Identification of an Immunoglobulin Paratope Binding to Keratan Sulfate and Expression of a Single-Chain Derivative for Imaging
Abstract
1. Introduction
2. Materials and Methods
2.1. Cultivation of MZ15 Hybridoma Cells and Production of MZ15 mAb
2.2. Identification, Cloning, and Sequencing of the MZ15 Variable Regions
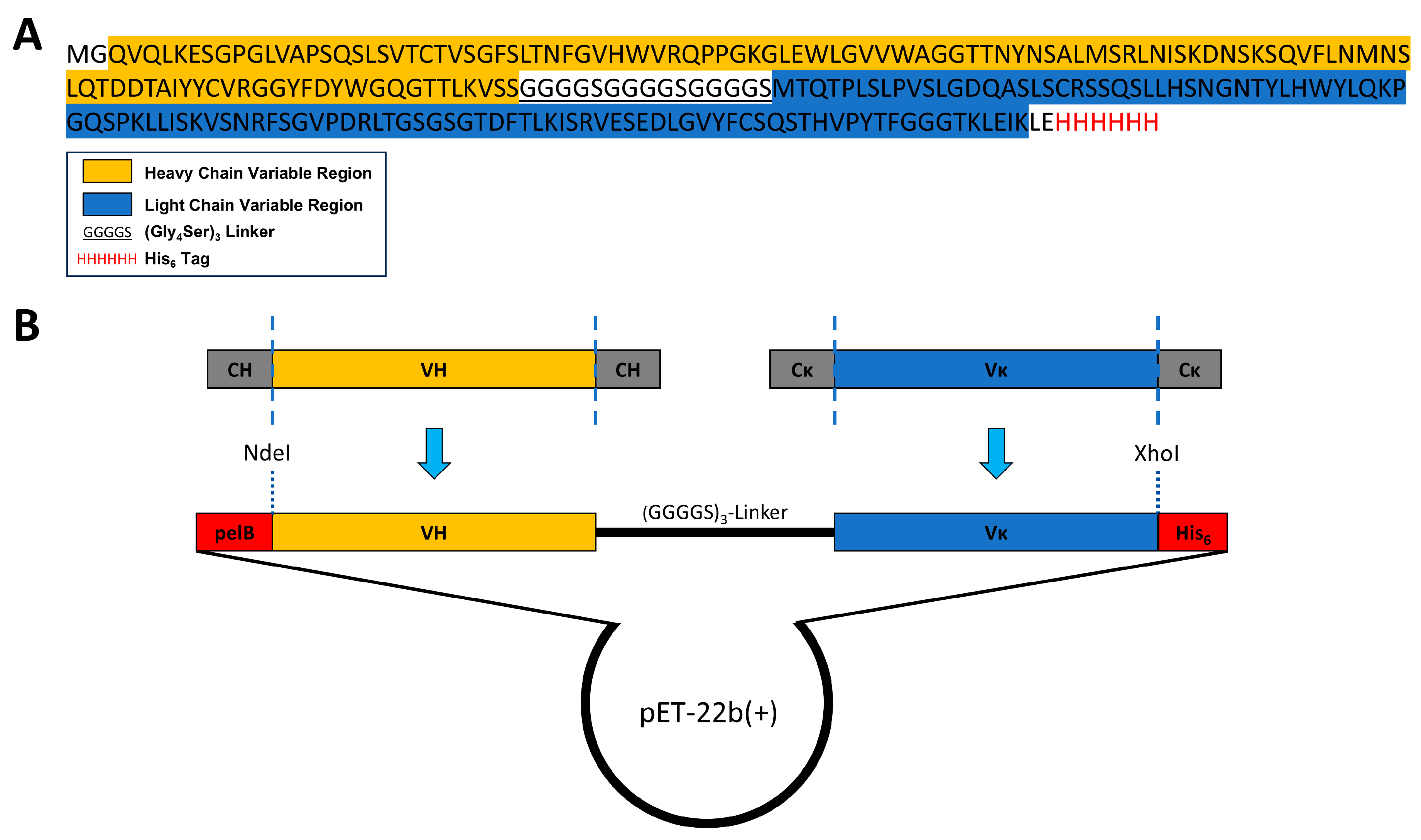
2.3. Construction of a ScFv Expression Vector and Protein Expression
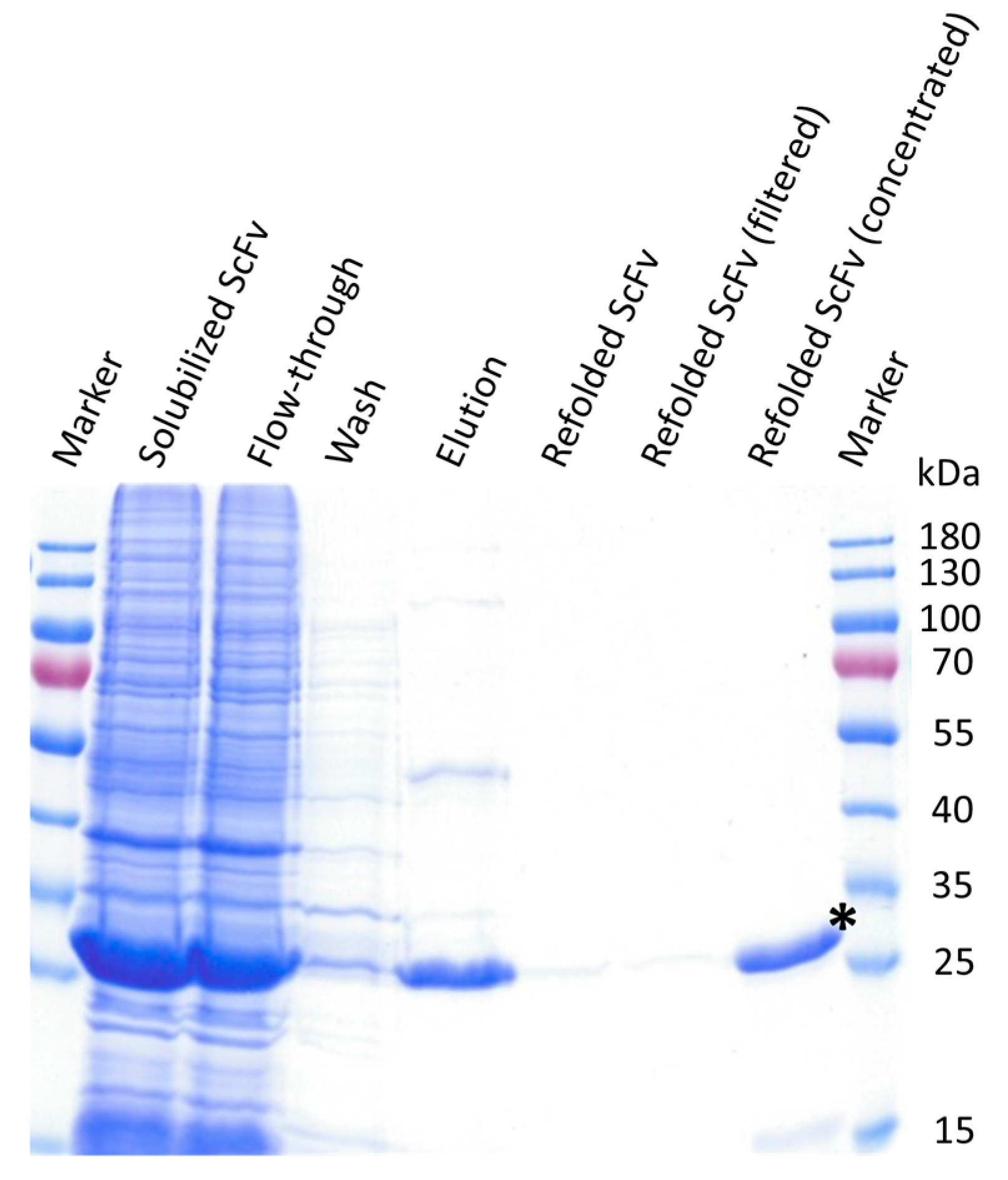
2.4. Solubilization, Functional Refolding, and Purification of the ScFv
2.5. KS Staining of Rat Eye Cryosections with the Single-Chain Variable Fragment
3. Results
3.1. Identification of the VH and VL Genes from the Anti-Keratan Sulfate mAb MZ15
3.2. Construction of a ScFv Expression Vector Based on the Anti-KS mAb MZ15
3.3. ScFv Was Successfully Expressed and Purified from Inclusion Bodies
3.4. ScFv Specifically Detects KS in Rat Eye Tissue Sections
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cox, T.R.; Erler, J.T. Remodeling and homeostasis of the extracellular matrix: Implications for fibrotic diseases and cancer. Dis. Models Mech. 2011, 4, 165–178. [Google Scholar] [CrossRef]
- Frantz, C.; Stewart, K.M.; Weaver, V.M. The extracellular matrix at a glance. J. Cell Sci. 2010, 123, 4195–4200. [Google Scholar] [CrossRef]
- Theocharis, A.D.; Skandalis, S.S.; Gialeli, C.; Karamanos, N.K. Extracellular matrix structure. Adv. Drug Deliv. Rev. 2016, 97, 4–27. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Zhang, F.; Li, L.; Li, G.; He, W.; Linhardt, R.J. Compositional analysis and structural elucidation of glycosaminoglycans in chicken eggs. Glycoconj. J. 2014, 31, 593–602. [Google Scholar] [CrossRef]
- Rabenstein, D.L. Heparin and heparan sulfate: Structure and function. Nat. Prod. Rep. 2002, 19, 312–331. [Google Scholar] [CrossRef]
- Funderburgh, J.L. MINI REVIEW Keratan sulfate: Structure, biosynthesis, and function. Glycobiology 2000, 10, 951–958. [Google Scholar] [CrossRef] [PubMed]
- Pomin, V.H.; Mulloy, B. Glycosaminoglycans and Proteoglycans. Pharmaceuticals 2018, 11, 27. [Google Scholar] [CrossRef]
- Sugahara, K.; Mikami, T.; Uyama, T.; Mizuguchi, S.; Nomura, K.; Kitagawa, H. Recent advances in the structural biology of chondroitin sulfate and dermatan sulfate. Curr. Opin. Struct. Biol. 2003, 13, 612–620. [Google Scholar] [CrossRef]
- Fallacara, A.; Baldini, E.; Manfredini, S.; Vertuani, S. Hyaluronic Acid in the Third Millennium. Polymers 2018, 10, 701. [Google Scholar] [CrossRef]
