PPARs Activity Affects the Hatchability Through Lipid Metabolism Regulation in Silkworm, Bombyx mori L.
Abstract
1. Introduction
2. Materials and Methods
2.1. Silkworm Strain and Cell Line
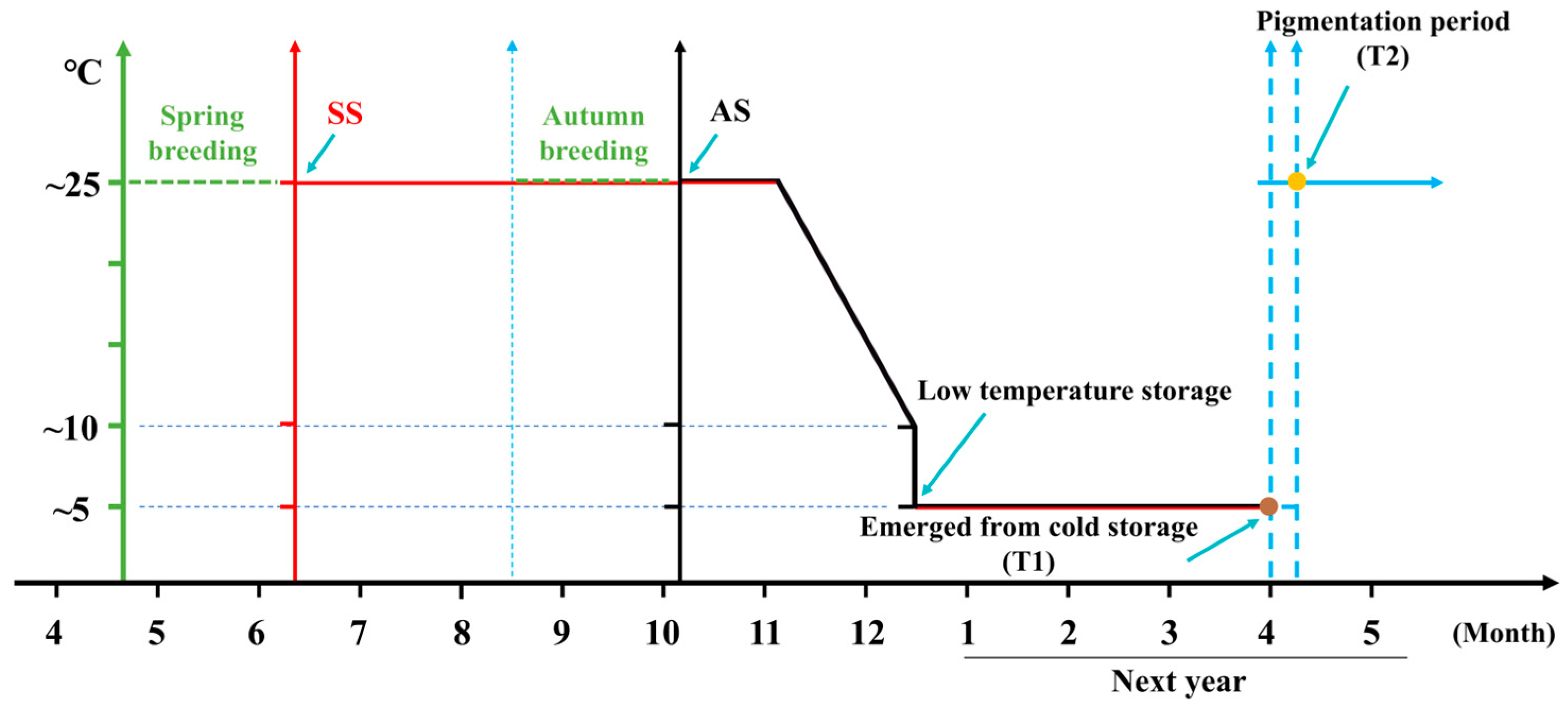
2.2. Hatchability Statistics
2.3. Embryo Observation
2.4. RNA-Seq Analysis
2.5. RNA Isolation, cDNA Synthesis, and qPCR Analysis
2.6. Phylogenetic Analysis
2.7. Determination of Fatty Acids
2.8. Artificial Regulation of PPARs Activity
2.9. mRNA Synthesis and Cell Transfection
2.10. Cell Viability Assay and Oil Red Staining
2.11. Statistical Analysis
3. Results
3.1. Hatching Rate and Embryo Development of QiufengA
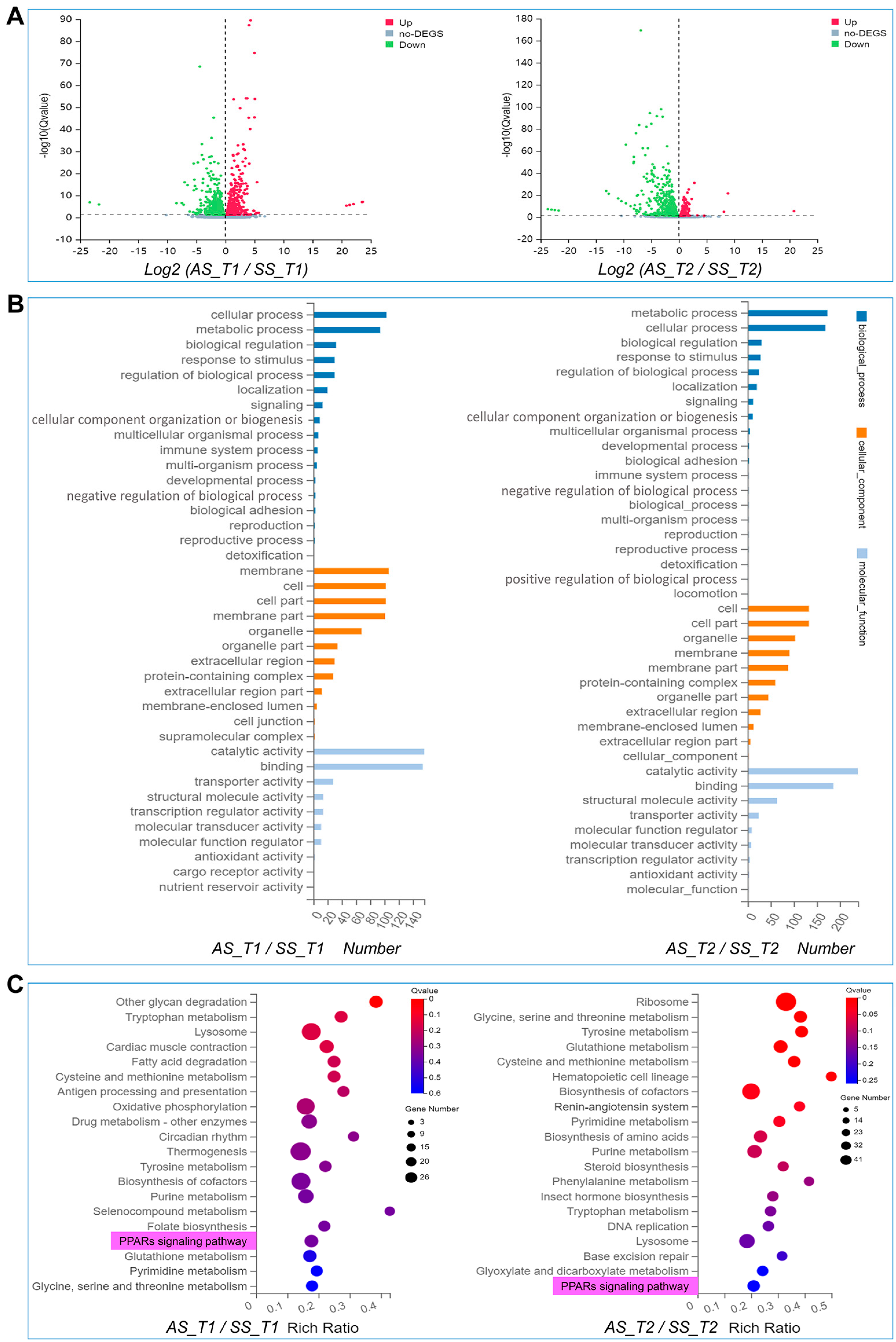
3.2. Differentially Expressed Genes of AS and SS
3.3. Phylogenetic Identification of PPARs Protein
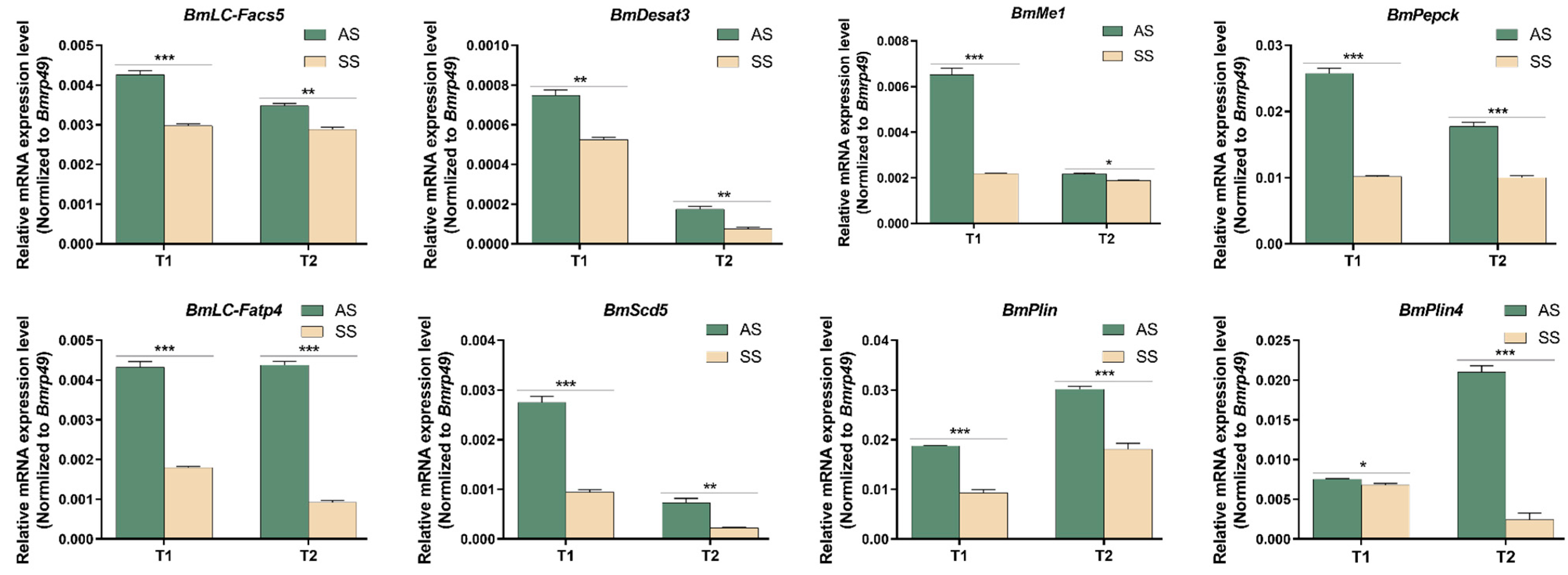
3.4. The Key Gene Expression Pattern of PPARs Pathway
3.5. The Contents of Main Fatty Acids in AS and SS Were Changed
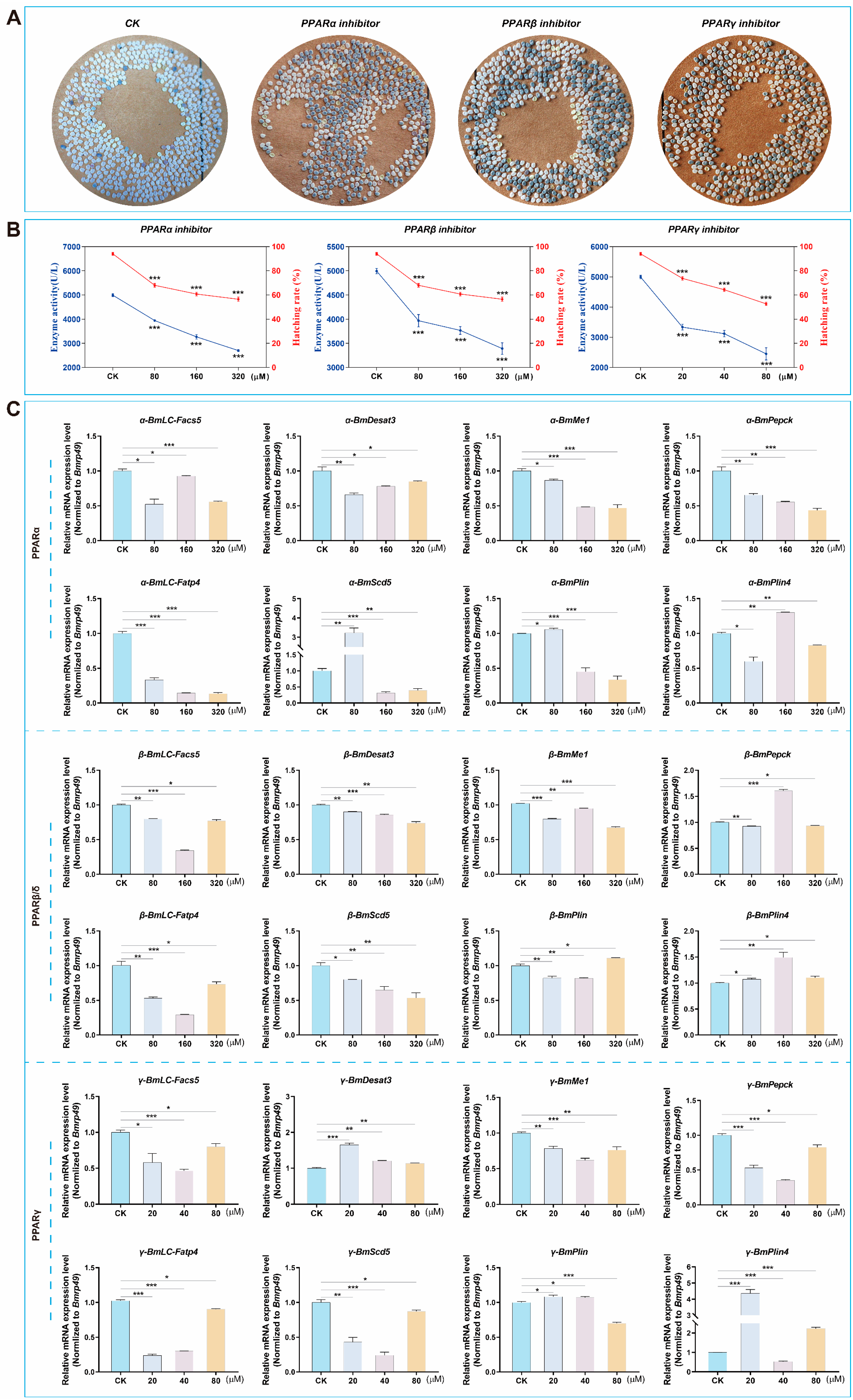
3.6. Decreased PPARs Activity Affected Hatching of QiufengA Strain
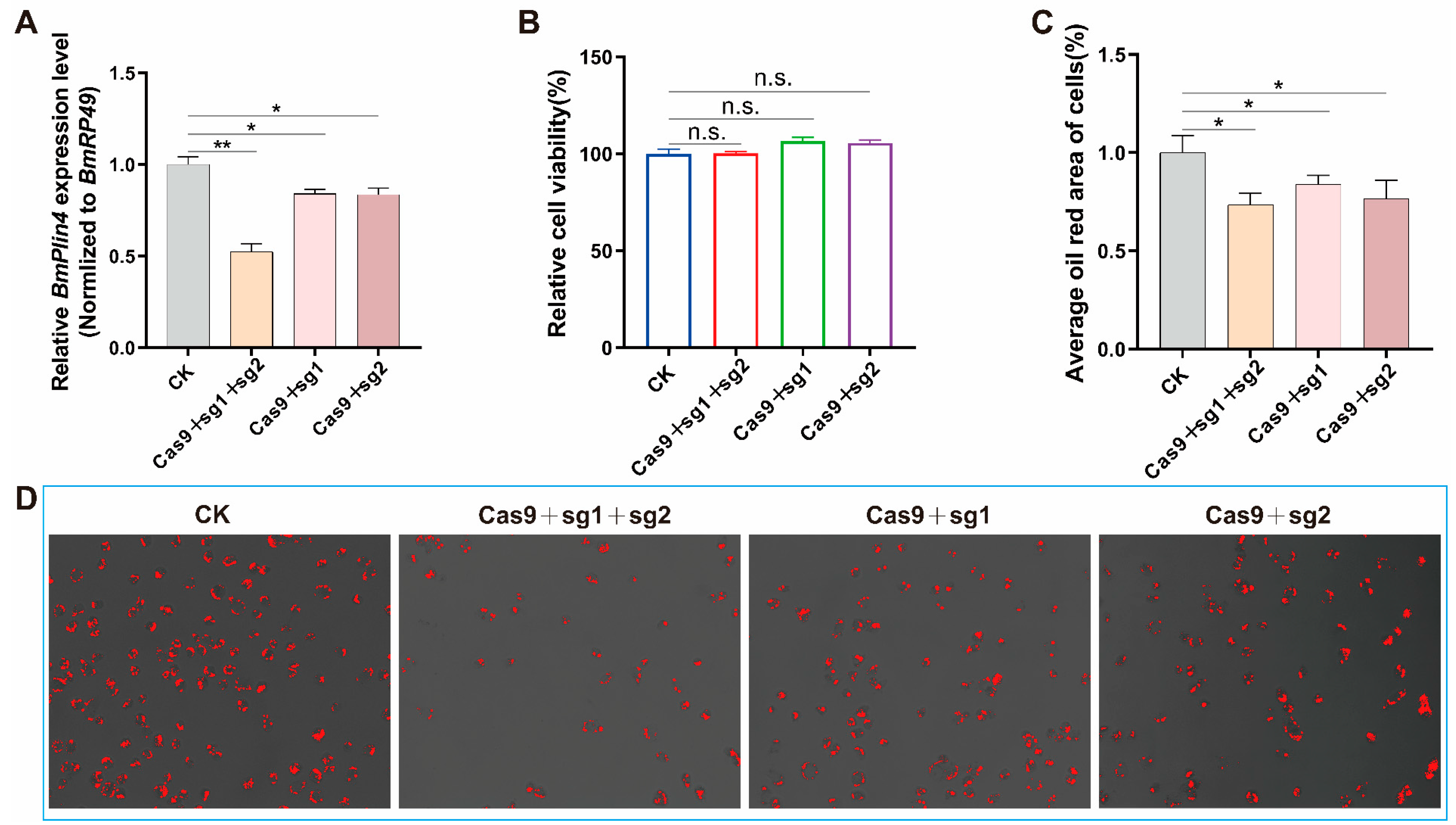
3.7. Knockdown the Key Gene BmPlin4 Affected Lipid Homeostasis in BmN Cells
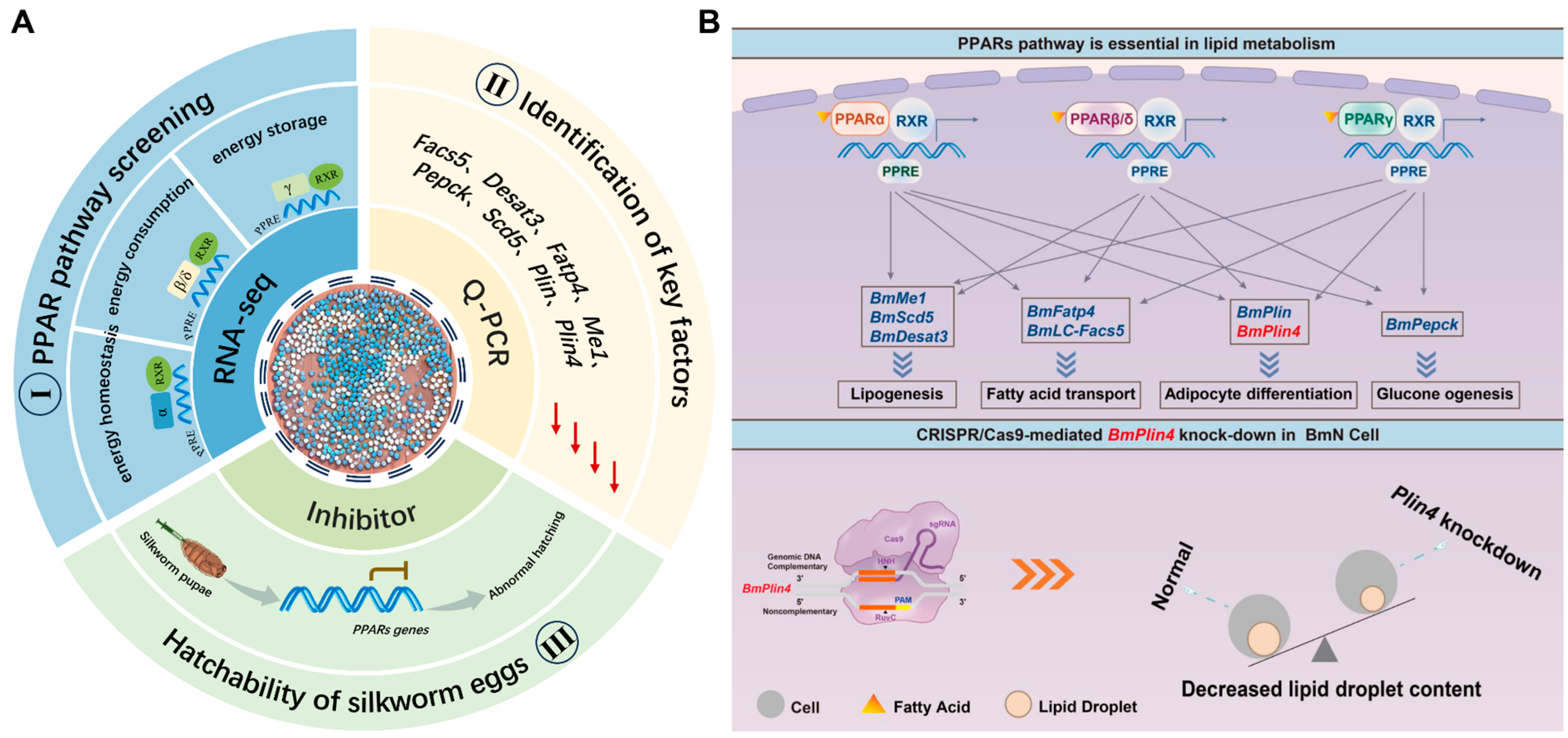
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kawamoto, M.; Jouraku, A.; Toyoda, A.; Yokoi, K.; Minakuchi, Y.; Katsuma, S.; Fujiyama, A.; Kiuchi, T.; Yamamoto, K.; Shimada, T. High-Quality Genome Assembly of the Silkworm, Bombyx mori. Insect Biochem. Mol. Biol. 2019, 107, 53–62. [Google Scholar] [CrossRef] [PubMed]
- Nakao, H. Early Embryonic Development of Bombyx. Dev. Genes Evol. 2021, 231, 95–107. [Google Scholar] [CrossRef]
- He, P.; Wei, E.; Wang, R.; Wang, Q.; Zhang, Y.; Tang, X.; Zhu, F.; Shen, Z. The Spirotetramat Inhibits Growth and Reproduction of Silkworm by Interfering with the Fatty Acid Metabolism. Pestic. Biochem. Physiol. 2022, 188, 105282. [Google Scholar] [CrossRef] [PubMed]
- Tong, X.; Han, M.J.; Lu, K.; Tai, S.; Liang, S.; Liu, Y.; Hu, H.; Shen, J.; Long, A.; Zhan, C.; et al. High-Resolution Silkworm Pan-Genome Provides Genetic Insights into Artificial Selection and Ecological Adaptation. Nat. Commun. 2022, 13, 5619. [Google Scholar] [CrossRef]
- Yamashita, O.; Yaginuma, T. Silkworm Eggs at Low Temperatures: Implications for Sericulture. In Insects at Low Temperature; Lee, R.E., Denlinger, D.L., Eds.; Springer: Boston, MA, USA, 1991; pp. 424–445. ISBN 978-1-4757-0192-0. [Google Scholar]
- Chen, L.; Hou, Y.; Hu, W.; Qiu, X.; Lu, H.; Wei, J.; Yu, S.; He, N.; Zhang, H.; Shen, G. The Molecular Chaperon AKR2A Increases the Mulberry Chilling-Tolerant Capacity by Maintaining SOD Activity and Unsaturated Fatty Acids Composition. Sci. Rep. 2018, 8, 12120. [Google Scholar] [CrossRef]
- Jingade, A.H.; Lekha, G.; Srinivasa, B.G.K.; Manjula, A.; Sivaprasad, V.A. Feasibility Study towards Cryopreservation of Silkworm Eggs: Response of Non-Diapause Silkworm Eggs to Low Temperature. Cryo Lett. 2015, 36, 19–24. [Google Scholar]
- Zhao, C.; Guo, Y.; Liu, Z.; Xia, Y.; Li, Y.; Song, Z.; Zhang, B.; Li, D. Temperature and Photoperiodic Response of Diapause Induction in Anastatus japonicus, an Egg Parasitoid of Stink Bugs. Insects 2021, 12, 872. [Google Scholar] [CrossRef] [PubMed]
