Indoleacetylglutamine Pathway Is a Potential Biomarker for Cardiovascular Diseases
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Source and Study Participants
2.2. Metabolomics
2.3. Statistical Analysis
3. Results
3.1. General Characteristics of Participants
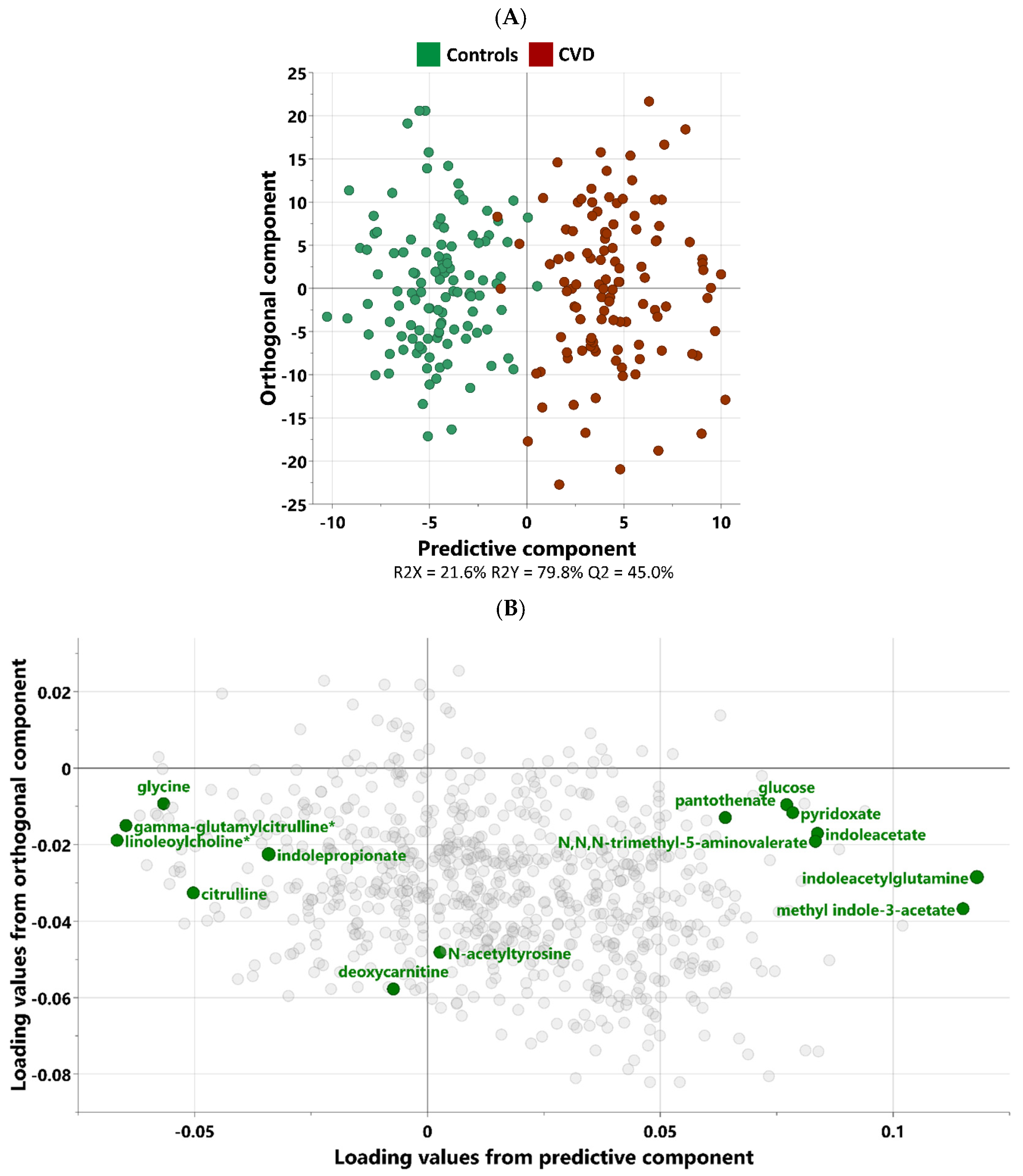
3.2. Multivariate Analysis
3.3. Univariate Analysis
3.4. Spearman’s Correlation Analysis
4. Discussion
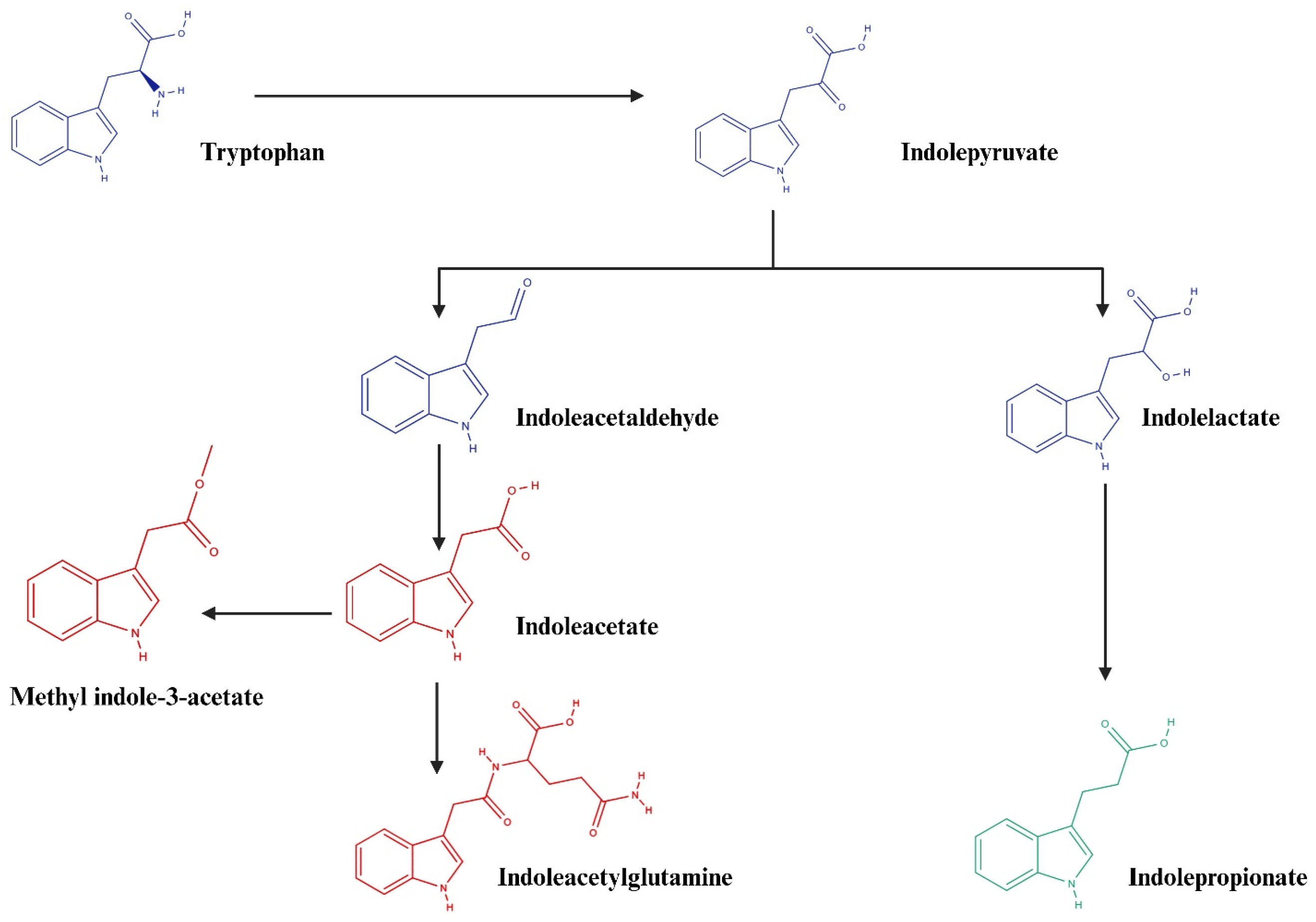
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Martin, S.S.; Aday, A.W.; Almarzooq, Z.I.; Anderson, C.A.; Arora, P.; Avery, C.L.; Baker-Smith, C.M.; Gibbs, B.B.; Beaton, A.Z.; Boehme, A.K.; et al. 2024 Heart Disease and Stroke Statistics: A Report of US and Global Data from the American Heart Association. Circulation 2024, 149, e347–e913. [Google Scholar] [PubMed]
- Luo, Y.; Liu, J.; Zeng, J.; Pan, H. Global burden of cardiovascular diseases attributed to low physical activity: An analysis of 204 countries and territories between 1990 and 2019. Am. J. Prev. Cardiol. 2024, 17, 100633. [Google Scholar] [CrossRef] [PubMed]
- Mendis, S.; Graham, I.; Narula, J. Addressing the Global Burden of Cardiovascular Diseases; Need for Scalable and Sustainable Frameworks. Glob. Heart 2022, 17, 48. [Google Scholar] [CrossRef]
- Perticone, M.; Molfino, A.; Maio, R. Editorial: Classical and Novel Biomarkers for Cardiovascular Disease. Front. Cardiovasc. Med. 2022, 9, 943227. [Google Scholar] [CrossRef]
- Wang, J.; Tan, G.-J.; Han, L.-N.; Bai, Y.-Y.; He, M.; Liu, H.-B. Novel biomarkers for cardiovascular risk prediction. J. Geriatr. Cardiol. 2017, 14, 135–150. [Google Scholar] [PubMed]
- Dambrova, M.; Makrecka-Kuka, M.; Kuka, J.; Vilskersts, R.; Nordberg, D.; Attwood, M.M.; Smesny, S.; Sen, Z.D.; Guo, A.C.; Oler, E.; et al. Acylcarnitines: Nomenclature, Biomarkers, Therapeutic Potential, Drug Targets, and Clinical Trials. Pharmacol. Rev. 2022, 74, 506–551. [Google Scholar] [CrossRef]
