Bioinformatics Strategies in Breast Cancer Research
Abstract
1. Introduction
2. Bioinformatics Tools and Techniques for Biomarker Discovery
2.1. Data Acquisition
2.2. Data Collection and Organization
- (1)
- Sequence databases. These encompass both primary and secondary collections of DNA, RNA, or protein sequences. Besides the above-mentioned GenBank collection, other online available databases include the following: (i) EMBL (EMBL Nucleotide Sequence Database), a comprehensive repository of nucleotide sequences and annotations maintained by EMBL’s European Bioinformatics Institute (EMBL-EBI), drawing data from public databases [34]; (ii) RNAcentral, a public resource that provides integrated access to a continually updated, extensive collection of non-coding RNA sequences [35]; (iii) UniProt, a freely accessible resource for protein sequences and their functional annotations. The UniProt Knowledgebase (UniProtKB), containing over 227 million sequences, is continuously updated by the UniProt team using machine learning and data extracted from scientific literature [36].
- (2)
- Gene expression databases. These store information about gene expression patterns across various cell types, tissues, or organisms at different times or under specific conditions. These databases enable, for example, the comparison of gene expression levels in healthy versus tumor tissues or in tissues treated with a placebo versus those treated with a drug. Gene Expression Omnibus (GEO) is a public functional genomics data repository, enabling researchers to explore and analyze gene expression data, including both raw and processed data; it also offers web tools that allow users to analyze and interpret data [37]. The Cancer Genome Atlas (TCGA) is a comprehensive database, containing over 20,000 tumor and matched normal samples across 33 of the most prevalent forms of cancer, all molecularly characterized at the DNA (copy number changes, epigenetic modifications), RNA (messenger RNA and microRNA), and protein levels [38].
- (3)
- Genetic databases. These contain information on genetic variants, including mutations, SNPs, and other genomic modifications linked to genetic diseases and pathological conditions. Examples include the following: (i) Clinical Genome Resource (ClinVar) catalogs various types of structural variants (SNPs, copy number variations, inversions, and translocations) together with their association with diseases [39]. Clinical relevance information for these variants is contributed by clinical testing labs, research institutions, and expert groups [39]. (ii) Single-Nucleotide Polymorphism Database (dbSNP) stores information on single nucleotide variants, microsatellites, insertions, and deletions that are prevalent in the human genome, useful for cancer research and genetic association studies [40,41].
- (4)
- Molecular structure databases. These provide access to the three-dimensional (3D) structures of biological molecules. Understanding these structures is critical for elucidating their functions and roles within cells. Among the most significant databases deserve mention: (i) Protein Data Bank (PDB), an open-access repository that houses over 210,000 experimentally validated 3D structures of proteins and nucleic acids [42]. The database is weekly updated with relevant functional annotations sourced from various external biodata resources [43]; (ii) Structural Classification of Proteins (SCOP), a database that organizes proteins with known 3D structures based on their evolutionary and structural relationships [44].
- (5)
- Molecular interaction databases. These focus on biomolecular interactions, particularly protein–protein interaction (PPIs). By identifying biological pathways, molecular patterns, and discovering new protein functions, these databases can elucidate the molecular basis of various pathologies, making them valuable tools for prevention, diagnosis, and therapy [45]. Databases in this category include the following: (i) Biological General Repository for Interaction Datasets (BioGRID), which provides comprehensive information on protein and genetic interactions across multiple species (including yeast, mice, and humans), thus allowing users to create intricate network graphs [46]; (ii) STRING, a key resource for studying physical and functional PPIs, deriving from experimental interaction databases, scientific literature, and computational predictions based on co-expression [47]; (iii) IntAct, a curated database system and analysis tool for investigating molecular interactions derived from scientific literature and direct data submissions. IntAct features over one million binary interactions and is continuously updated, with annotations that detail how even minor sequence changes can affect protein interactions [48].
- (6)
- Biological Pathway Databases. These provide valuable insights into the biological roles of molecules and the metabolic pathways they participate in. The most common functional database is Kyoto Encyclopedia of Genes and Genomes (KEGG), a comprehensive database designed to assign functional meanings to genes and genomes at both molecular and broader biological levels. This integrated resource combines 15 manually curated databases with one computationally generated database, organized into four main categories (systems, genomic, information, and health information). KEGG serves as a vital tool for studying metabolism, genetic pathways, organismal functions, and human diseases [49].
| Database | Name | Details | Website (10 June 2025) |
|---|---|---|---|
| Sequence | GenBank | DNA sequences | https://www.ncbi.nlm.nih.gov/genbank/ |
| EMBL | Nucleotide sequences and annotations | https://www.ebi.ac.uk/embl/ | |
| RNAcentral | Non-coding RNA sequences and annotations | https://rnacentral.org/ | |
| UniProt | Protein sequences and annotations | https://www.uniprot.org/ | |
| Gene expression | GEO | Multi-omics data | https://www.ncbi.nlm.nih.gov/geo/ |
| TCGA | Multi-omics data | https://www.cancer.gov/ccg/research/genome-sequencing/tcga | |
