Polysome Profiling Proves Impaired IL-10 and Caspase-8 Translation in PBMCs of Hemodialysis Patients
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Population
2.2. Absolute Cell Count Determination
2.3. PBMC Isolation
2.4. Stimulation of PBMCs
2.5. Preparation of Cytosolic Lysates
2.6. Preparation of Linear Sucrose Gradients
2.7. RNA/cDNA/qPCR
2.8. Cytokine Analysis
2.9. Statistics
3. Results
3.1. Demographic Data
3.2. Cellular Parameters
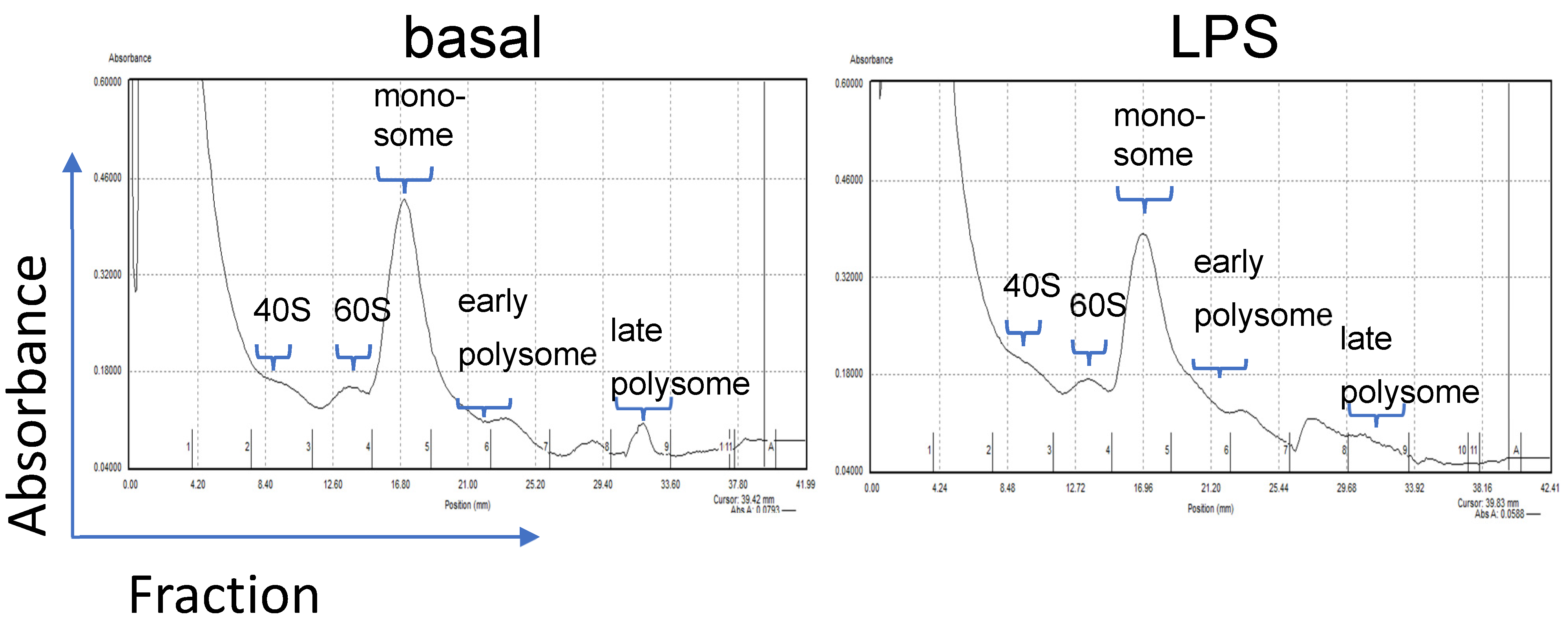
3.3. Translational Efficiency Measured by Polysome Profiling
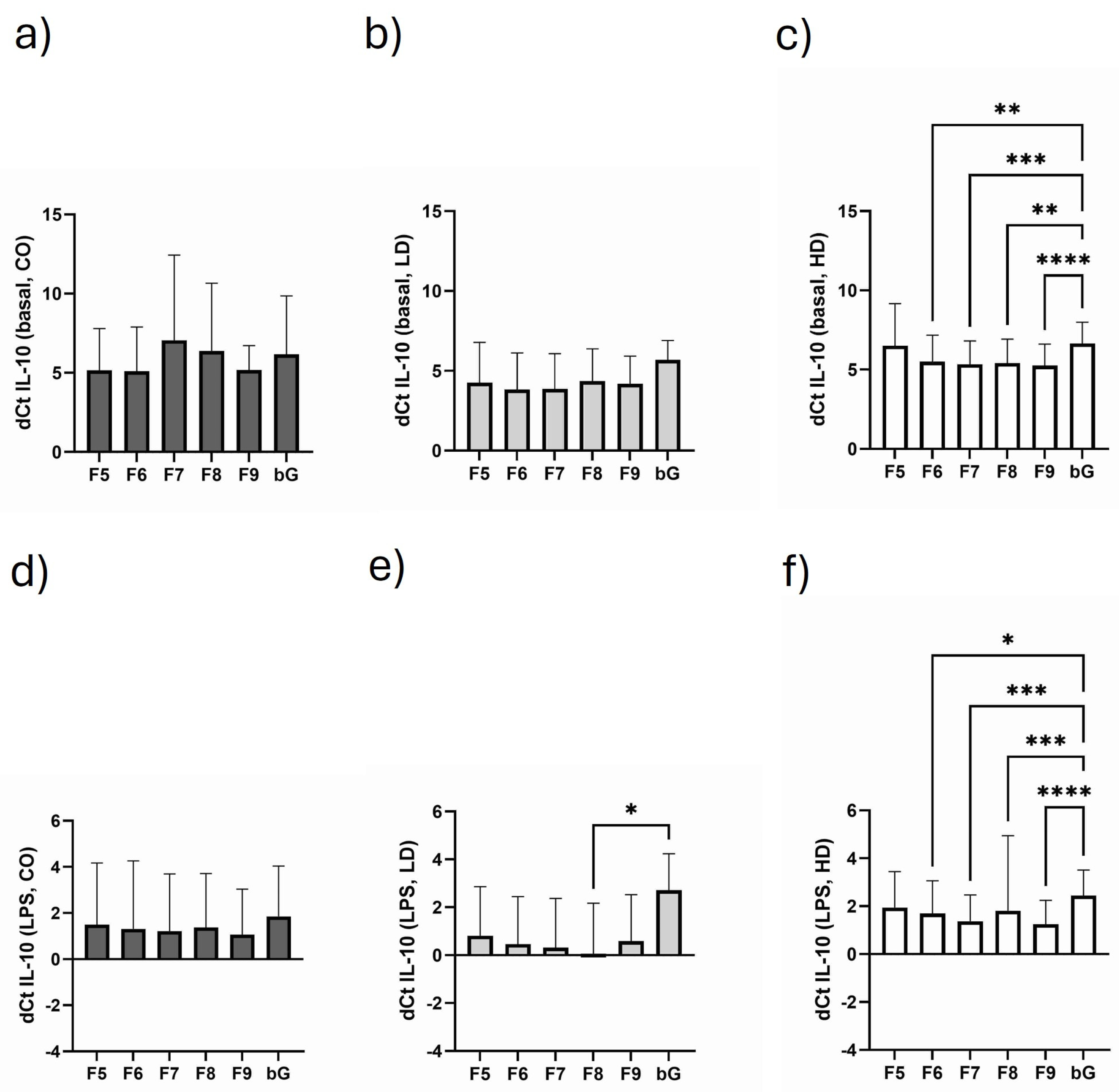
3.4. Course of IL-10 Expression in the Different Monosomal/Polysomal Fractions
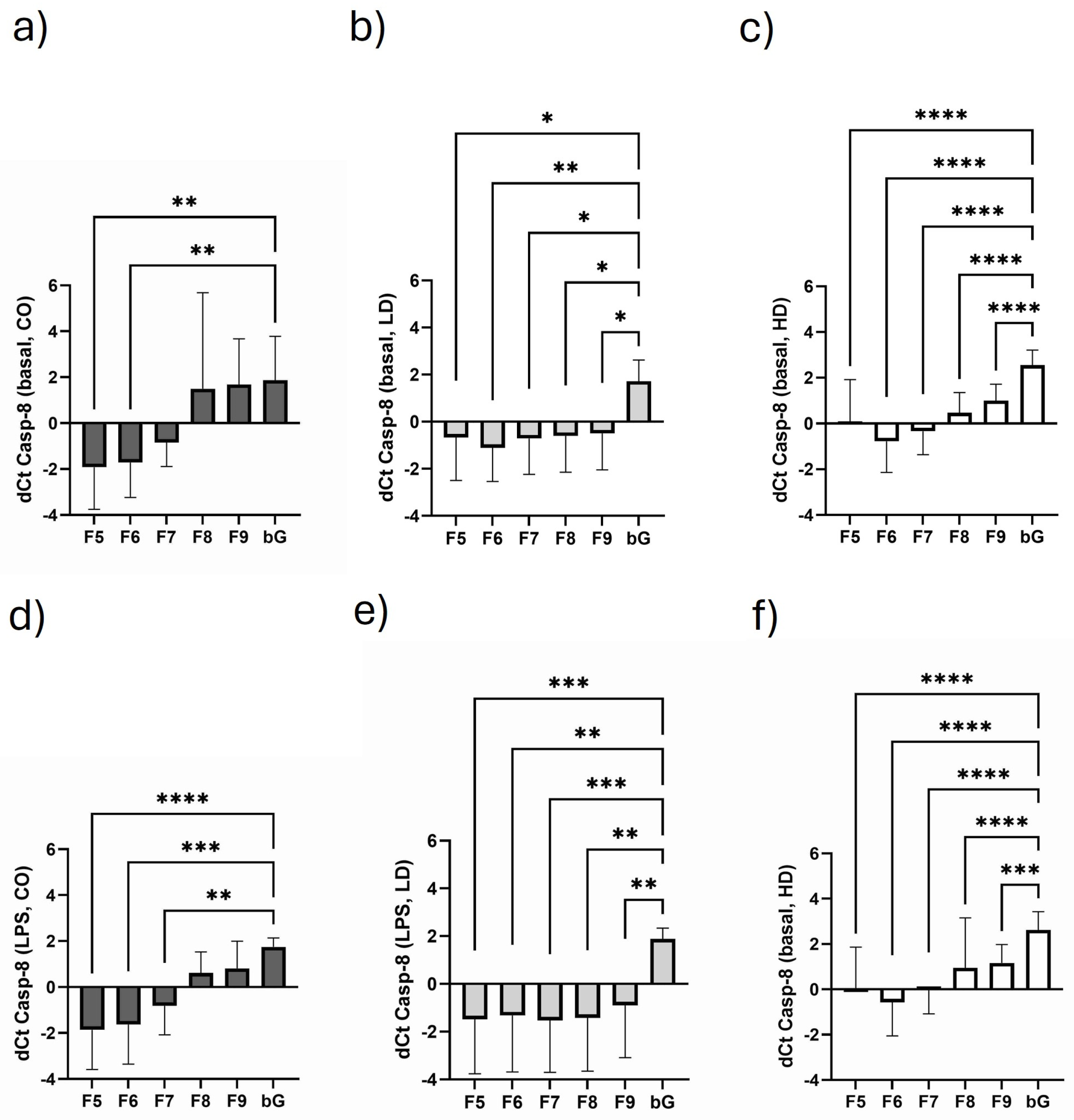
3.5. Course of Casp-8 Expression in the Different Monosomal/Polysomal Fractions
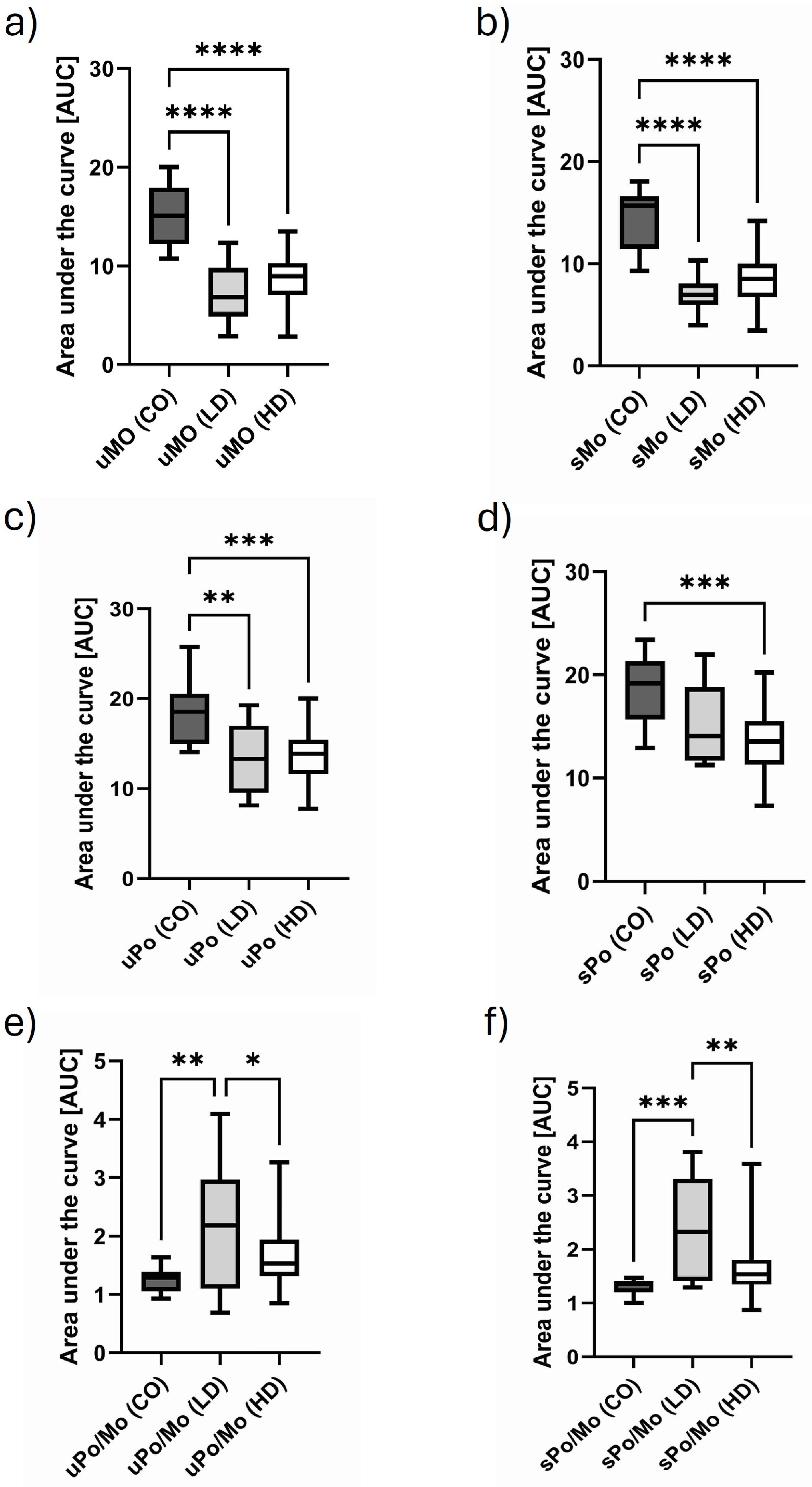
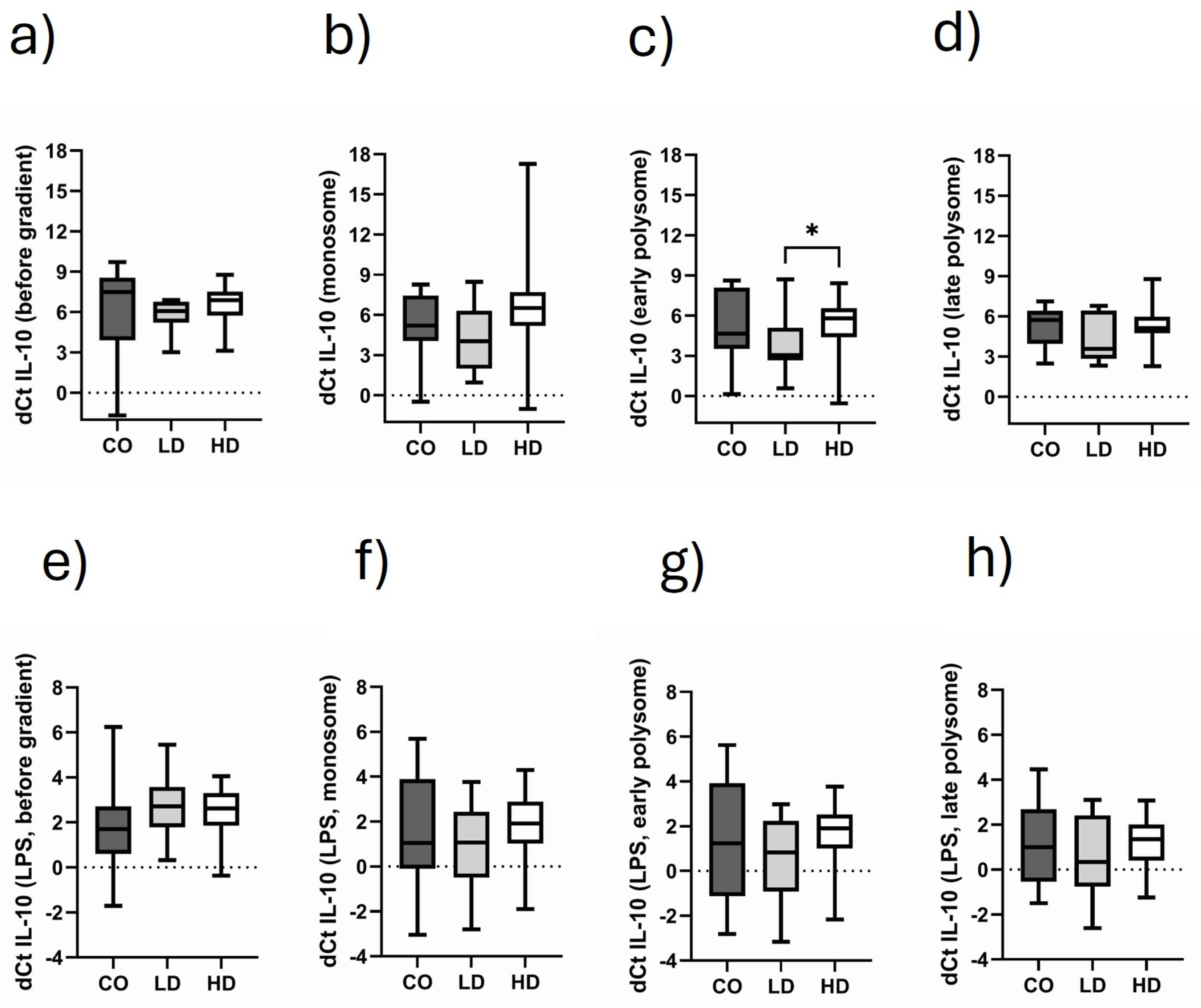
3.6. Comparison of Monosomal and Polysomal IL-10 Translational Activity in Different Cohorts
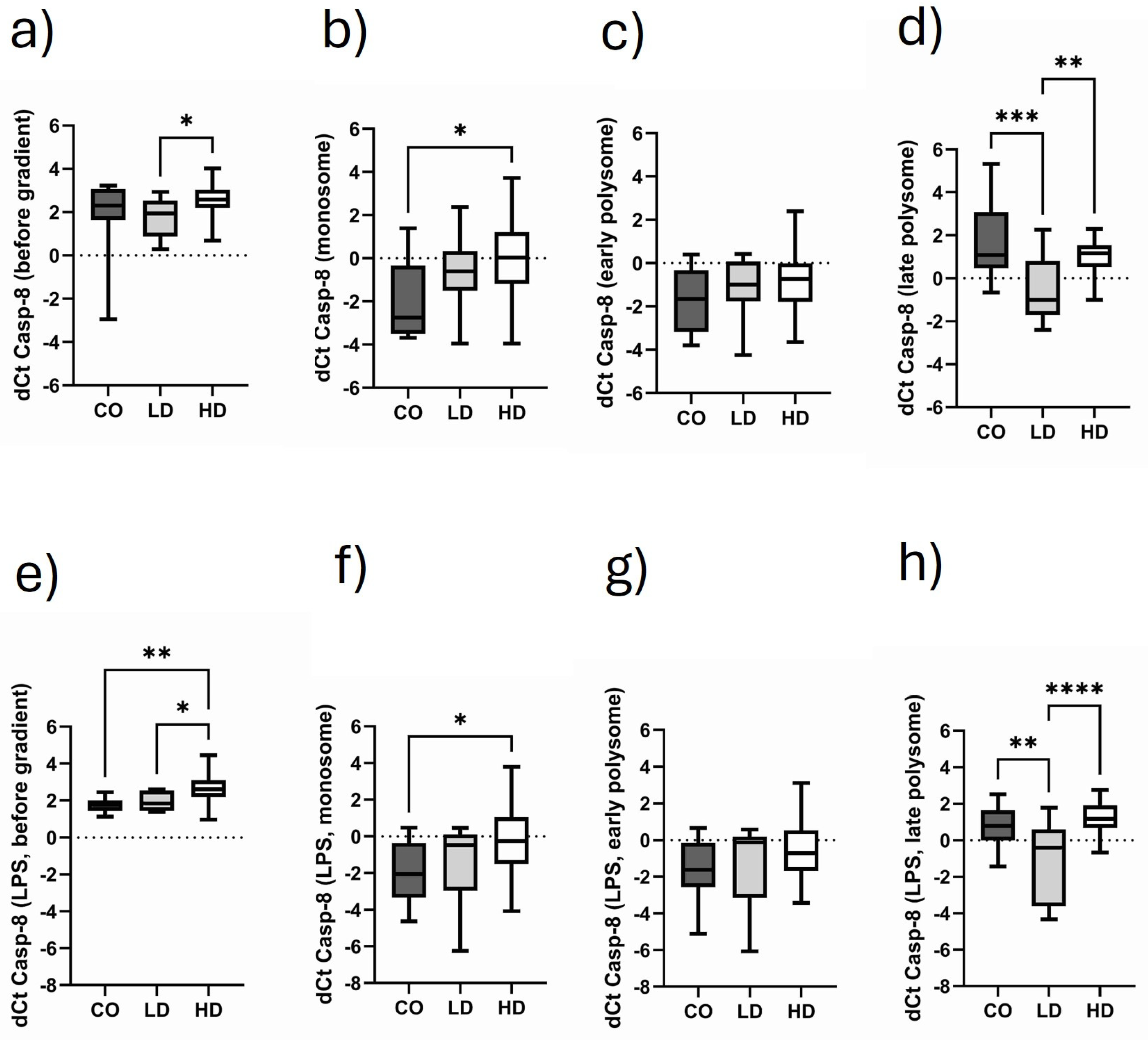
3.7. Comparison of Monosomal and Polysomal Casp-8 Translational Activity in Different Cohorts
3.8. Influence of Dialysis-Related Parameters with Regard to Monosomal/Polysomal IL-10 and Casp-8 Expression
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Inna, S.; Ilia, B.; Zhan, A.; Judith, S. Role of IL-10 in the progression of kidney disease. World J. Transplant. 2013, 3, 91–98. [Google Scholar] [CrossRef]
