A Potential Role of EFR3A in Human Disease States
Abstract
1. Introduction
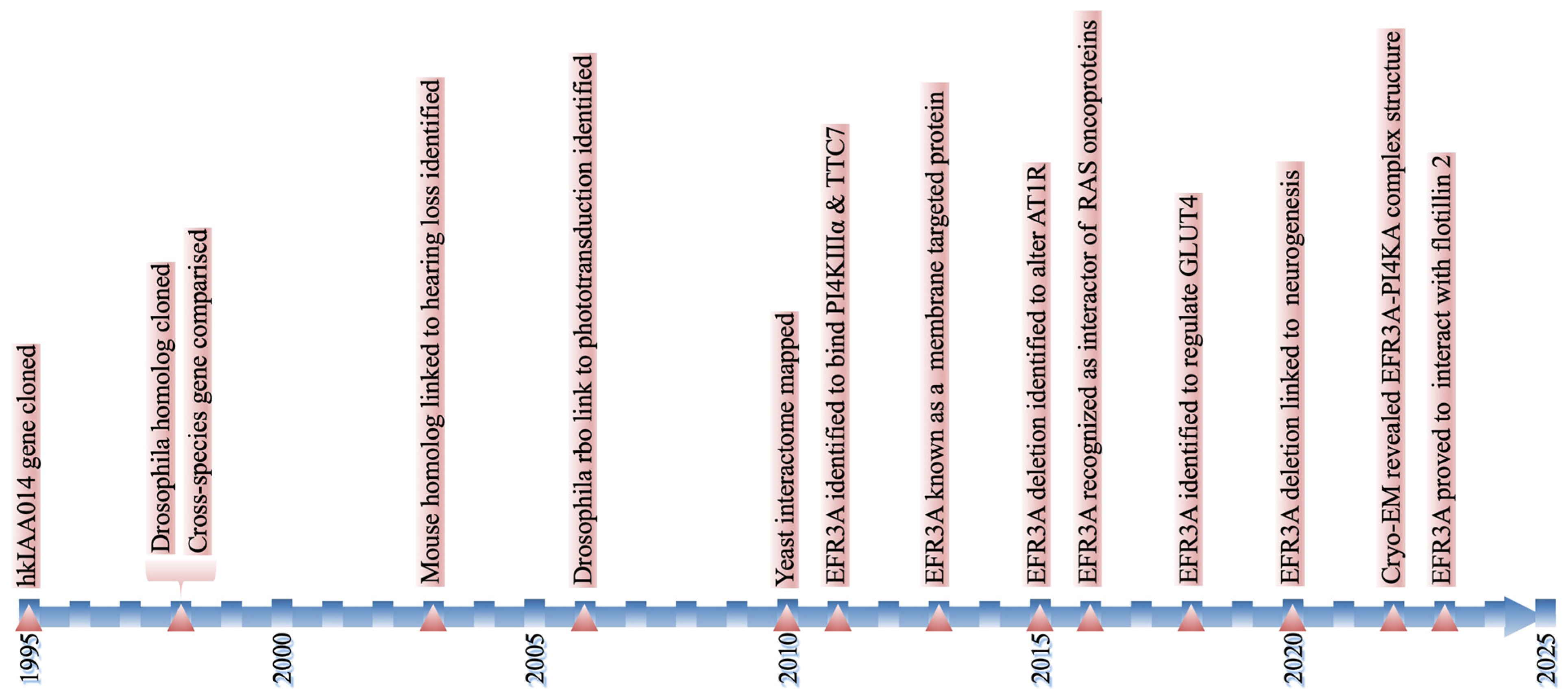
2. EFR3A—History in a Nutshell
3. Mutations and Changes in EFR3A Expression Levels Are Linked to Human Pathologies
3.1. Neurological Disorders
3.2. Cardiovascular Diseases
3.3. Colorectal Cancer
3.4. Pancreatic Ductal Adenocarcinoma
3.5. Nasopharyngeal Cancer
3.6. Brain Tumors
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| ACS | acute coronary syndrome |
| ASD | autism spectrum disorder |
| AT1R | angiotensin II receptor type 1 |
| BDNF | brain-derived neurotrophic factor |
| CAD | coronary artery disease |
| CRC | colorectal cancer |
| EFR3A | eighty five requiring 3 |
| ET | essential tremor |
| HCC | hypomyelination and congenital cataract |
| HDX-MS | hydrogen deuterium exchange mass spectrometry |
| MACE | major adverse cardiovascular events |
| NPC | nasopharyngeal carcinoma |
| PDAC | pancreatic ductal adenocarcinoma |
| PI(4,5)P2 | phosphatidyl inositol 4,5 bisphosphate |
| PI4KA | phosphatidylinositol 4-kinase alpha |
| PLC | phospholipase C |
| PM | plasma membrane |
| SNP | single nucleotide polymorphism |
| TRP | transient receptor potential |
| TTC7 | tetratricopeptide repeat domain 7 |
References
- Dornan, G.L.; Dalwadi, U.; Hamelin, D.J.; Hoffmann, R.M.; Yip, C.K.; Burke, J.E. Probing the Architecture, Dynamics, and Inhibition of the PI4KIIIalpha/TTC7/FAM126 Complex. J. Mol. Biol. 2018, 430 Pt B, 3129–3142. [Google Scholar] [CrossRef]
- Lees, J.A.; Zhang, Y.; Oh, M.S.; Schauder, C.M.; Yu, X.; Baskin, J.M.; Dobbs, K.; Notarangelo, L.D.; De Camilli, P.; Walz, T.; et al. Architecture of the human PI4KIIIalpha lipid kinase complex. Proc. Natl. Acad. Sci. USA 2017, 114, 13720–13725. [Google Scholar] [CrossRef]
- Suresh, S.; Shaw, A.L.; Pemberton, J.G.; Scott, M.K.; Harris, N.J.; Parson, M.A.H.; Jenkins, M.L.; Rohilla, P.; Alvarez-Prats, A.; Balla, T.; et al. Molecular basis for plasma membrane recruitment of PI4KA by EFR3. Sci. Adv. 2024, 10, eadp6660. [Google Scholar] [CrossRef] [PubMed]
- Trybus, M.; Hryniewicz-Jankowska, A.; Czogalla, A.; Sikorski, A.F. EFR3A, an intriguing gene, and protein with a scaffolding function. Cells 2025, 14, 445. [Google Scholar] [CrossRef]
- Baba, T.; Balla, T. Emerging roles of phosphatidylinositol 4-phosphate and phosphatidylinositol 4,5-bisphosphate as regulators of multiple steps in autophagy. J. Biochem. 2020, 168, 329–336. [Google Scholar] [CrossRef] [PubMed]
