Proteomic Profiling of Inflammatory Protein Dysregulation in HLA-B27-Positive Ankylosing Spondylitis: Molecular Signatures and Potential Biomarkers
Abstract
1. Introduction
2. Materials and Methods
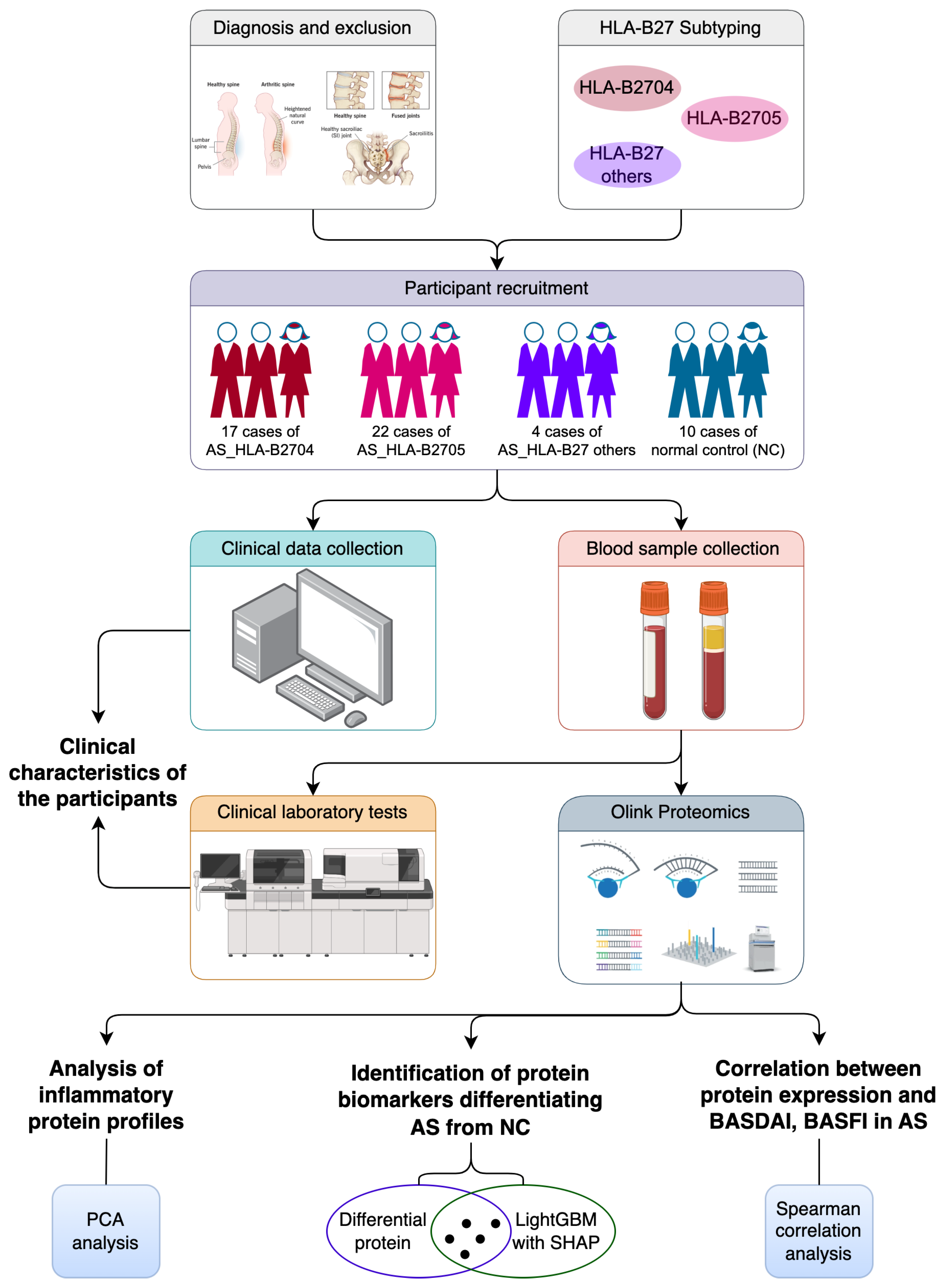
2.1. Study Design
2.2. Participant Recruitment
2.3. Data Collection
2.4. Sample Collection and Processing
2.5. Clinical Laboratory Test
2.6. HLA-B27 Subtyping
2.7. Inflammatory Protein Analysis
2.8. Statistical Analysis
3. Results
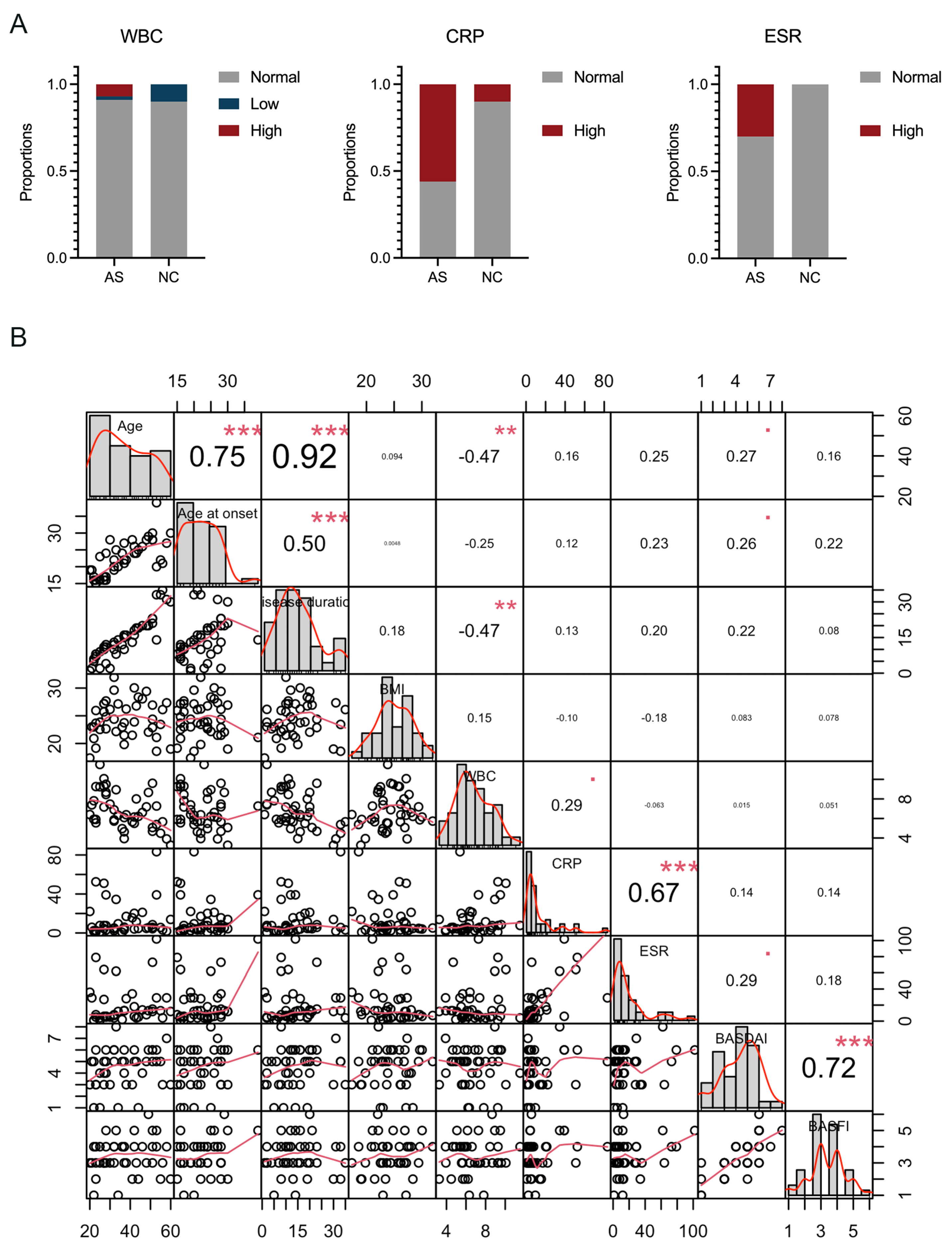
3.1. Clinical Characteristics of the Participants
3.2. Analysis of Inflammatory Protein Profiles in AS Patients and NC Subjects
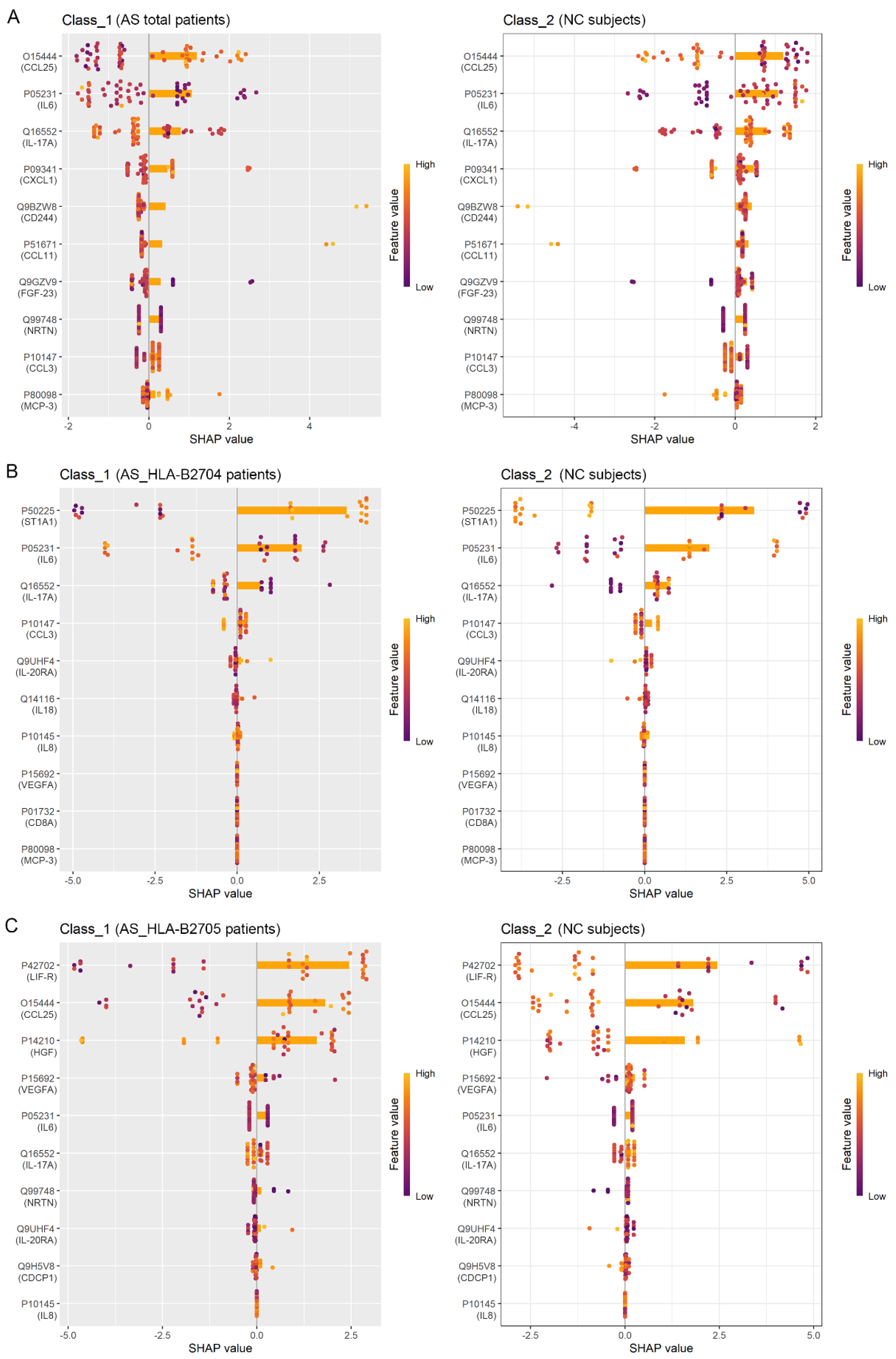
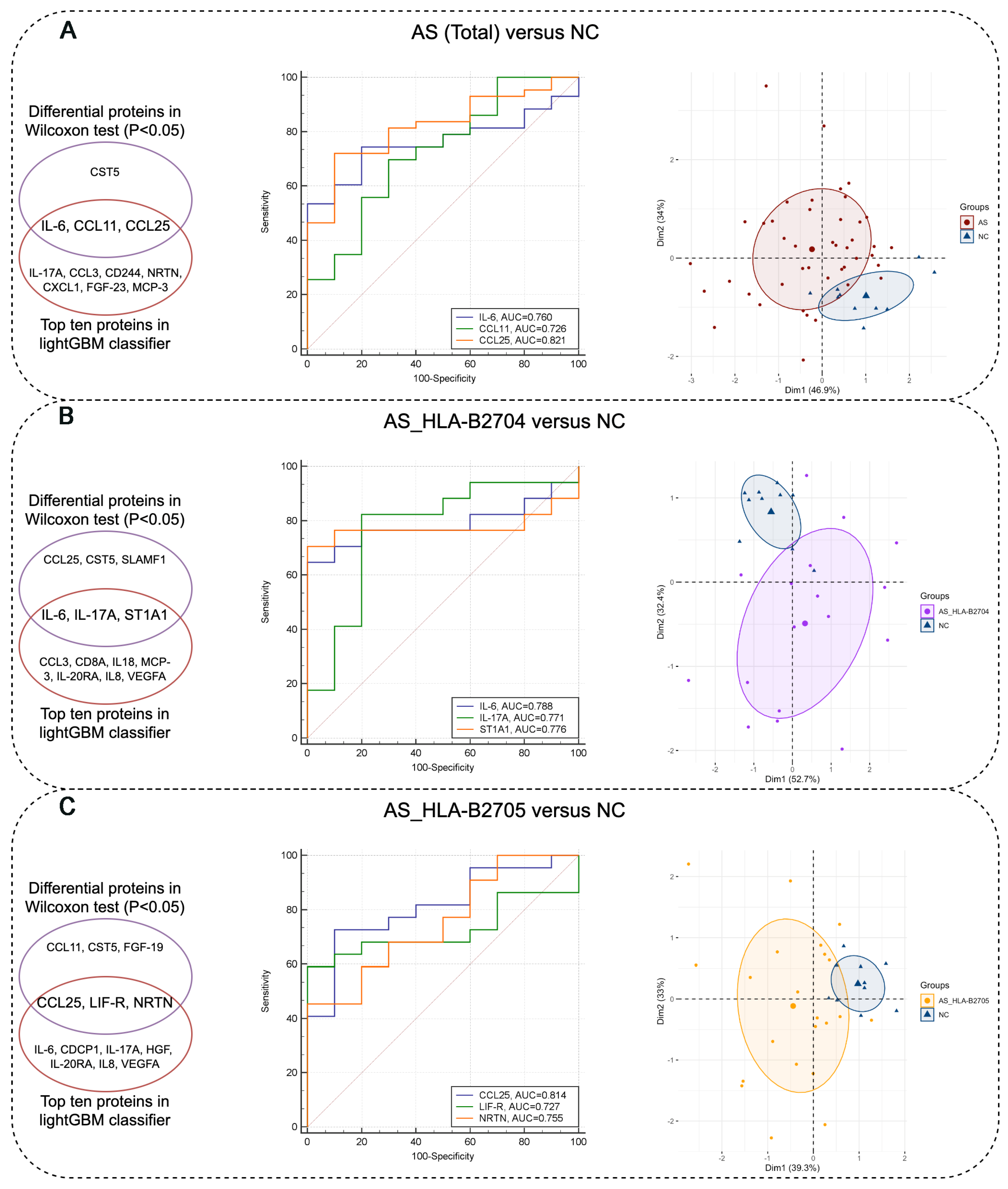
3.3. Identification of Protein Biomarkers Differentiating AS Patients from NC Subjects
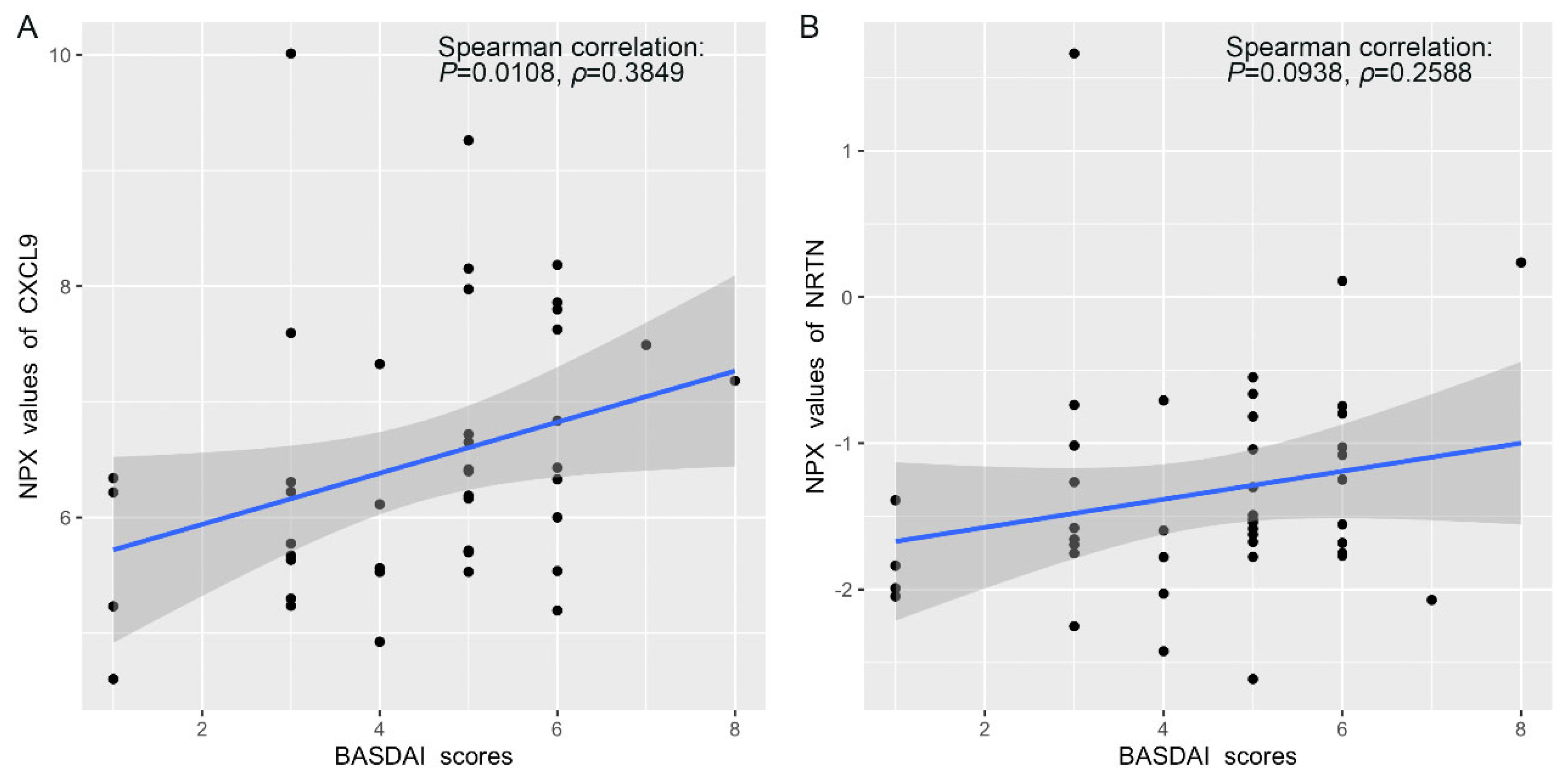
3.4. Correlation Between Protein Expression and BASDAI and BASFI Scores in AS Patients
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| AS | Ankylosing spondylitis |
| r-axSpA | Adiographic axial spondyloarthritis |
| HLA-B27 | Human leukocyte antigen-B*27 |
| NSAIDs | Non-steroidal anti-inflammatory drugs |
| TNF | Tumor necrosis factor |
| PEA | Proximity extension assay |
| ELISA | Enzyme-linked immunosorbent assay |
| NC | Normal control |
| BMI | Body mass index |
| WBC | White blood cell |
| CRP | C-reactive protein |
| ESR | Erythrocyte sedimentation rate |
| BASDAI | Bath Ankylosing Spondylitis Disease Activity Index |
| BASFI | Bath Ankylosing Spondylitis Functional Index |
| PCA | Principal component analysis |
| NPX | Normalized protein expression |
| SHAP | Shapley Additive Explanations |
| LightGBM | Light gradient boosting machine |
| ROC | Receiver operating characteristic |
| AUC | Area under the curve |
| PBMCs | Peripheral blood mononuclear cells |
| TNFi | TNF inhibitor |
| PsA | Psoriatic arthritis |