| Genetic | ClinVar | Genetic variants and associations with diseases | https://www.ncbi.nlm.nih.gov/clinvar/ |
| dbSNP | Small genetic variations | https://www.ncbi.nlm.nih.gov/snp/ | |
| Molecular structure | PDB | 3D structure of proteins, nucleic acids and complexes with functional annotations | https://www.rcsb.org/ |
| SCOP | Evolutionary structure and structural relationships of proteins | https://scop.mrc-lmb.cam.ac.uk/ | |
| Molecular interactions | BioGRID | Protein, genetic and chemical interactions | https://thebiogrid.org/ |
| STRING | Protein–protein interactions | https://string-db.org/ | |
| IntAct | Molecular interactions for macromolecular complexes | https://www.ebi.ac.uk/intact/ | |
| Biological Pathways | KEGG | High-level functions of biological systems | https://www.kegg.jp/ |
2.3. Data Analysis
2.4. Machine Learning and AI
3. Bioinformatics in BC Research
3.1. Diagnostic and Prognostic Biomarkers
3.2. Harnessing Bioinformatics for BC Therapy
| Drug | Class | Original FDA-Approved Use | Repurposing for BC (Approval Status) | Refs |
|---|---|---|---|---|
| Anastrozole | Aromatase inhibitor | Therapy in postmenopausal women with advanced HR+ BC Adjuvant therapy in early HR+ BC | Postmenopausal women at high risk of developing BC (UK-approved) | [124] |
| Azelastine | Histamine receptor antagonist | Allergy | HR+, HER2+ and TNBC subtypes (not yet approved, pre-clinical study) | [125] |
| Diclofenac | COX inhibitor | NSAID for pain and inflammation | TNBC (not yet approved, pre-clinical study) | [126] |
| Metformin | Mitochondrial complex I inhibitor | Type-2 diabetes mellitus | HR+, HER2+ and TNBC subtypes (not yet approved, pre-clinical and clinical studies) | [122] |
| Nebivolol | β-adrenergic receptor antagonist | Hypertension | TNBC (not yet approved, pre-clinical studies) | [127,128] |
| Olaparib | PARP inhibitor | Advanced BRCA-mutated ovarian cancer | early and metastatic BRCA-mutated BC (FDA-approved) | [129] |
| Ruxolitinib | JAK inhibitor | Bone marrow and blood cancers | HR+ metastatic BC and TNBC (not yet approved, clinical study) | [130] |
| Trametinib | MEK inhibitor | BRAF-mutated melanoma, NSCLC, thyroid cancer, and low-grade gliomas | TNBC (not yet approved, clinical study) | [131] |
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kim, J.; Harper, A.; McCormack, V.; Sung, H.; Houssami, N.; Morgan, E.; Mutebi, M.; Garvey, G.; Soerjomataram, I.; Fidler-Benaoudia, M.M. Global Patterns and Trends in Breast Cancer Incidence and Mortality across 185 Countries. Nat. Med. 2025, 31, 1154–1162. [Google Scholar] [CrossRef] [PubMed]
- Fumagalli, C.; Barberis, M. Breast Cancer Heterogeneity. Diagnostics 2021, 11, 1555. [Google Scholar] [CrossRef]
- Lüönd, F.; Tiede, S.; Christofori, G. Breast Cancer as an Example of Tumour Heterogeneity and Tumour Cell Plasticity during Malignant Progression. Br. J. Cancer 2021, 125, 164–175. [Google Scholar] [CrossRef]
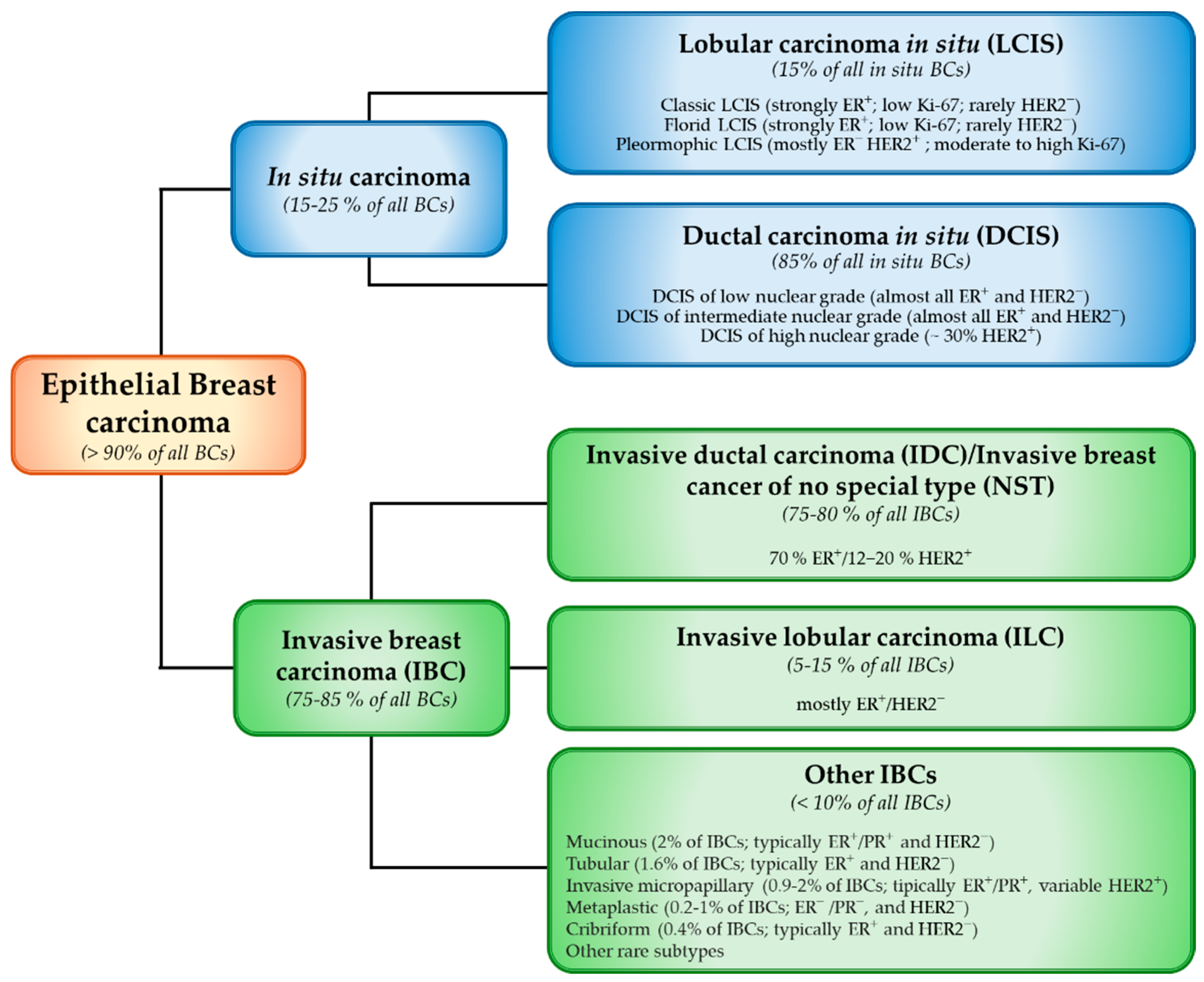
- Tan, P.H.; Ellis, I.; Allison, K.; Brogi, E.; Fox, S.B.; Lakhani, S.; Lazar, A.J.; Morris, E.A.; Sahin, A.; Salgado, R.; et al. The 2019 World Health Organization Classification of Tumours of the Breast. Histopathology 2020, 77, 181–185. [Google Scholar] [CrossRef]
- Radu, I.; Scripcariu, V.; Panuța, A.; Rusu, A.; Afrăsânie, V.-A.; Cojocaru, E.; Aniței, M.G.; Alexa-Stratulat, T.; Terinte, C.; Șerban, C.F.; et al. Breast Sarcomas—How Different Are They from Breast Carcinomas? Clinical, Pathological, Imaging and Treatment Insights. Diagnostics 2023, 13, 1370. [Google Scholar] [CrossRef]
- Orrantia-Borunda, E.; Anchondo-Nuñez, P.; Acuña-Aguilar, L.E.; Gómez-Valles, F.O.; Ramírez-Valdespino, C.A. Subtypes of Breast Cancer. In Breast Cancer; Exon Publications: Brisbane, QLD, Australia, 2022; pp. 31–42. [Google Scholar]
- Breast Cancer Association Consortium. Breast Cancer Risk Genes—Association Analysis in More than 113,000 Women. N. Engl. J. Med. 2021, 384, 428–439. [Google Scholar] [CrossRef] [PubMed]
- Taylor, A.; Brady, A.F.; Frayling, I.M.; Hanson, H.; Tischkowitz, M.; Turnbull, C.; Side, L. Consensus for Genes to Be Included on Cancer Panel Tests Offered by UK Genetics Services: Guidelines of the UK Cancer Genetics Group. J. Med. Genet. 2018, 55, 372–377. [Google Scholar] [CrossRef]
- McDevitt, T.; Durkie, M.; Arnold, N.; Burghel, G.J.; Butler, S.; Claes, K.B.M.; Logan, P.; Robinson, R.; Sheils, K.; Wolstenholme, N.; et al. EMQN Best Practice Guidelines for Genetic Testing in Hereditary Breast and Ovarian Cancer. Eur. J. Hum. Genet. 2024, 32, 479–488. [Google Scholar] [CrossRef]
- Mavaddat, N.; Dorling, L.; Carvalho, S.; Allen, J.; González-Neira, A.; Keeman, R.; Bolla, M.K.; Dennis, J.; Wang, Q.; Ahearn, T.U.; et al. Pathology of Tumors Associated With Pathogenic Germline Variants in 9 Breast Cancer Susceptibility Genes. JAMA Oncol. 2022, 8, e216744. [Google Scholar] [CrossRef]
- Feng, Y.; Spezia, M.; Huang, S.; Yuan, C.; Zeng, Z.; Zhang, L.; Ji, X.; Liu, W.; Huang, B.; Luo, W.; et al. Breast Cancer Development and Progression: Risk Factors, Cancer Stem Cells, Signaling Pathways, Genomics, and Molecular Pathogenesis. Genes Dis. 2018, 5, 77–106. [Google Scholar] [CrossRef] [PubMed]
- Koboldt, D.C.; Fulton, R.S.; McLellan, M.D.; Schmidt, H.; Kalicki-Veizer, J.; McMichael, J.F.; Fulton, L.L.; Dooling, D.J.; Ding, L.; Mardis, E.R.; et al. Comprehensive Molecular Portraits of Human Breast Tumours. Nature 2012, 490, 61–70. [Google Scholar] [CrossRef]
- Goetz, L.H.; Schork, N.J. Personalized Medicine: Motivation, Challenges, and Progress. Fertil. Steril. 2018, 109, 952–963. [Google Scholar] [CrossRef]
- Clark, A.J.; Lillard, J.W. A Comprehensive Review of Bioinformatics Tools for Genomic Biomarker Discovery Driving Precision Oncology. Genes 2024, 15, 1036. [Google Scholar] [CrossRef]
- Bagger, F.O.; Borgwardt, L.; Jespersen, A.S.; Hansen, A.R.; Bertelsen, B.; Kodama, M.; Nielsen, F.C. Whole Genome Sequencing in Clinical Practice. BMC Med. Genom. 2024, 17, 39. [Google Scholar] [CrossRef]