- Cockcroft, S. Expanding functions of the phosphatidylinositol/phosphatidate lipid transporter, PITPNC1 in physiology and in pathology. Adv. Biol. Regul. 2025, 95, 101056. [Google Scholar] [CrossRef]
- Li, F.L.; Fu, V.; Liu, G.; Tang, T.; Konradi, A.W.; Peng, X.; Kemper, E.; Cravatt, B.F.; Franklin, J.M.; Wu, Z.; et al. Hippo pathway regulation by phosphatidylinositol transfer protein and phosphoinositides. Nat. Chem. Biol. 2022, 18, 1076–1086. [Google Scholar] [CrossRef]
- Kagan, J.C.; Medzhitov, R. Phosphoinositide-mediated adaptor recruitment controls Toll-like receptor signaling. Cell 2006, 125, 943–955. [Google Scholar] [CrossRef]
- Ostuni, R.; Zanoni, I.; Granucci, F. Deciphering the complexity of Toll-like receptor signaling. Cell Mol. Life Sci. 2010, 67, 4109–4134. [Google Scholar] [CrossRef]
- Cockcroft, S. The expanding roles of PI4P and PI(4,5)P(2) at the plasma membrane: Role of phosphatidylinositol transfer proteins. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2024, 1869, 159394. [Google Scholar] [CrossRef]
- Ubeysinghe, S.; Wijayaratna, D.; Kankanamge, D.; Karunarathne, A. Molecular regulation of PLCbeta signaling. Methods Enzymol. 2023, 682, 17–52. [Google Scholar] [CrossRef] [PubMed]
- Barlow-Busch, I.; Shaw, A.L.; Burke, J.E. PI4KA and PIKfyve: Essential phosphoinositide signaling enzymes involved in myriad human diseases. Curr. Opin. Cell Biol. 2023, 83, 102207. [Google Scholar] [CrossRef] [PubMed]
- Suresh, S.; Burke, J.E. Structural basis for the conserved roles of PI4KA and its regulatory partners and their misregulation in disease. Adv. Biol. Regul. 2023, 90, 100996. [Google Scholar] [CrossRef]
- Krieger, E.; Vriend, G. YASARA View—molecular graphics for all devices—from smartphones to workstations. Bioinformatics 2014, 30, 2981–2982. [Google Scholar] [CrossRef]
- Nagase, T.; Seki, N.; Tanaka, A.; Ishikawa, K.; Nomura, N. Prediction of the coding sequences of unidentified human genes. IV. The coding sequences of 40 new genes (KIAA0121-KIAA0160) deduced by analysis of cDNA clones from human cell line KG-1. DNA Res. 1995, 2, 167–174. [Google Scholar] [CrossRef]
- Fagerberg, L.; Hallstrom, B.M.; Oksvold, P.; Kampf, C.; Djureinovic, D.; Odeberg, J.; Habuka, M.; Tahmasebpoor, S.; Danielsson, A.; Edlund, K.; et al. Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Mol. Cell Proteom. 2014, 13, 397–406. [Google Scholar] [CrossRef]