- Soares da Costa, D.; Reis, R.L.; Pashkuleva, I. Sulfation of Glycosaminoglycans and Its Implications in Human Health and Disorders. Annu. Rev. Biomed. Eng. 2017, 19, 1–26. [Google Scholar] [CrossRef] [PubMed]
- Klaver, D.W.; Wilce, M.C.J.; Gasperini, R.; Freeman, C.; Juliano, J.P.; Parish, C.; Foa, L.; Aguilar, M.-I.; Small, D.H. Glycosaminoglycan-induced activation of the β-secretase (BACE1) of Alzheimer’s disease. J. Neurochem. 2010, 112, 1552–1561. [Google Scholar] [CrossRef] [PubMed]
- Bonnans, C.; Chou, J.; Werb, Z. Remodelling the extracellular matrix in development and disease. Nat. Rev. Mol. Cell Biol. 2014, 15, 786–801. [Google Scholar] [CrossRef] [PubMed]
- Winkler, J.; Abisoye-Ogunniyan, A.; Metcalf, K.J.; Werb, Z. Concepts of extracellular matrix remodelling in tumour progression and metastasis. Nat. Commun. 2020, 11, 5120. [Google Scholar] [CrossRef] [PubMed]
- Sorokin, L. The impact of the extracellular matrix on inflammation. Nat. Rev. Immunol. 2010, 10, 712–723. [Google Scholar] [CrossRef]
- Meyer, K.; Linker, A.; Davidson, E.A.; Weissmann, B. The mucopolysaccharides of bovine cornea. J. Biol. Chem. 1953, 205, 611–616. [Google Scholar] [CrossRef] [PubMed]
- Hirano, S.; Hoffman, P.; Meyer, K. The Structure of Keratosulfate of Bovine Cornea1. J. Org. Chem. 1961, 26, 5064–5069. [Google Scholar] [CrossRef]
- Imagama, S.; Sakamoto, K.; Tauchi, R.; Shinjo, R.; Ohgomori, T.; Ito, Z.; Zhang, H.; Nishida, Y.; Asami, N.; Takeshita, S.; et al. Keratan Sulfate Restricts Neural Plasticity after Spinal Cord Injury. J. Neurosci. 2011, 31, 17091–17102. [Google Scholar] [CrossRef] [PubMed]
- Chakravarti, S.; Petroll, W.M.; Hassell, J.R.; Jester, J.V.; Lass, J.H.; Paul, J.; Birk, D.E. Corneal opacity in lumican-null mice: Defects in collagen fibril structure and packing in the posterior stroma. Investig. Ophthalmol. Vis. Sci. 2000, 41, 3365–3373. [Google Scholar]
- Chen, S.; Oldberg, A.; Chakravarti, S.; Birk, D.E. Fibromodulin regulates collagen fibrillogenesis during peripheral corneal development. Dev. Dyn. 2010, 239, 844–854. [Google Scholar] [CrossRef]
- Hassell, J.R.; Cintron, C.; Kublin, C.; Newsome, D.A. Proteoglycan changes during restoration of transparency in corneal scars. Arch. Biochem. Biophys. 1983, 222, 362–369. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, T.; Otake, H.; Hiramatsu, N.; Yamamoto, N.; Taga, A.; Nagai, N. A Proteomic Approach for Understanding the Mechanisms of Delayed Corneal Wound Healing in Diabetic Keratopathy Using Diabetic Model Rat. Int. J. Mol. Sci. 2018, 19, 3635. [Google Scholar] [CrossRef] [PubMed]