- Apostolos, P.; Karim, H.; Konstantinos, V.; Vikash, R.; Nagy, H.; Vassilios, P. The Effect of Interleukin-10 Immunotherapy on Renal Ischemia-Reperfusion Injury: A Systematic Review and Meta-Analysis of Preclinical Studies. Int. J. Mol. Sci. 2024, 25, 6231. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Sun, B.; Wang, H.; Yin, C.; Wang, X.; Ji, X. Increased serum IL-10 in lupus patients promotes apoptosis of T cell subsets via the caspase 8 pathway initiated by Fas signaling. J. Biomed. Res. 2015, 29, 232–240. [Google Scholar] [CrossRef] [PubMed]
- Han, J.-H.; Park, J.; Kang, T.-B.; Lee, K.-H. Regulation of Caspase-8 Activity at the Crossroads of Pro-Inflammation and Anti-Inflammation. Int. J. Mol. Sci. 2021, 22, 3318. [Google Scholar] [CrossRef] [PubMed]
- Arici, M.; Walls, J. End-stage renal disease, atherosclerosis, and cardiovascular mortality: Is C-reactive protein the missing link? Kidney Int. 2001, 59, 407–414. [Google Scholar] [CrossRef] [PubMed]
- Girndt, M.; Seibert, E. Premature cardiovascular disease in chronic renal failure (CRF): A model for an advanced ageing process. Exp. Gerontol. 2010, 45, 797–800. [Google Scholar] [CrossRef]
- Syed-Ahmed, M.; Narayanan, M. Immune Dysfunction and Risk of Infection in Chronic Kidney Disease. Adv. Chronic Kidney Dis. 2019, 26, 8–15. [Google Scholar] [CrossRef] [PubMed]
- Ross, R. Atherosclerosis—An inflammatory disease. N. Engl. J. Med. 1999, 340, 115–126. [Google Scholar] [CrossRef]
- Ulrich, C.; Wilke, A.; Schleicher, N.; Girndt, M.; Fiedler, R. Hypervolemia-Induced Immune Disturbances Do Not Involve IL-1ß but IL-6 and IL-10 Activation in Haemodialysis Patients. Toxins 2020, 12, 159. [Google Scholar] [CrossRef]
