Oncogenic Role of SAMD4B in Breast Cancer Progression by Activating Wnt/β-Catenin Pathway
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Lines and Cell Culture
2.2. Construction of Stable Breast Cancer Cell Lines
2.3. Reverse Transcription (RT)-qPCR
2.4. Western Blotting
2.5. MTT Assay
2.6. Plate Colony Formation Assay
2.7. Cell Cycle Analysis
2.8. Dual-Luciferase Reporter Gene Assay
2.9. Wound Healing Assay
2.10. Transwell Assay
2.11. Immunofluorescence Analysis
2.12. mRNA Stability Analysis
2.13. RNA Immunoprecipitation (RIP) Assay
2.14. Bioinformatics Analysis
2.15. Statistical Analysis
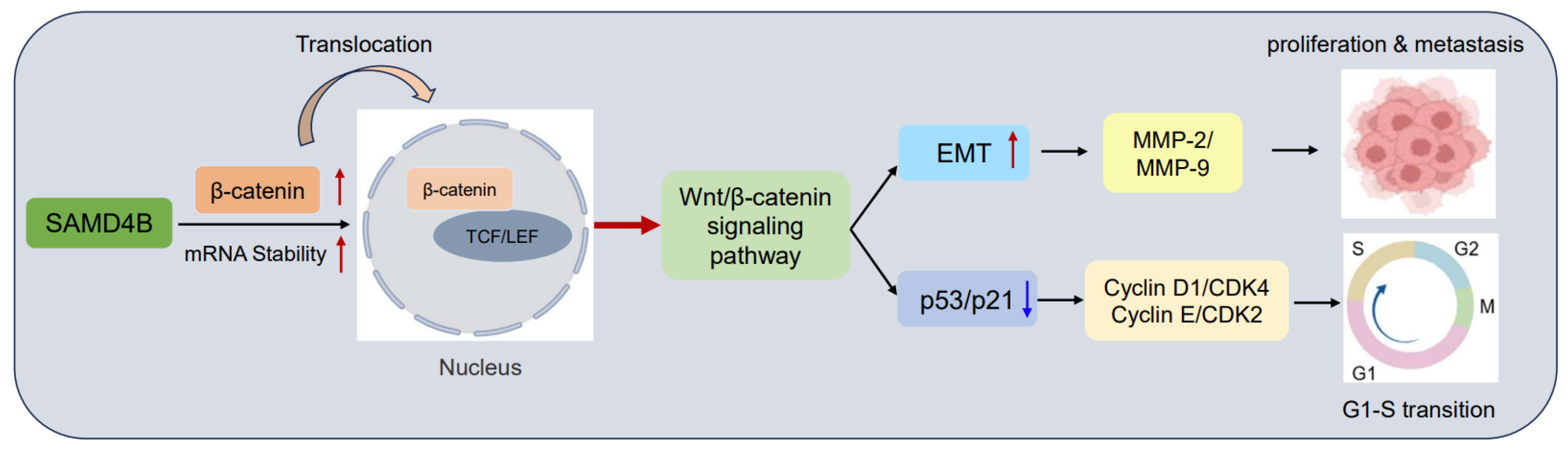
3. Results
3.1. SAMD4B Expression Is Upregulated in Breast Cancer
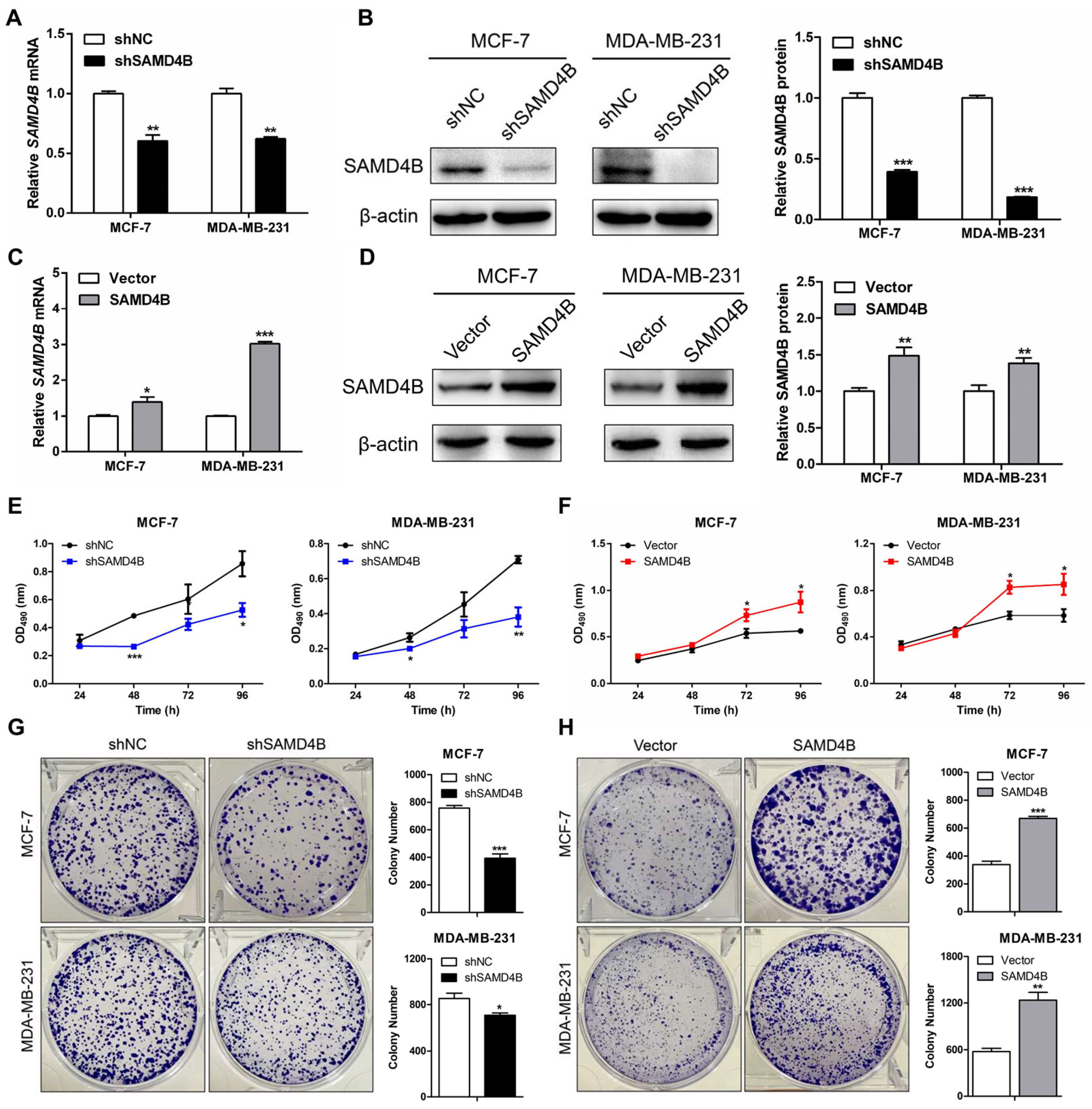
3.2. SAMD4B Promotes the Growth and Proliferation of Breast Cancer Cells
3.3. SAMD4B Promotes G1-S Phase Transition of Breast Cancer Cells
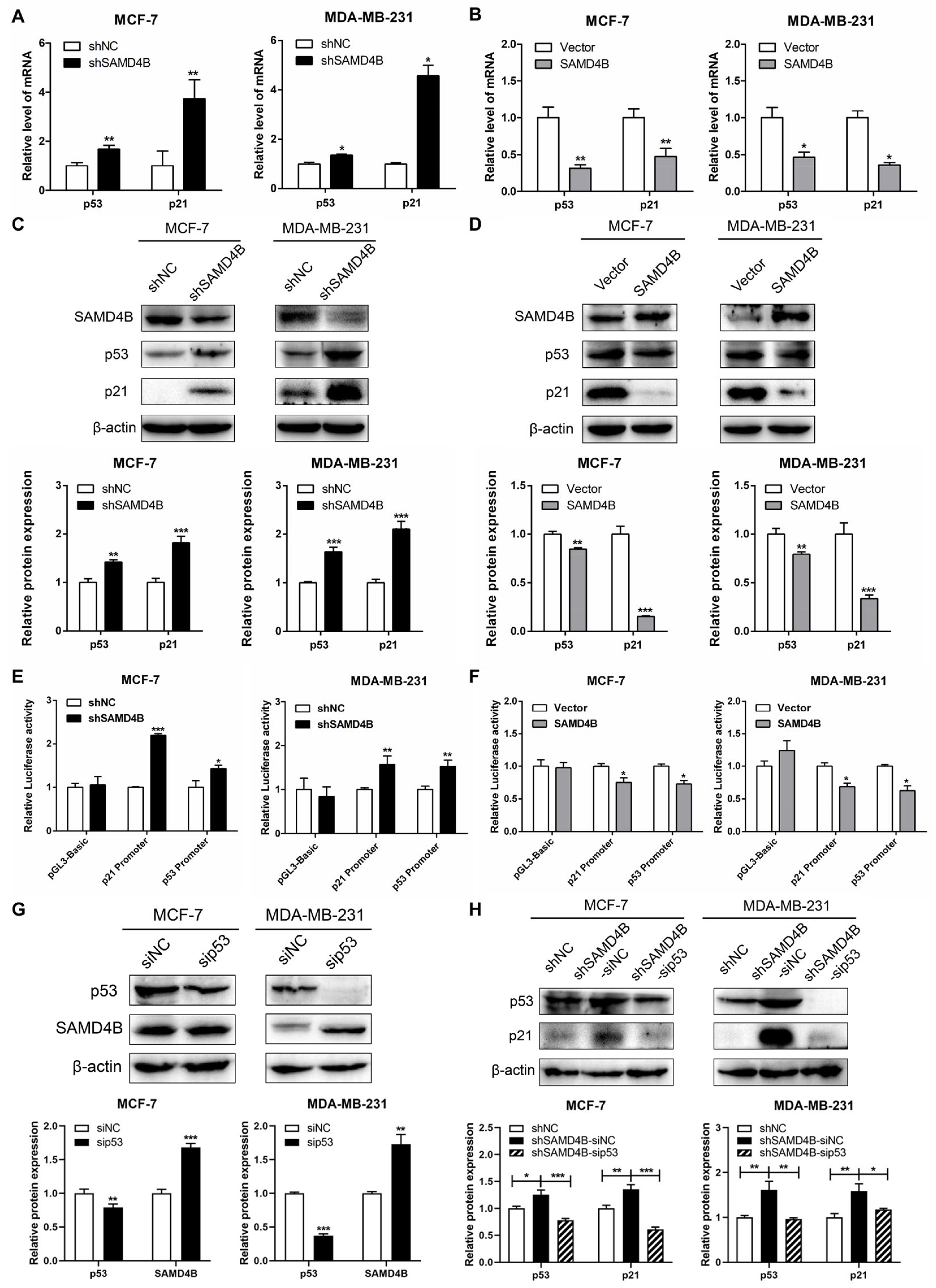
3.4. SAMD4B Affects Breast Cancer Cell Cycle Through Regulating p53 Expression
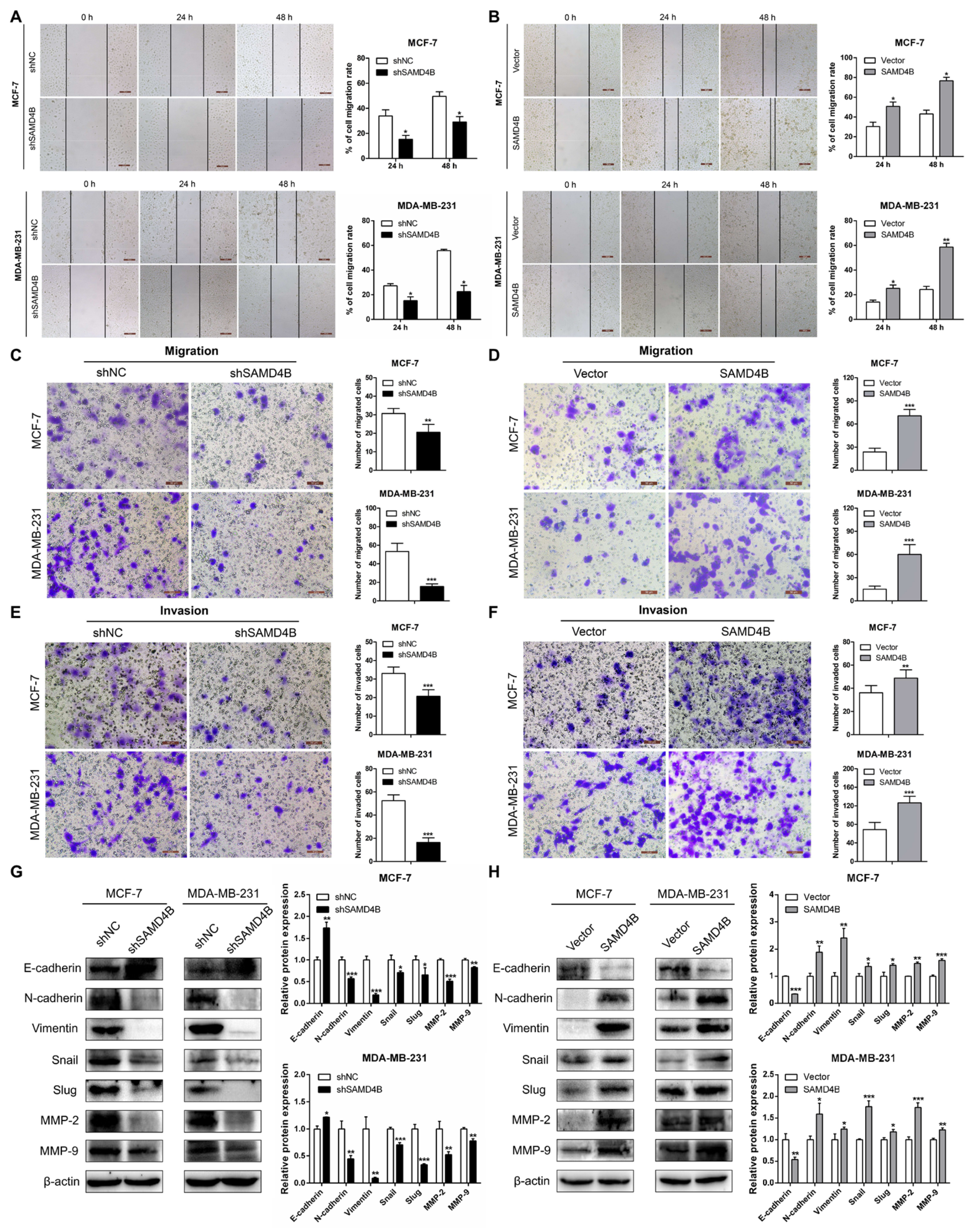
3.5. SAMD4B Promotes Migration and Invasion of Breast Cancer Cells Through Inducing EMT
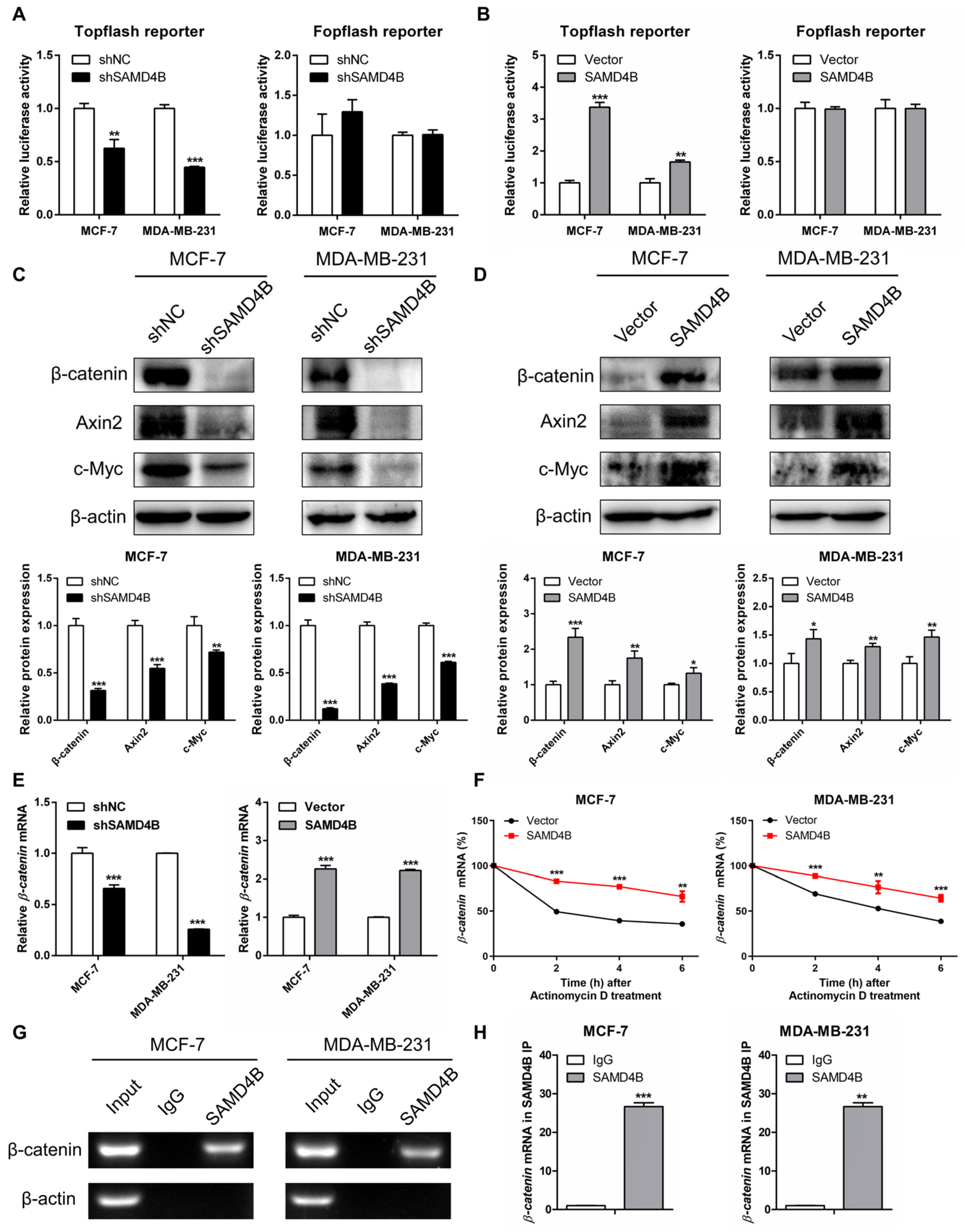
3.6. SAMD4B Activates the Wnt/β-Catenin Pathway in Breast Cancer Cells
3.7. Inhibition of Wnt/β-Catenin Pathway Reverses the Promoting Effects of SAMD4B Overexpression on Breast Cancer Progression
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| SAMD4B | Sterile alpha motif domain-containing protein 4B |
| EMT | Epithelial–mesenchymal transition |
| MMP | Matrix metalloproteinase |
| CDK | Cyclin-dependent kinases |
| RIP | RNA immunoprecipitation |
| HBV | Human hepatitis B virus |
| UTR | Untranslated region |
| CCR4 | Carbon Catabolite Repressor 4 |
| POP2 | Poly(A) polymerase-associated protein 2 |
| KDM2B | Lysine (K)-specific demethylase 2B |
| FGF2 | Fibroblast growth factor 2 |