- Zhao, E.Y.; Jones, M.; Jones, S.J.M. Whole-Genome Sequencing in Cancer. Cold Spring Harb. Perspect. Med. 2019, 9, a034579. [Google Scholar] [CrossRef]
- Hong, M.; Tao, S.; Zhang, L.; Diao, L.T.; Huang, X.; Huang, S.; Xie, S.J.; Xiao, Z.D.; Zhang, H. RNA Sequencing: New Technologies and Applications in Cancer Research. J. Hematol. Oncol. 2020, 13, 166. [Google Scholar] [CrossRef]
- Rabbani, B.; Tekin, M.; Mahdieh, N. The Promise of Whole-Exome Sequencing in Medical Genetics. J. Hum. Genet. 2014, 59, 5–15. [Google Scholar] [CrossRef]
- Lelieveld, S.H.; Spielmann, M.; Mundlos, S.; Veltman, J.A.; Gilissen, C. Comparison of Exome and Genome Sequencing Technologies for the Complete Capture of Protein-Coding Regions. Hum. Mutat. 2015, 36, 815–822. [Google Scholar] [CrossRef]
- Pei, X.M.; Yeung, M.H.Y.; Wong, A.N.N.; Tsang, H.F.; Yu, A.C.S.; Yim, A.K.Y.; Wong, S.C.C. Targeted Sequencing Approach and Its Clinical Applications for the Molecular Diagnosis of Human Diseases. Cells 2023, 12, 493. [Google Scholar] [CrossRef] [PubMed]
- Tang, F.; Barbacioru, C.; Wang, Y.; Nordman, E.; Lee, C.; Xu, N.; Wang, X.; Bodeau, J.; Tuch, B.B.; Siddiqui, A.; et al. MRNA-Seq Whole-Transcriptome Analysis of a Single Cell. Nat. Methods 2009, 6, 377–382. [Google Scholar] [CrossRef] [PubMed]
- Wei, E.; Reisinger, A.; Li, J.; French, L.E.; Clanner-Engelshofen, B.; Reinholz, M. Integration of ScRNA-Seq and TCGA RNA-Seq to Analyze the Heterogeneity of HPV+ and HPV- Cervical Cancer Immune Cells and Establish Molecular Risk Models. Front. Oncol. 2022, 12, 860900. [Google Scholar] [CrossRef]
- Lopes, R.; Agami, R.; Korkmaz, G. GRO-Seq, a Tool for Identification of Transcripts Regulating Gene Expression. Methods Mol. Biol. 2017, 1543, 43–55. [Google Scholar] [CrossRef]
- Chen, X.; Xu, H.; Shu, X.; Song, C.-X. Mapping Epigenetic Modifications by Sequencing Technologies. Cell Death Differ. 2025, 32, 56–65. [Google Scholar] [CrossRef]
- Guanzon, D.; Ross, J.P.; Ma, C.; Berry, O.; Liew, Y.J. Comparing Methylation Levels Assayed in GC-Rich Regions with Current and Emerging Methods. BMC Genom. 2024, 25, 741. [Google Scholar] [CrossRef]
- Mehrmohamadi, M.; Sepehri, M.H.; Nazer, N.; Norouzi, M.R. A Comparative Overview of Epigenomic Profiling Methods. Front. Cell Dev. Biol. 2021, 9, 714687. [Google Scholar] [CrossRef]
- Caldera, M.; Buphamalai, P.; Müller, F.; Menche, J. Interactome-Based Approaches to Human Disease. Curr. Opin. Syst. Biol. 2017, 3, 88–94. [Google Scholar] [CrossRef]
- Li, G.; Cai, L.; Chang, H.; Hong, P.; Zhou, Q.; Kulakova, E.V.; Kolchanov, N.A.; Ruan, Y. Chromatin Interaction Analysis with Paired-End Tag (ChIA-PET) Sequencing Technology and Application. BMC Genom. 2014, 15, S11. [Google Scholar] [CrossRef]
- Cui, M.; Cheng, C.; Zhang, L. High-Throughput Proteomics: A Methodological Mini-Review. Lab. Investig. 2022, 102, 1170–1181. [Google Scholar] [CrossRef]
- Meng, X.; Liu, Y.; Xu, S.; Yang, L.; Yin, R. Review on Analytical Technologies and Applications in Metabolomics. BIOCELL 2024, 48, 65–78. [Google Scholar] [CrossRef]
- Al-Harazi, O.; El Allali, A.; Colak, D. Biomolecular Databases and Subnetwork Identification Approaches of Interest to Big Data Community: An Expert Review. Omi. A J. Integr. Biol. 2019, 23, 138–151. [Google Scholar] [CrossRef]
- Diniz, W.J.S.; Canduri, F. REVIEW-ARTICLE Bioinformatics: An Overview and Its Applications. Genet. Mol. Res. 2017, 16, 17. [Google Scholar] [CrossRef]
- Benson, D.A.; Cavanaugh, M.; Clark, K.; Karsch-Mizrachi, I.; Lipman, D.J.; Ostell, J.; Sayers, E.W. GenBank. Nucleic Acids Res. 2012, 41, D36–D42. [Google Scholar] [CrossRef]
- Stoesser, G. The EMBL Nucleotide Sequence Database. Nucleic Acids Res. 2002, 30, 21–26. [Google Scholar] [CrossRef] [PubMed]
- Sweeney, B.A.; Petrov, A.I.; Burkov, B.; Finn, R.D.; Bateman, A.; Szymanski, M.; Karlowski, W.M.; Gorodkin, J.; Seemann, S.E.; Cannone, J.J.; et al. RNAcentral: A Hub of Information for Non-Coding RNA Sequences. Nucleic Acids Res. 2019, 47, D221–D229. [Google Scholar] [CrossRef]
- Bateman, A.; Martin, M.-J.; Orchard, S.; Magrane, M.; Ahmad, S.; Alpi, E.; Bowler-Barnett, E.H.; Britto, R.; Bye-A-Jee, H.; Cukura, A.; et al. UniProt: The Universal Protein Knowledgebase in 2023. Nucleic Acids Res. 2023, 51, D523–D531. [Google Scholar] [CrossRef]
- Clough, E.; Barrett, T. The Gene Expression Omnibus Database. Methods Mol Biol. 2016, 1418, 93–110. [Google Scholar] [CrossRef]
- Ma, C.X.; Ellis, M.J. The Cancer Genome Atlas: Clinical Applications for Breast Cancer. Oncology 2013, 27, 1263–1269. [Google Scholar]
- Landrum, M.J.; Lee, J.M.; Benson, M.; Brown, G.R.; Chao, C.; Chitipiralla, S.; Gu, B.; Hart, J.; Hoffman, D.; Jang, W.; et al. ClinVar: Improving Access to Variant Interpretations and Supporting Evidence. Nucleic Acids Res. 2018, 46, D1062–D1067. [Google Scholar] [CrossRef]
- Phan, L.; Zhang, H.; Wang, Q.; Villamarin, R.; Hefferon, T.; Ramanathan, A.; Kattman, B. The Evolution of DbSNP: 25 Years of Impact in Genomic Research. Nucleic Acids Res. 2025, 53, D925–D931. [Google Scholar] [CrossRef]
- Pettersson, E. Tri-Nucleotide Threading for Parallel Amplification of Minute Amounts of Genomic DNA. Nucleic Acids Res. 2006, 34, e49. [Google Scholar] [CrossRef]
- Burley, S.K.; Piehl, D.W.; Vallat, B.; Zardecki, C. RCSB Protein Data Bank: Supporting Research and Education Worldwide through Explorations of Experimentally Determined and Computationally Predicted Atomic Level 3D Biostructures. IUCrJ 2024, 11, 279–286. [Google Scholar] [CrossRef] [PubMed]
- Burley, S.K.; Bhikadiya, C.; Bi, C.; Bittrich, S.; Chao, H.; Chen, L.; Craig, P.A.; Crichlow, G.V.; Dalenberg, K.; Duarte, J.M.; et al. RCSB Protein Data Bank (RCSB.Org): Delivery of Experimentally-Determined PDB Structures alongside One Million Computed Structure Models of Proteins from Artificial Intelligence/Machine Learning. Nucleic Acids Res. 2023, 51, D488–D508. [Google Scholar] [CrossRef]
