NEK Family Kinases: Structure, Function, and Role in Disease
Abstract
1. Introduction
2. Structure-Function Relationships Among NEKs
2.1. The NEK Catalytic Domains
2.2. Accessory Domains in NEK Family Members
2.2.1. Coiled-Coil Domain
2.2.2. PEST Sequences
2.2.3. DEAD-Box Domains
2.2.4. Regulator of Chromosome Condensation 1 (RCC1)-like Domain
2.2.5. Armadillo Repeat Domains
3. Regulation of NEKs
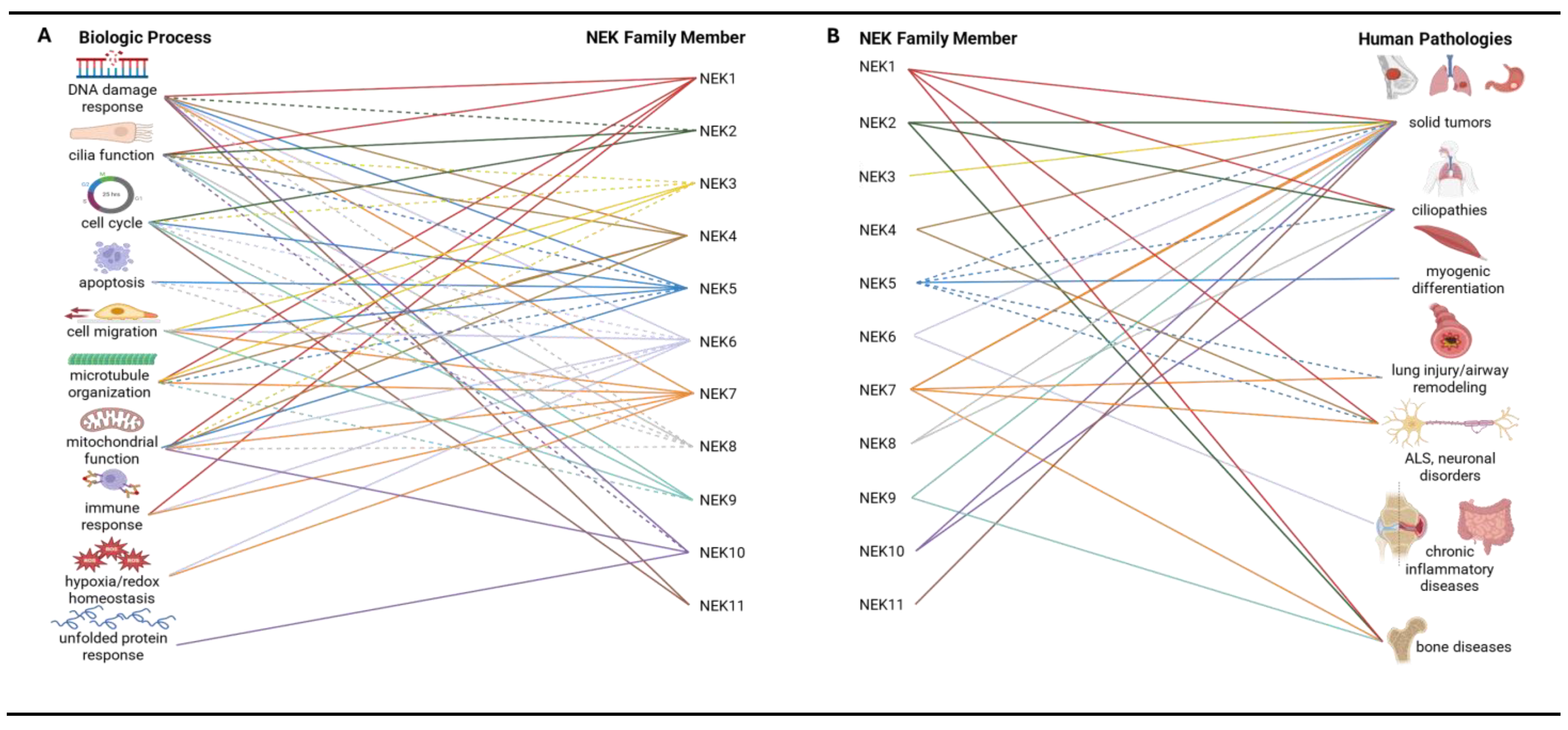
4. Functions of Specific NEK Family Members
4.1. NEK1
4.2. NEK2
4.3. NEK3
4.4. NEK4
4.5. NEK5
4.6. NEK6
4.7. NEK7
4.8. NEK8
4.9. NEK9
4.10. NEK10
4.11. NEK11
5. Perspectives and Future Directions
Future Directions
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Fry, A.M.; O’Regan, L.; Sabir, S.R.; Bayliss, R. Cell cycle regulation by the NEK family of protein kinases. J. Cell Sci. 2012, 125, 4423–4433. [Google Scholar] [CrossRef]
- Osmani, A.H.; McGuire, S.L.; Osmani, S.A. Parallel activation of the NIMA and p34cdc2 cell cycle-regulated protein kinases is required to initiate mitosis in A. nidulans. Cell 1991, 67, 283–291. [Google Scholar] [CrossRef]
- Moniz, L.S.; Dutt, P.; Haider, N.; Stambolic, V. Nek family of kinases in cell cycle, checkpoint control and cancer. Cell Div. 2011, 6, 18. [Google Scholar] [CrossRef]
- Fry, A.M.; Meraldi, P.; Nigg, E.A. A centrosomal function for the human Nek2 protein kinase, a member of the NIMA family of cell cycle regulators. EMBO J. 1998, 17, 470–481. [Google Scholar] [CrossRef]
- Belham, C.; Roig, J.; Caldwell, J.A.; Aoyama, Y.; Kemp, B.E.; Comb, M.; Avruch, J. A mitotic cascade of NIMA family kinases. Nercc1/Nek9 activates the Nek6 and Nek7 kinases. J. Biol. Chem. 2003, 278, 34897–34909. [Google Scholar] [CrossRef]
- Moniz, L.S.; Stambolic, V. Nek10 mediates G2/M cell cycle arrest and MEK autoactivation in response to UV irradiation. Mol. Cell. Biol. 2011, 31, 30–42. [Google Scholar] [CrossRef] [PubMed]
- Fry, A.M.; Schultz, S.J.; Bartek, J.; Nigg, E.A. Substrate specificity and cell cycle regulation of the Nek2 protein kinase, a potential human homolog of the mitotic regulator NIMA of Aspergillus nidulans. J. Biol. Chem. 1995, 270, 12899–12905. [Google Scholar] [CrossRef] [PubMed]
- Parker, J.D.K.; Bradley, B.A.; Mooers, A.O.; Quarmby, L.M. Phylogenetic analysis of the Neks reveals early diversification of ciliary-cell. PLoS ONE 2007, 2, e1076. [Google Scholar] [CrossRef] [PubMed]
- Pavan, I.; Peres de Oliveira, A.; Dias, P.; Basei, F.; Issayama, L.; Ferezin, C.; Silva, F.; Rodrigues de Oliveira, A.; Moura, L.A.; Martins, M.; et al. On Broken Ne(c)ks and Broken DNA: The role of human NEKs in the DNA damage response. Cells. 2021, 10, 507. [Google Scholar] [CrossRef]