| ATF4 | Activating transcription factor 4 |
| hnRNA | Heterogeneous nuclear RNA |
| BAG2 | Bcl2-associated athanogene 2 |
| RACK1 | Receptor for activated C kinase 1 |
References
- Bray, F.; Laversanne, M.; Sung, H.; Ferlay, J.; Siegel, R.L.; Soerjomataram, I.; Jemal, A. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2024, 74, 229–263. [Google Scholar] [CrossRef]
- Montazeri Aliabadi, H. Molecular Targets for Breast Cancer Therapy. Biomolecules 2024, 14, 1219. [Google Scholar] [CrossRef] [PubMed]
- Ye, F.; Dewanjee, S.; Li, Y.; Jha, N.K.; Chen, Z.S.; Kumar, A.; Vishakha; Behl, T.; Jha, S.K.; Tang, H. Advancements in clinical aspects of targeted therapy and immunotherapy in breast cancer. Mol. Cancer 2023, 22, 105. [Google Scholar] [CrossRef]
- Lancaster, C.L.; Moberg, K.H.; Corbett, A.H. Post-Transcriptional Regulation of Gene Expression and the Intricate Life of Eukaryotic mRNAs. Wiley Interdiscip. Rev. RNA 2025, 16, e70007. [Google Scholar] [CrossRef]
- Gerstberger, S.; Hafner, M.; Tuschl, T. A census of human RNA-binding proteins. Nat. Rev. Genet. 2014, 15, 829–845. [Google Scholar] [CrossRef]
- Gebauer, F.; Schwarzl, T.; Valcárcel, J.; Hentze, M.W. RNA-binding proteins in human genetic disease. Nat. Rev. Genet. 2021, 22, 185–198. [Google Scholar] [CrossRef]
- Wolozin, B.; Ivanov, P. Stress granules and neurodegeneration. Nat. Rev. Neurosci. 2019, 20, 649–666. [Google Scholar] [CrossRef]
- Li, W.; Deng, X.; Chen, J. RNA-binding proteins in regulating mRNA stability and translation: Roles and mechanisms in cancer. Semin. Cancer Biol. 2022, 86 Pt 2, 664–677. [Google Scholar] [CrossRef]
- Wang, S.; Sun, H.; Chen, G.; Wu, C.; Sun, B.; Lin, J.; Lin, D.; Zeng, D.; Lin, B.; Huang, G.; et al. RNA-binding proteins in breast cancer: Biological implications and therapeutic opportunities. Crit. Rev. Oncol./Hematol. 2024, 195, 104271. [Google Scholar] [CrossRef]