- Tang, C.L.; Zhou, H.H.; Zhu, Y.W.; Huang, J.; Wang, G.B. Glutathione S-Transferase Influences the Fecundity of Schistosoma japonicum. Acta Trop. 2019, 191, 8–12. [Google Scholar] [CrossRef]
- Goldsmith, M.R.; Shimada, T.; Abe, H. The Genetics and Genomics of the Silkworm, Bombyx mori. Annu. Rev. Entomol. 2005, 50, 71–100. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Meng, Y.; Wang, Y.X.; Luo, J.; Katsuma, S.; Yang, C.W.; Banno, Y.; Kusakabe, T.; Shimada, T.; Xia, Q.Y. Vitellogenin Receptor Mutation Leads to the Oogenesis Mutant Phenotype “Scanty Vitellin” of the Silkworm, Bombyx mori. J. Biol. Chem. 2013, 288, 13345–13355. [Google Scholar] [CrossRef]
- Kim, T.; Yang, Q. Peroxisome-Proliferator-Activated Receptors Regulate Redox Signaling in the Cardiovascular System. World J. Cardiol. 2013, 5, 164–174. [Google Scholar] [CrossRef]
- Peng, L.; Yang, H.; Ye, Y.; Ma, Z.; Kuhn, C.; Rahmeh, M.; Mahner, S.; Makrigiannakis, A.; Jeschke, U.; von Schönfeldt, V. Role of Peroxisome Proliferator-Activated Receptors (PPARs) in Trophoblast Functions. Int. J. Mol. Sci. 2021, 22, 433. [Google Scholar] [CrossRef]
- Nakken, B.; Varga, T.; Szatmari, I.; Szeles, L.; Gyongyosi, A.; Illarionov, P.A.; Dezso, B.; Gogolak, P.; Rajnavolgyi, E.; Nagy, L. Peroxisome Proliferator-Activated Receptor γ-Regulated Cathepsin D Is Required for Lipid Antigen Presentation by Dendritic Cells. J. Immunol. 2011, 187, 240–247. [Google Scholar] [CrossRef]
- Kersten, S. Integrated Physiology and Systems Biology of PPARα. Mol. Metab. 2014, 3, 354–371. [Google Scholar] [CrossRef] [PubMed]
- Cheng, H.S.; Tan, W.R.; Low, Z.S.; Marvalim, C.; Lee, J.Y.H.; Tan, N.S. Exploration and Development of PPAR Modulators in Health and Disease: An Update of Clinical Evidence. Int. J. Mol. Sci. 2019, 20, 5055. [Google Scholar] [CrossRef] [PubMed]
- Zoete, V.; Grosdidier, A.; Michielin, O. Peroxisome Proliferator-Activated Receptor Structures: Ligand Specificity, Molecular Switch and Interactions with Regulators. Biochim. Biophys. Acta 2007, 1771, 915–925. [Google Scholar] [CrossRef] [PubMed]
- Tyagi, S.; Gupta, P.; Saini, A.S.; Kaushal, C.; Sharma, S. The Peroxisome Proliferator-Activated Receptor: A Family of Nuclear Receptors Role in Various Diseases. J. Adv. Pharm. Technol. Res. 2011, 2, 236–240. [Google Scholar] [CrossRef]
- Finck, J.; Berdan, E.L.; Mayer, F.; Ronacher, B.; Geiselhardt, S. Divergence of Cuticular Hydrocarbons in Two Sympatric Grasshopper Species and the Evolution of Fatty Acid Synthases and Elongases across Insects. Sci. Rep. 2016, 6, 33695. [Google Scholar] [CrossRef]
- Fabjanowska, J.; Kowalczuk-Vasilev, E.; Klebaniuk, R.; Milewski, S.; Gümüş, H. N-3 Polyunsaturated Fatty Acids as a Nutritional Support of the Reproductive and Immune System of Cattle—A Review. Animals 2023, 13, 3589. [Google Scholar] [CrossRef]
- Chen, J.; Liu, H. Nutritional Indices for Assessing Fatty Acids: A Mini-Review. Int. J. Mol. Sci. 2020, 21, 5695. [Google Scholar] [CrossRef]