- de Castro Ferezin, C.; Lim Kam Sian, T.C.C.; Wu, Y.; Ma, X.; Chüeh, A.C.; Huang, C.; Schittenhelm, R.B.; Kobarg, J.; Daly, R.J. Identification of biological pathways and processes regulated by NEK5 in breast epithelial cells via an integrated proteomic approach. Cell Commun. Signal. 2022, 20, 197. [Google Scholar] [CrossRef]
- Chen, Y.; Jiang, L.; Zhang, L.; Chi, H.; Wang, Q. Immune microenvironment and molecular mechanisms in endometrial cancer: Implications for resistance and innovative treatments. Discov. Oncol. 2025, 16, 532. [Google Scholar] [CrossRef]
- Taylor, S.S.; Kornev, A.P. Protein kinases: Evolution of dynamic regulatory proteins. Trends Biochem. Sci. 2011, 36, 65–77. [Google Scholar] [CrossRef] [PubMed]
- Melo-Hanchuk, T.D.; Slepicka, P.F.; Meirelles, G.V.; Basei, F.L.; Lovato, D.V.; Granato, D.C.; Pauletti, B.A.; Kobarg, J.; Palmieri, L.; Leme, A.F.P.; et al. NEK1 kinase domain structure and its dynamic protein interactome after exposure to cisplatin. Sci. Rep. 2017, 7, 5445. [Google Scholar] [CrossRef] [PubMed]
- Bayliss, R.; Fry, A.; Haq, T.; Yeoh, S. On the molecular mechanisms of mitotic kinase activation. Open Biol. 2012, 2, 120136. [Google Scholar] [CrossRef]
- Rellos, P.; Ivins, F.J.; Baxter, J.E.; Pike, A.; Nott, T.J.; Parkinson, D.M.; Das, S.; Howell, S.; Fedorov, O.; Shen, Q.Y.; et al. Structure and regulation of the human Nek2 centrosomal kinase. J Biol Chem. 2007, 282, 6833. [Google Scholar] [CrossRef]
- van de Kooij, B.; Creixell, P.; van Vlimmeren, A.; Joughin, B.A.; Miller, C.J.; Haider, N.; Simpson, C.D.; Linding, R.; Stambolic, V.; Turk, B.E.; et al. Comprehensive substrate specificity profiling of the human Nek kinome reveals unexpected signaling outputs. eLife 2019, 8, 1–29. [Google Scholar] [CrossRef]
- Richards, M.W.; O’Regan, L.; Mas-Droux, C.; Blot, J.M.Y.; Cheung, J.; Hoelder, S.; Fry, A.M.; Bayliss, R. An autoinhibitory tyrosine motif in the cell-cycle-regulated Nek7 kinase is released through binding of Nek 9. Mol. Cell 2009, 36, 560–570. [Google Scholar] [CrossRef]
- Haq, T.; Richards, M.W.; Burgess, S.G.; Gallego, P.; Yeoh, S.; O’Regan, L.; Reverter, D.; Roig, J.; Fry, A.M.; Bayliss, R. Mechanistic basis of Nek7 activation through Nek9 binding and induced dimerization. Nat. Commun. 2015, 6, 8771. [Google Scholar] [CrossRef]
- Lupas, A.N.; Bassler, J.; Dunin-Horkawicz, S. The structure and topology of α-helical coiled coils. In Fibrous Proteins: Structures and Mechanisms; Subcellular Biochemistry; Springer: Cham, Switzerland, 2017; Volume 82, pp. 95–129. [Google Scholar] [CrossRef]
- Croasdale, R.; Ivins, F.J.; Muskett, F.; Daviter, T.; Scott, D.J.; Hardy, T.; Smerdon, S.J.; Fry, A.M.; Pfuhl, M.; Chin, J.W.; et al. An undecided coiled coil: The leucine zipper of Nek2 kinase exhibits atypical conformational exchange dynamics. J. Biol. Chem. 2011, 286, 27537–27547. [Google Scholar] [CrossRef]