- Chassé, H.; Boulben, S.; Costache, V.; Cormier, P.; Morales, J. Analysis of translation using polysome profiling. Nucleic Acids Res. 2017, 45, e15. [Google Scholar] [CrossRef]
- Lokdarshi, A.; von Arnim, A. A Miniature Sucrose Gradient for Polysome Profiling. Bio-Protoc. 2023, 13, e4622. [Google Scholar] [CrossRef]
- Liu, B.; Qian, S.-B. Characterizing inactive ribosomes in translational profiling. Translation 2016, 4, e1138018. [Google Scholar] [CrossRef]
- Mishra, J.; Dent, C.; Tarabishi, R.; Mitsnefes, M.M.; Ma, Q.; Kelly, C.; Ruff, S.M.; Zahedi, K.; Shao, M.; Bean, J.; et al. Neutrophil gelatinase-associated lipocalin (NGAL) as a biomarker for acute renal injury after cardiac surgery. Lancet 2005, 365, 1231–1238. [Google Scholar] [CrossRef] [PubMed]
- Malyszko, J.; Malyszko, J.S.; Koc-Zorawska, E.; Kozminski, P.; Mysliwiec, M. Neutrophil gelatinase-associated lipocalin in dialyzed patients is related to residual renal function, type of renal replacement therapy and inflammation. Kidney Blood Press. Res. 2009, 32, 464–469. [Google Scholar] [CrossRef] [PubMed]
- Jang, Y.; Lee, J.H.; Wang, Y.; Sweeney, G. Emerging clinical and experimental evidence for the role of lipocalin-2 in metabolic syndrome. Clin. Exp. Pharmacol. Physiol. 2012, 39, 194–199. [Google Scholar] [CrossRef] [PubMed]
