li, Y. Revealing epigenetic factors of circRNA expression by machine learning in various cellular contexts. Iscience 2020, 23, 101842. [Google Scholar] [CrossRef] [PubMed]
- Bai, C.; Hao, X.; Zhou, L.; Sun, Y.; Song, L.; Wang, F.; Yang, L.; Liu, J.; Chen, J. Machine learning-based identification of the novel circRNAs circERBB2 and circCHST12 as potential biomarkers of intracerebral hemorrhage. Front. Neurosci. 2022, 16, 1002590. [Google Scholar] [CrossRef]
- Dhuri, K.; Bechtold, C.; Quijano, E.; Pham, H.; Gupta, A.; Vikram, A.; Bahal, R. Antisense oligonucleotides: An emerging area in drug discovery and development. J. Clin. Med. 2020, 9, 2004. [Google Scholar] [CrossRef]
- Yan, Y.; Su, M.; Qin, B. CircHIPK3 promotes colorectal cancer cells proliferation and metastasis via modulating of miR-1207-5p/FMNL2 signal. Biochem. Biophys. Res. Commun. 2020, 524, 839–846. [Google Scholar] [CrossRef] [PubMed]
- Roberts, T.C.; Langer, R.; Wood, M.J. Advances in oligonucleotide drug delivery. Nat. Rev. Drug Discov. 2020, 19, 673–694. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Tao, Y.; Shan, L.; Chen, R.; Jiang, H.; Qian, Z.; Cai, F.; Ma, L.; Yu, Y. The role of MicroRNAs in hepatocellular carcinoma. J. Cancer 2018, 9, 3557. [Google Scholar] [CrossRef] [PubMed]
- Ngue, H.; Shrestha, J.; Kim, H.; Vasudevan, S. Circular RNA expression is enriched in breast cancer extracellular vesicles and is associated with chemotherapy resistance. Cancer Res. 2023, 83 (Suppl. S7), 3789. [Google Scholar]
- Zhang, X.O.; Dong, R.; Zhang, Y.; Zhang, J.L.; Luo, Z.; Zhang, J.; Chen, L.L.; Yang, L. Diverse alternative back-splicing and alternative splicing landscape of circular RNAs. Genome Res. 2016, 26, 1277–1287. [Google Scholar] [PubMed]
- Zhang, Y.; Nguyen, T.M.; Zhang, X.O.; Wang, L.; Phan, T.; Clohessy, J.G.; Pandolfi, P.P. Optimized RNA-targeting CRISPR/Cas13d technology outperforms shRNA in identifying functional circRNAs. Genome Biol. 2021, 22, 41. [Google Scholar]
- Damnernsawad, A.; Bottomly, D.; Kurtz, S.E.; Eide, C.A.; McWeeney, S.K.; Tyner, J.W.; Nechiporuk, T. A genome-wide CRISPR screen identifies regulators of MAPK and MTOR pathways that mediate resistance to sorafenib in acute myeloid leukemia. Haematologica 2020, 107, 77. [Google Scholar] [CrossRef]
- Palaz, F.; Kalkan, A.K.; Can, O.; Demir, A.N.; Tozluyurt, A.; Ozcan, A.; Ozsoz, M. CRISPR-Cas13 system as a promising and versatile tool for cancer diagnosis, therapy, and research. ACS Synth. Biol. 2021, 10, 1245–1267. [Google Scholar] [PubMed]
- Byrnes, A.E.; Dominguez, S.L.; Yen, C.W.; Laufer, B.I.; Foreman, O.; Reichelt, M.; Lin, H.; Sagolla, M.; Hötzel, K.; Ngu, H.; et al. Lipid nanoparticle delivery limits antisense oligonucleotide activity and cellular distribution in the brain after intracerebroventricular injection. Mol. Ther.-Nucleic Acids 2023, 32, 773–793. [Google Scholar] [CrossRef] [PubMed]
- Adamus, T.; Hung, C.Y.; Yu, C.; Kang, E.; Hammad, M.; Flores, L.; Nechaev, S.; Zhang, Q.; Gonzaga, J.M.; Muthaiyah, K.; et al. Glioma-targeted delivery of exosome-encapsulated antisense oligonucleotides using neural stem cells. Mol. Ther.-Nucleic Acids 2022, 27, 611–620. [Google Scholar] [CrossRef] [PubMed]