| Pso | Psoriasis without arthritis |
| AI | Artificial intelligence |
| GDNF | Glial cell-derived neurotrophic factor |
| GFLs | GDNF family ligands |
| DMARDs | Disease-modifying antirheumatic drugs |
| PCR-SSP | Polymerase chain reaction with sequence-specific primers |
References
- Braun, J.; Sieper, J. Ankylosing spondylitis. Lancet 2007, 369, 1379–1390. [Google Scholar] [CrossRef]
- Wenker, K.J.; Quint, J.M. Ankylosing Spondylitis; StatPearls: Treasure Island, FL, USA, 2024. [Google Scholar]
- Baraliakos, X.; Saffore, C.D.; Collins, E.B.; Parikh, B.; Ye, X.; Walsh, J.A. Comparative Efficacy of Advanced Therapies in the Treatment of Radiographic Axial Spondyloarthritis or Ankylosing Spondylitis as Evaluated by the ASDAS Low Disease Activity Criteria. Rheumatol. Ther. 2024, 11, 989–999. [Google Scholar] [PubMed]
- Zlatkovic-Svenda, M.; Djokic, A.; Perunicic, A.; Zdravkovic, M.; Dolijanovic, S.P.; Thorpe, J.; Dudok, D.; Milicevic, J.; Petrovic, D.; Radunovic, G. Translation, cross-cultural adaptation and psychometric evaluation of the Serbian Ankylosing Spondylitis Quality of Life (ASQoL) Questionnaire (refers to r-axSpA) and its relations with disease activity and functional status indices. J. Patient Rep. Outcomes 2025, 9, 8. [Google Scholar] [PubMed]
- Walsh, J.; Hunter, T.; Schroeder, K.; Sandoval, D.; Bolce, R. Trends in diagnostic prevalence and treatment patterns of male and female ankylosing spondylitis patients in the United States, 2006–2016. BMC Rheumatol 2019, 3, 39. [Google Scholar] [CrossRef] [PubMed]
- Dean, L.E.; Jones, G.T.; MacDonald, A.G.; Downham, C.; Sturrock, R.D.; Macfarlane, G.J. Global prevalence of ankylosing spondylitis. Rheumatology 2014, 53, 650–657. [Google Scholar]
- Stolwijk, C.; van Onna, M.; Boonen, A.; van Tubergen, A. Global Prevalence of Spondyloarthritis: A Systematic Review and Meta-Regression Analysis. Arthritis Care Res. 2016, 68, 1320–1331. [Google Scholar]