- Jofré, R.; Rodriguez-Benitez, P.; López-Gómez, J.M.; Pérez-Garcia, R. Inflammatory syndrome in patients on hemodialysis. J. Am. Soc. Nephrol. 2006, 17 (Suppl. 3), S274–S280. [Google Scholar] [CrossRef]
- Caglar, K.; Peng, Y.; Pupim, L.B.; Flakoll, P.J.; Levenhagen, D.; Hakim, R.M.; Ikizler, T.A. Inflammatory signals associated with hemodialysis. Kidney Int. 2002, 62, 1408–1416. [Google Scholar] [CrossRef]
- Taraz, M.; Khatami, M.; Hajiseyedjavadi, M.; Farrokhian, A.; Amini, M.; Khalili, H.; Abdollahi, A.; Dashti-Khavidaki, S. Association between antiinflammatory cytokine, IL-10, and sleep quality in patients on maintenance hemodialysis. Hemodial. International. Int. Symp. Home Hemodial. 2013, 17, 382–390. [Google Scholar] [CrossRef]
- Bale, B.F.; Doneen, A.L.; Leimgruber, P.P.; Vigerust, D.J. The critical issue linking lipids and inflammation: Clinical utility of stopping oxidative stress. Front. Cardiovasc. Med. 2022, 9, 1042729. [Google Scholar] [CrossRef]
- Wazir, M.; Olanrewaju, O.A.; Yahya, M.; Kumari, J.; Kumar, N.; Singh, J.; Al-Itbi, A.Y.A.; Kumari, K.; Ahmed, A.; Islam, T.; et al. Lipid Disorders and Cardiovascular Risk: A Comprehensive Analysis of Current Perspectives. Cureus 2023, 15, e51395. [Google Scholar] [CrossRef] [PubMed]
- Bhargava, S.; de la Puente-Secades, S.; Schurgers, L.; Jankowski, J. Lipids and lipoproteins in cardiovascular diseases: A classification. Trends Endocrinol. Metab. 2022, 33, 409–423. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, S.; Kaur, S.; Guha, S.; Batra, S.K. The Multifaceted Roles of Neutrophil Gelatinase Associated Lipocalin (NGAL) In Inflammation and Cancer. Biochim. Biophys. Acta 2012, 1826, 129–169. [Google Scholar] [CrossRef]