- Caterson, B.; Melrose, J. Keratan sulfate, a complex glycosaminoglycan with unique functional capability. Glycobiology 2018, 28, 182–206. [Google Scholar] [CrossRef] [PubMed]
- Pomin, V.H. Keratan sulfate: An up-to-date review. Int. J. Biol. Macromol. 2015, 72, 282–289. [Google Scholar] [CrossRef]
- Funderburgh, J.L.; Caterson, B.; Conrad, G.W. Distribution of proteoglycans antigenically related to corneal keratan sulfate proteoglycan. J. Biol. Chem. 1987, 262, 11634–11640. [Google Scholar] [CrossRef]
- Scott, J.E.; Haigh, M. ‘Small’-proteoglycan:collagen interactions: Keratan sulphate proteoglycan associates with rabbit corneal collagen fibrils at the ‘a’ and ‘c’ bands. Biosci. Rep. 1985, 5, 765–774. [Google Scholar] [CrossRef]
- Saito, T.; Nishida, K.; Nakayama, J.; Akama, T.O.; Fukuda, M.N.; Watanabe, K.; Quantock, A.J.; Maeda, N.; Watanabe, H.; Tano, Y. Sulfation patterns of keratan sulfate in different macular corneal dystrophy immunophenotypes using three different probes. Br. J. Ophthalmol. 2008, 92, 1434–1436. [Google Scholar] [CrossRef] [PubMed]
- Foyez, T.; Takeda-Uchimura, Y.; Ishigaki, S.; Narentuya; Zhang, Z.; Sobue, G.; Kadomatsu, K.; Uchimura, K. Microglial keratan sulfate epitope elicits in central nervous tissues of transgenic model mice and patients with amyotrophic lateral sclerosis. Am. J. Pathol. 2015, 185, 3053–3065. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Takeda-Uchimura, Y.; Foyez, T.; Ohtake-Niimi, S.; Narentuya; Akatsu, H.; Nishitsuji, K.; Michikawa, M.; Wyss-Coray, T.; Kadomatsu, K.; et al. Deficiency of a sulfotransferase for sialic acid-modified glycans mitigates Alzheimer’s pathology. Proc. Natl. Acad. Sci. USA 2017, 114, E2947–E2954. [Google Scholar] [CrossRef] [PubMed]
- Wakitani, S.; Nawata, M.; Kawaguchi, A.; Okabe, T.; Takaoka, K.; Tsuchiya, T.; Nakaoka, R.; Masuda, H.; Miyazaki, K. Serum keratan sulfate is a promising marker of early articular cartilage breakdown. Rheumatology 2007, 46, 1652–1656. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Leiphrakpam, P.D.; Patil, P.P.; Remmers, N.; Swanson, B.; Grandgenett, P.M.; Qiu, F.; Yu, F.; Radhakrishnan, P. Role of keratan sulfate expression in human pancreatic cancer malignancy. Sci. Rep. 2019, 9, 9665. [Google Scholar] [CrossRef] [PubMed]