- Faulkner, D.L.; Dockendorff, T.C.; Jongens, T.A. Clonal analysis of cmp44E, which encodes a conserved putative transmembrane protein, indicates a requirement for cell viability in Drosophila. Dev. Genet. 1998, 23, 264–274. [Google Scholar] [CrossRef]
- Huang, F.D.; Matthies, H.J.; Speese, S.D.; Smith, M.A.; Broadie, K. Rolling blackout, a newly identified PIP2-DAG pathway lipase required for Drosophila phototransduction. Nat. Neurosci. 2004, 7, 1070–1078. [Google Scholar] [CrossRef] [PubMed]
- Huang, F.D.; Woodruff, E.; Mohrmann, R.; Broadie, K. Rolling blackout is required for synaptic vesicle exocytosis. J. Neurosci. 2006, 26, 2369–2379. [Google Scholar] [CrossRef]
- RCSB Protein Data Bank. Available online: https://www.rcsb.org/ (accessed on 1 December 2024).
- Nagase, T.; Ishikawa, K.; Suyama, M.; Kikuno, R.; Hirosawa, M.; Miyajima, N.; Tanaka, A.; Kotani, H.; Nomura, N.; Ohara, O. Prediction of the coding sequences of unidentified human genes. XIII. The complete sequences of 100 new cDNA clones from brain which code for large proteins in vitro. DNA Res. 1999, 6, 63–70. [Google Scholar] [CrossRef]
- Bojjireddy, N.; Guzman-Hernandez, M.L.; Reinhard, N.R.; Jovic, M.; Balla, T. EFR3s are palmitoylated plasma membrane proteins that control responsiveness to G-protein-coupled receptors. J. Cell Sci. 2015, 128, 118–128. [Google Scholar] [CrossRef]
- Munemoto, Y.; Houtani, T.; Kase, M.; Sakuma, S.; Baba, K.; Yamashita, T.; Sugimoto, T. Mouse homolog of KIAA0143 protein: Hearing deficit induces specific changes of expression in auditory brainstem neurons. Brain Res. Mol. Brain Res. 2004, 128, 131–140. [Google Scholar] [CrossRef]
- Vijayakrishnan, N.; Phillips, S.E.; Broadie, K. Drosophila rolling blackout displays lipase domain-dependent and -independent endocytic functions downstream of dynamin. Traffic 2010, 11, 1567–1578. [Google Scholar] [CrossRef] [PubMed]
- Vijayakrishnan, N.; Woodruff, E.A., 3rd; Broadie, K. Rolling blackout is required for bulk endocytosis in non-neuronal cells and neuronal synapses. J. Cell Sci. 2009, 122 Pt 1, 114–125. [Google Scholar] [CrossRef]
- Shyngle, J.; Sharma, R.P. Studies on paralysis and development of second chromosome temperature sensitive paralytic mutants of Drosophila melanogaster. Indian. J. Exp. Biol. 1985, 23, 235–240. [Google Scholar] [PubMed]
- Tarassov, K.; Messier, V.; Landry, C.R.; Radinovic, S.; Serna Molina, M.M.; Shames, I.; Malitskaya, Y.; Vogel, J.; Bussey, H.; Michnick, S.W. An in vivo map of the yeast protein interactome. Science 2008, 320, 1465–1470. [Google Scholar] [CrossRef]