- Romejko, K.; Markowska, M.; Niemczyk, S. The Review of Current Knowledge on Neutrophil Gelatinase-Associated Lipocalin (NGAL). Int. J. Mol. Sci. 2023, 24, 10470. [Google Scholar] [CrossRef]
- Ulrich, C.; Fiedler, R.; Herberger, E.; Canim, Z.; Markau, S.; Girndt, M. Hypervolemia in Dialysis Patients Impairs STAT3 Signaling and Upregulates miR-142-3p: Effects on IL-10 and IL-6. Int. J. Mol. Sci. 2024, 25, 3719. [Google Scholar] [CrossRef]
- Magno, A.L.; Herat, L.Y.; Carnagarin, R.; Schlaich, M.P.; Matthews, V.B. Current Knowledge of IL-6 Cytokine Family Members in Acute and Chronic Kidney Disease. Biomedicines 2019, 7, 19. [Google Scholar] [CrossRef] [PubMed]
- Yasukawa, H.; Ohishi, M.; Mori, H.; Murakami, M.; Chinen, T.; Aki, D.; Hanada, T.; Takeda, K.; Akira, S.; Hoshijima, M.; et al. IL-6 induces an anti-inflammatory response in the absence of SOCS3 in macrophages. Nat. Immunol. 2003, 4, 551–556. [Google Scholar] [CrossRef] [PubMed]
- Komili, S.; Silver, P.A. Coupling and coordination in gene expression processes: A systems biology view. Nat. Rev. Genet. 2008, 9, 38–48. [Google Scholar] [CrossRef]
- Cohen, G. Immune Dysfunction in Uremia. Toxins 2020, 12, 439. [Google Scholar] [CrossRef]
- Girndt, M.; Ulrich, C.; Kaul, H.; Sester, U.; Sester, M.; Köhler, H. Uremia-associated immune defect: The IL-10-CRP axis. Kidney Int. 2003, 63 (Suppl. 84), S76–S79. [Google Scholar] [CrossRef] [PubMed]