- Zanetti, M.; Ratcliffe, A.; Watt, F.M. Two subpopulations of differentiated chondrocytes identified with a monoclonal antibody to keratan sulfate. J. Cell Biol. 1985, 101, 53–59. [Google Scholar] [CrossRef] [PubMed]
- Mehmet, H.; Scudder, P.; Tang, P.W.; Hounsell, E.F.; Caterson, B.; Feizi, T. The antigenic determinants recognized by three monoclonal antibodies to keratan sulphate involve sulphated hepta- or larger oligosaccharides of the poly(N-acetyllactosamine) series. Eur. J. Biochem. 1986, 157, 385–391. [Google Scholar] [CrossRef] [PubMed]
- Boyraz, B.; Saatz, J.; Pompös, I.-M.; Gad, M.; Dernedde, J.; Maier, A.-K.B.; Moscovitz, O.; Seeberger, P.H.; Traub, H.; Tauber, R. Imaging Keratan Sulfate in Ocular Tissue Sections by Immunofluorescence Microscopy and LA-ICP-MS. ACS Appl. Bio Mater. 2022, 5, 853–861. [Google Scholar] [CrossRef] [PubMed]
- Robbins, R.A.; Wagner, W.D.; Sawyer, L.M.; Caterson, B. Immunolocalization of proteoglycan types in aortas of pigeons with spontaneous or diet-induced atherosclerosis. Am. J. Pathol. 1989, 134, 615–626. [Google Scholar] [PubMed]
- Funderburgh, J.L.; Funderburgh, M.L.; Rodrigues, M.M.; Krachmer, J.H.; Conrad, G.W. Altered antigenicity of keratan sulfate proteoglycan in selected corneal diseases. Investig. Ophthalmol. Vis. Sci. 1990, 31, 419–428. [Google Scholar]
- Hasegawa, N.; Torii, T.; Kato, T.; Miyajima, H.; Furuhata, A.; Nakayasu, K.; Kanai, A.; Habuchi, O. Decreased GlcNAc 6-O-Sulfotransferase Activity in the Cornea with Macular Corneal Dystrophy. Investig. Ophthalmol. Vis. Sci. 2000, 41, 3670–3677. [Google Scholar]
- Matsui, H.; Ohgomori, T.; Natori, T.; Miyamoto, K.; Kusunoki, S.; Sakamoto, K.; Ishiguro, N.; Imagama, S.; Kadomatsu, K. Keratan sulfate expression in microglia is diminished in the spinal cord in experimental autoimmune neuritis. Cell Death Dis. 2013, 4, e946. [Google Scholar] [CrossRef]
- Colcher, D.; Bird, R.; Roselli, M.; Hardman, K.D.; Johnson, S.; Pope, S.; Dodd, S.W.; Pantoliano, M.W.; Milenic, D.E.; Schlom, J. In Vivo Tumor Targeting of a Recombinant Single-Chain Antigen-Binding Protein. JNCI J. Natl. Cancer Inst. 1990, 82, 1191–1197. [Google Scholar] [CrossRef]
- Ahmad, Z.A.; Yeap, S.K.; Ali, A.M.; Ho, W.Y.; Alitheen, N.B.; Hamid, M. scFv antibody: Principles and clinical application. Clin. Dev. Immunol. 2012, 2012, 980250. [Google Scholar] [CrossRef] [PubMed]