- Andreeva, A.; Kulesha, E.; Gough, J.; Murzin, A.G. The SCOP Database in 2020: Expanded Classification of Representative Family and Superfamily Domains of Known Protein Structures. Nucleic Acids Res. 2020, 48, D376–D382. [Google Scholar] [CrossRef]
- Safari-Alighiarloo, N.; Taghizadeh, M.; Rezaei-Tavirani, M.; Goliaei, B.; Peyvandi, A.A. Protein-Protein Interaction Networks (PPI) and Complex Diseases. Gastroenterol. Hepatol. Bed Bench 2014, 7, 17–31. [Google Scholar]
- Oughtred, R.; Rust, J.; Chang, C.; Breitkreutz, B.; Stark, C.; Willems, A.; Boucher, L.; Leung, G.; Kolas, N.; Zhang, F.; et al. The BioGRID Database: A Comprehensive Biomedical Resource of Curated Protein, Genetic, and Chemical Interactions. Protein Sci. 2021, 30, 187–200. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Kirsch, R.; Koutrouli, M.; Nastou, K.; Mehryary, F.; Hachilif, R.; Gable, A.L.; Fang, T.; Doncheva, N.T.; Pyysalo, S.; et al. The STRING Database in 2023: Protein–Protein Association Networks and Functional Enrichment Analyses for Any Sequenced Genome of Interest. Nucleic Acids Res. 2023, 51, D638–D646. [Google Scholar] [CrossRef]
- del Toro, N.; Shrivastava, A.; Ragueneau, E.; Meldal, B.; Combe, C.; Barrera, E.; Perfetto, L.; How, K.; Ratan, P.; Shirodkar, G.; et al. The IntAct Database: Efficient Access to Fine-Grained Molecular Interaction Data. Nucleic Acids Res. 2022, 50, D648–D653. [Google Scholar] [CrossRef]
- Kanehisa, M.; Furumichi, M.; Tanabe, M.; Sato, Y.; Morishima, K. KEGG: New Perspectives on Genomes, Pathways, Diseases and Drugs. Nucleic Acids Res. 2017, 45, D353–D361. [Google Scholar] [CrossRef]
- Chowdhury, B.; Garai, G. A Review on Multiple Sequence Alignment from the Perspective of Genetic Algorithm. Genomics 2017, 109, 419–431. [Google Scholar] [CrossRef] [PubMed]
- Dayhoff, M.O.; Schwartz, R.M.; Orcutt, B.C. A Model of Evolutionary Change. In Atlas of Protein Sequence and Structure; National Biomedical Research Foundation: Waltham, MA, USA, 1978; pp. 345–352. [Google Scholar]
- Henikoff, S.; Henikoff, J.G. Amino Acid Substitution Matrices from Protein Blocks. Proc. Natl. Acad. Sci. USA 1992, 89, 10915–10919. [Google Scholar] [CrossRef] [PubMed]
- Bernardi, G.; Bernardi, G. Compositional Constraints and Genome Evolution. J. Mol. Evol. 1986, 24, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.; Duan, T.; Ye, P.; Chen, K.; Zhang, G.; Lai, M.; Zhang, H. TSVdb: A Web-Tool for TCGA Splicing Variants Analysis. BMC Genom. 2018, 19, 405. [Google Scholar] [CrossRef]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative Analysis of Complex Cancer Genomics and Clinical Profiles Using the CBioPortal. Sci. Signal. 2013, 6, pl1. [Google Scholar] [CrossRef]
- Achawanantakun, R.; Chen, J.; Sun, Y.; Zhang, Y. LncRNA-ID: Long Non-Coding RNA IDentification Using Balanced Random Forests. Bioinformatics 2015, 31, 3897–3905. [Google Scholar] [CrossRef]
- Li, Z.; Liu, L.; Feng, C.; Qin, Y.; Xiao, J.; Zhang, Z.; Ma, L. LncBook 2.0: integrating human long non-coding RNAs with multi-omics annotations. Nucleic Acids Res. 2023, 51, D186–D191. [Google Scholar] [CrossRef]
- Chang, L.; Xia, J. MicroRNA Regulatory Network Analysis Using MiRNet 2.0. Methods Mol Biol. 2023, 2594, 185–204. [Google Scholar] [CrossRef]
- Kern, F.; Aparicio-Puerta, E.; Li, Y.; Fehlmann, T.; Kehl, T.; Wagner, V.; Ray, K.; Ludwig, N.; Lenhof, H.-P.; Meese, E.; et al. MiRTargetLink 2.0—Interactive MiRNA Target Gene and Target Pathway Networks. Nucleic Acids Res. 2021, 49, W409–W416. [Google Scholar] [CrossRef]
- Chen, Y.; Wang, X. MiRDB: An Online Database for Prediction of Functional MicroRNA Targets. Nucleic Acids Res. 2020, 48, D127–D131. [Google Scholar] [CrossRef] [PubMed]
- Tomczak, A.; Mortensen, J.M.; Winnenburg, R.; Liu, C.; Alessi, D.T.; Swamy, V.; Vallania, F.; Lofgren, S.; Haynes, W.; Shah, N.H.; et al. Interpretation of Biological Experiments Changes with Evolution of the Gene Ontology and Its Annotations. Sci. Rep. 2018, 8, 5115. [Google Scholar] [CrossRef] [PubMed]
- The Gene Ontology Consortium. The Gene Ontology Resource: 20 Years and Still GOing Strong. Nucleic Acids Res. 2019, 47, D330–D338. [Google Scholar] [CrossRef]
- Pomaznoy, M.; Ha, B.; Peters, B. GOnet: A Tool for Interactive Gene Ontology Analysis. BMC Bioinform. 2018, 19, 470. [Google Scholar] [CrossRef]
- Ge, S.X.; Jung, D.; Yao, R. ShinyGO: A Graphical Gene-Set Enrichment Tool for Animals and Plants. Bioinformatics 2020, 36, 2628–2629. [Google Scholar] [CrossRef]
- Cheng, X.; Yan, J.; Liu, Y.; Wang, J.; Taubert, S. EVITTA: A Web-Based Visualization and Inference Toolbox for Transcriptome Analysis. Nucleic Acids Res. 2021, 49, W207–W215. [Google Scholar] [CrossRef] [PubMed]
- Ge, X. IDEP Web Application for RNA-Seq Data Analysis. Methods Mol. Biol. 2021, 2284, 417–443. [Google Scholar] [PubMed]
- Reimand, J.; Kull, M.; Peterson, H.; Hansen, J.; Vilo, J. G:Profiler—A Web-Based Toolset for Functional Profiling of Gene Lists from Large-Scale Experiments. Nucleic Acids Res. 2007, 35, W193–W200. [Google Scholar] [CrossRef] [PubMed]
- Abramson, J.; Adler, J.; Dunger, J.; Evans, R.; Green, T.; Pritzel, A.; Ronneberger, O.; Willmore, L.; Ballard, A.J.; Bambrick, J.; et al. Accurate Structure Prediction of Biomolecular Interactions with AlphaFold 3. Nature 2024, 630, 493–500. [Google Scholar] [CrossRef]
- Agu, P.C.; Afiukwa, C.A.; Orji, O.U.; Ezeh, E.M.; Ofoke, I.H.; Ogbu, C.O.; Ugwuja, E.I.; Aja, P.M. Molecular Docking as a Tool for the Discovery of Molecular Targets of Nutraceuticals in Diseases Management. Sci. Rep. 2023, 13, 13398. [Google Scholar] [CrossRef]
- Paggi, J.M.; Pandit, A.; Dror, R.O. The Art and Science of Molecular Docking. Annu. Rev. Biochem. 2024, 93, 389–410. [Google Scholar] [CrossRef]
- Sugiki, T.; Kobayashi, N.; Fujiwara, T. Modern Technologies of Solution Nuclear Magnetic Resonance Spectroscopy for Three-Dimensional Structure Determination of Proteins Open Avenues for Life Scientists. Comput. Struct. Biotechnol. J. 2017, 15, 328–339. [Google Scholar] [CrossRef]