- Baird, D.; Stefan, C.; Audhya, A.; Weys, S.; Emr, S.D. Assembly of the PtdIns 4-kinase Stt4 complex at the plasma membrane requires Ypp1 and Efr3. J. Cell Biol. 2008, 183, 1061–1074. [Google Scholar] [CrossRef] [PubMed]
- Zara, F.; Biancheri, R.; Bruno, C.; Bordo, L.; Assereto, S.; Gazzerro, E.; Sotgia, F.; Wang, X.B.; Gianotti, S.; Stringara, S.; et al. Deficiency of hyccin, a newly identified membrane protein, causes hypomyelination and congenital cataract. Nat. Genet. 2006, 38, 1111–1113. [Google Scholar] [CrossRef]
- Baskin, J.M.; Wu, X.; Christiano, R.; Oh, M.S.; Schauder, C.M.; Gazzerro, E.; Messa, M.; Baldassari, S.; Assereto, S.; Biancheri, R.; et al. The leukodystrophy protein FAM126A (hyccin) regulates PtdIns(4)P synthesis at the plasma membrane. Nat. Cell Biol. 2016, 18, 132–138. [Google Scholar] [CrossRef]
- Zhu, L.; Plow, E.F.; Qin, J. Initiation of focal adhesion assembly by talin and kindlin: A dynamic view. Protein Sci. 2021, 30, 531–542. [Google Scholar] [CrossRef]
- Dong, J.M.; Tay, F.P.; Swa, H.L.; Gunaratne, J.; Leung, T.; Burke, B.; Manser, E. Proximity biotinylation provides insight into the molecular composition of focal adhesions at the nanometer scale. Sci. Signal 2016, 9, rs4. [Google Scholar] [CrossRef]
- Adhikari, H.; Kattan, W.E.; Kumar, S.; Zhou, P.; Hancock, J.F.; Counter, C.M. Oncogenic KRAS is dependent upon an EFR3A-PI4KA signaling axis for potent tumorigenic activity. Nat. Commun. 2021, 12, 5248. [Google Scholar] [CrossRef]
- Koester, A.M.; Geiser, A.; Laidlaw, K.M.E.; Morris, S.; Cutiongco, M.F.A.; Stirrat, L.; Gadegaard, N.; Boles, E.; Black, H.L.; Bryant, N.J.; et al. EFR3 and phosphatidylinositol 4-kinase IIIalpha regulate insulin-stimulated glucose transport and GLUT4 dispersal in 3T3-L1 adipocytes. Biosci. Rep. 2022, 42, BSR20221181. [Google Scholar] [CrossRef] [PubMed]
- Koester, A.M.; Geiser, A.; Bowman, P.R.T.; van de Linde, S.; Gadegaard, N.; Bryant, N.J.; Gould, G.W. GLUT4 translocation and dispersal operate in multiple cell types and are negatively correlated with cell size in adipocytes. Sci. Rep. 2022, 12, 20535. [Google Scholar] [CrossRef] [PubMed]
- Qian, Q.; Liu, Q.; Zhou, D.; Pan, H.; Liu, Z.; He, F.; Ji, S.; Wang, D.; Bao, W.; Liu, X.; et al. Brain-specific ablation of Efr3a promotes adult hippocampal neurogenesis via the brain-derived neurotrophic factor pathway. FASEB J. 2017, 31, 2104–2113. [Google Scholar] [CrossRef] [PubMed]