- Messina, C.M.; Gaglio, R.; Morghese, M.; Tolone, M.; Arena, R.; Moschetti, G.; Santulli, A.; Francesca, N.; Settanni, L. Microbiological Profile and Bioactive Properties of Insect Powders Used in Food and Feed Formulations. Foods 2019, 8, 400. [Google Scholar] [CrossRef] [PubMed]
- Pietrzak-Fiećko, R.; Kamelska-Sadowska, A.M. The Comparison of Nutritional Value of Human Milk with Other Mammals’ Milk. Nutrients 2020, 12, 1404. [Google Scholar] [CrossRef]
- Ma, J.; Sun, S.; Whelan, J.; Shou, H. CRISPR/Cas9-Mediated Knockout of GmFATB1 Significantly Reduced the Amount of Saturated Fatty Acids in Soybean Seeds. Int. J. Mol. Sci. 2021, 22, 3877. [Google Scholar] [CrossRef] [PubMed]
- Tutunchi, H.; Ostadrahimi, A.; Saghafi-Asl, M. The Effects of Diets Enriched in Monounsaturated Oleic Acid on the Management and Prevention of Obesity: A Systematic Review of Human Intervention Studies. Adv. Nutr. 2020, 11, 864–877. [Google Scholar] [CrossRef] [PubMed]
- Shrestha, N.; Vidimce, J.; Holland, O.J.; Cuffe, J.S.M.; Beck, B.R.; Perkins, A.V.; McAinch, A.J.; Hryciw, D.H. Maternal and Postnatal High Linoleic Acid Diet Impacts Lipid Metabolism in Adult Rat Offspring in a Sex-Specific Manner. Int. J. Mol. Sci. 2021, 22, 2946. [Google Scholar] [CrossRef]
- Xu, X.; Bi, H.; Wang, Y.; Li, X.; Xu, J.; Liu, Z.; He, L.; Li, K.; Huang, Y. Disruption of the Ovarian Serine Protease (Osp) Gene Causes Female Sterility in Bombyx mori and Spodoptera litura. Pest. Manag. Sci. 2020, 76, 1245–1255. [Google Scholar] [CrossRef]
- Pan, M.H.; Cai, X.J.; Liu, M.; Lv, J.; Tang, H.; Tan, J.; Lu, C. Establishment and Characterization of an Ovarian Cell Line of the Silkworm, Bombyx mori. Tissue Cell. 2010, 42, 42–46. [Google Scholar] [CrossRef]
- Xu, X.; Wang, Y.H.; Liu, Z.L.; Wang, Y.Q.; He, L.; Li, K.; Huang, Y.P. Disruption of Egg-Specific Protein Causes Female Sterility in Bombyx mori. Insect Sci. 2022, 29, 128–138. [Google Scholar] [CrossRef]
- Teng, X.; Zhang, Z.; He, G.; Yang, L.; Li, F. Validation of Reference Genes for Quantitative Expression Analysis by Real-Time RT-PCR in Four Lepidopteran Insects. J. Insect Sci. 2012, 12, 60. [Google Scholar] [CrossRef]
- Felsenstein, J. Confidence Limits on Phylogenies: An Approach Using the Bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef]
- Saitou, N.; Nei, M. The Neighbor-Joining Method: A New Method for Reconstructing Phylogenetic Trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Li, Z.; Xu, J.; Zeng, B.; Ling, L.; You, L.; Chen, Y.; Huang, Y.; Tan, A. The CRISPR/Cas System Mediates Efficient Genome Engineering in Bombyx mori. Cell Res. 2013, 23, 1414–1416. [Google Scholar] [CrossRef] [PubMed]
- Li, H.L.; Wang, X.Y.; Zheng, X.L.; Lu, W. Research Progress on Oviposition-Related Genes in Insects. J. Insect Sci. 2020, 20, 36. [Google Scholar] [CrossRef]
- Lamichane, S.; Dahal Lamichane, B.; Kwon, S.M. Pivotal Roles of Peroxisome Proliferator-Activated Receptors (PPARs) and Their Signal Cascade for Cellular and Whole-Body Energy Homeostasis. Int. J. Mol. Sci. 2018, 19, 949. [Google Scholar] [CrossRef]