- Rama Ballesteros, A.R.; Quiñonero Muñoz, F.J.; Mesas Hernández, C.; Melguizo Alonso, C.; Prados Salazar, J.C. Synthetic Circular miR-21 Sponge as Tool for Lung Cancer Treatment. Int. J. Mol. Sci. 2022, 23, 2963. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Yu, S.; Ding, L.; Ma, L.; Chen, H.; Zhou, H.; Zou, Y.; Yu, M.; Lin, J.; Cui, Q. The dual role of circular RNAs as miRNA sponges in breast cancer and colon cancer. Biomedicines 2021, 9, 1590. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Shin, S.H.; Lim, C.G.; Heo, Y.H.; Choi, I.Y.; Kim, H.H. Synthetic RNA therapeutics in cancer. J. Pharmacol. Exp. Ther. 2023, 386, 212–223. [Google Scholar] [CrossRef] [PubMed]
- Obi, P.; Chen, Y.G. The design and synthesis of circular RNAs. Methods 2021, 196, 85–103. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Zhang, Y.; Zhou, S.; Dain, L.; Mei, L.; Zhu, G. Circular RNA: An emerging frontier in RNA therapeutic targets, RNA therapeutics, and mRNA vaccines. J. Control. Release 2022, 348, 84–94. [Google Scholar] [CrossRef]
- Sarkar, S.; Moitra, P.; Bera, S.; Bhattacharya, S. Antisense Oligonucleotide Embedded Context Responsive Nanoparticles Derived from Synthetic Ionizable Lipids for lncRNA Targeted Therapy of Breast Cancer. ACS Appl. Mater. Interfaces 2024, 16, 45871–45887. [Google Scholar] [CrossRef]
- Kim, H.; Jang, H.; Cho, H.; Choi, J.; Hwang, K.Y.; Choi, Y.; Kim, S.H.; Yang, Y. Recent advances in exosome-based drug delivery for cancer therapy. Cancers 2021, 13, 4435. [Google Scholar] [CrossRef] [PubMed]
- Khorasani, A.B.; Hafezi, N.; Sanaei, M.J.; Jafari-Raddani, F.; Pourbagheri-Sigaroodi, A.; Bashash, D. The PI3K/AKT/mTOR signaling pathway in breast cancer: Review of clinical trials and latest advances. Cell Biochem. Funct. 2024, 42, e3998. [Google Scholar] [CrossRef]
- Han, R.; Rao, X.; Zhou, H.; Lu, L. Synergistic Immunoregulation: Harnessing CircRNAs and PiRNAs to Amplify PD-1/PD-L1 Inhibition Therapy. Int. J. Nanomed. 2024, 19, 4803–4834. [Google Scholar] [CrossRef]
- Wang, B.; Shen, W.; Yan, L.; Li, X.; Zhang, L.; Zhao, S.; Jin, X. Reveal the potential molecular mechanism of circRNA regulating immune-related mRNA through sponge miRNA in the occurrence and immune regulation of papillary thyroid cancer. Ann. Med. 2023, 55, 2244515. [Google Scholar] [PubMed]
- Antonangeli, F.; Natalini, A.; Garassino, M.C.; Sica, A.; Santoni, A.; Di Rosa, F. Regulation of PD-L1 Expression by NF-κB in Cancer. Front. Immunol. 2020, 11, 584626. [Google Scholar]
- Bejugam, P.R.; Das, A.; Panda, A.C. Seeing is believing: Visualizing circular RNAs. Non-Coding RNA 2020, 6, 45. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.O.; Wang, H.B.; Zhang, Y.; Lu, X.; Chen, L.L.; Yang, L. Complementary sequence-mediated exon circularization. Cell 2014, 159, 134–147. [Google Scholar] [CrossRef]