- Zhang, S.; Peng, L.; Li, Q.; Zhao, J.; Xu, D.; Zhao, J.; Wang, Q.; Li, M.; Zhang, W.; Tian, X.; et al. Spectrum of Spondyloarthritis Among Chinese Populations. Curr. Rheumatol. Rep. 2022, 24, 247–258. [Google Scholar]
- Nelson, D.A.; Kaplan, R.M.; Kurina, L.M.; Weisman, M.H. Incidence of Ankylosing Spondylitis Among Male and Female United States Army Personnel. Arthritis Care Res. 2023, 75, 332–339. [Google Scholar]
- Braun, J.; Sieper, J. Fifty years after the discovery of the association of HLA B27 with ankylosing spondylitis. RMD Open 2023, 9, e003102. [Google Scholar]
- Brewerton, D.A.; Hart, F.D.; Nicholls, A.; Caffrey, M.; James, D.C.; Sturrock, R.D. Ankylosing spondylitis and HL-A 27. Lancet 1973, 1, 904–907. [Google Scholar] [CrossRef]
- Schlosstein, L.; Terasaki, P.I.; Bluestone, R.; Pearson, C.M. High association of an HL-A antigen, W27, with ankylosing spondylitis. N. Engl. J. Med. 1973, 288, 704–706. [Google Scholar] [CrossRef]
- Khan, M.A. Polymorphism of HLA-B27: 105 subtypes currently known. Curr. Rheumatol. Rep. 2013, 15, 362. [Google Scholar] [PubMed]
- Montserrat, V.; Galocha, B.; Marcilla, M.; Vazquez, M.; Lopez de Castro, J.A. HLA-B*2704, an allotype associated with ankylosing spondylitis, is critically dependent on transporter associated with antigen processing and relatively independent of tapasin and immunoproteasome for maturation, surface expression, and T cell recognition: Relationship to B*2705 and B*2706. J. Immunol. 2006, 177, 7015–7023. [Google Scholar]
- Pimentel-Santos, F.M.; Ligeiro, D.; Matos, M.; Mourao, A.F.; Costa, J.; Santos, H.; Barcelos, A.; Godinho, F.; Pinto, P.; Cruz, M.; et al. Whole blood transcriptional profiling in ankylosing spondylitis identifies novel candidate genes that might contribute to the inflammatory and tissue-destructive disease aspects. Arthritis Res. Ther. 2011, 13, R57. [Google Scholar] [CrossRef]