- Fang, M.; Webster, T.F.; Ferguson, P.L.; Stapleton, H.M. Characterizing the Peroxisome Proliferator-Activated Receptor (PPARγ) Ligand Binding Potential of Several Major Flame Retardants, Their Metabolites, and Chemical Mixtures in House Dust. Environ. Health Perspect. 2015, 123, 166–172. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.Q.; Deng, W.; Xiao, Y.; Chen, J.J.; Liu, C.; Wang, J.; Guo, Y.; Duan, M.; Cai, Z.; Xie, S.; et al. The 5-Lipoxygenase Inhibitor Zileuton Protects Pressure Overload-Induced Cardiac Remodeling via Activating PPARα. Oxid. Med. Cell. Longev. 2019, 2019, 7536803. [Google Scholar] [CrossRef]
- Song, L.; Gao, Y.; Li, J.; Ban, L. iTRAQ-Based Comparative Proteomic Analysis Reveals Molecular Mechanisms Underlying Wing Dimorphism of the Pea Aphid, Acyrthosiphon pisum. Front. Physiol. 2018, 9, 1016. [Google Scholar] [CrossRef]
- Han, L.; Shen, W.J.; Bittner, S.; Kraemer, F.B.; Azhar, S. PPARs: Regulators of Metabolism and as Therapeutic Targets in Cardiovascular Disease. Part I: PPAR-α. Future Cardiol. 2017, 13, 259–278. [Google Scholar] [CrossRef] [PubMed]
- Han, L.; Shen, W.J.; Bittner, S.; Kraemer, F.B.; Azhar, S. PPARs: Regulators of Metabolism and as Therapeutic Targets in Cardiovascular Disease. Part II: PPAR-β/δ and PPAR-γ. Future Cardiol. 2017, 13, 279–296. [Google Scholar] [CrossRef]
- Matsumoto, S.; Yoshiga, T.; Yokoyama, N.; Iwanaga, M.; Koshiba, S.; Kigawa, T.; Hirota, H.; Yokoyama, S.; Okano, K.; Mita, K.; et al. Characterization of Acyl-CoA-Binding Protein (ACBP) in the Pheromone Gland of the Silkworm, Bombyx mori. Insect Biochem. Mol. Biol. 2001, 31, 603–609. [Google Scholar] [CrossRef]
- Bowman, T.A.; O’Keeffe, K.R.; D’Aquila, T.; Yan, Q.W.; Griffin, J.D.; Killion, E.A.; Salter, D.M.; Mashek, D.G.; Buhman, K.K.; Greenberg, A.S. Acyl CoA Synthetase 5 (ACSL5) Ablation in Mice Increases Energy Expenditure and Insulin Sensitivity and Delays Fat Absorption. Mol. Metab. 2016, 5, 210–220. [Google Scholar] [CrossRef] [PubMed]
- Yoshiga, T.; Okano, K.; Mita, K.; Shimada, T.; Matsumoto, S. cDNA Cloning of Acyl-CoA Desaturase Homologs in the Silkworm, Bombyx mori. Gene 2000, 246, 339–345. [Google Scholar] [CrossRef] [PubMed]
- Simmen, F.A.; Alhallak, I.; Simmen, R.C.M. Malic Enzyme 1 (ME1) in the Biology of Cancer: It Is Not Just Intermediary Metabolism. J. Mol. Endocrinol. 2020, 65, R77–R90. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.F.; Duan, C.C.; Yang, Z.Q.; Wang, Y.S.; Yue, Z.P.; Guo, B. Malic Enzyme 1 Is Important for Uterine Decidualization in Response to Progesterone/cAMP/PKA/HB-EGF Pathway. FASEB J. 2020, 34, 3820–3837. [Google Scholar] [CrossRef]
- Hanson, R.W.; Patel, Y.M. Phosphoenolpyruvate Carboxykinase (GTP): The Gene and the Enzyme. Adv. Enzymol. Relat. Areas Mol. Biol. 1994, 69, 203–281. [Google Scholar] [CrossRef]
- Hanson, R.W.; Reshef, L. Regulation of Phosphoenolpyruvate Carboxykinase (GTP) Gene Expression. Annu. Rev. Biochem. 1997, 66, 581–611. [Google Scholar] [CrossRef]
- Okamura, T.; Shimizu, H.; Nagao, T.; Ueda, R.; Ishii, S. ATF-2 Regulates Fat Metabolism in Drosophila. Mol. Biol. Cell. 2007, 18, 1519–1529. [Google Scholar] [CrossRef]