- Jeck, W.R.; Sorrentino, J.A.; Wang, K.; Slevin, M.K.; Burd, C.E.; Liu, J.; Marzluff, W.F.; Sharpless, N.E. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 2013, 19, 141–157. [Google Scholar] [CrossRef] [PubMed]
- Szabo, L.; Salzman, J. Detecting circular RNAs: Bioinformatic and experimental challenges. Nat. Rev. Genet. 2016, 17, 679–692. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Tao, C.; Li, S.; Du, M.; Bai, Y.; Hu, X.; Li, Y.; Chen, J.; Yang, E. circFL-seq reveals full-length circular RNAs with rolling circular reverse transcription and nanopore sequencing. eLife 2021, 10, e69457. [Google Scholar] [CrossRef] [PubMed]
- Micheel, J.; Safrastyan, A.; Wollny, D. Advances in non-coding RNA sequencing. Non-Coding RNA 2021, 7, 70. [Google Scholar] [CrossRef]
- Zhang, J.; Luo, Z.; Zheng, Y.; Duan, M.; Qiu, Z.; Huang, C. CircRNA as an Achilles heel of cancer: Characterization, biomarker and therapeutic modalities. J. Transl. Med. 2024, 22, 752. [Google Scholar] [CrossRef] [PubMed]
- Thwala, L.N.; Ndlovu, S.C.; Mpofu, K.T.; Lugongolo, M.Y.; Mthunzi-Kufa, P. Nanotechnology-Based Diagnostics for Diseases Prevalent in Developing Countries: Current Advances in Point-of-Care Tests. Nanomaterials 2023, 13, 1247. [Google Scholar] [CrossRef] [PubMed]
- Salzman, J.; Gawad, C.; Wang, P.L.; Lacayo, N.; Brown, P.O. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS ONE 2012, 7, e30733. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, H.; Zuo, Y.; Wang, J.; Zhang, M.Q.; Malhotra, A.; Mayeda, A. Characterization of RNase R-digested cellular RNA source that consists of lariat and circular RNAs from pre-mRNA splicing. Nucleic Acids Res. 2006, 34, e63. [Google Scholar] [CrossRef]
- Ma, X.K.; Gao, X.; Cao, M.; Yang, L. Base-editor-mediated circRNA knockout by targeting predominantly back-splice sites. In Circular RNAs; Springer: New York, NY, USA, 2024; pp. 193–208. [Google Scholar]
- Li, Z.; Huang, C.; Bao, C.; Chen, L.; Lin, M.; Wang, X.; Zhong, G.; Yu, B.; Hu, W.; Dai, L.; et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat. Struct. Mol. Biol. 2015, 22, 256–264. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Deng, Y.; Liu, Q.; Ye, B.; Dai, Z.; Chen, Y.; Dai, X. Identifying circular RNA and predicting its regulatory interactions by machine learning. Front. Genetics 2020, 11, 655. [Google Scholar] [CrossRef]
- Xu, Y. An overview of the main circRNA databases. Non-Coding Coding Coding RNA Investig. 2017, 1, 22. [Google Scholar] [CrossRef]
- Glažar, P.; Papavasileiou, P.; Rajewsky, N. circBase: A database for circular RNAs. RNA 2014, 20, 1666–1670. [Google Scholar] [CrossRef] [PubMed]
- Vromman, M.; Vandesompele, J.; Volders, P.J. Closing the circle: Current state and perspectives of circular RNA databases. Brief. Bioinform. 2021, 22, 288–297. [Google Scholar] [CrossRef] [PubMed]
- Cao, X.; Zhou, X.; Hou, F.; Huang, Y.E.; Yuan, M.; Long, M.; Chen, S.; Lei, W.; Zhu, J.; Chen, J.; et al. ncRNADrug: A database for validated and predicted ncRNAs associated with drug resistance and targeted by drugs. Nucleic Acids Res. 2024, 52, D1393–D1399. [Google Scholar] [CrossRef]