- Gander, J.; Carrard, J.; Gallart-Ayala, H.; Borreggine, R.; Teav, T.; Infanger, D.; Colledge, F.; Streese, L.; Wagner, J.; Klenk, C.; et al. Metabolic Impairment in Coronary Artery Disease: Elevated Serum Acylcarnitines Under the Spotlights. Front. Cardiovasc. Med. 2021, 8, 792350. [Google Scholar] [CrossRef]
- Gao, C.; Hou, L. Branched chain amino acids metabolism in heart failure. Front. Nutr. 2023, 10, 1279066. [Google Scholar] [CrossRef]
- Zhang, B.-C.; Chen, J.-H.; Xiang, C.-H.; Su, M.-Y.; Zhang, X.-S.; Ma, Y.-F. Increased serum bile acid level is associated with high-risk coronary artery plaques in an asymptomatic population detected by coronary computed tomography angiography. J. Thorac. Dis. 2019, 11, 5063–5070. [Google Scholar] [CrossRef]
- Shalon, D.; Culver, R.N.; Grembi, J.A.; Folz, J.; Treit, P.V.; Shi, H.; Rosenberger, F.A.; Dethlefsen, L.; Meng, X.; Yaffe, E.; et al. Profiling the human intestinal environment under physiological conditions. Nature 2023, 617, 581–591. [Google Scholar] [CrossRef]
- Iliou, A.; Mikros, E.; Karaman, I.; Elliott, F.; Griffin, J.L.; Tzoulaki, I.; Elliott, P. Metabolic phenotyping and cardiovascular disease: An overview of evidence from epidemiological settings. Heart 2021, 107, 1123–1129. [Google Scholar] [CrossRef]
- Albert, C.L.; Tang, W.H.W. Metabolic Biomarkers in Heart Failure. Heart Fail. Clin. 2018, 14, 109–118. [Google Scholar] [CrossRef] [PubMed]
- Lv, J.; Pan, C.; Cai, Y.; Han, X.; Wang, C.; Ma, J.; Pang, J.; Xu, F.; Wu, S.; Kou, T.; et al. Plasma metabolomics reveals the shared and distinct metabolic disturbances associated with cardiovascular events in coronary artery disease. Nat. Commun. 2024, 15, 5729. [Google Scholar] [CrossRef]
- Cavus, E.; Karakas, M.; Ojeda, F.M.; Kontto, J.; Veronesi, G.; Ferrario, M.M.; Linneberg, A.; Jorgensen, T.; Meisinger, C.; Thorand, B.; et al. Association of Circulating Metabolites With Risk of Coronary Heart Disease in a European Population: Results from the Biomarkers for Cardiovascular Risk Assessment in Europe (BiomarCaRE) Consortium. JAMA Cardiol. 2019, 4, 1270–1279. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.L.; Lin, W.; Di, C.; Hou, H.; Chen, H.; Zhou, J.; Yang, Q.; He, G. Metabolomics and Biomarkers for Paroxysmal and Persistent Atrial Fibrillation. J. Am. Heart Assoc. 2024, 13, e032153. [Google Scholar] [CrossRef]