- Trybus, M.; Hryniewicz-Jankowska, A.; Wojtowicz, K.; Trombik, T.; Czogalla, A.; Sikorski, A.F. EFR3A: A new raft domain organizing protein? Cell Mol. Biol. Lett. 2023, 28, 86. [Google Scholar] [CrossRef]
- Jiao, D.; Xu, Y.; Tian, F.; Zhou, Y.; Chen, D.; Wang, Y. Establishment of animal models and behavioral studies for autism spectrum disorders. J. Int. Med. Res. 2024, 52, 3000605241245293. [Google Scholar] [CrossRef]
- Gupta, A.R.; Pirruccello, M.; Cheng, F.; Kang, H.J.; Fernandez, T.V.; Baskin, J.M.; Choi, M.; Liu, L.; Ercan-Sencicek, A.G.; Murdoch, J.D.; et al. Rare deleterious mutations of the gene EFR3A in autism spectrum disorders. Mol. Autism. 2014, 5, 31. [Google Scholar] [CrossRef]
- Voineagu, I.; Wang, X.; Johnston, P.; Lowe, J.K.; Tian, Y.; Horvath, S.; Mill, J.; Cantor, R.M.; Blencowe, B.J.; Geschwind, D.H. Transcriptomic analysis of autistic brain reveals convergent molecular pathology. Nature 2011, 474, 380–384. [Google Scholar] [CrossRef]
- Zhao, K.; Bai, X.; Wang, X.; Cao, Y.; Zhang, L.; Li, W.; Wang, S. Insight on the hub gene associated signatures and potential therapeutic agents in epilepsy and glioma. Brain Res. Bull. 2023, 199, 110666. [Google Scholar] [CrossRef]
- Gao, Y.; Ding, L.; Liu, J.; Wang, X.; Meng, Q. Exploring the diagnostic markers of essential tremor: A study based on machine learning algorithms. Open Life Sci. 2023, 18, 20220622. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Wei, M.; Wu, Y.; Qin, H.; Li, W.; Ma, X.; Cheng, J.; Ren, J.; Shen, Y.; Chen, Z.; et al. Amyloid beta oligomers suppress excitatory transmitter release via presynaptic depletion of phosphatidylinositol-4,5-bisphosphate. Nat. Commun. 2019, 10, 1193. [Google Scholar] [CrossRef] [PubMed]
- Wei, X.; Wang, J.; Yang, E.; Zhang, Y.; Qian, Q.; Li, X.; Huang, F.; Sun, B. Efr3b is essential for social recognition by modulating the excitability of CA2 pyramidal neurons. Proc. Natl. Acad. Sci. USA 2024, 121, e2314557121. [Google Scholar] [CrossRef] [PubMed]
- Frak, W.; Wojtasinska, A.; Lisinska, W.; Mlynarska, E.; Franczyk, B.; Rysz, J. Pathophysiology of Cardiovascular Diseases: New Insights into Molecular Mechanisms of Atherosclerosis, Arterial Hypertension, and Coronary Artery Disease. Biomedicines 2022, 10, 1938. [Google Scholar] [CrossRef]