- Chen, Y.; Wang, J.; Wang, C.; Zou, Q. AutoEdge-CCP: A novel approach for predicting cancer-associated circRNAs and drugs based on automated edge embedding. PLoS Comput. Biol. 2024, 20, e1011851. [Google Scholar] [CrossRef]
- Chen, Y.; Yao, L.; Tang, Y.; Jhong, J.H.; Wan, J.; Chang, J.; Cui, S.; Luo, Y.; Cai, X.; Li, W.; et al. CircNet 2.0: An updated database for exploring circular RNA regulatory networks in cancers. Nucleic Acids Res. 2022, 50, D93–D101. [Google Scholar] [PubMed]
- Ji, P.; Wu, W.; Chen, S.; Zheng, Y.; Zhou, L.; Zhang, J.; Cheng, H.; Yan, J.; Zhang, S.; Yang, P.; et al. Expanded expression landscape and prioritization of circular RNAs in mammals. Cell Rep. 2019, 26, 3444–3460. [Google Scholar] [CrossRef]
- Chen, X.; Han, P.; Zhou, T.; Guo, X.; Song, X.; Li, Y. circRNADb: A comprehensive database for human circular RNAs with protein-coding annotations. Sci. Rep. 2016, 6, 34985. [Google Scholar] [CrossRef] [PubMed]
- Ruan, H.; Xiang, Y.; Ko, J.; Li, S.; Jing, Y.; Zhu, X.; Ye, Y.; Zhang, Z.; Mills, T.; Feng, J.; et al. Comprehensive characterization of circular RNAs in~ 1000 human cancer cell lines. Genome Med. 2019, 11, 55. [Google Scholar]
- Chen, X.; Yan, C.C.; Zhang, X.; Zhang, X.; Dai, F.; Yin, J.; Zhang, Y. Drug-target interaction prediction: Databases, web servers and computational models. Brief. Bioinform. 2016, 17, 696–712. [Google Scholar]
- Yin, S.; Xu, P.; Jiang, Y.; Yang, X.; Lin, Y.; Zheng, M.; Hu, J.; Zhao, Q. Predicting the potential associations between circRNA and drug sensitivity using a multisource feature-based approach. J. Cell. Mol. Med. 2024, 28, e18591. [Google Scholar] [PubMed]
- Drula, R.; Braicu, C.; Berindan-Neagoe, I. Current advances in circular RNA detection and investigation methods: Are we running in circles? Wiley Interdiscip. Rev. RNA 2024, 15, e1850. [Google Scholar] [CrossRef] [PubMed]
- He, A.T.; Liu, J.; Li, F.; Yang, B.B. Targeting circular RNAs as a therapeutic approach: Current strategies and challenges. Signal Transduct. Target. Ther. 2021, 6, 185. [Google Scholar] [CrossRef] [PubMed]
- Santer, L.; Bär, C.; Thum, T. Circular RNAs: A novel class of functional RNA molecules with a therapeutic perspective. Mol. Ther. 2019, 27, 1350–1363. [Google Scholar] [CrossRef] [PubMed]
- Lai, Y.; Wei, X.; Lin, S.; Qin, L.; Cheng, L.; Li, P. Current status and perspectives of patient-derived xenograft models in cancer research. J. Hematol. Oncol. 2017, 10, 106. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Wu, W.; Cai, C.; Zhang, H.; Shen, H.; Han, Y. Patient-derived xenograft models in cancer therapy: Technologies and applications. Signal Transduct. Target. Ther. 2023, 8, 160. [Google Scholar] [CrossRef] [PubMed]
- Karkampouna, S.; La Manna, F.; Benjak, A.; Kiener, M.; De Menna, M.; Zoni, E.; Grosjean, J.; Klima, I.; Garofoli, A.; Bolis, M.; et al. Patient-derived xenografts and organoids model therapy response in prostate cancer. Nat. Commun. 2021, 12, 1117. [Google Scholar] [CrossRef]