- Huston, J.S.; Levinson, D.; Mudgett-Hunter, M.; Tai, M.S.; Novotný, J.; Margolies, M.N.; Ridge, R.J.; Bruccoleri, R.E.; Haber, E.; Crea, R.; et al. Protein engineering of antibody binding sites: Recovery of specific activity in an anti-digoxin single-chain Fv analogue produced in Escherichia coli. Proc. Natl. Acad. Sci. USA 1988, 85, 5879–5883. [Google Scholar] [CrossRef] [PubMed]
- Muñoz-López, P.; Ribas-Aparicio, R.M.; Becerra-Báez, E.I.; Fraga-Pérez, K.; Flores-Martínez, L.F.; Mateos-Chávez, A.A.; Luria-Pérez, R. Single-Chain Fragment Variable: Recent Progress in Cancer Diagnosis and Therapy. Cancers 2022, 14, 4206. [Google Scholar] [CrossRef] [PubMed]
- Lensen, J.F.M.; Rops, A.L.W.M.M.; Wijnhoven, T.J.M.; Hafmans, T.; Feitz, W.F.J.; Oosterwijk, E.; Banas, B.; Bindels, R.J.M.; van den Heuvel, L.P.W.J.; van der Vlag, J.; et al. Localization and Functional Characterization of Glycosaminoglycan Domains in the Normal Human Kidney as Revealed by Phage Display-Derived Single Chain Antibodies. J. Am. Soc. Nephrol. 2005, 16, 1279–1288. [Google Scholar] [CrossRef]
- van der Steen, S.C.H.A.; van Tilborg, A.A.G.; Vallen, M.J.E.; Bulten, J.; van Kuppevelt, T.H.; Massuger, L.F.A.G. Prognostic significance of highly sulfated chondroitin sulfates in ovarian cancer defined by the single chain antibody GD3A11. Gynecol. Oncol. 2016, 140, 527–536. [Google Scholar] [CrossRef] [PubMed]
- Smetsers, T.F.C.M.; van de Westerlo, E.M.A.; ten Dam, G.B.; Overes, I.M.; Schalkwijk, J.; van Muijen, G.N.P.; van Kuppevelt, T.H. Human Single-Chain Antibodies Reactive with Native Chondroitin Sulfate Detect Chondroitin Sulfate Alterations in Melanoma and Psoriasis. J. Investig. Dermatol. 2004, 122, 707–716. [Google Scholar] [CrossRef]
- van Kuppevelt, T.H.; Dennissen, M.A.B.A.; van Venrooij, W.J.; Hoet, R.M.A.; Veerkamp, J.H. Generation and Application of Type-specific Anti-Heparan Sulfate Antibodies Using Phage Display Technology: Further evidence for heparan sulfate heterogeneity in the kidney. J. Biol. Chem. 1998, 273, 12960–12966. [Google Scholar] [CrossRef]
- Damen, L.A.A.; van de Westerlo, E.M.A.; Versteeg, E.M.M.; van Wessel, T.; Daamen, W.F.; van Kuppevelt, T.H. Construction and evaluation of an antibody phage display library targeting heparan sulfate. Glycoconj. J. 2020, 37, 445–455. [Google Scholar] [CrossRef]
- Rohatgi, S.; Ganju, P.; Sehgal, D. Systematic design and testing of nested (RT-)PCR primers for specific amplification of mouse rearranged/expressed immunoglobulin variable region genes from small number of B cells. J. Immunol. Methods 2008, 339, 205–219. [Google Scholar] [CrossRef] [PubMed]