- Girndt, M.; Sester, M.; Sester, U.; Kaul, H.; Köhler, H. Molecular aspects of T- and B-cell function in uremia. Kidney Int. 2001, 59 (Suppl. 78), S206–S211. [Google Scholar] [CrossRef]
- Girndt, M.; Sester, U.; Sester, M.; Kaul, H.; Köhler, H. Impaired cellular immune function in patients with end-stage renal failure. Nephrol. Dial. Transplant. 1999, 14, 2807–2810. [Google Scholar] [CrossRef] [PubMed]
- Pai, A.B.; Pai, M.P. Optimizing antimicrobial therapy for gram-positive bloodstream infections in patients on hemodialysis. Adv. Chronic Kidney Dis. 2006, 13, 259–270. [Google Scholar] [CrossRef] [PubMed]
- Rteil, A.; Kazma, J.M.; El Sawda, J.; Gharamti, A.; Koubar, S.H.; Kanafani, Z.A. Clinical characteristics, risk factors and microbiology of infections in patients receiving chronic hemodialysis. J. Infect. Public Health 2020, 13, 1166–1171. [Google Scholar] [CrossRef] [PubMed]
- Mandal, R.; Barrón, J.C.; Kostova, I.; Becker, S.; Strebhardt, K. Caspase-8: The double-edged sword. Biochim. Biophys. Acta 2020, 1873, 188357. [Google Scholar] [CrossRef]
- Orning, P.; Lien, E. Multiple roles of caspase-8 in cell death, inflammation, and innate immunity. J. Leukoc. Biol. 2021, 109, 121–141. [Google Scholar] [CrossRef] [PubMed]
- Sanz, A.B.; Sanchez-Niño, M.D.; Ramos, A.M.; Ortiz, A. Regulated cell death pathways in kidney disease. Nat. Rev. Nephrol. 2023, 19, 281–299. [Google Scholar] [CrossRef]