- Lee, C.M.; Chan, E.R.; Schueller, D.; Breitman, M.; Haghiac, M.; Magrey, M. IL-17-producing cells in ankylosing spondylitis patients show gender-based differences in gene expression. Clin. Exp. Rheumatol. 2024, 42, 1057–1066. [Google Scholar]
- Deng, Y.; Xu, W.; Ni, M.; Sun, X.; Wang, X.; Zhang, T.; Pan, F. DNA methylation and expression of LGR6 gene in ankylosing spondylitis: A case-control study. Hum. Immunol. 2023, 84, 110719. [Google Scholar] [PubMed]
- Saevik, A.B.; Ueland, G.; Akerman, A.K.; Methlie, P.; Quinkler, M.; Jorgensen, A.P.; Hoybye, C.; Debowska, A.W.J.; Nedrebo, B.G.; Dahle, A.L.; et al. Altered biomarkers for cardiovascular disease and inflammation in autoimmune Addison’s disease—A cross-sectional study. Eur. J. Endocrinol. 2023, 189, 438–447. [Google Scholar] [PubMed]
- Jia, X.; He, X.; Huang, C.; Li, J.; Dong, Z.; Liu, K. Protein translation: Biological processes and therapeutic strategies for human diseases. Signal Transduct. Target. Ther. 2024, 9, 44. [Google Scholar]
- Al-Janabi, A.; Martin, P.; Khan, A.R.; Foulkes, A.C.; Smith, C.H.; Griffiths, C.E.M.; Morris, A.P.; Eyre, S.; Warren, R.B.; Group, B.S.; et al. Integrated proteomics and genomics analysis of paradoxical eczema in psoriasis patients treated with biologics. J. Allergy Clin. Immunol. 2023, 152, 1237–1246. [Google Scholar] [CrossRef]
- Fredriksson, T.; Brudin, L.; Henningsson, A.J.; Skogman, B.H.; Tjernberg, I. Diagnostic patterns of serum inflammatory protein markers in children with Lyme neuroborreliosis. Ticks Tick. Borne Dis. 2024, 15, 102349. [Google Scholar]
- Hoyer, A.; Chakraborty, S.; Lilienthal, I.; Konradsen, J.R.; Katayama, S.; Soderhall, C. The functional role of CST1 and CCL26 in asthma development. Immun. Inflamm. Dis. 2024, 12, e1162. [Google Scholar] [CrossRef] [PubMed]