- Ye, J.; Ma, N.; Madden, T.L.; Ostell, J.M. IgBLAST: An immunoglobulin variable domain sequence analysis tool. Nucleic Acids Res. 2013, 41, W34–W40. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Zaro, J.L.; Shen, W.C. Fusion protein linkers: Property, design and functionality. Adv. Drug Deliv. Rev. 2013, 65, 1357–1369. [Google Scholar] [CrossRef] [PubMed]
- Kipriyanov, S.M.; Moldenhauer, G.; Little, M. High level production of soluble single chain antibodies in small-scale Escherichia coli cultures. J. Immunol. Methods 1997, 200, 69–77. [Google Scholar] [CrossRef]
- Dewi, K.S.; Retnoningrum, D.S.; Riani, C.; Fuad, A.M. Construction and Periplasmic Expression of the Anti-EGFRvIII ScFv Antibody Gene in Escherichia coli. Sci. Pharm. 2016, 84, 141–152. [Google Scholar] [CrossRef] [PubMed]
- Sarker, A.; Rathore, A.S.; Gupta, R.D. Evaluation of scFv protein recovery from E. coli by in vitro refolding and mild solubilization process. Microb. Cell Factories 2019, 18, 5. [Google Scholar] [CrossRef]
- Song, H.N.; Jang, J.H.; Kim, Y.W.; Kim, D.H.; Park, S.G.; Lee, M.K.; Paek, S.H.; Woo, E.J. Refolded scFv antibody fragment against myoglobin shows rapid reaction kinetics. Int. J. Mol. Sci. 2014, 15, 23658–23671. [Google Scholar] [CrossRef] [PubMed]
- Umetsu, M.; Tsumoto, K.; Hara, M.; Ashish, K.; Goda, S.; Adschiri, T.; Kumagai, I. How Additives Influence the Refolding of Immunoglobulin-folded Proteins in a Stepwise Dialysis System: Spectroscopic evidence for highly efficient refolding of a single-chain Fv fragment. J. Biol. Chem. 2003, 278, 8979–8987. [Google Scholar] [CrossRef]
- Ozaki, C.Y.; Silveira, C.R.; Andrade, F.B.; Nepomuceno, R.; Silva, A.; Munhoz, D.D.; Yamamoto, B.B.; Luz, D.; Abreu, P.A.; Horton, D.S.; et al. Single Chain Variable Fragments Produced in Escherichia coli against Heat-Labile and Heat-Stable Toxins from Enterotoxigenic E. coli. PLoS ONE 2015, 10, e0131484. [Google Scholar] [CrossRef] [PubMed]
- Koçer, İ.; Cox, E.C.; DeLisa, M.P.; Çelik, E. Effects of variable domain orientation on anti-HER2 single-chain variable fragment antibody expressed in the Escherichia coli cytoplasm. Biotechnol. Prog. 2021, 37, e3102. [Google Scholar] [CrossRef] [PubMed]
- Baneyx, F.; Mujacic, M. Recombinant protein folding and misfolding in Escherichia coli. Nat. Biotechnol. 2004, 22, 1399–1408. [Google Scholar] [CrossRef]