- Lu, X.; Zhong, J.; Liu, L.; Zhang, W.; Zhao, S.; Chen, L.; Wei, Y.; Zhang, H.; Wu, J.; Chen, W.; et al. The function and regulatory mechanism of RNA-binding proteins in breast cancer and their future clinical treatment prospects. Front. Oncol. 2022, 12, 929037. [Google Scholar] [CrossRef]
- Zou, H.; Luo, J.; Guo, Y.; Liu, Y.; Wang, Y.; Deng, L.; Li, P. RNA-binding protein complex LIN28/MSI2 enhances cancer stem cell-like properties by modulating Hippo-YAP1 signaling and independently of Let-7. Oncogene 2022, 41, 1657–1672. [Google Scholar] [CrossRef] [PubMed]
- Shen, M.; Zhang, R.; Jia, W.; Zhu, Z.; Zhao, L.; Huang, G.; Liu, J. RNA-binding protein p54(nrb)/NONO potentiates nuclear EGFR-mediated tumorigenesis of triple-negative breast cancer. Cell Death Dis. 2022, 13, 42. [Google Scholar] [CrossRef]
- Wang, X.Y.; Zhang, L.N. RNA binding protein SAMD4: Current knowledge and future perspectives. Cell Biosci. 2023, 13, 21. [Google Scholar] [CrossRef]
- Aviv, T.; Lin, Z.; Lau, S.; Rendl, L.M.; Sicheri, F.; Smibert, C.A. The RNA-binding SAM domain of Smaug defines a new family of post-transcriptional regulators. Nat. Struct. Biol. 2003, 10, 614–621. [Google Scholar] [CrossRef]
- Dahanukar, A.; Walker, J.A.; Wharton, R.P. Smaug, a novel RNA-binding protein that operates a translational switch in Drosophila. Mol. Cell 1999, 4, 209–218. [Google Scholar] [CrossRef]
- Green, J.B.; Gardner, C.D.; Wharton, R.P.; Aggarwal, A.K. RNA recognition via the SAM domain of Smaug. Mol. Cell 2003, 11, 1537–1548. [Google Scholar] [CrossRef]
- Semotok, J.L.; Cooperstock, R.L.; Pinder, B.D.; Vari, H.K.; Lipshitz, H.D.; Smibert, C.A. Smaug recruits the CCR4/POP2/NOT deadenylase complex to trigger maternal transcript localization in the early Drosophila embryo. Curr. Biol. 2005, 15, 284–294. [Google Scholar] [CrossRef]