- Szabo, I.; Badii, M.; Gaal, I.O.; Szabo, R.; Sirbe, C.; Humita, O.; Joosten, L.A.B.; Crisan, T.O.; Rednic, S. Immune Profiling of Patients with Systemic Sclerosis through Targeted Proteomic Analysis. Int. J. Mol. Sci. 2023, 24, 17601. [Google Scholar] [CrossRef] [PubMed]
- van der Linden, S.; Valkenburg, H.A.; Cats, A. Evaluation of diagnostic criteria for ankylosing spondylitis. A proposal for modification of the New York criteria. Arthritis Rheum. 1984, 27, 361–368. [Google Scholar] [CrossRef]
- Silva, C.F.O.; Obara, K.; Paixao, L.; Santos, E.H.; Santos, A.I.Z.; Cardoso, J.R. Use of posturography in patients with ankylosing spondylitis: A systematic review. S. Afr. J. Physiother. 2024, 80, 1953. [Google Scholar] [CrossRef]
- Lundberg, M.; Eriksson, A.; Tran, B.; Assarsson, E.; Fredriksson, S. Homogeneous antibody-based proximity extension assays provide sensitive and specific detection of low-abundant proteins in human blood. Nucleic Acids Res. 2011, 39, e102. [Google Scholar] [CrossRef]
- Sode, J.; Bank, S.; Vogel, U.; Andersen, P.S.; Sorensen, S.B.; Bojesen, A.B.; Andersen, M.R.; Brandslund, I.; Dessau, R.B.; Hoffmann, H.J.; et al. Genetically determined high activities of the TNF-alpha, IL23/IL17, and NFkB pathways were associated with increased risk of ankylosing spondylitis. BMC Med. Genet. 2018, 19, 165. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.I.; Kim, H.J.; Jo, S.; Shim, S.C.; Kim, T.H.; Won, E.J.; Kim, T.J. IL-6 activates pathologic Th17 cell via STAT 3 phosphorylation in inflammatory joint of Ankylosing Spondylitis. Biochem. Biophys. Res. Commun. 2022, 620, 69–75. [Google Scholar] [CrossRef]
- Schinocca, C.; Rizzo, C.; Fasano, S.; Grasso, G.; La Barbera, L.; Ciccia, F.; Guggino, G. Role of the IL-23/IL-17 Pathway in Rheumatic Diseases: An Overview. Front. Immunol. 2021, 12, 637829. [Google Scholar] [CrossRef]