- Choi, J.H.; Lee, S.Y. Secretory and extracellular production of recombinant proteins using Escherichia coli. Appl. Microbiol. Biotechnol. 2004, 64, 625–635. [Google Scholar] [CrossRef]
- Bach, H.; Mazor, Y.; Shaky, S.; Shoham-Lev, A.; Berdichevsky, Y.; Gutnick, D.L.; Benhar, I. Escherichia coli maltose-binding protein as a molecular chaperone for recombinant intracellular cytoplasmic single-chain antibodies. J. Mol. Biol. 2001, 312, 79–93. [Google Scholar] [CrossRef] [PubMed]
- Hira, V.V.V.; de Jong, A.L.; Ferro, K.; Khurshed, M.; Molenaar, R.J.; Van Noorden, C.J.F. Comparison of different methodologies and cryostat versus paraffin sections for chromogenic immunohistochemistry. Acta Histochem. 2019, 121, 125–134. [Google Scholar] [CrossRef]
- Quantock, A.J.; Young, R.D.; Akama, T.O. Structural and biochemical aspects of keratan sulphate in the cornea. Cell. Mol. Life Sci. 2010, 67, 891–906. [Google Scholar] [CrossRef]
- Puri, S.; Coulson-Thomas, Y.M.; Gesteira, T.F.; Coulson-Thomas, V.J. Distribution and Function of Glycosaminoglycans and Proteoglycans in the Development, Homeostasis and Pathology of the Ocular Surface. Front. Cell Dev. Biol. 2020, 8, 731. [Google Scholar] [CrossRef] [PubMed]
- Akhtar, S.; Kerr, B.C.; Hayes, A.J.; Hughes, C.E.; Meek, K.M.; Caterson, B. Immunochemical Localization of Keratan Sulfate Proteoglycans in Cornea, Sclera, and Limbus Using a Keratanase-Generated Neoepitope Monoclonal Antibody. Investig. Ophthalmol. Vis. Sci. 2008, 49, 2424–2431. [Google Scholar] [CrossRef] [PubMed]
- Azuma, N.; Hirakata, A.; Hida, T.; Kohsaka, S. Histochemical and immunohistochemical studies on keratan sulfate in the anterior segment of the developing human eye. Exp. Eye Res. 1994, 58, 277–286. [Google Scholar] [CrossRef] [PubMed]
- Brown, G.M.; Huckerby, T.N.; Morris, H.G.; Abram, B.L.; Nieduszynski, I.A. Oligosaccharides derived from bovine articular cartilage keratan sulfates after keratanase II digestion: Implications for keratan sulfate structural fingerprinting. Biochemistry 1994, 33, 4836–4846. [Google Scholar] [CrossRef]
- Brown, G.M.; Huckerby, T.N.; Nieduszynski, I.A. Oligosaccharides derived by keratanase II digestion of bovine articular cartilage keratan sulphates. Eur. J. Biochem. 1994, 224, 281–308. [Google Scholar] [CrossRef]