- Ong, K.T.; Mashek, M.T.; Davidson, N.O.; Mashek, D.G. Hepatic ATGL Mediates PPAR-α Signaling and Fatty Acid Channeling through an L-FABP Independent Mechanism. J. Lipid Res. 2014, 55, 808–815. [Google Scholar] [CrossRef]
- Sim, C.; Denlinger, D.L. Transcription Profiling and Regulation of Fat Metabolism Genes in Diapausing Adults of the Mosquito Culex pipiens. Physiol. Genom. 2009, 39, 202–209. [Google Scholar] [CrossRef]
- Li, H.; Herrmann, T.; Seeßle, J.; Liebisch, G.; Merle, U.; Stremmel, W.; Chamulitrat, W. Role of Fatty Acid Transport Protein 4 in Metabolic Tissues: Insights into Obesity and Fatty Liver Disease. Biosci. Rep. 2022, 42, BSR20211854. [Google Scholar] [CrossRef]
- Wang, Z.; Zou, L.; Zhang, Y.; Zhu, M.; Zhang, S.; Wu, D.; Lan, J.; Zang, X.; Wang, Q.; Zhang, H.; et al. ACS-20/FATP4 Mediates the Anti-Ageing Effect of Dietary Restriction in C. elegans. Nat. Commun. 2023, 14, 7683. [Google Scholar] [CrossRef]
- Yamamoto, H.; Hattori, M.; Chamulitrat, W.; Ohno, Y.; Kihara, A. Skin Permeability Barrier Formation by the Ichthyosis-Causative Gene FATP4 through Formation of the Barrier Lipid ω-O-Acylceramide. Proc. Natl. Acad. Sci. USA 2020, 117, 2914–2922. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, S.; Stanley, D.; Kim, Y. An Insect Prostaglandin E2 Synthase Acts in Immunity and Reproduction. Front. Physiol. 2018, 9, 1231. [Google Scholar] [CrossRef]
- Xu, G.; Lyu, H.; Yi, Y.; Peng, Y.; Feng, Q.; Song, Q.; Gong, C.; Peng, X.; Palli, S.R.; Zheng, S. Intragenic DNA Methylation Regulates Insect Gene Expression and Reproduction through the MBD/Tip60 Complex. iScience 2021, 24, 102040. [Google Scholar] [CrossRef] [PubMed]
- Teixeira, L.; Rabouille, C.; Rørth, P.; Ephrussi, A.; Vanzo, N.F. Drosophila Perilipin/ADRP Homologue Lsd2 Regulates Lipid Metabolism. Mech. Dev. 2003, 120, 1071–1081. [Google Scholar] [CrossRef]
- Griseti, E.; Bello, A.A.; Bieth, E.; Sabbagh, B.; Iacovoni, J.S.; Bigay, J.; Laurell, H.; Čopič, A. Molecular Mechanisms of Perilipin Protein Function in Lipid Droplet Metabolism. FEBS Lett. 2024, 598, 1170–1198. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Chang, B.; Wu, X.; Li, L.; Sleeman, M.; Chan, L. Inactivation of Plin4 Downregulates Plin5 and Reduces Cardiac Lipid Accumulation in Mice. Am. J. Physiol. Endocrinol. Metab. 2013, 304, E770–E779. [Google Scholar] [CrossRef]
- Zipper, L.; Jassmann, D.; Burgmer, S.; Görlich, B.; Reiff, T. Ecdysone Steroid Hormone Remote Controls Intestinal Stem Cell Fate Decisions via the PPARγ-Homolog Eip75B in Drosophila. eLife 2020, 9, e55795. [Google Scholar] [CrossRef]
- Waite, L.L.; Person, E.C.; Zhou, Y.; Lim, K.H.; Scanlan, T.S.; Taylor, R.N. Placental Peroxisome Proliferator-Activated Receptor-Gamma Is Up-Regulated by Pregnancy Serum. J. Clin. Endocrinol. Metab. 2000, 85, 3808–3814. [Google Scholar] [CrossRef][Green Version]
- Hong, J.-W.; Park, K.W. Further Understanding of Fat Biology: Lessons from a Fat Fly. Exp. Mol. Med. 2010, 42, 12–20. [Google Scholar] [CrossRef]
- Hoedjes, K.M.; Kostic, H.; Flatt, T.; Keller, L. A Single Nucleotide Variant in the PPARγ-Homolog Eip75B Affects Fecundity in Drosophila. Mol. Biol. Evol. 2023, 40, msad018. [Google Scholar] [CrossRef]