- Zhao, W.B.; Lin, K.R.; Xu, Q.F. Correlation of serum IL-6, TNF-alpha levels and disease activity in patients with ankylosing spondylitis. Eur. Rev. Med. Pharmacol. Sci. 2024, 28, 80–89. [Google Scholar]
- Hu, Y.; Lou, B.; Jiang, Z.; Yu, C. Predictive Values of Blood Type I and Type II Interferon Production for Disease Activity and Clinical Response to TNF-alpha Blocking Therapy in Patients with Ankylosing Spondylitis. Tohoku J. Exp. Med. 2023, 260, 263–271. [Google Scholar] [CrossRef]
- Leijten, E.; Tao, W.; Pouw, J.; van Kempen, T.; Olde Nordkamp, M.; Balak, D.; Tekstra, J.; Munoz-Elias, E.; DePrimo, S.; Drylewicz, J.; et al. Broad proteomic screen reveals shared serum proteomic signature in patients with psoriatic arthritis and psoriasis without arthritis. Rheumatology 2021, 60, 751–761. [Google Scholar] [PubMed]
- Bitkina, O.V.; Park, J.; Kim, H.K. Application of artificial intelligence in medical technologies: A systematic review of main trends. Digit. Health 2023, 9, 20552076231189331. [Google Scholar] [CrossRef] [PubMed]
- da Silva, R.G.L. The advancement of artificial intelligence in biomedical research and health innovation: Challenges and opportunities in emerging economies. Glob. Health 2024, 20, 44. [Google Scholar]
- Hanna, M.G.; Pantanowitz, L.; Dash, R.; Harrison, J.H.; Deebajah, M.; Pantanowitz, J.; Rashidi, H.H. Future of Artificial Intelligence-Machine Learning Trends in Pathology and Medicine. Mod. Pathol. 2025, 38, 100705. [Google Scholar]
- Loos, T.; Dekeyzer, L.; Struyf, S.; Schutyser, E.; Gijsbers, K.; Gouwy, M.; Fraeyman, A.; Put, W.; Ronsse, I.; Grillet, B.; et al. TLR ligands and cytokines induce CXCR3 ligands in endothelial cells: Enhanced CXCL9 in autoimmune arthritis. Lab. Investig. 2006, 86, 902–916. [Google Scholar] [PubMed]