- Eberhardt, J.; Santos-Martins, D.; Tillack, A.F.; Forli, S. AutoDock Vina 1.2.0: New Docking Methods, Expanded Force Field, and Python Bindings. J. Chem. Inf. Model. 2021, 61, 3891–3898. [Google Scholar] [CrossRef]
- Liu, Y.; Yang, X.; Gan, J.; Chen, S.; Xiao, Z.-X.; Cao, Y. CB-Dock2: Improved Protein–Ligand Blind Docking by Integrating Cavity Detection, Docking and Homologous Template Fitting. Nucleic Acids Res. 2022, 50, W159–W164. [Google Scholar] [CrossRef]
- Greener, J.G.; Kandathil, S.M.; Moffat, L.; Jones, D.T. A Guide to Machine Learning for Biologists. Nat. Rev. Mol. Cell Biol. 2022, 23, 40–55. [Google Scholar] [CrossRef]
- Singh, A.; Singh, A.; Bhattacharya, S. Research Trends on AI in Breast Cancer Diagnosis, and Treatment over Two Decades. Discov. Oncol. 2024, 15, 772. [Google Scholar] [CrossRef]
- Yan, S.; Yue, S. Identification of Early Diagnostic Biomarkers for Breast Cancer through Bioinformatics Analysis. Medicine 2023, 102, e35273. [Google Scholar] [CrossRef]
- Wang, N.; Zhang, H.; Li, D.; Jiang, C.; Zhao, H.; Teng, Y. Identification of Novel Biomarkers in Breast Cancer via Integrated Bioinformatics Analysis and Experimental Validation. Bioengineered 2021, 12, 12431–12446. [Google Scholar] [CrossRef]
- Golestan, A.; Tahmasebi, A.; Maghsoodi, N.; Faraji, S.N.; Irajie, C.; Ramezani, A. Unveiling Promising Breast Cancer Biomarkers: An Integrative Approach Combining Bioinformatics Analysis and Experimental Verification. BMC Cancer 2024, 24, 155. [Google Scholar] [CrossRef]
- Xu, K.; Wang, R.; Xie, H.; Hu, L.; Wang, C.; Xu, J.; Zhu, C.; Liu, Y.; Gao, F.; Li, X.; et al. Single-Cell RNA Sequencing Reveals Cell Heterogeneity and Transcriptome Profile of Breast Cancer Lymph Node Metastasis. Oncogenesis 2021, 10, 66. [Google Scholar] [CrossRef]
- Wang, Y.; Yi, K.; Chen, B.; Zhang, B.; Jidong, G. Elucidating the Susceptibility to Breast Cancer: An in-Depth Proteomic and Transcriptomic Investigation into Novel Potential Plasma Protein Biomarkers. Front. Mol. Biosci. 2024, 10, 1340917. [Google Scholar] [CrossRef] [PubMed]
- Gambacurta, A.; Tullio, V.; Savini, I.; Mauriello, A.; Catani, M.V.; Gasperi, V. Identification of the EBF1/ETS2/KLF2-MiR-126-Gene Feed-Forward Loop in Breast Carcinogenesis and Stemness. Int. J. Mol. Sci. 2025, 26, 328. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Y.; Ma, D.; Yang, Y.-S.; Yang, F.; Ding, J.-H.; Gong, Y.; Jiang, L.; Ge, L.-P.; Wu, S.-Y.; Yu, Q.; et al. Comprehensive Metabolomics Expands Precision Medicine for Triple-Negative Breast Cancer. Cell Res. 2022, 32, 477–490. [Google Scholar] [CrossRef] [PubMed]
- An, R.; Yu, H.; Wang, Y.; Lu, J.; Gao, Y.; Xie, X.; Zhang, J. Integrative Analysis of Plasma Metabolomics and Proteomics Reveals the Metabolic Landscape of Breast Cancer. Cancer Metab. 2022, 10, 13. [Google Scholar] [CrossRef]
- Asleh, K.; Negri, G.L.; Spencer Miko, S.E.; Colborne, S.; Hughes, C.S.; Wang, X.Q.; Gao, D.; Gilks, C.B.; Chia, S.K.L.; Nielsen, T.O.; et al. Proteomic Analysis of Archival Breast Cancer Clinical Specimens Identifies Biological Subtypes with Distinct Survival Outcomes. Nat. Commun. 2022, 13, 896. [Google Scholar] [CrossRef] [PubMed]
- Jeon, Y.; Lee, G.; Jeong, H.; Gong, G.; Kim, J.; Kim, K.; Jeong, J.H.; Lee, H.J. Proteomic Analysis of Breast Cancer Based on Immune Subtypes. Clin. Proteom. 2024, 21, 17. [Google Scholar] [CrossRef] [PubMed]
- Azevedo, A.L.K.; Gomig, T.H.B.; Batista, M.; Marchini, F.K.; Spautz, C.C.; Rabinovich, I.; Sebastião, A.P.M.; Oliveira, J.C.; Gradia, D.F.; Cavalli, I.J.; et al. High-Throughput Proteomics of Breast Cancer Subtypes: Biological Characterization and Multiple Candidate Biomarker Panels to Patients’ Stratification. J. Proteom. 2023, 285, 104955. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.M.; Chae, H. MoBRCA-Net: A Breast Cancer Subtype Classification Framework Based on Multi-Omics Attention Neural Networks. BMC Bioinform. 2023, 24, 169. [Google Scholar] [CrossRef]
- Smith-Byrne, K.; Hedman, Å.; Dimitriou, M.; Desai, T.; Sokolov, A.V.; Schioth, H.B.; Koprulu, M.; Pietzner, M.; Langenberg, C.; Atkins, J.; et al. Identifying Therapeutic Targets for Cancer among 2074 Circulating Proteins and Risk of Nine Cancers. Nat. Commun. 2024, 15, 3621. [Google Scholar] [CrossRef]
- Kalocsay, M.; Berberich, M.J.; Everley, R.A.; Nariya, M.K.; Chung, M.; Gaudio, B.; Victor, C.; Bradshaw, G.A.; Eisert, R.J.; Hafner, M.; et al. Proteomic Profiling across Breast Cancer Cell Lines and Models. Sci. Data 2023, 10, 514. [Google Scholar] [CrossRef]
- Shenoy, A.; Belugali Nataraj, N.; Perry, G.; Loayza Puch, F.; Nagel, R.; Marin, I.; Balint, N.; Bossel, N.; Pavlovsky, A.; Barshack, I.; et al. Proteomic Patterns Associated with Response to Breast Cancer Neoadjuvant Treatment. Mol. Syst. Biol. 2020, 16, e9443. [Google Scholar] [CrossRef]
- Zheng, B.; Du, P.; Zeng, Z.; Cao, P.; Ma, X.; Jiang, Y. Propranolol Inhibits EMT and Metastasis in Breast Cancer through MiR-499-5p-Mediated Sox6. J. Cancer Res. Clin. Oncol. 2024, 150, 59. [Google Scholar] [CrossRef]
- Alam, M.S.; Rahaman, M.M.; Sultana, A.; Wang, G.; Mollah, M.N.H. Statistics and Network-Based Approaches to Identify Molecular Mechanisms That Drive the Progression of Breast Cancer. Comput. Biol. Med. 2022, 145, 105508. [Google Scholar] [CrossRef]
- Lord, S.R.; Collins, J.M.; Cheng, W.-C.; Haider, S.; Wigfield, S.; Gaude, E.; Fielding, B.A.; Pinnick, K.E.; Harjes, U.; Segaran, A.; et al. Transcriptomic Analysis of Human Primary Breast Cancer Identifies Fatty Acid Oxidation as a Target for Metformin. Br. J. Cancer 2020, 122, 258–265. [Google Scholar] [CrossRef]
- Mujawar, T.; Tare, H.; Deshmukh, N.; Udugade, B.; Thube, U. Repurposing FDA-Approved Anastrozole-Based Drugs for Breast Cancer through Drug-Drug Transcriptomic Similarity and Cavity Detection Guided Blind Docking. Int. J. DRUG Deliv. Technol. 2023, 13, 1172–1177. [Google Scholar] [CrossRef]
- Firoozbakht, F.; Rezaeian, I.; Rueda, L.; Ngom, A. Computationally Repurposing Drugs for Breast Cancer Subtypes Using a Network-Based Approach. BMC Bioinform. 2022, 23, 143. [Google Scholar] [CrossRef] [PubMed]