- Sun, J.; Shen, C.; Jin, X.; Li, X.; Wu, D. Mir-367 is downregulated in coronary artery disaese and its overexpression exerts anti-inflammatory effect via inhibition of the NF-κB-activated inflammatory pathway. Int. J. Clin. Exp. Patholol. 2017, 10, 4047–4057. [Google Scholar]
- Yuan, B.; Shen, H.; Lin, L.; Su, T.; Zhong, L.; Yang, Z. MicroRNA367 negatively regulates the inflammatory response of microglia by targeting IRAK4 in intracerebral hemorrhage. J. Neuroinflammation 2015, 12, 206. [Google Scholar] [CrossRef]
- Botey-Bataller, J.; Vrijmoeth, H.D.; Ursinus, J.; Kullberg, B.J.; van den Wijngaard, C.C.; Ter Hofstede, H.; Alaswad, A.; Gupta, M.K.; Roesner, L.M.; Huehn, J.; et al. A comprehensive genetic map of cytokine responses in Lyme borreliosis. Nat. Commun. 2024, 15, 3795. [Google Scholar] [CrossRef]
- Florek, K.; Kubler, M.; Gorka, M.; Kubler, P. New Modifiable Risk Factors Influencing Coronary Artery Disease Severity. Int. J. Mol. Sci. 2024, 25, 7766. [Google Scholar] [CrossRef]
- Liu, X.; Xu, H.; Xu, H.; Geng, Q.; Mak, W.H.; Ling, F.; Su, Z.; Yang, F.; Zhang, T.; Chen, J.; et al. New genetic variants associated with major adverse cardiovascular events in patients with acute coronary syndromes and treated with clopidogrel and aspirin. Pharmacogen. J. 2021, 21, 664–672. [Google Scholar] [CrossRef]
- Zhou, D.; Yang, L.; Zheng, L.; Ge, W.; Li, D.; Zhang, Y.; Hu, X.; Gao, Z.; Xu, J.; Huang, Y.; et al. Exome capture sequencing of adenoma reveals genetic alterations in multiple cellular pathways at the early stage of colorectal tumorigenesis. PLoS ONE 2013, 8, e53310. [Google Scholar] [CrossRef]
- Lin, S.H.; Raju, G.S.; Huff, C.; Ye, Y.; Gu, J.; Chen, J.S.; Hildebrandt, M.A.T.; Liang, H.; Menter, D.G.; Morris, J.; et al. The somatic mutation landscape of premalignant colorectal adenoma. Gut 2018, 67, 1299–1305. [Google Scholar] [CrossRef]
- Li, S.; Han, T. Frequent loss of FAM126A expression in colorectal cancer results in selective FAM126B dependency. iScience 2024, 27, 109646. [Google Scholar] [CrossRef]
- Thompson, N.A.; Ranzani, M.; van der Weyden, L.; Iyer, V.; Offord, V.; Droop, A.; Behan, F.; Goncalves, E.; Speak, A.; Iorio, F.; et al. Combinatorial CRISPR screen identifies fitness effects of gene paralogues. Nat. Commun. 2021, 12, 1302. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Topatana, W.; Juengpanich, S.; Cao, J.; Hu, J.; Zhang, B.; Ma, D.; Cai, X.; Chen, M. Development of synthetic lethality in cancer: Molecular and cellular classification. Signal Transduct. Target. Ther. 2020, 5, 241. [Google Scholar] [CrossRef]