- Chen, L.; Dumelie, J.G.; Li, X.; Cheng, M.H.; Yang, Z.; Laver, J.D.; Siddiqui, N.U.; Westwood, J.T.; Morris, Q.; Lipshitz, H.D.; et al. Global regulation of mRNA translation and stability in the early Drosophila embryo by the Smaug RNA-binding protein. Genome Biol. 2014, 15, R4. [Google Scholar] [CrossRef]
- Tadros, W.; Goldman, A.L.; Babak, T.; Menzies, F.; Vardy, L.; Orr-Weaver, T.; Hughes, T.R.; Westwood, J.T.; Smibert, C.A.; Lipshitz, H.D. SMAUG is a major regulator of maternal mRNA destabilization in Drosophila and its translation is activated by the PAN GU kinase. Dev. Cell 2007, 12, 143–155. [Google Scholar] [CrossRef]
- Semotok, J.L.; Luo, H.; Cooperstock, R.L.; Karaiskakis, A.; Vari, H.K.; Smibert, C.A.; Lipshitz, H.D. Drosophila maternal Hsp83 mRNA destabilization is directed by multiple SMAUG recognition elements in the open reading frame. Mol. Cell. Biol. 2008, 28, 6757–6772. [Google Scholar] [CrossRef]
- Nelson, M.R.; Leidal, A.M.; Smibert, C.A. Drosophila Cup is an eIF4E-binding protein that functions in Smaug-mediated translational repression. EMBO J. 2004, 23, 150–159. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Fan, X.; Song, Y.; Liu, Y.; Liu, R.; Wu, J.; Li, X.; Yuan, Q.; Fu, G.; Xia, N.; et al. SAMD4 family members suppress human hepatitis B virus by directly binding to the Smaug recognition region of viral RNA. Cell. Mol. Immunol. 2021, 18, 1032–1044. [Google Scholar] [CrossRef] [PubMed]
- Niu, N.; Xiang, J.F.; Yang, Q.; Wang, L.; Wei, Z.; Chen, L.L.; Yang, L.; Zou, W. RNA-binding protein SAMD4 regulates skeleton development through translational inhibition of Mig6 expression. Cell Discov. 2017, 3, 16050. [Google Scholar] [CrossRef] [PubMed]
- Yi, N.; Wang, H.R.; Zhu, Y.P.; Xiao, T.; Lin, Q.; Liu, H.; Meng, Y.L.; Sun, Y.Z.; Lin, F.; Hu, S.Y.; et al. RNA-binding protein SAMD4A targets FGF2 to regulate cardiomyocyte lineage specification from human embryonic stem cells. Stem Cell Res. Ther. 2025, 16, 144. [Google Scholar] [CrossRef]