- Neagu, A.-N.; Whitham, D.; Buonanno, E.; Jenkins, A.; Alexa-Stratulat, T.; Tamba, B.I.; Darie, C.C. Proteomics and Its Applications in Breast Cancer. Am. J. Cancer Res. 2021, 11, 4006–4049. [Google Scholar]
- Huang, Y.; Zeng, P.; Zhong, C. Classifying Breast Cancer Subtypes on Multi-Omics Data via Sparse Canonical Correlation Analysis and Deep Learning. BMC Bioinform. 2024, 25, 132. [Google Scholar] [CrossRef]
- Li, M.; Guo, Y.; Feng, Y.-M.; Zhang, N. Identification of Triple-Negative Breast Cancer Genes and a Novel High-Risk Breast Cancer Prediction Model Development Based on PPI Data and Support Vector Machines. Front. Genet. 2019, 10, 180. [Google Scholar] [CrossRef]
- Kuilman, M.M.; Ellappalayam, A.; Barcaru, A.; Haan, J.C.; Bhaskaran, R.; Wehkamp, D.; Menicucci, A.R.; Audeh, W.M.; Mittempergher, L.; Glas, A.M. BluePrint Breast Cancer Molecular Subtyping Recognizes Single and Dual Subtype Tumors with Implications for Therapeutic Guidance. Breast Cancer Res. Treat. 2022, 195, 263–274. [Google Scholar] [CrossRef]
- Loo, S.K.; Yates, M.E.; Yang, S.; Oesterreich, S.; Lee, A.V.; Wang, X.-S. Fusion-Associated Carcinomas of the Breast: Diagnostic, Prognostic, and Therapeutic Significance. Genes. Chromosomes Cancer 2022, 61, 261–273. [Google Scholar] [CrossRef]
- Roy, S.; Gupta, D. Analysis of Breast Cancer Next-Generation Sequencing Datasets for Identifying Fusion Genes Responsible for the Cancer Progression. Inform. Med. Unlocked 2023, 41, 101306. [Google Scholar] [CrossRef]
- Elia, I.; Broekaert, D.; Christen, S.; Boon, R.; Radaelli, E.; Orth, M.F.; Verfaillie, C.; Grünewald, T.G.P.; Fendt, S.-M. Proline Metabolism Supports Metastasis Formation and Could Be Inhibited to Selectively Target Metastasizing Cancer Cells. Nat. Commun. 2017, 8, 15267. [Google Scholar] [CrossRef] [PubMed]
- Karsli-Ceppioglu, S.; Dagdemir, A.; Judes, G.; Lebert, A.; Penault-Llorca, F.; Bignon, Y.J.; Bernard-Gallon, D. The Epigenetic Landscape of Promoter Genome-Wide Analysis in Breast Cancer. Sci. Rep. 2017, 7, 6597. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Q.Y.; Lei, P.J.; Zhang, X.; Zheng, J.Y.; Wang, H.Y.; Zhao, J.; Li, Y.M.; Ye, M.; Li, L.; Wei, G.; et al. Global Histone Modification Profiling Reveals the Epigenomic Dynamics during Malignant Transformation in a Four-Stage Breast Cancer Model. Clin. Epigenet. 2016, 8, 34. [Google Scholar] [CrossRef] [PubMed]
- Cortellesi, E.; Savini, I.; Veneziano, M.; Gambacurta, A.; Catani, M.V.; Gasperi, V. Decoding the Epigenome of Breast Cancer. Int. J. Mol. Sci. 2025, 26, 2605. [Google Scholar] [CrossRef] [PubMed]
- Argelaguet, R.; Arnol, D.; Bredikhin, D.; Deloro, Y.; Velten, B.; Marioni, J.C.; Stegle, O. MOFA+: A Statistical Framework for Comprehensive Integration of Multi-Modal Single-Cell Data. Genome Biol. 2020, 21, 111. [Google Scholar] [CrossRef]
- Sharma, A.; Debik, J.; Naume, B.; Ohnstad, H.O.; Sahlber, K.K.; Borgen, E.; Børresen-Dale, A.-L.; Engebråten, O.; Fritzman, B.; Garred, Ø.; et al. Comprehensive Multi-Omics Analysis of Breast Cancer Reveals Distinct Long-Term Prognostic Subtypes. Oncogenesis 2024, 13, 22. [Google Scholar] [CrossRef]
- Malighetti, F.; Villa, M.; Villa, A.M.; Pelucchi, S.; Aroldi, A.; Cortinovis, D.L.; Canova, S.; Capici, S.; Cazzaniga, M.E.; Mologni, L.; et al. Prognostic Biomarkers in Breast Cancer via Multi-Omics Clustering Analysis. Int. J. Mol. Sci. 2025, 26, 1943. [Google Scholar] [CrossRef]
- Zhang, S.; Liu, K.; Liu, Y.; Hu, X.; Gu, X. The Role and Application of Bioinformatics Techniques and Tools in Drug Discovery. Front. Pharmacol. 2025, 16, 1547131. [Google Scholar] [CrossRef] [PubMed]
- Savage, P.; Pacis, A.; Kuasne, H.; Liu, L.; Lai, D.; Wan, A.; Dankner, M.; Martinez, C.; Muñoz-Ramos, V.; Pilon, V.; et al. Chemogenomic Profiling of Breast Cancer Patient-Derived Xenografts Reveals Targetable Vulnerabilities for Difficult-to-Treat Tumors. Commun. Biol. 2020, 3, 310. [Google Scholar] [CrossRef]
- Meisel, J.L.; Venur, V.A.; Gnant, M.; Carey, L. Evolution of Targeted Therapy in Breast Cancer: Where Precision Medicine Began. Am. Soc. Clin. Oncol. Educ. B. 2018, 38, 78–86. [Google Scholar] [CrossRef]
- Nielsen, T.O.; Parker, J.S.; Leung, S.; Voduc, D.; Ebbert, M.; Vickery, T.; Davies, S.R.; Snider, J.; Stijleman, I.J.; Reed, J.; et al. A Comparison of PAM50 Intrinsic Subtyping with Immunohistochemistry and Clinical Prognostic Factors in Tamoxifen-Treated Estrogen Receptor–Positive Breast Cancer. Clin. Cancer Res. 2010, 16, 5222–5232. [Google Scholar] [CrossRef]
- Ohara, A.M.; Naoi, Y.; Shimazu, K.; Kagara, N.; Shimoda, M.; Tanei, T.; Miyake, T.; Kim, S.J.; Noguchi, S. PAM50 for Prediction of Response to Neoadjuvant Chemotherapy for ER-Positive Breast Cancer. Breast Cancer Res. Treat. 2019, 173, 533–543. [Google Scholar] [CrossRef]
- Rodríguez-Bejarano, O.H.; Parra-López, C.; Patarroyo, M.A. A Review Concerning the Breast Cancer-Related Tumour Microenvironment. Crit. Rev. Oncol. Hematol. 2024, 199, 104389. [Google Scholar] [CrossRef]
- Le, T.; Aronow, R.A.; Kirshtein, A.; Shahriyari, L. A Review of Digital Cytometry Methods: Estimating the Relative Abundance of Cell Types in a Bulk of Cells. Brief. Bioinform. 2021, 22, bbaa219. [Google Scholar] [CrossRef]
- Fernández, E.A.; Mahmoud, Y.D.; Veigas, F.; Rocha, D.; Miranda, M.; Merlo, J.; Balzarini, M.; Lujan, H.D.; Rabinovich, G.A.; Girotti, M.R. Unveiling the Immune Infiltrate Modulation in Cancer and Response to Immunotherapy by MIXTURE—An Enhanced Deconvolution Method. Brief. Bioinform. 2021, 22, bbaa317. [Google Scholar] [CrossRef]
- Zerdes, I.; Matikas, A.; Mezheyeuski, A.; Manikis, G.; Acs, B.; Johansson, H.; Boyaci, C.; Boman, C.; Poncet, C.; Ignatiadis, M.; et al. Machine Learning-Based Spatial Characterization of Tumor-Immune Microenvironment in the EORTC 10994/BIG 1-00 Early Breast Cancer Trial. NPJ Breast Cancer 2025, 11, 23. [Google Scholar] [CrossRef]
- National Cancer Institute. Drugs Approved for Breast Cancer. Available online: https://www.cancer.gov/about-cancer/treatment/drugs/breast (accessed on 12 May 2025).