- Helps, N.R.; Luo, X.; Barker, H.M.; Cohen, P.T. NIMA-related kinase 2 (Nek2), a cell-cycle-regulated protein kinase localized to centrosomes, is complexed to protein phosphatase 1. Biochem. J. 2000, 349, 509–518. [Google Scholar] [CrossRef] [PubMed]
- Hames, R.S.; Wattam, S.L.; Yamano, H.; Bacchieri, R.; Fry, A.M. APC/C-mediated destruction of the centrosomal kinase Nek2A occurs in early mitosis and depends upon a cyclin A-type D-box. EMBO J. 2001, 20, 7117–7127. [Google Scholar] [CrossRef]
- Rechsteiner, M.; Rogers, S.W. PEST sequences and regulation by proteolysis. Trends Biochem. Sci. 1996, 21, 267–271. [Google Scholar] [CrossRef] [PubMed]
- Mendenhall, M.D.; Hodge, A.E. Regulation of Cdc28 cyclin-dependent protein kinase activity during the cell cycle of the yeast Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 1998, 62, 1191–1243. [Google Scholar] [CrossRef]
- Xu, G.; Bernaudo, S.; Fu, G.; Lee, D.Y.; Yang, B.B.; Peng, C. Cyclin G2 is degraded through the ubiquitin-proteasome pathway and mediates the antiproliferative effect of activin receptor-like kinase 7. Mol. Biol. Cell 2008, 19, 4968–4979. [Google Scholar] [CrossRef]
- Li, Y.; Zhu, J.; Zhai, F.; Kong, L.; Li, H.; Jin, X. Advances in the understanding of nuclear pore complexes in human diseases. J. Cancer Res. Clin. Oncol. 2024, 150, 374. [Google Scholar] [CrossRef]
- Sekhar, K.R.; Freeman, M.L. PEST sequences in proteins involved in cyclic nucleotide signalling pathways. J. Recept. Signal Transduct. Res. 1998, 18, 113–132. [Google Scholar] [CrossRef]
- Burton, J.L.; Solomon, M.J. D box and KEN box motifs in budding yeast Hsl1p are required for APC-mediated degradation and direct binding to Cdc20p and Cdh1p. Genes Dev. 2001, 15, 2381–2395. [Google Scholar] [CrossRef]
- Bachus, S.; Graves, D.; Fulham, L.; Akkerman, N.; Stephanson, C.; Shieh, J.; Pelka, P. In mitosis you are not: The NIMA family of kinases in Aspergillus, yeast, and mammals. Int. J. Mol. Sci. 2022, 23, 4041. [Google Scholar] [CrossRef] [PubMed]
- Chang, J.; Baloh, R.H.; Milbrandt, J. The NIMA-family kinase Nek3 regulates microtubule acetylation in neurons. J. Cell Sci. 2009, 122, 2274–2282. [Google Scholar] [CrossRef] [PubMed]
- Krien, M.J.E.; West, R.R.; John, U.P.; Koniaras, K.; McIntosh, J.R.; O’Connell, M.J. The fission yeast NIMA kinase Fin1p is required for spindle function and nuclear envelope integrity. EMBO J. 2002, 21, 1713–1722. [Google Scholar] [CrossRef][Green Version]
- Fuller-Pace, F.V. DEAD box RNA helicase functions in cancer. RNA Biol. 2013, 10, 121–132. [Google Scholar] [CrossRef]