- Duftner, C.; Dejaco, C.; Kullich, W.; Klauser, A.; Goldberger, C.; Falkenbach, A.; Schirmer, M. Preferential type 1 chemokine receptors and cytokine production of CD28- T cells in ankylosing spondylitis. Ann. Rheum. Dis. 2006, 65, 647–653. [Google Scholar] [CrossRef]
- Morel, L.; Domingues, O.; Zimmer, J.; Michel, T. Revisiting the Role of Neurotrophic Factors in Inflammation. Cells 2020, 9, 865. [Google Scholar] [CrossRef]

| AS Patients (Number = 43) | NC (Number = 10) | |
|---|---|---|
| Basic clinical information | ||
| Age, years, (mean ± SD [range]) | 37.0 ± 11.8 (20~60) | 40.1 ± 9.0 (28~53) |
| Age at onset, years, (mean ± SD [range]) | 22.5 ± 5.3 (15~39) | ––– |
| Disease duration, years, (mean ± SD, range) | 14.5 ± 8.8 (1~35) | ––– |
| Gender (number of Male/Female) | 30/13 | 7/3 |
| BMI, kg/m2, (mean ± SD [range]) | 24.4 ± 3.3 (17.4~32.0) | 23.5 ± 4.4 (18.0~33.9) |
| Drug intervention (number of Yes/No) | 32/11 | ––– |
| Clinical laboratory tests | ||
| HLA-B27+ (number, %) | 43, 100% | ––– |
| HLA-B27 Subtypes | ––– | |
| HLA-B2704 (number, %) | 17, 39.5% | ––– |
| HLA-B2705 (number, %) | 22, 51.2% | ––– |
| Others (number, %) | 4, 9.3% | ––– |
| WBC, 10^9, (mean ± SD [range]) | 6.8 ± 1.9 (3.26~11.52) | 5.9 ± 1.8 (3.03~8.43) |
| CRP, mg/L, (mean ± SD [range]) | 12.8 ± 17.2 (0.50~83.32) ** | 2.3 ± 2.0 (0.20~5.75) |
| ESR, mm/H, (mean ± SD [range]) | 19.4 ± 23.3 (1~102) | 8.4 ± 4.1 (1~14) |
| Index scores | ||
| BASDAI (mean ± SD [range]) | 4.4 ± 1.7 (1~8) | ––– |
| BASFI (mean ± SD [range]) | 3.4 ± 1.1 (1~6) | ––– |
| Assay | OlinkID | UniProt | Disease (Mean) | NC (Mean) | p_Value | |
|---|---|---|---|---|---|---|
| AS (Total) vs. NC | CCL25 | OID00551 | O15444 | 5.52 | 6.17 | 0.0011 |
| IL-6 | OID00482 | P05231 | 2.80 | 1.58 | 0.0097 | |
| CST5 | OID00491 | P28325 | 5.88 | 6.29 | 0.0266 | |
| CCL11 | OID00505 | P51671 | 6.55 | 6.93 | 0.0266 | |
| AS_HLA-B2704 vs. NC | CCL25 | OID00551 | O15444 | 5.44 | 6.17 | 0.0022 |