- Kulkarni, V.S.; Alagarsamy, V.; Solomon, V.R.; Jose, P.A.; Murugesan, S. Drug Repurposing: An Effective Tool in Modern Drug Discovery. Russ. J. Bioorg. Chem. 2023, 49, 157–166. [Google Scholar] [CrossRef]
- Correia, A.S.; Gärtner, F.; Vale, N. Drug Combination and Repurposing for Cancer Therapy: The Example of Breast Cancer. Heliyon 2021, 7, e05948. [Google Scholar] [CrossRef] [PubMed]
- Hernández-Lemus, E.; Martínez-García, M. Pathway-Based Drug-Repurposing Schemes in Cancer: The Role of Translational Bioinformatics. Front. Oncol. 2021, 10, 605680. [Google Scholar] [CrossRef]
- Corleto, K.A.; Strandmo, J.L.; Giles, E.D. Metformin and Breast Cancer: Current Findings and Future Perspectives from Preclinical and Clinical Studies. Pharmaceuticals 2024, 17, 396. [Google Scholar] [CrossRef] [PubMed]
- Lord, S.R.; Cheng, W.-C.; Liu, D.; Gaude, E.; Haider, S.; Metcalf, T.; Patel, N.; Teoh, E.J.; Gleeson, F.; Bradley, K.; et al. Integrated Pharmacodynamic Analysis Identifies Two Metabolic Adaption Pathways to Metformin in Breast Cancer. Cell Metab. 2018, 28, 679–688.e4. [Google Scholar] [CrossRef] [PubMed]
- Cuzick, J.; Sestak, I.; Forbes, J.F.; Dowsett, M.; Cawthorn, S.; Mansel, R.E.; Loibl, S.; Bonanni, B.; Evans, D.G.; Howell, A. Use of Anastrozole for Breast Cancer Prevention (IBIS-II): Long-Term Results of a Randomised Controlled Trial. Lancet 2020, 395, 117–122. [Google Scholar] [CrossRef]
- Moraca, F.; Arciuolo, V.; Marzano, S.; Napolitano, F.; Castellano, G.; D’Aria, F.; Di Porzio, A.; Landolfi, L.; Catalanotti, B.; Randazzo, A.; et al. Repurposing FDA-Approved Drugs to Target G-Quadruplexes in Breast Cancer. Eur. J. Med. Chem. 2025, 285, 117245. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Li, J.; Li, Y.; Zhou, Y.; Wang, Z.; Zhang, D.; Liu, J.; Zhang, X. Diclofenac Impairs the Proliferation and Glucose Metabolism of Triple-Negative Breast Cancer Cells by Targeting the c-Myc Pathway. Exp. Ther. Med. 2021, 21, 584. [Google Scholar] [CrossRef]
- Kim, Y.J.; Jang, S.-K.; Kim, G.; Hong, S.-E.; Park, C.S.; Seong, M.-K.; Kim, H.-A.; Kim, K.S.; Kim, C.-H.; Park, K.S.; et al. Nebivolol Sensitizes BT-474 Breast Cancer Cells to FGFR Inhibitors. Anticancer. Res. 2023, 43, 1973–1980. [Google Scholar] [CrossRef]
- Nuevo-Tapioles, C.; Santacatterina, F.; Stamatakis, K.; Núñez de Arenas, C.; Gómez de Cedrón, M.; Formentini, L.; Cuezva, J.M. Coordinate β-Adrenergic Inhibition of Mitochondrial Activity and Angiogenesis Arrest Tumor Growth. Nat. Commun. 2020, 11, 3606. [Google Scholar] [CrossRef]
- Tutt, A.N.J.; Garber, J.E.; Kaufman, B.; Viale, G.; Fumagalli, D.; Rastogi, P.; Gelber, R.D.; de Azambuja, E.; Fielding, A.; Balmaña, J.; et al. Adjuvant Olaparib for Patients with BRCA1- or BRCA2- Mutated Breast Cancer. N. Engl. J. Med. 2021, 384, 2394–2405. [Google Scholar] [CrossRef] [PubMed]
- Makhlin, I.; McAndrew, N.P.; Wileyto, E.P.; Clark, A.S.; Holmes, R.; Bottalico, L.N.; Mesaros, C.; Blair, I.A.; Jeschke, G.R.; Fox, K.R.; et al. Ruxolitinib and Exemestane for Estrogen Receptor Positive, Aromatase Inhibitor Resistant Advanced Breast Cancer. NPJ Breast Cancer 2022, 8, 122. [Google Scholar] [CrossRef] [PubMed]
- Zhu, S.; Wu, Y.; Song, B.; Yi, M.; Yan, Y.; Mei, Q.; Wu, K. Recent Advances in Targeted Strategies for Triple-Negative Breast Cancer. J. Hematol. Oncol. 2023, 16, 100. [Google Scholar] [CrossRef]
- Bravaccini, S.; Mazza, M.; Maltoni, R. No More Disparities among Regions in Italy: Recent Approval of Genomic Test Reimbursability for Early Breast Cancer Patients in the Country. Breast Cancer Res. Treat. 2023, 201, 1–3. [Google Scholar] [CrossRef]
- Jamialahmadi, H.; Khalili-Tanha, G.; Nazari, E.; Rezaei-Tavirani, M. Artificial Intelligence and Bioinformatics: A Journey from Traditional Techniques to Smart Approaches. Gastroenterol. Hepatol. Bed Bench 2024, 17, 241–252. [Google Scholar] [CrossRef]
- Flores, J.E.; Claborne, D.M.; Weller, Z.D.; Webb-Robertson, B.-J.M.; Waters, K.M.; Bramer, L.M. Missing Data in Multi-Omics Integration: Recent Advances through Artificial Intelligence. Front. Artif. Intell. 2023, 6, 1098308. [Google Scholar] [CrossRef]
- Vaseghi, H.; Akrami, S.M.; Rashidi-Nezhad, A. The Challenges in the Interpretation of Genetic Variants Detected by Genomics Techniques in Patients with Congenital Anomalies. J. Clin. Lab. Anal. 2023, 37, e24967. [Google Scholar] [CrossRef] [PubMed]
- Thorn, C.F.; Klein, T.E.; Altman, R.B. PharmGKB: The Pharmacogenomics Knowledge Base. Methods Mol. Biol. 2013, 1015, 311–320. [Google Scholar] [CrossRef]
- Field, M.A. Bioinformatic Challenges Detecting Genetic Variation in Precision Medicine Programs. Front. Med. 2022, 9, 806696. [Google Scholar] [CrossRef]
- Roy, S.; Coldren, C.; Karunamurthy, A.; Kip, N.S.; Klee, E.W.; Lincoln, S.E.; Leon, A.; Pullambhatla, M.; Temple-Smolkin, R.L.; Voelkerding, K.V.; et al. Standards and Guidelines for Validating Next-Generation Sequencing Bioinformatics Pipelines. J. Mol. Diagn. 2018, 20, 4–27. [Google Scholar] [CrossRef]
- Guo, J.; Hu, J.; Zheng, Y.; Zhao, S.; Ma, J. Artificial Intelligence: Opportunities and Challenges in the Clinical Applications of Triple-Negative Breast Cancer. Br. J. Cancer 2023, 128, 2141–2149. [Google Scholar] [CrossRef] [PubMed]
- Ahn, J.S.; Shin, S.; Yang, S.-A.; Park, E.K.; Kim, K.H.; Cho, S.I.; Ock, C.-Y.; Kim, S. Artificial Intelligence in Breast Cancer Diagnosis and Personalized Medicine. J. Breast Cancer 2023, 26, 405. [Google Scholar] [CrossRef] [PubMed]
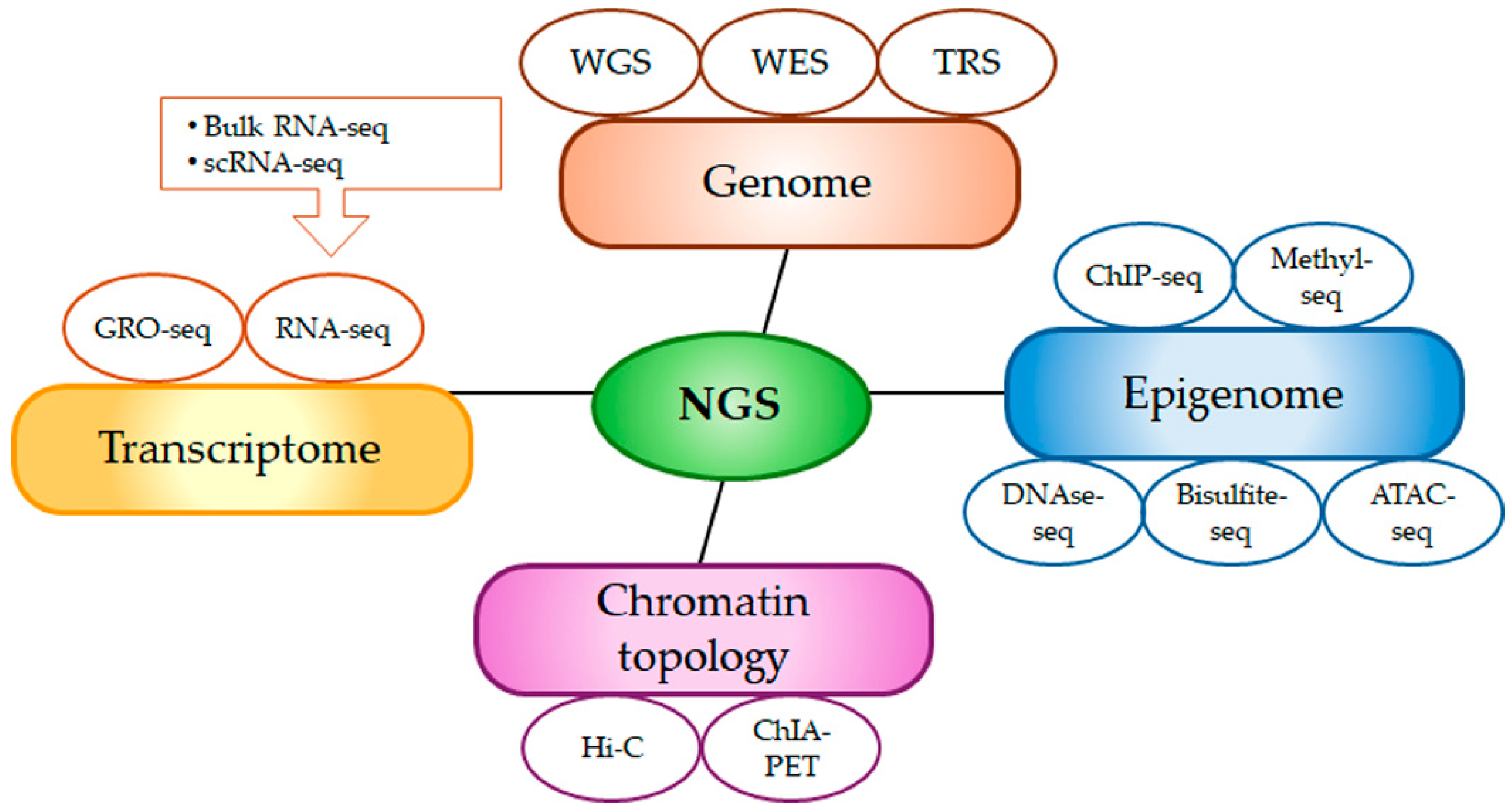
| Database | Analysis | Website (10 June 2025) |
|---|---|---|
| AlphaFold 3 | Prediction of protein, DNA and RNA structure and modeling of structural complexes | https://alphafold.ebi.ac.uk/ |
| AutoDock Vina 1.2.0 | Protein–ligand docking | https://vina.scripps.edu/ |
| CB-Dock2 | Protein–ligand blind docking | https://cadd.labshare.cn/cb-dock2/ |
| cBioPortal for Cancer Genomics | Multi-omics cancer genomics data | http://cbioportal.org/ |
| eVITTA | Transcriptome functional characterization | https://tau.cmmt.ubc.ca/eVITTA/ |
| g:Profiler | Functional enrichment analysis at gene level | https://biit.cs.ut.ee/gprofiler/gost |
| GO | Gene functions, cellular processes and subcellular localization of proteins | http://www.geneontology.org/ |
| GOnet | GO term annotation and enrichment analysis | https://tools.dice-database.org/GOnet/ |