- Xing, Z.; Ma, W.K.; Tran, E.J. The DDX5/Dbp2 subfamily of DEAD-box RNA helicases. Wiley Interdiscip. Rev. RNA 2019, 10, e1519. [Google Scholar] [CrossRef]
- Hirth, A.; Fatti, E.; Netz, E.; Acebron, S.P.; Papageorgiou, D.; Švorinić, A.; Cruciat, C.-M.; Karaulanov, E.; Gopanenko, A.; Zhu, T.; et al. DEAD box RNA helicases are pervasive protein kinase interactors and activators. Genome Res. 2024, 34, 952–966. [Google Scholar] [CrossRef]
- Basei, F.L.; Silva, I.R.E.; Firmino Dias, P.R.; Ferezin, C.C.; Peres de Oliveira, A.; Issayama, L.K.; Moura, L.A.R.; Riback da Silva, F.; Kobarg, J. The mitochondrial connection: The Nek kinases’ new functional axis in mitochondrial homeostasis. Cells 2024, 13, 473. [Google Scholar] [CrossRef]
- Cargill, M.; Venkataraman, R.; Lee, S. DEAD-box RNA helicases and genome stability. Genes 2021, 12, 1471. [Google Scholar] [CrossRef]
- England, J.R.; Huang, J.; Jennings, M.J.; Makde, R.D.; Tan, S. RCC1 uses a conformationally diverse loop region to interact with the nucleosome: A model for the RCC1-nucleosome complex. J. Mol. Biol. 2010, 398, 518–529. [Google Scholar] [CrossRef][Green Version]
- Seki, T.; Hayashi, N.; Nishimoto, T. RCC1 in the Ran pathway. J. Biochem. 1996, 120, 207–214. [Google Scholar] [CrossRef] [PubMed]
- Hadjebi, O.; Casas-Terradellas, E.; Garcia-Gonzalo, F.R.; Rosa, J.L. The RCC1 superfamily: From genes, to function, to disease. Biochim. Biophys. Acta 2008, 1783, 1467–1479. [Google Scholar] [CrossRef] [PubMed]
- Roig, J. NEK8, a NIMA-family protein kinase at the core of the ciliary INV complex. Cell Commun. Signal. 2025, 23, 170. [Google Scholar] [CrossRef] [PubMed]
- Bertran, M.T.; Sdelci, S.; Regué, L.; Avruch, J.; Caelles, C.; Roig, J. Nek9 is a Plk1-activated kinase that controls early centrosome separation through Nek6/7 and Eg 5. EMBO J. 2011, 30, 2634–2647. [Google Scholar] [CrossRef]
- Renault, L.; Kuhlmann, J.; Henkel, A.; Wittinghofer, A. Structural basis for guanine nucleotide exchange on Ran by the regulator of chromosome condensation (RCC1). Cell 2001, 105, 245–255. [Google Scholar] [CrossRef] [PubMed]
- Roig, J.; Mikhailov, A.; Belham, C.; Avruch, J. Nercc1, a mammalian NIMA-family kinase, binds the Ran GTPase and regulates mitotic progression. Genes Dev. 2002, 16, 1640–1658. [Google Scholar] [CrossRef]
- Yang, S.W.; Gao, C.; Chen, L.; Song, Y.L.; Zhu, J.L.; Qi, S.T.; Jiang, Z.Z.; Li, Y.Q.; Han, C.S.; Sun, Q.Y. Nek9 regulates spindle organization and cell cycle progression during mouse oocyte meiosis and its location in early embryo mitosis. Cell Cycle 2012, 11, 4366–4377. [Google Scholar] [CrossRef]
- Li, H.; Li, J.; Zhang, Y.; Cao, R.; Guo, C.; Jiao, M. The NIMA-related kinase family and cancer. Front. Oncol. 2025, 15, 1556917. [Google Scholar] [CrossRef]