- Jian, M.; Ren, L.; He, G.; Lin, Q.; Tang, W.; Chen, Y.; Chen, J.; Liu, T.; Ji, M.; Wei, Y.; et al. A novel patient-derived organoids-based xenografts model for preclinical drug response testing in patients with colorectal liver metastases. J. Transl. Med. 2020, 18, 234. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, A.F.; Bindereif, A.; Bozzoni, I.; Hanan, M.; Hansen, T.B.; Irimia, M.; Kadener, S.; Kristensen, L.S.; Legnini, I.; Morlando, M.; et al. Best practice standards for circular RNA research. Nat. Methods 2022, 19, 1208–1220. [Google Scholar] [CrossRef]
- Xie, F.; Zhao, N.; Zhang, H.; Xie, D. Circular RNA CircHIPK3 promotes gemcitabine sensitivity in bladder cancer. J. Cancer 2020, 11, 1907. [Google Scholar] [CrossRef]
- Shang, Z.; Luo, Z.; Wang, Y.; Liu, Q.; Xin, Y.; Zhang, M.; Li, X.; Zeng, S.; Yu, L.; Zhang, X.; et al. CircHIPK3 contributes to cisplatin resistance in gastric cancer by blocking autophagy-dependent ferroptosis. J. Cell Physiol. 2023, 238, 2407–2424. [Google Scholar] [CrossRef] [PubMed]
- Sun, M.; Liu, X.; Zhao, W.; Zhang, B.; Deng, P. Circ_0058063 contributes to cisplatin-resistance of bladder cancer cells by upregulating B2M through acting as RNA sponges for miR-335-5p. BMC Cancer 2022, 22, 313. [Google Scholar] [CrossRef]
- Su, Y.; Lin, T. MP17-01 circmorc3 contributes to cisplatin-resistance by affecting m6a modification on dna damage response related genes in bladder cancer. J. Urol. 2020, 203 (Suppl. S4), e225. [Google Scholar]
- Kun-Peng, Z.; Xiao-Long, M.; Chun-Lin, Z. Overexpressed circPVT1, a potential new circular RNA biomarker, contributes to doxorubicin and cisplatin resistance of osteosarcoma cells by regulating ABCB1. Int. J. Biol. Sci. 2018, 14, 321. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Gong, Q.; Yu, Y.; Zhu, J.; Li, W. Knockdown of circ-ABCB10 promotes sensitivity of lung cancer cells to cisplatin via miR-556-3p/AK4 axis. BMC Pulm. Med. 2020, 20, 10. [Google Scholar] [CrossRef]
- Xie, X.; Sun, F.K.; Huang, X.; Wang, C.H.; Dai, J.; Zhao, J.P.; Fang, C.; He, W. A circular RNA, circSMARCA5, inhibits prostate cancer proliferative, migrative, and invasive capabilities via the miR-181b-5p/miR-17-3p-TIMP3 axis. Aging 2021, 13, 19908. [Google Scholar] [CrossRef]
- Gopikrishnan, M.; Ashour, H.M.; Pintus, G.; Hammad, M.; Kashyap, M.K.; Zayed, H. Therapeutic and diagnostic applications of exosomal circRNAs in breast cancer. Funct. Integr. Genom. 2023, 23, 184. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Luo, J.; Yang, W.; Ye, W.C. CircRNAs in colorectal cancer: Potential biomarkers and therapeutic targets. Cell Death Dis. 2023, 14, 353. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wang, Y.; Su, X.; Wang, P.; Lin, W. The value of circulating circular RNA in cancer diagnosis, monitoring, prognosis, and guiding treatment. Front. Oncol. 2021, 11, 736546. [Google Scholar] [CrossRef]
- Yang, Z.; Guan, F.; Bronk, L.; Zhao, L. Multi-omics approaches for biomarker discovery in predicting the response of esophageal cancer to neoadjuvant therapy: A multidimensional perspective. Pharmacol. Ther. 2024, 254, 108591. [Google Scholar] [CrossRef]