- Yamagishi, K.; Suzuki, K.; Imai, K.; Mochizuki, H.; Morikawa, K.; Kyogashima, M.; Kimata, K.; Watanabe, H. Purification, Characterization, and Molecular Cloning of a Novel Keratan Sulfate Hydrolase, Endo-β-N-acetylglucosaminidase, from Bacillus circulans. J. Biol. Chem. 2003, 278, 25766–25772. [Google Scholar] [CrossRef]
- Wang, H.; He, W.; Jiang, P.; Yu, Y.; Lin, L.; Sun, X.; Koffas, M.; Zhang, F.; Linhardt, R.J. Construction and functional characterization of truncated versions of recombinant keratanase II from Bacillus circulans. Glycoconj. J. 2017, 34, 643–649. [Google Scholar] [CrossRef]
- Caterson, B.; Christner, J.E.; Baker, J.R. Identification of a monoclonal antibody that specifically recognizes corneal and skeletal keratan sulfate. Monoclonal antibodies to cartilage proteoglycan. J. Biol. Chem. 1983, 258, 8848–8854. [Google Scholar] [CrossRef]
- Hayashida, Y.; Akama, T.O.; Beecher, N.; Lewis, P.; Young, R.D.; Meek, K.M.; Kerr, B.; Hughes, C.E.; Caterson, B.; Tanigami, A.; et al. Matrix morphogenesis in cornea is mediated by the modification of keratan sulfate by GlcNAc 6-O-sulfotransferase. Proc. Natl. Acad. Sci. USA 2006, 103, 13333–13338. [Google Scholar] [CrossRef]
- Kawabe, K.; Tateyama, D.; Toyoda, H.; Kawasaki, N.; Hashii, N.; Nakao, H.; Matsumoto, S.; Nonaka, M.; Matsumura, H.; Hirose, Y.; et al. A novel antibody for human induced pluripotent stem cells and embryonic stem cells recognizes a type of keratan sulfate lacking oversulfated structures. Glycobiology 2012, 23, 322–336. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Boyraz, B.; Tauber, R.; Dernedde, J. Identification of an Immunoglobulin Paratope Binding to Keratan Sulfate and Expression of a Single-Chain Derivative for Imaging. Biomolecules 2025, 15, 178. https://doi.org/10.3390/biom15020178
Boyraz B, Tauber R, Dernedde J. Identification of an Immunoglobulin Paratope Binding to Keratan Sulfate and Expression of a Single-Chain Derivative for Imaging. Biomolecules. 2025; 15(2):178. https://doi.org/10.3390/biom15020178
Chicago/Turabian StyleBoyraz, Burak, Rudolf Tauber, and Jens Dernedde. 2025. "Identification of an Immunoglobulin Paratope Binding to Keratan Sulfate and Expression of a Single-Chain Derivative for Imaging" Biomolecules 15, no. 2: 178. https://doi.org/10.3390/biom15020178
APA StyleBoyraz, B., Tauber, R., & Dernedde, J. (2025). Identification of an Immunoglobulin Paratope Binding to Keratan Sulfate and Expression of a Single-Chain Derivative for Imaging. Biomolecules, 15(2), 178. https://doi.org/10.3390/biom15020178