- Al Thani, A.; Fthenou, E.; Paparrodopoulos, S.; Al Marri, A.; Shi, Z.; Qafoud, F.; Afifi, N. Qatar Biobank Cohort Study: Study Design and First Results. Am. J. Epidemiol. 2019, 188, 1420–1433. [Google Scholar] [CrossRef] [PubMed]
- Al-Khelaifi, F.; Diboun, I.; Donati, F.; Botrè, F.; Alsayrafi, M.; Georgakopoulos, C.; Suhre, K.; Yousri, N.A.; Elrayess, M.A. A pilot study comparing the metabolic profiles of elite-level athletes from different sporting disciplines. Sports Med. Open 2018, 4, 2. [Google Scholar] [CrossRef] [PubMed]
- Evans, A.; Bridgewater, B.; Liu, Q.; Mitchell, M.W.; Robinson, R.J.; Dai, H.; Stewart, S.J.; DeHaven, C.D.; Miller, L.A.D. High Resolution Mass Spectrometry Improves Data Quantity and Quality as Compared to Unit Mass Resolution Mass Spectrometry in High-Throughput Profiling Metabolomics. Metabolomics 2014, 4, 1000132. [Google Scholar]
- Roager, H.M.; Licht, T.R. Microbial tryptophan catabolites in health and disease. Nat. Commun. 2018, 9, 3294. [Google Scholar] [CrossRef]
- Gao, J.; Xu, K.; Liu, H.; Liu, G.; Bai, M.; Peng, C.; Li, T.; Yin, Y. Impact of the Gut Microbiota on Intestinal Immunity Mediated by Tryptophan Metabolism. Front. Cell. Infect. Microbiol. 2018, 8, 13. [Google Scholar] [CrossRef]
- Dou, L.; Sallée, M.; Cerini, C.; Poitevin, S.; Gondouin, B.; Jourde-Chiche, N.; Fallague, K.; Brunet, P.; Calaf, R.; Dussol, B.; et al. The cardiovascular effect of the uremic solute indole-3 acetic acid. J. Am. Soc. Nephrol. 2015, 26, 876–887. [Google Scholar] [CrossRef] [PubMed]
- Wu, P.-H.; Lin, Y.-T.; Wu, P.-Y.; Lee, H.-H.; Lee, S.-C.; Hung, S.-C.; Chen, S.-C.; Kuo, M.-C.; Chiu, Y.-W. Association between Circulation Indole-3-Acetic Acid Levels and Stem Cell Factor in Maintenance Hemodialysis Patients: A Cross-Sectional Study. J. Clin. Med. 2020, 9, 124. [Google Scholar] [CrossRef]