- Huber, A.H.; Nelson, W.J.; Weis, W.I. Three-dimensional structure of the armadillo repeat region of β-catenin. Cell. 1997, 90, 871–882. [Google Scholar] [CrossRef]
- Arrías, P.N.; Osmanli, Z.; Paralta, E.; Chinestrad, P.M.; Monzon, A.M.; Tosatto, S.C.E. Diversity and structural-functional insights of alpha-solenoid proteins. Prot. Sci. 2024, 33, e5189. [Google Scholar] [CrossRef] [PubMed]
- Power, K.M.; Akella, J.S.; Gu, A.; Walsh, J.D.; Bellotti, S.; Morash, M.; Zhang, W.; Ramadan, Y.H.; Ross, N.; Golden, A.; et al. Mutation of NEKL-4/NEK10 and TTLL genes suppress neuronal ciliary degeneration caused by loss of CCPP-1 deglutamylase function. PLoS Genet. 2020, 16, e1009052. [Google Scholar] [CrossRef]
- Haider, N.; Dutt, P.; van de Kooij, B.; Ho, J.; Palomero, L.; Pujana, M.A.; Yaffe, M.; Stambolic, V. NEK10 tyrosine phosphorylates p53 and controls its transcriptional activity. Oncogene 2020, 39, 5252–5266. [Google Scholar] [CrossRef]
- Meirelles, G.V.; Perez, A.M.; de Souza, E.E.; Basei, F.L.; Papa, P.F.; Melo Hanchuk, T.D.; Cardoso, V.B.; Kobarg, J. “Stop Ne(c)king around”: How interactomics contributes to functionally characterize Nek family kinases. World J. Biol. Chem. 2014, 5, 141–160. [Google Scholar] [CrossRef] [PubMed]
- Maier, B.D.; Petursson, B.; Lussana, A.; Petsalaki, E. Data-driven extraction of human kinase-substrate relationships from Omics datasets. Mol. Cell Proteomics. 2025, 24, 100994. [Google Scholar] [CrossRef]
- Chivukula, R.R.; Montoro, D.T.; Leung, H.M.; Yang, J.; Shamseldin, H.E.; Taylor, M.S.; Dougherty, G.W.; Zariwala, M.A.; Carson, J.; Daniels, M.L.A.; et al. A human ciliopathy reveals essential functions for NEK10 in airway mucociliary clearance. Nat. Med. 2020, 26, 378–388. [Google Scholar] [CrossRef]
- Fry, A.M.; Arnaud, L.; Nigg, E.A. Activity of the human centrosomal kinase, Nek2, depends on an unusual leucine zipper dimerization motif. ” J. Biol. Chem. 1999, 274, 38863–38873. [Google Scholar] [CrossRef]
- Dutt, P.; Haider, N.; Mouaaz, S.; Stambolic, V. β-Catenin turnover is regulated by Nek10-mediated tyrosine phosphorylation in A549 lung adenocarcinoma cells. Proc. Natl. Acad. Sci. USA 2024, 121, e2300606121. [Google Scholar] [CrossRef]
- Fry, A.M.; Bayliss, R.; Roig, J. Mitotic regulation by NEK kinase networks. Front. Cell Dev. Biol. 2017, 5, 102. [Google Scholar] [CrossRef]
- Freund, I.; Hehlgans, S.; Martin, D.; Ensminger, M.; Fokas, E.; Rödel, C.; Löbrich, M.; Rödel, F. Fractionation-dependent radiosensitization by molecular targeting of Nek1. Cells 2020, 9, 1235. [Google Scholar] [CrossRef] [PubMed]
- Singh, V.; Connelly, Z.M.; Shen, X.; De Benedetti, A. Identification of the proteome complement of human TLK1 reveals it binds and phosphorylates NEK1 regulating its activity. Cell Cycle 2017, 16, 915–926. [Google Scholar] [CrossRef] [PubMed]