- Chen, Q.; Liu, S.; Zhou, H.; Wang, J.; Xiao, X.; Chen, G.; Du, J.; Zhong, L.; Song, H.; Huang, X. SAMD4A inhibits abdominal aortic aneurysm development and VSMC phenotypic transformation through targeting KDM2B. J. Adv. Res. 2025, in press. [Google Scholar] [CrossRef]
- Zhou, M.; Wang, B.; Li, H.; Han, J.; Li, A.; Lu, W. RNA-binding protein SAMD4A inhibits breast tumor angiogenesis by modulating the balance of angiogenesis program. Cancer Sci. 2021, 112, 3835–3845. [Google Scholar] [CrossRef]
- Li, D.; Qi, T.; Chen, J. SAMD4A serves as a negative prognostic marker for gastric cancer patients. Tissue Cell 2023, 84, 102167. [Google Scholar] [CrossRef]
- Luo, N.; Li, G.; Li, Y.; Fan, X.; Wang, Y.; Ye, X.; Mo, X.; Zhou, J.; Yuan, W.; Tan, M.; et al. SAMD4B, a novel SAM-containing protein, inhibits AP-1-, p53- and p21-mediated transcriptional activity. BMB Rep. 2010, 43, 355–361. [Google Scholar] [CrossRef]
- Wu, C.; Liu, X.; Li, B.; Sun, G.; Peng, C.; Xiang, D. miR-451 suppresses the malignant characteristics of colorectal cancer via targeting SAMD4B. Mol. Med. Rep. 2021, 24, 557. [Google Scholar] [CrossRef]
- Chen, M.Y.; Sun, C.Y.; Zhao, R.; Guan, X.L.; Li, M.L.; Zhang, F.; Wan, Z.H.; Feng, J.X.; Yin, M.; Lei, Q.Y.; et al. BAG2 releases SAMD4B upon sensing of arginine deficiency to promote tumor cell survival. Mol. Cell 2025, 85, 2581–2596.e6. [Google Scholar] [CrossRef]
- Huang, Y.; Hong, W.; Wei, X. The molecular mechanisms and therapeutic strategies of EMT in tumor progression and metastasis. J. Hematol. Oncol. 2022, 15, 129. [Google Scholar] [CrossRef]
- Lamouille, S.; Xu, J.; Derynck, R. Molecular mechanisms of epithelial-mesenchymal transition. Nat. Rev. Mol. Cell Biol. 2014, 15, 178–196. [Google Scholar] [CrossRef]