- Nayak, S.; Boopathi, S.; Chandrasekar, M.; Panda, S.P.; Manikandan, K.; Chitra, V.; Almutairi, B.O.; Arokiyaraj, S.; Guru, A.; Arockiaraj, J. Indole-3-acetic acid exposure leads to cardiovascular inflammation and fibrosis in chronic kidney disease rat model. Food Chem. Toxicol. 2024, 192, 114917. [Google Scholar] [CrossRef]
- Stoll, M.L.; Kumar, R.; Lefkowitz, E.J.; Cron, R.Q.; Morrow, C.D.; Barnes, S. Fecal metabolomics in pediatric spondyloarthritis implicate decreased metabolic diversity and altered tryptophan metabolism as pathogenic factors. Genes Immun. 2016, 17, 400–405. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Zhang, H.; Ren, S.; Ding, Y.; Remex, N.S.; Bhuiyan, S.; Qu, J.; Tang, X. Gut microbiota and microbiota-derived metabolites in cardiovascular diseases. Chin. Med. J. 2023, 136, 2269–2284. [Google Scholar] [CrossRef]
- Almeida, C.; Goncalves-Nobre, J.G.; Costa, D.A.; Barata, P. The potential links between human gut microbiota and cardiovascular health and disease—Is there a gut-cardiovascular axis? Front. Gastroenterol. 2023, 2, 1235126. [Google Scholar] [CrossRef]
- Talmor-Barkan, Y.; Bar, N.; Shaul, A.A.; Shahaf, N.; Godneva, A.; Bussi, Y.; Lotan-Pompan, M.; Weinberger, A.; Shechter, A.; Chezar-Azerrad, C.; et al. Metabolomic and microbiome profiling reveals personalized risk factors for coronary artery disease. Nat. Med. 2022, 28, 295–302. [Google Scholar] [CrossRef]
- Ettinger, G.; Macdonald, K.; Reid, G.; Burton, J.P. The influence of the human microbiome and probiotics on cardiovascular health. Gut Microbes 2014, 5, 719–728. [Google Scholar] [CrossRef]
- Song, Y.; Wei, H.; Zhou, Z.; Wang, H.; Hang, W.; Wu, J.; Wang, D.W. Gut microbiota-dependent phenylacetylglutamine in cardiovascular disease: Current knowledge and new insights. Front. Med. 2024, 18, 31–45. [Google Scholar] [CrossRef]
- Saha, P.P.; Gogonea, V.; Sweet, W.; Mohan, M.L.; Singh, K.D.; Anderson, J.T.; Mallela, D.; Witherow, C.; Kar, N.; Stenson, K.; et al. Gut microbe-generated phenylacetylglutamine is an endogenous allosteric modulator of β2-adrenergic receptors. Nat. Commun. 2024, 15, 6696. [Google Scholar] [CrossRef]