- Kimmel, A.R.; Brasaemle, D.L.; McAndrews-Hill, M.; Sztalryd, C.; Londos, C. Adoption of PERILIPIN as a unifying nomenclature for the mammalian PAT-family of intracellular lipid storage droplet proteins. J. Lipid Res. 2010, 51, 468–471. [Google Scholar] [CrossRef] [PubMed]
- Pei, J.; Zou, D.; Li, L.; Kang, L.; Sun, M.; Li, X.; Chen, Q.; Chen, D.; Qu, B.; Gao, X.; et al. Senp7 Deficiency Impairs Lipid Droplets Maturation in White Adipose Tissues via Plin4 deSUMOylation. J. Biol. Chem. 2024, 300, 107319. [Google Scholar] [CrossRef]
- Olzmann, J.A.; Carvalho, P. Dynamics and functions of lipid droplets. Nat. Rev. Mol. Cell Biol. 2019, 20, 37–155. [Google Scholar] [CrossRef]
- Dunning, K.R.; Anastasi, M.R.; Zhang, V.J.; Russell, D.L.; Robker, R.L. Regulation of fatty acid oxidation in mouse cumulus-oocyte complexes during maturation and modulation by PPAR agonists. PLoS ONE 2014, 9, e87327. [Google Scholar] [CrossRef]
- Li, Z.; Thiel, K.; Thul, P.J.; Beller, M.; Kühnlein, R.P.; Welte, M.A. Lipid droplets control the maternal histone supply of Drosophila embryos. Curr. Biol. 2012, 22, 2104–2113. [Google Scholar] [CrossRef]
- Li, T.; Jin, Y.; Wu, J.; Ren, Z. Beyond energy provider: Multifunction of lipid droplets in embryonic development. Biol. Res. 2023, 56, 38. [Google Scholar] [CrossRef]
- Aizawa, R.; Ibayashi, M.; Tatsumi, T.; Yamamoto, A.; Kokubo, T.; Miyasaka, N.; Sato, K.; Ikeda, S.; Minami, N.; Tsukamoto, S. Synthesis and Maintenance of Lipid Droplets Are Essential for Mouse Preimplantation Embryonic Development. Development 2019, 146, dev181925. [Google Scholar] [CrossRef]
- de la Rosa Rodriguez, M.A.; Kersten, S. Regulation of Lipid Droplet-Associated Proteins by Peroxisome Proliferator-Activated Receptors. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2017, 1862, 1212–1220. [Google Scholar] [CrossRef]
- Echeverría, F.; Ortiz, M.; Valenzuela, R.; Videla, L.A. Long-Chain Polyunsaturated Fatty Acids Regulation of PPARs, Signaling: Relationship to Tissue Development and Aging. Prostaglandins Leukot. Essent. Fatty Acids 2016, 114, 28–34. [Google Scholar] [CrossRef]
- Leroy, J.L.M.R.; Vanholder, T.; Mateusen, B.; Christophe, A.; Opsomer, G.; de Kruif, A.; Genicot, G.; Van Soom, A. Non-Esterified Fatty Acids in Follicular Fluid of Dairy Cows and Their Effect on Developmental Capacity of Bovine Oocytes In Vitro. Reproduction 2005, 130, 485–495. [Google Scholar] [CrossRef]
- Prades, J.; Funari, S.S.; Escribá, P.V.; Barceló, F. Effects of Unsaturated Fatty Acids and Triacylglycerols on Phosphatidylethanolamine Membrane Structure. J. Lipid Res. 2003, 44, 1720–1727. [Google Scholar] [CrossRef]
- Bashan, M.; Cakmak, O. Changes in Composition of Phospholipid and Triacylglycerol Fatty Acids Prepared from Prediapausing and Diapausing Individuals of Dolycoris baccarum and Piezodorus lituratus (Heteroptera: Pentatomidae). Ann. Entomol. Soc. Am. 2005, 98, 575–579. [Google Scholar] [CrossRef]
- Khani, A.; Moharramipour, S.; Barzegar, M. Cold Tolerance and Trehalose Accumulation in Overwintering Larvae of the Codling Moth, Cydia pomonella (Lepidoptera: Tortricidae). Eur. J. Entomol. 2007, 104, 385–392. [Google Scholar] [CrossRef]