| iDEP 2.0 | RNA-seq data analysis | https://bioinformatics.sdstate.edu/idep/ |
| LncBook 2.0 | human lncRNAs integration with multi-omics annotations | https://ngdc.cncb.ac.cn/lncbook/ |
| LncRNA-ID | lncRNA identification | https://github.com/zhangy72/LncRNA-ID |
| miRNet 2.0 | miRNA functions and interaction networks with genes, diseases, compounds, transcription factors. | https://www.mirnet.ca/ |
| miRTargetLink 2.0 | miRNA–mRNA interactions | https://ccb-compute.cs.uni-saarland.de/mirtargetlink2/ |
| miRDB | miRNA–mRNA interactions and functional annotations | https://mirdb.org/mirdb/index.html |
| ShinyGO 0.82 | Graphical gene-set enrichment | https://bioinformatics.sdstate.edu/go/ |
| TSVdb | TCGA splicing variants | https://github.com/wenjie1991/TSVdb |
| BC Samples | Integrated Strategy | Main Findings | Refs |
|---|---|---|---|
| GEO and TCGA databases | Transcriptomic profiling PPI network construction Survival analysis | 10 hub genes (PBK, CCNA2, CDCA8, MELK, NUSAP1, BIRC5, CCNB2, HMMR, MAD2L1, and PRC1) strongly associated with BC evolution. | [76] |
| GEO and TCGA databases | Transcriptomic profiling PPI network construction Survival analysis | 23 hub genes negatively correlated with BC overall survival. Increased cell cycle gene (CDK1, CDC20, AURKA and MCM4) expression as predictive biomarker for poor prognosis. | [77] |
| TCGA and METABRIC databases | Transcriptomic profiling | Genes involved in cell communication (CACNG4 and CHRNA6), cell cycle regulation and DNA replication (PKMYT1) pathways, and invasion and metastasis (EPYC) as diagnostic and prognostic markers. | [78] |
| Fresh tissues and axillary lymph nodes | Single cell transcriptomic profiling | CD44 +/ALDH2 +/ALDH6A1+ cluster in BC stem cells. PTMA, STC2, CST3, and RAMP3 genes involved in lymph node metastasis. | [79] |
| ARIC study | Transcriptomic and proteomic profiling | Five plasma proteins with strong and causal links to BC: PEX14 and CTSF positively associated; SNUPN, CSK, and PARK7 negatively associated. | [80] |
| Fresh tissues TCGA and GEO databases | Epigenomic and transcriptomic profiling PPI network construction | Identification of a TFs/miR-126/gene FFL regulating cell identity/stemness. FFL disruption promotes oncogenic transformation and BC progression. | [81] |
| Fresh tissues | Genomic, transcriptomic, metabolomic and lipidomic profiling. Gene–protein–reaction relationship construction | Subclassification of TNBCs in three metabolomic subtypes with prognostic value. N-acetyl-aspartyl-glutamate as potential therapeutic target for high-risk tumors. | [82] |
| Plasma samples | Metabolomic and proteomic profiling PPI network construction Machine learning models for diagnostic efficacy evaluation | Downregulation of metabolism of specific amino acids. Among the 31 DEPs, four enzymes (GOT1, LDHB, GSS, GPX3) linked to deregulated metabolic pathways. Identification of plasma metabolic signature for BC. | [83] |
| FFPE samples | Proteomic profiling Survival analysis | Subclassification of basal-like, HER2-enriched and TNBCs based on immune responses and clinical outcomes | [84] |
| FFPE samples TCGA database | Proteomic profiling Survival analysis | Coronin-1A and α-1-antitrypsin as markers for immune subtype-stratification | [85] |
| Fresh tissues NCG database | Proteomic profiling PPI network construction | Classification of BC subtypes based on oncoproteins/tumor suppressor DEPs. | [86] |
| TCGA database | Transcriptomic and epigenomic profiling | Development of a BC subtype classification framework (moBRCA-net). | [87] |
| Public protein-GWAS studies | Proteomic profiling/disease causal relationship construction | Genetically predicted concentrations of circulating AOC2, SPN1, CD160, RALB, GDI2, CPNE1, ULK3, CTSF, and PLAUR associated with BC risk and subtypes. | [88] |
| Cell lines | Proteomic profiling | ~13,000 cell type-specific proteins correlated with HR status and molecular signatures. RB1 and CB2X as strong predictors of palbociclib response. | [89] |
| FFPE samples TCGA database Cell lines | Proteome and metabolome profiling PPI network construction Survival analysis | PYCR1 and ALDH18A1 associated with NAT resistance, tumor relapse and poor prognosis. PYCR1 KO: increased glutamine catabolism and chemotherapy-sensitivity in ER+ cells, decreased integrin and laminin expression in ER+ and TNBC. | [90] |
| Cell lines Murine models | Transcriptomic profiling miRNA target gene prediction | EMT and metastasis inhibition by propranolol. | [91] |
| GEO and TCGA databases PDB and PubChem databases | Transcriptomic profiling PPI network construction Survival analysis TFs/miRNA/genes network construction Drug sensitivity analysis Molecular modeling and docking | Seven key genes (BUB1, CCNB1, ASPM, TTK, CCNA2, CENPF, and RFC4), regulated by specific TFs and miRNAs, involved in BC progression with prognostic value. Trametinib, selumetinib, and refametinib repurposing for BCs. | [92] |
| Fresh tissues | Transcriptomic and lipidomic profiling | Upregulation of fatty acid oxidation genes depending on metformin resistance or sensitivity. | [93] |
| CMap database | Transcriptomic profiling Molecular modeling and docking | Dolasetron and granisetron repurposing as aromatase inhibitors | [94] |
| METABRIC and LINCS databases | Genomic and transcriptomic profiling Drug–drug interaction analysis | Novel network-based approach for drug repurposing. BC subtype-specific ruxolitinib repurposing. | [95] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Veneziano, M.; Savini, I.; Cortellesi, E.; Gasperi, V.; Gambacurta, A.; Catani, M.V. Bioinformatics Strategies in Breast Cancer Research. Biomolecules 2025, 15, 1409. https://doi.org/10.3390/biom15101409
Veneziano M, Savini I, Cortellesi E, Gasperi V, Gambacurta A, Catani MV. Bioinformatics Strategies in Breast Cancer Research. Biomolecules. 2025; 15(10):1409. https://doi.org/10.3390/biom15101409
Chicago/Turabian StyleVeneziano, Matteo, Isabella Savini, Elisa Cortellesi, Valeria Gasperi, Alessandra Gambacurta, and Maria Valeria Catani. 2025. "Bioinformatics Strategies in Breast Cancer Research" Biomolecules 15, no. 10: 1409. https://doi.org/10.3390/biom15101409
APA StyleVeneziano, M., Savini, I., Cortellesi, E., Gasperi, V., Gambacurta, A., & Catani, M. V. (2025). Bioinformatics Strategies in Breast Cancer Research. Biomolecules, 15(10), 1409. https://doi.org/10.3390/biom15101409