- Romano, K.A.; Nemet, I.; Saha, P.P.; Haghikia, A.; Li, X.S.; Mohan, M.L.; Lovano, B.; Castel, L.; Witkowski, M.; Buffa, J.A.; et al. Gut Microbiota-Generated Phenylacetylglutamine and Heart Failure. Circ. Heart Fail. 2023, 16, e009972. [Google Scholar] [CrossRef] [PubMed]
- Heianza, Y.; Tiwari, S.; Wang, X.; Watrous, J.D.; Rexrode, K.M.; Hu, F.B.; Alotaibi, M.; Jain, M.; Sun, Q.; Manson, J.E.; et al. Gut-Microbiota-Related Metabolite Phenylacetylglutamine and Risk of Incident Coronary Heart Disease Among Women. J. Clin. Endocrinol. Metab. 2024, 30, dgae525. [Google Scholar] [CrossRef] [PubMed]
- Tang, W.H.W.; Nemet, I.; Li, X.S.; Wu, Y.; Haghikia, A.; Witkowski, M.; Koeth, R.A.; Demuth, I.; König, M.; Steinhagen-Thiessen, E.; et al. Prognostic value of gut microbe-generated metabolite phenylacetylglutamine in patients with heart failure. Eur. J. Heart Fail. 2024, 26, 233–241. [Google Scholar] [CrossRef]
- Nemet, I.; Saha, P.P.; Gupta, N.; Zhu, W.; Romano, K.A.; Skye, S.M.; Cajka, T.; Mohan, M.L.; Li, L.; Wu, Y.; et al. A Cardiovascular Disease-Linked Gut Microbial Metabolite Acts via Adrenergic Receptors. Cell 2020, 180, 862–877.e22. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Wang, S.; Zhang, Q.; He, C.; Fu, C.; Wei, Q. The role of the gut microbiota in health and cardiovascular diseases. Mol. Biomed. 2022, 3, 30. [Google Scholar] [CrossRef]
- Lee, A.M.; Xu, Y.; Hooper, S.R.; Abraham, A.G.; Hu, J.; Xiao, R.; Matheson, M.B.; Brunson, C.; Rhee, E.P.; Coresh, J.; et al. Circulating Metabolomic Associations with Neurocognitive Outcomes in Pediatric CKD. Clin. J. Am. Soc. Nephrol. 2023, 19, 13–25. [Google Scholar] [CrossRef]
- Nemet, I.; Li, X.S.; Haghikia, A.; Li, L.; Wilcox, J.; Romano, K.A.; Buffa, J.A.; Witkowski, M.; Demuth, I.; König, M.; et al. Atlas of gut microbe-derived products from aromatic amino acids and risk of cardiovascular morbidity and mortality. Eur. Heart J. 2023, 44, 3085–3096. [Google Scholar] [CrossRef]