- Xue, W.; Yang, L.; Chen, C.; Ashrafizadeh, M.; Tian, Y.; Sun, R. Wnt/β-catenin-driven EMT regulation in human cancers. Cell. Mol. Life Sci. CMLS 2024, 81, 79. [Google Scholar] [CrossRef] [PubMed]
- Parichha, A.; Suresh, V.; Chatterjee, M.; Kshirsagar, A.; Ben-Reuven, L.; Olender, T.; Taketo, M.M.; Radosevic, V.; Bobic-Rasonja, M.; Trnski, S.; et al. Constitutive activation of canonical Wnt signaling disrupts choroid plexus epithelial fate. Nat. Commun. 2022, 13, 633. [Google Scholar] [CrossRef] [PubMed]
- Bertoli, C.; Skotheim, J.M.; de Bruin, R.A. Control of cell cycle transcription during G1 and S phases. Nat. Rev. Mol. Cell Biol. 2013, 14, 518–528. [Google Scholar] [CrossRef]
- Chen, J. The Cell-Cycle Arrest and Apoptotic Functions of p53 in Tumor Initiation and Progression. Cold Spring Harb. Perspect. Med. 2016, 6, a026104. [Google Scholar] [CrossRef]
- Mello, S.S.; Attardi, L.D. Deciphering p53 signaling in tumor suppression. Curr. Opin. Cell Biol. 2018, 51, 65–72. [Google Scholar] [CrossRef]
- Bakir, B.; Chiarella, A.M.; Pitarresi, J.R.; Rustgi, A.K. EMT, MET, Plasticity, and Tumor Metastasis. Trends Cell Biol. 2020, 30, 764–776. [Google Scholar] [CrossRef]
- Xu, X.; Zhang, M.; Xu, F.; Jiang, S. Wnt signaling in breast cancer: Biological mechanisms, challenges and opportunities. Mol. Cancer 2020, 19, 165. [Google Scholar] [CrossRef]
- Qin, H.; Ni, H.; Liu, Y.; Yuan, Y.; Xi, T.; Li, X.; Zheng, L. RNA-binding proteins in tumor progression. J. Hematol. Oncol. 2020, 13, 90. [Google Scholar] [CrossRef]
- Pereira, B.; Billaud, M.; Almeida, R. RNA-Binding Proteins in Cancer: Old Players and New Actors. Trends Cancer 2017, 3, 506–528. [Google Scholar] [CrossRef]
- Smibert, C.A.; Lie, Y.S.; Shillinglaw, W.; Henzel, W.J.; Macdonald, P.M. Smaug, a novel and conserved protein, contributes to repression of nanos mRNA translation in vitro. RNA 1999, 5, 1535–1547. [Google Scholar] [CrossRef] [PubMed]