- World Health Organization Colorectal Cancer. Available online: https://www.who.int/news-room/fact-sheets/detail/colorectal-cancer (accessed on 11 February 2025).
- Leguay, K.; Kent, O.A. Dynamic Coupling of MAPK Signaling to the Guanine Nucleotide Exchange Factor GEF-H1. Onco Targets Ther. 2025, 18, 147–159. [Google Scholar] [CrossRef] [PubMed]
- Luo, J. KRAS mutation in pancreatic cancer. Semin. Oncol. 2021, 48, 10–18. [Google Scholar] [CrossRef]
- Mark, J.K.K.; Teh, A.H.; Yap, B.K. Epstein-Barr virus-infected nasopharyngeal carcinoma therapeutics: Oncoprotein targets and clinical implications. Med. Oncol. 2025, 42, 59. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Gong, Y.; Jiang, Q.; Liu, L.; Li, S.; Zhou, Q.; Huang, F.; Liu, Z. Circular RNA Expression Profiles in Nasopharyngeal Carcinoma by Sequence Analysis. Front. Oncol. 2020, 10, 601. [Google Scholar] [CrossRef] [PubMed]
- Jiang, P.; Xia, B. Circular RNA EFR3A promotes nasopharyngeal carcinoma progression through modulating the miR-654-3p/EFR3A axis. Cell Mol. Biol. 2023, 69, 111–117. [Google Scholar] [CrossRef]
- Conn, V.M.; Chinnaiyan, A.M.; Conn, S.J. Circular RNA in cancer. Nat. Rev. Cancer 2024, 24, 597–613. [Google Scholar] [CrossRef]
- Latif, G.; Al Anezi, F.Y.; Iskandar, D.; Bashar, A.; Alghazo, J. Recent Advances in Classification of Brain Tumor from MR Images—State of the Art Review from 2017 to 2021. Curr. Med. Imaging 2022, 18, 903–918. [Google Scholar] [CrossRef]
- Cacciotti, C.; Fleming, A.; Ramaswamy, V. Advances in the molecular classification of pediatric brain tumors: A guide to the galaxy. J. Pathol. 2020, 251, 249–261. [Google Scholar] [CrossRef] [PubMed]
- Hoellerbauer, P.; Biery, M.C.; Arora, S.; Rao, Y.; Girard, E.J.; Mitchell, K.; Dighe, P.; Kufeld, M.; Kuppers, D.A.; Herman, J.A.; et al. Functional genomic analysis of adult and pediatric brain tumor isolates. bioRxiv 2023. [Google Scholar] [CrossRef]
- Zhao, B.; Rao, Y.; Leighow, S.; O’Brien, E.P.; Gilbert, L.; Pritchard, J.R. A pan-CRISPR analysis of mammalian cell specificity identifies ultra-compact sgRNA subsets for genome-scale experiments. Nat. Commun. 2022, 13, 625. [Google Scholar] [CrossRef]
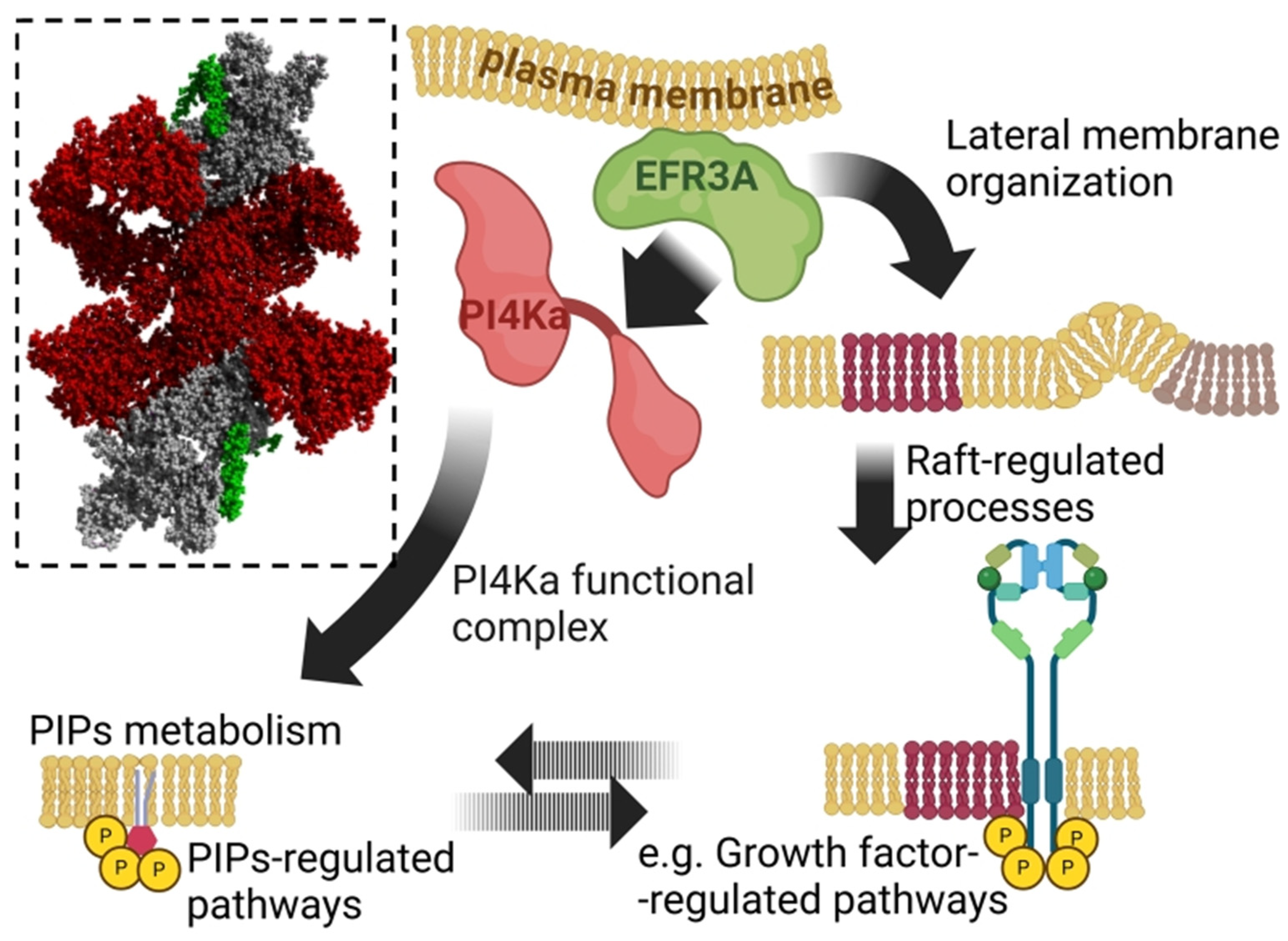
| Disease Category | Associated Findings | References |
|---|---|---|
| Neurological Disorders | EFR3A mutations linked to autism spectrum disorder (ASD). | [39] |