- Gesper, M.; Nonnast, A.B.H.; Kumowski, N.; Stoehr, R.; Schuett, K.; Marx, N.; Kappel, B.A. Gut-Derived Metabolite Indole-3-Propionic Acid Modulates Mitochondrial Function in Cardiomyocytes and Alters Cardiac Function. Front. Med. 2021, 8, 648259. [Google Scholar] [CrossRef]
- Xue, H.; Yu, C.; Deng, Y.; Zhang, Y.; Chen, S.; Chen, X.; Chen, K.; Yang, Y.; Ling, W. Gut Microbially Produced Indole-3-Propionic Acid Inhibits Atherosclerosis by Promoting Reverse Cholesterol Transport and Its Deficiency Is Causally Related to Atherosclerotic Cardiovascular Disease. Circ. Res. 2022, 131, 404–420. [Google Scholar] [CrossRef]
- Wang, Y.C.; Koay, Y.C.; Pan, C.; Zhou, Z.; Tang, W.; Wilcox, J.; Li, X.S.; Zagouras, A.; Marques, F.; Allayee, H.; et al. Indole-3-Propionic Acid Protects Against Heart Failure With Preserved Ejection Fraction. Circ. Res. 2024, 134, 371–389. [Google Scholar] [CrossRef] [PubMed]


| Test | Variable | Control Group (n = 112) | CVDs Group (n = 112) | p Value |
|---|---|---|---|---|
| General characteristics | Gender (M/F) | 68/44 | 67/45 | 0.99 |
| Age | 47 (42–51) | 53 (46–55) | 0.000 | |
| Waist size (cm) | 91.5 (85.7–99.2) | 98 (91.7–106) | 0.000 | |
| Hips size (cm) | 104.5 (100–113) | 107 (100–115) | 0.527 | |
| BMI (kg/m2) | 29.2 (25.9–33.2) | 30.8 (27.5–34.1) | 0.029 | |
| Systolic blood pressure (mmHg) | 116 (108–123) | 126 (119–136) | 0.000 | |
| Diastolic blood pressure (mmHg) | 75 (69–82) | 79 (73–87) | 0.006 | |
| Glycemic profile | Fasting blood glucose (mmol/L) | 5.1 (4.8–5.6) | 6.4 (5.4–9.2) | 0.000 |
| HbA1C (%) | 5.5 (5.2–5.8) | 6.5 (5.9–7.8) | 0.000 | |
| C-peptide (ng/mL) | 2.4 (1.8–3.2) | 3.1 (2.2–4.0) | 0.001 | |
| Insulin (uU/mL) | 9.7 (6.7–14) | 15 (9.9–23.1) | 0.000 | |
| Lipid profile | Total cholesterol (mmol/L) | 5.2 (4.8–5.7) | 4.9 (4.2–5.6) | 0.014 |
| HDL-cholesterol (mmol/L) | 1.3 (1.1–1.5) | 1.1 (0.9–1.4) | 0.009 | |
| LDL-cholesterol (mmol/L) | 3.2 (2.7–3.9) | 3.0 (2.1–3.5) | 0.007 | |
| Non-HDL-cholesterol (mmol/L) | 3.9 (3.3–4.5) | 3.7 (2.9–4.4) | 0.182 | |
| Triglyceride (mmol/L) | 1.21 (0.9–1.7) | 1.68 (1.2–2.4) | 0.000 | |
| Kidney function | Creatinine (µmol/L) | 69.5 (60–81) | 74 (64–84.2) | 0.020 |