- Soppert, J.; Lehrke, M.; Marx, N.; Jankowski, J.; Noels, H. Lipoproteins and lipids in cardiovascular disease: From mechanistic insights to therapeutic targeting. Adv. Drug Deliv. Rev. 2020, 159, 4–33. [Google Scholar] [CrossRef] [PubMed]
- Schieweck, R.; Ciccopiedi, G.; Klau, K.; Popper, B. Monosomes buffer translational stress to allow for active ribosome elongation. Front. Mol. Biosci. 2023, 10, 1158043. [Google Scholar] [CrossRef]
| Basal | bG | F5 | F6 | F7 | F8 | F9 |
|---|---|---|---|---|---|---|
| CO | 1.7 ± 0.2 | 1.9 ± 0.1 | 1.8 ± 0.1 | 1.7 ± 0.1 | 1.7 ± 0.2 | 1.6 ± 0.2 |
| LD | 1.8 ± 0.6 | 1.9 ± 0.1 | 1.8 ± 0.1 | 1.7 ± 0.2 | 1.7 ± 0.2 | 1.6 ± 0.2 |
| HD | 1.7 ± 0.1 | 1.9 ± 0.1 | 1.8 ± 0.2 | 1.7 ± 0.2 | 1.7 ± 0.1 | 1.6 ± 0.3 |
| Stimulated | ||||||
| CO | 1.6 ± 0.2 | 1.9 ± 0.1 | 1.8 ± 0.1 | 1.7 ± 0.1 | 1.6 ± 0.1 | 1.7 ± 0.1 |
| LD | 1.6 ± 0.3 | 1.9 ± 0.1 | 1.8 ± 0.2 | 1.7 ± 0.2 | 1.8 ± 0.5 | 1.7 ± 0.4 |
| HD | 1.7 ± 0.3 | 1.8 ± 0.1 | 1.7 ± 0.2 | 1.6 ± 0.2 | 1.6 ± 0.2 | 1.6 ± 0.3 |
| CO (n = 9) | LD (n = 10) | HD (n = 42) | Statistics | |
|---|---|---|---|---|
| Age (years) | 37.0 ± 16.8 | 55.0 ± 7.8 | 59.6 ± 15.0 | * CO vs. LD *** CO vs. HD |
| Gender (female, %) | 33.3 | 50.0 | 41.5 | n.s. |
| Diabetes (%) | 0 | 30.0 | 28.6 | n.s. |
| Sys. BP (mm Hg) | - | 147 ± 23 | 149 ± 18 | n.s. |
| Diast. BP (mm Hg) | - | 89 ± 9 | 79 ± 12 | * LD vs. HD |
| CRP (mg/L) | 0.9 ± 0.6 | 12.5 ± 6.7 | 23.1 ± 15.3 | **** CO vs. HD * CO vs. LD |
| NGAL (ng/L) | 219.0 ± 76.5 | 270.0 ± 145.4 | 2333.0 ± 742.6 | **** CO vs. HD **** LD vs. HD |