- Maréchal, A.; Zou, L. DNA damage sensing by the ATM and ATR kinases. Cold Spring Harb Perspect Biol. 2013, 5, a012716. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Ho, C.K.; Ouyang, J.; Zou, L. Nek1 kinase associates with ATR-ATRIP and primes ATR for efficient DNA damage signaling. Proc. Natl. Acad. Sci. USA 2013, 110, 2175–2180. [Google Scholar] [CrossRef] [PubMed]
- Cannavo, E.; Giordano Reginato, M.; Cejka, P. Stepwise 5′ DNA end-specific resection of DNA breaks by the Mre11-Rad50-Xrs2 and Sae2 nuclease ensemble. Proc. Natl. Acad. Sci. USA 2019, 116, 5505–5513. [Google Scholar] [CrossRef]
- Spies, J.; Waizenegger, A.; Barton, O.; Sürder, M.; Wright, W.D.; Heyer, W.D.; Löbrich, M. Nek1 regulates Rad54 to orchestrate homologous recombination and replication fork stability. Mol. Cell 2016, 62, 903–917. [Google Scholar] [CrossRef]
- Gregorczyk, M.; Pastore, G.; Muñoz, I.; Carroll, T.; Streubel, J.; Munro, M.; Lis, P.; Lange, S.; Lamoliatte, F.; Macartney, T.; et al. ; et al. Functional characterization of C21ORF2 association with the NEK1 kinase mutated in human diseases. Life Sci. Alliance 2023, 6, e202201740. [Google Scholar] [CrossRef]
- Watanabe, Y.; Nakagawa, T.; Akiyama, T.; Nakagawa, M.; Suzuki, N.; Warita, H.; Aoki, M.; Nakayama, K. An amyotrophic lateral sclerosis-associated mutant of C21ORF2 is stabilized by NEK1-mediated hyperphosphorylation and the inability to bind FBXO3. iScience 2020, 23, 101491. [Google Scholar] [CrossRef]
- Hardy, T.; Lee, M.; Hames, R.S.; Prosser, S.L.; Cheary, D.M.; Samant, M.D.; Schultz, F.; Baxter, J.E.; Rhee, K.; Fry, A.M. Multisite phosphorylation of C-Nap1 releases it from Cep135 to trigger centrosome disjunction. J. Cell Sci. 2014, 127, 2493–2506. [Google Scholar] [CrossRef]
- Bahe, S.; Stierhof, Y.D.; Wilkinson, C.J.; Leiss, F.; Nigg, E.A. Rootletin forms centriole-associated filaments and functions in centrosome cohesion. J. Cell Biol. 2005, 171, 27–33. [Google Scholar] [CrossRef]
- Wei, R.; Ngo, B.; Wu, G.; Lee, W.H. Phosphorylation of the Ndc80 complex protein, HEC1, by Nek2 kinase modulates chromosome alignment and signaling of the spindle assembly checkpoint. Mol. Biol. Cell. 2011, 22, 3584–3595. [Google Scholar] [CrossRef] [PubMed]
- Fang, Y.; Zhang, X. Targeting NEK2 as a promising therapeutic approach for cancer treatment. Cell Cycle 2016, 15, 895–907. [Google Scholar] [CrossRef]
- Park, J.; Rhee, K. NEK2 phosphorylation antagonizes the microtubule stabilizing activity of centrobin. Biochem. Biophys. Res. Commun. 2013, 431, 302–308. [Google Scholar] [CrossRef] [PubMed]