| Chloride (mmol/L) | 69.5 (60–81) | 74 (64–84.2) | 0.001 | |
| Urea (mmol/L) | 101 (100–102) | 100 (99–102) | 0.004 | |
| Bicarbonate (mmol/L) | 4.7 (3.8–5.2) | 5 (4.1–6.5) | 0.370 | |
| Total protein (g/L) | 27 (26–28) | 27 (25–28) | 0.445 | |
| Cardiac function | NT-proBNP (pg/mL) | 72.9 (4.1) | 73.3 (3.9) | 0.057 |
| Homocysteine (µmol/L) | 20.6 (12.6–38.8) | 28.7 (15.4–51.1) | 0.099 | |
| Liver function | Albumin (g/L) | 8.7 (7.2–10.2) | 9.6 (7.4–11.4) | 0.427 |
| ALT (U/L) | 45.1(2.7) | 44.8 (3.0) | 0.100 | |
| AST (U/L) | 21 (15–28) | 22 (18–30.2) | 0.256 | |
| GGT (U/L) | 18 (15–20.2) | 18 (15.7–22) | 0.001 |
| Metabolite | Sub Pathway | Super-Pathway | Estimate | SE | p-Value | FDR |
|---|---|---|---|---|---|---|
| Indoleacetylglutamine | Tryptophan Metabolism | Amino Acid | 0.836 | 0.127 | 4.91 × 10−10 | 4.14 × 10−7 |
| Methyl indole-3-acetate | Food Component/Plant | Xenobiotics | 0.564 | 0.093 | 6.42 × 10−9 | 2.71 × 10−6 |
| Indolepropionate | Tryptophan Metabolism | Amino Acid | −0.645 | 0.132 | 1.85 × 10−6 | 5.21 × 10−4 |
| Indoleacetate | Tryptophan Metabolism | Amino Acid | 0.405 | 0.094 | 2.32 × 10−5 | 4.89 × 10−3 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Naja, K.; Anwardeen, N.; Al-Shafai, M.; Elrayess, M.A. Indoleacetylglutamine Pathway Is a Potential Biomarker for Cardiovascular Diseases. Biomolecules 2025, 15, 377. https://doi.org/10.3390/biom15030377
Naja K, Anwardeen N, Al-Shafai M, Elrayess MA. Indoleacetylglutamine Pathway Is a Potential Biomarker for Cardiovascular Diseases. Biomolecules. 2025; 15(3):377. https://doi.org/10.3390/biom15030377
Chicago/Turabian StyleNaja, Khaled, Najeha Anwardeen, Mashael Al-Shafai, and Mohamed A. Elrayess. 2025. "Indoleacetylglutamine Pathway Is a Potential Biomarker for Cardiovascular Diseases" Biomolecules 15, no. 3: 377. https://doi.org/10.3390/biom15030377
APA StyleNaja, K., Anwardeen, N., Al-Shafai, M., & Elrayess, M. A. (2025). Indoleacetylglutamine Pathway Is a Potential Biomarker for Cardiovascular Diseases. Biomolecules, 15(3), 377. https://doi.org/10.3390/biom15030377