- Rao, D.; Yu, C.; Sheng, J.; Lv, E.; Huang, W. The emerging roles of circFOXO3 in cancer. Front. Cell Dev. Biol. 2021, 9, 659417. [Google Scholar] [CrossRef]
- Pfafenrot, C.; Preußer, C. Establishing essential quality criteria for the validation of circular RNAs as biomarkers. Biomol. Detect. Quantif. 2019, 17, 100085. [Google Scholar] [CrossRef] [PubMed]
- Cai, J.; Qiu, Z.; Chi-Shing Cho, W.; Liu, Z.; Chen, S.; Li, H.; Chen, K.; Li, Y.; Zuo, C.; Qiu, M. Synthetic circRNA therapeutics: Innovations, strategies, and future horizons. MedComm 2024, 5, e720. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Zhang, K.; Tan, S.; Xin, J.; Yuan, Q.; Xu, H.; Xu, X.; Liang, Q.; Christiani, D.C.; Wang, M.; et al. Circular RNAs in body fluids as cancer biomarkers: The new frontier of liquid biopsies. Mol. Cancer 2021, 20, 13. [Google Scholar] [CrossRef] [PubMed]
- Newsham, I.; Sendera, M.; Jammula, S.G.; Samarajiwa, S.A. Early detection and diagnosis of cancer with interpretable machine learning to uncover cancer-specific DNA methylation patterns. Biol. Methods Protoc. 2024, 9, bpae028. [Google Scholar] [CrossRef] [PubMed]
- Jiang, W.; Pan, S.; Chen, X.; Wang, Z.W.; Zhu, X. The role of lncRNAs and circRNAs in the PD-1/PD-L1 pathway in cancer immunotherapy. Mol. Cancer 2021, 20, 116. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Wen, Y.; Yu, W.; Lu, L.; Yang, Y.; Liu, C.; Hu, Z.; Fang, Z.; Huang, S. Optimized circular RNA vaccines for superior cancer immunotherapy. Theranostics 2025, 15, 1420. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Liu, W.R.; Tan, S.; Zhou, J.K.; Xu, X.; Ming, Y.; Cheng, J.; Li, J.; Zeng, Z.; Zuo, Y.; et al. Characterization of distinct circular RNA signatures in solid tumors. Mol. Cancer 2022, 21, 63. [Google Scholar] [CrossRef] [PubMed]
- Long, G.; Ma, S.; Shi, R.; Sun, Y.; Hu, Z.; Chen, K. Circular RNAs and drug resistance in genitourinary cancers: A literature review. Cancers 2022, 14, 866. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Xie, F.; Lin, J.; Zhao, Y.; Zhang, Q.; Liao, Z.; Wei, P. Diagnostic and prognostic value of circulating CircRNAs in cancer. Front. Med. 2021, 8, 649383. [Google Scholar] [CrossRef]
- Bonelli, P.; Borrelli, A.; Tuccillo, F.M.; Buonaguro, F.M.; Tornesello, M.L. The role of circRNAs in human papillomavirus (HPV)-associated cancers. Cancers 2021, 13, 1173. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Fan, Q.; Wang, Y.; Liu, Y.; Xu, X.; Liang, Y.; Xie, J.; Li, J.; Ai, F.; Cao, Y.; et al. CircRNAs in colorectal cancer: Potential roles, clinical applications, and natural product-based regulation. Front. Oncol. 2025, 15, 1525779. [Google Scholar] [CrossRef] [PubMed]
- Kristensen, L.S.; Hansen, T.B.; Venø, M.T.; Kjems, J. Circular RNAs in cancer: Opportunities and challenges in the field. Oncogene 2018, 37, 555–565. [Google Scholar] [CrossRef] [PubMed]

| Database | Focus on Drug Response | Strengths | Limitations | Reference * |
|---|---|---|---|---|