- Mbom, B.C.; Siemers, K.A.; Ostrowski, M.A.; Nelson, W.J.; Barth, A.I. Nek2 phosphorylates and stabilizes β-catenin at mitotic centrosomes downstream of Plk1. Mol Biol Cell. 2014, 25, 977–991. [Google Scholar] [CrossRef] [PubMed]
- Miller, S.L.; Antico, G.; Raghunath, P.N.; Tomaszewski, J.E.; Clevenger, C.V. Nek3 kinase regulates prolactin-mediated cytoskeletal reorganization and motility of breast cancer cells. Oncogene 2007, 26, 4668–4678. [Google Scholar] [CrossRef]
- Nguyen, C.L.; Possemato, R.; Bauerlein, E.L.; Xie, A.; Scully, R.; Hahn, W.C. Nek4 regulates entry into replicative senescence and the response to DNA damage in human fibroblasts. Mol. Cell. Biol. 2012, 32, 3963–3977. [Google Scholar] [CrossRef]
- Ferezin, C.D.C.; Basei, F.L.; Melo-Hanchuk, T.D.; de Oliveira, A.L.; Peres de Oliveira, A.; Mori, M.P.; de Souza-Pinto, N.C.; Kobarg, J. NEK5 interacts with LonP1 and its kinase activity is essential for the regulation. FEBS Open Bio. 2021, 11, 546–563. [Google Scholar] [CrossRef]
- Adib, R.; Montgomery, J.M.; Atherton, J.; O’Regan, L.; Richards, M.W.; Straatman, K.R.; Roth, D.; Straube, A.; Bayliss, R.; Moores, C.A.; et al. Mitotic phosphorylation by NEK6 and NEK7 reduces the microtubule affinity of EML4 to promote chromosome congression. Sci. Signal. 2019, 12, eaaw2939. [Google Scholar] [CrossRef]
- Rapley, J.; Nicolàs, M.; Groen, A.; Regué, L.; Bertran, M.T.; Caelles, C.; Avruch, J.; Roig, J. The NIMA-family kinase Nek6 phosphorylates the kinesin Eg5 at a novel site necessary for mitotic spindle formation. J. Cell Sci. 2008, 121, 3912–3921. [Google Scholar] [CrossRef]
- Hashimoto, Y.; Akita, H.; Hibino, M.; Kohri, K.; Nakanishi, M. Identification and characterization of Nek6 protein kinase, a potential human homolog of NIMA histone H3 kinase. Biochem. Biophys. Res. Commun. 2002, 293, 753–758. [Google Scholar] [CrossRef] [PubMed]
- O’Regan, L.; Sampson, J.; Richards, M.W.; Knebel, A.; Roth, D.; Hood, F.E.; Straube, A.; Royle, S.J.; Bayliss, R.; Fry, A.M. Hsp72 is targeted to the mitotic spindle by Nek6 to promote K-fiber assembly and mitotic progression. J. Cell Biol. 2015, 209, 349–358. [Google Scholar] [CrossRef] [PubMed]
- Takatani, S.; Ozawa, S.; Yagi, N.; Hotta, T.; Hashimoto, T.; Takahashi, Y.; Takahashi, T.; Motose, H. Directional cell expansion requires NIMA-related kinase 6 (NEK6)-mediated cortical microtubule destabilization. Sci. Rep. 2017, 7, 7826. [Google Scholar] [CrossRef]
- Belham, C.; Comb, M.J.; Avruch, J. Identification of the NIMA family kinases NEK6/7 as regulators of the p70 ribosomal S6 kinase. Curr. Biol. 2001, 11, 1155–1167. [Google Scholar] [CrossRef] [PubMed]
- Jeon, Y.J.; Lee, K.Y.; Cho, Y.Y.; Pugliese, A.; Kim, H.G.; Jeong, C.H.; Bode, A.M.; Dong, Z. Role of NEK6 in tumor promoter-induced transformation in JB6 C141 mouse skin epidermal cells. J. Biol. Chem. 2010, 285, 28126–28133. [Google Scholar] [CrossRef]