| EFR3A identified as a hub gene predicting seizures in primary glioma patients. | [41] | |
| Association with essential tremor. | [42] | |
| Reducing EFR3A gene expression prevents the SC-CA1 synapse dysfunction and rescues synaptic and spatial learning and memory deficits in Alzheimer’s disease mouse model. | [43] | |
| Depletion of the EFR3B isoform in the CA2/CA3 regions of pyramidal neurons (PNs) led to impaired excitability and deficits in the recognition of social novelty in mice. | [44] | |
| Cardiovascular Diseases | Overexpression of EFR3A linked to coronary artery disease (CAD), regulated by miR-367 and affecting inflammatory pathways. | [46] |
| SNP in EFR3A associated with major adverse cardiovascular events (MACEs) in acute coronary syndrome patients. | [50] | |
| Oncological Diseases | Colorectal Cancer (CRC): | |
| Somatic mutation identified in adenoma during adenoma-to-carcinoma progression. | [51,52] | |
| Protective synthetic lethality mechanism involving EFR3A in CRC. | [53,54,55] | |
| Pancreatic Ductal Adenocarcinoma (PDAC): | ||
| EFR3A identified as a player in KRAS-mutant PDAC, affecting survival and tumor progression. | [33] | |
| Nasopharyngeal Carcinoma (NPC): | ||
| circEFR3A overexpression promotes NPC growth; silencing circEFR3A shows anti-oncogenic effects. | [60,61] | |
| Brain Tumors: | ||
| Strong genetic dependency between EFR3A and EFR3B, affecting cell viability in glioblastoma cell lines. | [65] | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Marek-Bukowiec, K.; Trybus, M.; Hryniewicz-Jankowska, A.; Czogalla, A.; Sikorski, A.F. A Potential Role of EFR3A in Human Disease States. Biomolecules 2025, 15, 466. https://doi.org/10.3390/biom15040466
Marek-Bukowiec K, Trybus M, Hryniewicz-Jankowska A, Czogalla A, Sikorski AF. A Potential Role of EFR3A in Human Disease States. Biomolecules. 2025; 15(4):466. https://doi.org/10.3390/biom15040466
Chicago/Turabian StyleMarek-Bukowiec, Karolina, Magdalena Trybus, Anita Hryniewicz-Jankowska, Aleksander Czogalla, and Aleksander F. Sikorski. 2025. "A Potential Role of EFR3A in Human Disease States" Biomolecules 15, no. 4: 466. https://doi.org/10.3390/biom15040466
APA StyleMarek-Bukowiec, K., Trybus, M., Hryniewicz-Jankowska, A., Czogalla, A., & Sikorski, A. F. (2025). A Potential Role of EFR3A in Human Disease States. Biomolecules, 15(4), 466. https://doi.org/10.3390/biom15040466