- Ohtsu, T.; Katagiri, C.; Kimura, M.T.; Hori, S.H. Cold Adaptations in Drosophila. Qualitative Changes of Triacylglycerols with Relation to Overwintering. J. Biol. Chem. 1993, 268, 1830–1834. [Google Scholar] [CrossRef] [PubMed]
- Koštál, V.; Berková, P.; Šimek, P. Remodelling of Membrane Phospholipids during Transition to Diapause and Cold-Acclimation in the Larvae of Chymomyza costata (Drosophilidae). Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 2003, 135, 407–419. [Google Scholar] [CrossRef]
- V, K. Cell Structural Modifications in Insects at Low Temperature. Low Temp. Biol. Insects 2010, 116–140. [Google Scholar]
- Rozsypal, J.; Koštál, V.; Berková, P.; Zahradníčková, H.; Šimek, P. Seasonal Changes in the Composition of Storage and Membrane Lipids in Overwintering Larvae of the Codling Moth, Cydia pomonella. J. Therm. Biol. 2014, 45, 124–133. [Google Scholar] [CrossRef]
- Walkowiak-Nowicka, K.; Chowański, S.; Pacholska-Bogalska, J.; Adamski, Z.; Kuczer, M.; Rosiński, G. Effects of Alloferon and Its Analogues on Reproduction and Development of the Tenebrio Molitor Beetle. Sci. Rep. 2024, 14, 17016. [Google Scholar] [CrossRef]
- Atella, G.C.; Shahabuddin, M. Differential Partitioning of Maternal Fatty Acid and Phospholipid in Neonate Mosquito Larvae. J. Exp. Biol. 2002, 205, 3623–3630. [Google Scholar] [CrossRef]



| Primer Name | Primer Sequence (5′-3′) |
|---|---|
| qRT-PCR | |
| BmLC-Facs5-F | CGTTGTAGCTTCGATGTGCA |
| Bm LC-Facs5-R | CCAGCCTCCTTATATCGCCA |
| BmDesat3-F | CACAGACTTTGGGCACACAA |
| BmDesat3-R | GGCTTTGATTTCGGGATGCT |
| BmMe1-F | ACTGCACTTTCAACGACGAC |
| BmMe1-R | TTCTTGACGATGAGGCCCTT |
| BmPepck-F | AATCTCGCAATGATGACGCC |
| BmPepck-R | TTTTGAACACCGTCGCCATT |
| BmLC-Fatp4-F | TGTGATCAGGCCCAAGTTCT |
| BmLC-Fatp4-R | CCGGTCCACTATTTGTTGCC |
| BmScd5-F | TGAGTGACCTTCAAGCGGAT |
| BmScd5-R | CGCACTATTGACCAGCCAAG |
| BmPlin-F | TTGGACCACTATCTACCGCC |
| BmPlin-R | ACCCGAGTTTCTGAGTTCGT |
| BmPlin4-F | CGAAGACGCCAAAGAGAAGG |
| BmPlin4-R | TACTTGTCGGCGTCCTTGAT |
| BmRP49-F | TCAATCGGATCGCTATGACA |
| BmRP49-R | ATGACGGGTCTTCTTGTTGG |
| Plasmid construction | |
| sgRNA-F1 | TAATACGACTCACTATAGGCAGCCTTCTTGGTATTGTGTTTTAGAGCTAGAAATAGCAA |
| sgRNA-F2 | TAATACGACTCACTATAGGGGCGGCTACTGTTGAGAAGTTTTAGAGCTAGAAATAGCAA |
| sgRNA-R | AGCACCGACTCGGTGCCACTTTTTCAAGTTGATAACGGACTAGCCTTATTTTAACTTGCTATTTCTAGCT |
| Species Name | Sequence Number |
|---|---|
| Bombyx mori | XP_012550651.2, XP_012545279.1, XP_012549547.1, XP_037873833.1, XP_004929241.1, NP_001296524.1, XP_012545064.1, XP_021204279.2, |
| Trichoplusia ni | XP_026724857.1, XP_026732544.1, XP_026725998.1, XP_02672833.1, XP_02673812.1, XP_026744351, XP_026727208.1, XP_026739507.1, |
| Bicyclus anynana | XP_023938956.1, XP_023935398.1, XP_023943582.1, XP_023942610.2, XP_023953014.1, XP_052739993.1, XP_023948300.1, XP_052741276.1, |
| Helicoverpa armigera | XP_021190129.1, XP_02118361.3, XP_063893783.1, XP_063897571.1, XP_049706614.2, XP_021195974.3, XP_021195577.3, XP_063892118.1, |