| CO (n = 9) | LD (n = 10) | HD (n = 42) | Statistics | |
|---|---|---|---|---|
| Leukocytes (×106/mL) | 9.3 ± 2.4 | 9.8 ± 2.9 | 9.6 ± 3.0 | n.s. |
| Granulocytes (×106/mL) | 4.2 ± 0.4 | 5.7 ± 2.3 | 5.6 ± 2.2 | n.s. |
| Monocytes (×106/mL) | 0.6 ± 0.1 | 0.74 ± 0.08 | 0.71 ± 0.04 | n.s. |
| Lymphocytes (×106/mL) | 1.6 ± 0.4 | 1.7 ± 0.7 | 1.3 ± 0.6 | * LD vs. HD |
| CD4+ (×106/mL) | 0.9 ± 0.4 | 1.1 ± 0.4 | 0.7 ± 0.3 | * LD vs. HD |
| CD8+ (×106/mL) | 0.5 ± 0.3 | 0.5 ± 0.4 | 0.3 ± 0.3 | n.s. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dawood, A.; Fiedler, R.; Markau, S.; Girndt, M.; Ulrich, C. Polysome Profiling Proves Impaired IL-10 and Caspase-8 Translation in PBMCs of Hemodialysis Patients. Biomolecules 2025, 15, 335. https://doi.org/10.3390/biom15030335
Dawood A, Fiedler R, Markau S, Girndt M, Ulrich C. Polysome Profiling Proves Impaired IL-10 and Caspase-8 Translation in PBMCs of Hemodialysis Patients. Biomolecules. 2025; 15(3):335. https://doi.org/10.3390/biom15030335
Chicago/Turabian StyleDawood, Amanda, Roman Fiedler, Silke Markau, Matthias Girndt, and Christof Ulrich. 2025. "Polysome Profiling Proves Impaired IL-10 and Caspase-8 Translation in PBMCs of Hemodialysis Patients" Biomolecules 15, no. 3: 335. https://doi.org/10.3390/biom15030335
APA StyleDawood, A., Fiedler, R., Markau, S., Girndt, M., & Ulrich, C. (2025). Polysome Profiling Proves Impaired IL-10 and Caspase-8 Translation in PBMCs of Hemodialysis Patients. Biomolecules, 15(3), 335. https://doi.org/10.3390/biom15030335