- Baez, M.V.; Boccaccio, G.L. Mammalian Smaug is a translational repressor that forms cytoplasmic foci similar to stress granules. J. Biol. Chem. 2005, 280, 43131–43140. [Google Scholar] [CrossRef] [PubMed]
- Klejewski, A.; Świerczewska, M.; Zaorska, K.; Brązert, M.; Nowicki, M.; Zabel, M.; Januchowski, R. New and Old Genes Associated with Topotecan Resistance Development in Ovarian Cancer Cell Lines. Anticancer. Res. 2017, 37, 1625–1636. [Google Scholar] [CrossRef] [PubMed]
- Januchowski, R.; Sterzyńska, K.; Zawierucha, P.; Ruciński, M.; Świerczewska, M.; Partyka, M.; Bednarek-Rajewska, K.; Brązert, M.; Nowicki, M.; Zabel, M.; et al. Microarray-based detection and expression analysis of new genes associated with drug resistance in ovarian cancer cell lines. Oncotarget 2017, 8, 49944–49958. [Google Scholar] [CrossRef]
- Kayser, S.; Hills, R.K.; Langova, R.; Kramer, M.; Guijarro, F.; Sustkova, Z.; Estey, E.H.; Shaw, C.M.; Ráčil, Z.; Mayer, J.; et al. Characteristics and outcome of patients with acute myeloid leukaemia and t(8;16)(p11;p13): Results from an International Collaborative Study. Br. J. Haematol. 2021, 192, 832–842. [Google Scholar] [CrossRef]
- Liu, Y.; Su, Z.; Tavana, O.; Gu, W. Understanding the complexity of p53 in a new era of tumor suppression. Cancer Cell 2024, 42, 946–967. [Google Scholar] [CrossRef]
- Brabletz, S.; Schuhwerk, H.; Brabletz, T.; Stemmler, M.P. Dynamic EMT: A multi-tool for tumor progression. EMBO J. 2021, 40, e108647. [Google Scholar] [CrossRef]
- Hashemi, M.; Hasani, S.; Hajimazdarany, S.; Ghadyani, F.; Olyaee, Y.; Khodadadi, M.; Ziyarani, M.F.; Dehghanpour, A.; Salehi, H.; Kakavand, A.; et al. Biological functions and molecular interactions of Wnt/β-catenin in breast cancer: Revisiting signaling networks. Int. J. Biol. Macromol. 2023, 232, 123377. [Google Scholar] [CrossRef]