| IL-6 | OID00482 | P05231 | 2.72 | 1.58 | 0.0129 | |
| CST5 | OID00491 | P28325 | 5.85 | 6.29 | 0.0459 | |
| ST1A1 | OID00557 | P50225 | 7.42 | 8.91 | 0.0175 | |
| SLAMF1 | OID00502 | Q13291 | 1.00 | 0.63 | 0.0459 | |
| IL-17A | OID00485 | Q16552 | 0.98 | 0.43 | 0.0203 | |
| AS_HLA-B2705 vs. NC | CCL25 | OID00551 | O15444 | 5.56 | 6.17 | 0.0040 |
| FGF-19 | OID00545 | O95750 | 5.39 | 6.18 | 0.0385 | |
| CST5 | OID00491 | P28325 | 5.86 | 6.29 | 0.0385 | |
| LIF-R | OID00511 | P42702 | 2.78 | 3.00 | 0.0428 | |
| CCL11 | OID00505 | P51671 | 6.53 | 6.93 | 0.0249 | |
| NRTN | OID00548 | Q99748 | −1.31 | −1.80 | 0.0222 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yan, Y.; Wang, J.; Wang, Y.; Liu, J.; Yang, W.; Niu, M.; Yu, Y.; Zhao, H. Proteomic Profiling of Inflammatory Protein Dysregulation in HLA-B27-Positive Ankylosing Spondylitis: Molecular Signatures and Potential Biomarkers. Biomolecules 2025, 15, 516. https://doi.org/10.3390/biom15040516
Yan Y, Wang J, Wang Y, Liu J, Yang W, Niu M, Yu Y, Zhao H. Proteomic Profiling of Inflammatory Protein Dysregulation in HLA-B27-Positive Ankylosing Spondylitis: Molecular Signatures and Potential Biomarkers. Biomolecules. 2025; 15(4):516. https://doi.org/10.3390/biom15040516
Chicago/Turabian StyleYan, Yuzhu, Jihan Wang, Yangyang Wang, Junye Liu, Wenjuan Yang, Min Niu, Yan Yu, and Heping Zhao. 2025. "Proteomic Profiling of Inflammatory Protein Dysregulation in HLA-B27-Positive Ankylosing Spondylitis: Molecular Signatures and Potential Biomarkers" Biomolecules 15, no. 4: 516. https://doi.org/10.3390/biom15040516
APA StyleYan, Y., Wang, J., Wang, Y., Liu, J., Yang, W., Niu, M., Yu, Y., & Zhao, H. (2025). Proteomic Profiling of Inflammatory Protein Dysregulation in HLA-B27-Positive Ankylosing Spondylitis: Molecular Signatures and Potential Biomarkers. Biomolecules, 15(4), 516. https://doi.org/10.3390/biom15040516