| Papilio machaon | XP_014360141.1, XP_014362552.2, XP_045535389.1, XP_0455405334.1, XP_014362762.2, KPJ14531.1, XP_045535146.1, XP_045536721.1, |
| Spodoptera litura | XP_022822534.1, XP_022814607.1, XP_022825750.1, XP_022818283.1, XP_0228204601, XP_022825758.1, XP_022826143.1, XP_022834648.1, |
| Diatraea saccharalis | CAH0755737.1, CAG9785496.1, CAG9787336.1, CAG9792994.1, CAG9784543.1, CAG9787313.1, CAG9787424.1, CAH0751628.1, |
| Chilo suppressalis | RVE52369.1, CAH0407053.1, CAH0399100.1, RVE51794.1, RVE51223.1, RVE48477.1, CAH2981644.1, CAH0398210.1, |
| Drosophila melanogaster | NP_649067.2, NP_652731.1, NP_5248802, NP_611341.1, Np_001260354.1, CAB69054.1, NP_001036276.1, NP_001286253.1, |
| Anopheles maculipalpis | XP_050080276.1, XP_050070393.1, XP_05006733.1, XP_050069062.1, XP_050068915.1, XP_050069472.1, XP_050071398.1, XP_050067548.1, |
| Drosophila busckii | XP_017842548.1, ALC47890.1, ALC46605.1, XP_017838813.1, XP_017835074.2, XP_017846257.1, XP_017853279.1, ALC40480.1, |
| Mus musculus | NP_082252.1, AAA40103.1, NP_852072.2, EDL36290.1, NP_03611.1, 4YMK_A, BAC27409.1, NP_001397092.1, |
| Cricetulus griseus | XP_007650740.1, XP_027263037.1, XP_027263401.1, XP_003502116.1, XP_035292398.1, XP_003508493.1, XP_035297263.1, XP_027272679.1, |
| Microtus ochrogaster | XP_005352541.1, KAH0504555.1, KAH0500186.1, KAH0509637.1, XP_005346222.1, XP_005352393.1, KAH0505204.1, XP_005357949.2, |
| Sus scrofa | XP_005671765.1, NP_001107750.1, NP_001231187.1, NP_001155225.1, XP_013849357.2, NP_998946.1, AEZ36149.1, XP_020939716.1, |
| Bubalus bubalis | AIU41596.1, XP_025145597.1, XP_025141974.3, XP_025151907.1, Xp_006065433.2, NP_001277844.1, XP_044789244.1, XP_006067173.3, |
| Bos mutus | XP_005906616.1, XP_005892117.1, MXQ91848.1, XP_005904960.1, XP_005908530.1, XP_005892117.1, MXQ82814.1, MXQ83873.1), |
| Homo sapiens | KAI4077466.1, NP_001032671.2, AAC50613.1, NP_004554.3, NP_005085.2, CAA73998.1, KAI2575794.1, NP_001354797.1, |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, X.; Cui, C.; Du, X.; Chen, J.; He, X.; Zhu, L.; Hu, C.; Xu, F.; Ma, C.; Yu, S.; et al. PPARs Activity Affects the Hatchability Through Lipid Metabolism Regulation in Silkworm, Bombyx mori L. Biomolecules 2025, 15, 492. https://doi.org/10.3390/biom15040492
Xu X, Cui C, Du X, Chen J, He X, Zhu L, Hu C, Xu F, Ma C, Yu S, et al. PPARs Activity Affects the Hatchability Through Lipid Metabolism Regulation in Silkworm, Bombyx mori L. Biomolecules. 2025; 15(4):492. https://doi.org/10.3390/biom15040492
Chicago/Turabian StyleXu, Xia, Chunguang Cui, Xin Du, Jine Chen, Xiuling He, Linbao Zhu, Chengjie Hu, Fang Xu, Chenkai Ma, Shaofang Yu, and et al. 2025. "PPARs Activity Affects the Hatchability Through Lipid Metabolism Regulation in Silkworm, Bombyx mori L." Biomolecules 15, no. 4: 492. https://doi.org/10.3390/biom15040492
APA StyleXu, X., Cui, C., Du, X., Chen, J., He, X., Zhu, L., Hu, C., Xu, F., Ma, C., Yu, S., He, X., Song, H., & Wang, Y. (2025). PPARs Activity Affects the Hatchability Through Lipid Metabolism Regulation in Silkworm, Bombyx mori L. Biomolecules, 15(4), 492. https://doi.org/10.3390/biom15040492