- Buoso, E.; Ronfani, M.; Galasso, M.; Ventura, D.; Corsini, E.; Racchi, M. Cortisol-induced SRSF3 expression promotes GR splicing, RACK1 expression and breast cancer cells migration. Pharmacol. Res. 2019, 143, 17–26. [Google Scholar] [CrossRef]
- Tian, R.; Tian, J.; Zuo, X.; Ren, S.; Zhang, H.; Liu, H.; Wang, Z.; Cui, Y.; Niu, R.; Zhang, F. RACK1 facilitates breast cancer progression by competitively inhibiting the binding of β-catenin to PSMD2 and enhancing the stability of β-catenin. Cell Death Dis. 2023, 14, 685. [Google Scholar] [CrossRef]



| Gene | Forward Primer (5ʹ-3ʹ) | Reverse Primer (5ʹ -3ʹ) |
|---|---|---|
| SAMD4B p21 p53 β-catenin GAPDH | CCTGCCAAATCCACCCTAGC GGCGAGGCCGGGATGAGTTG GAGGTTGGCTCTGACTGTACC AGCTTCCAGACACGCTATCAT CGCTCTCTGCTCCTCCTGTT | TGATCGGGCGTGGTAAAAGC CTGCCGCCGTTTTCGACCCT TCCGTCCCAGTAGATTACCAC CGGTACAACGAGCTGTTTCTAC CCATGGTGTCTGAGCGATGT |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, J.-H.; Wang, X.-Y.; Song, H.-X.; Nie, X.-F.; Zhang, L.-N. Oncogenic Role of SAMD4B in Breast Cancer Progression by Activating Wnt/β-Catenin Pathway. Biomolecules 2025, 15, 1423. https://doi.org/10.3390/biom15101423
Li J-H, Wang X-Y, Song H-X, Nie X-F, Zhang L-N. Oncogenic Role of SAMD4B in Breast Cancer Progression by Activating Wnt/β-Catenin Pathway. Biomolecules. 2025; 15(10):1423. https://doi.org/10.3390/biom15101423
Chicago/Turabian StyleLi, Jia-Hui, Xin-Ya Wang, Huan-Xi Song, Xiao-Fei Nie, and Li-Na Zhang. 2025. "Oncogenic Role of SAMD4B in Breast Cancer Progression by Activating Wnt/β-Catenin Pathway" Biomolecules 15, no. 10: 1423. https://doi.org/10.3390/biom15101423
APA StyleLi, J.-H., Wang, X.-Y., Song, H.-X., Nie, X.-F., & Zhang, L.-N. (2025). Oncogenic Role of SAMD4B in Breast Cancer Progression by Activating Wnt/β-Catenin Pathway. Biomolecules, 15(10), 1423. https://doi.org/10.3390/biom15101423