| circBase | Does not directly focus on drug response but offers foundational circRNA annotations | Serves as a foundational resource consolidating circRNA data from multiple studies. | Lacks pharmacogenomic-specific annotations, limiting its utility for drug interaction studies. | [125,126] |
| ncRNADrug | Provides insights into ncRNA roles in drug resistance mechanisms | Catalogs both validated and predicted interactions between ncRNAs (including circRNAs) and drug resistance. | Computational predictions may introduce biases without experimental validation. | [128] |
| CircNet 2.0 | Facilitates study of circRNA-miRNA-mRNA interactions, relevant for drug response | Provides an interactive platform for circRNA regulatory networks, including circRNA-miRNA-mRNA interactions. | Does not explicitly focus on pharmacogenomics. | [129] |
| CircAtlas | Not specifically focused on drug response but may aid in exploring circRNA-drug interactions. | Provides detailed human circRNA data, including sequences, annotations and the potential circRNA–miRNA interactions | Not explicitly designed for drug response studies. | [130,131] |
| circRNADb | Provides annotations that can be used for exploring drug response mechanisms | Provides a comprehensive catalog of human exonic circRNAs with annotations. | Not specifically tailored for drug resistance mechanisms. | [127,131] |
| CircRiC | Cancer-specific circRNAs | Specializes in circRNA expression and drug sensitivity in cancer cell lines. | Focuses on cancer models, limiting its generalizability. | [132,133] |
| GATECDA Framework | Predicts circRNA influence on drug sensitivity | Leverages graph attention auto-encoder techniques to predict circRNA influence on drug sensitivity. | Computational framework may lack broad experimental validation. | [134] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alqahtani, S.; Alqahtani, T.; Venkatesan, K.; Sivadasan, D.; Ahmed, R.; Elfadil, H.; Paulsamy, P.; Periannan, K. Unveiling Pharmacogenomics Insights into Circular RNAs: Toward Precision Medicine in Cancer Therapy. Biomolecules 2025, 15, 535. https://doi.org/10.3390/biom15040535
Alqahtani S, Alqahtani T, Venkatesan K, Sivadasan D, Ahmed R, Elfadil H, Paulsamy P, Periannan K. Unveiling Pharmacogenomics Insights into Circular RNAs: Toward Precision Medicine in Cancer Therapy. Biomolecules. 2025; 15(4):535. https://doi.org/10.3390/biom15040535
Chicago/Turabian StyleAlqahtani, Saud, Taha Alqahtani, Krishnaraju Venkatesan, Durgaramani Sivadasan, Rehab Ahmed, Hassabelrasoul Elfadil, Premalatha Paulsamy, and Kalaiselvi Periannan. 2025. "Unveiling Pharmacogenomics Insights into Circular RNAs: Toward Precision Medicine in Cancer Therapy" Biomolecules 15, no. 4: 535. https://doi.org/10.3390/biom15040535
APA StyleAlqahtani, S., Alqahtani, T., Venkatesan, K., Sivadasan, D., Ahmed, R., Elfadil, H., Paulsamy, P., & Periannan, K. (2025). Unveiling Pharmacogenomics Insights into Circular RNAs: Toward Precision Medicine in Cancer Therapy. Biomolecules, 15(4), 535. https://doi.org/10.3390/biom15040535