- Tan, R.; Nakajima, S.; Wang, Q.; Sun, H.; Xue, J.; Wu, J.; Hellwig, S.; Zeng, X.; Yates, N.A.; Smithgall, T.E.; et al. Nek7 Protects Telomeres from Oxidative DNA Damage by Phosphorylation and Stabilization of TRF1. Mol. Cell 2017, 65, 715–729. [Google Scholar] [CrossRef]
- Xu, J.; Zhang, L.; Duan, Y.; Sun, F.; Odeh, N.; He, Y.; Núñez, G. NEK7 phosphorylation amplifies NLRP3 inflammasome activation downstream of potassium efflux and gasdermin D. Sci. Immunol. 2025, 10, eadl2993. [Google Scholar] [CrossRef]
- Freixo, F.; Martinez Delgado, P.; Manso, Y.; Sánchez-Huertas, C.; Lacasa, C.; Soriano, E.; Roig, J.; Lüders, J. NEK7 regulates dendrite morphogenesis in neurons via Eg5-dependent microtubule stabilization. Nat. Commun. 2018, 9, 2330. [Google Scholar] [CrossRef]
- Czarnecki, P.G.; Gabriel, G.C.; Manning, D.K.; Sergeev, M.; Lemke, K.; Klena, N.T.; Liu, X.; Chen, Y.; Li, Y.; San Agustin, J.T.; et al. ANKS6 is the critical activator of NEK8 kinase in embryonic situs determination and organ patterning. Nat. Commun. 2015, 6, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Cao, B.; Zhang, K.; Pan, C.; Dong, Y.; Lu, F. NEK8 regulates colorectal cancer progression via phosphorylating MYC. Cell Commun. Signal. 2023, 21, 209. [Google Scholar] [CrossRef]
- Holland, P.M.; Milne, A.; Garka, K.; Johnson, R.S.; Willis, C.; Sims, J.E.; Rauch, C.T.; Bird, T.A.; Virca, G.D. Purification, cloning, and characterization of Nek8, a novel NIMA-related kinase, and its candidate substrate Bicd2. J. Biol. Chem. 2002, 277, 16229–16240. [Google Scholar] [CrossRef]
- Sun, Z.; Wang, Q.; Sun, L.; Wu, M.; Li, S.; Hua, H.; Sun, Y.; Ni, T.; Zhou, C.; Huang, S.; et al. Acetaminophen-induced reduction of NIMA-related kinase 7 expression exacerbates acute liver injury. JHEP Rep. 2022, 4, 100545. [Google Scholar] [CrossRef]
- Sdelci, S.; Schütz, M.; Pinyol, R.; Bertran, M.T.; Regué, L.; Caelles, C.; Vernos, I.; Roig, J. Nek9 phosphorylation of NEDD1/GCP-WD contributes to Plk1 control of γ-tubulin recruitment to the mitotic centrosome. Curr. Biol. 2012, 22, 1516–1523. [Google Scholar] [CrossRef] [PubMed]
- Lu, G.; Du, R.; Dong, J.; Sun, Y.; Zhou, F.; Feng, F.; Feng, B.; Han, Y.; Shang, Y. Cancer-associated fibroblast derived SLIT2 drives gastric cancer cell metastasis by activating NEK9. Cell Death Dis. 2023, 14, 421. [Google Scholar] [CrossRef]
- Lu, G.; Tian, S.; Sun, Y.; Dong, J.; Wang, N.; Zeng, J.; Nie, Y.; Wu, K.; Han, Y.; Feng, B.; et al. NEK9, a novel effector of IL-6/STAT3, regulates metastasis of gastric cancer by targeting ARHGEF2 phosphorylation. Theranostics 2021, 11, 2460–2474. [Google Scholar] [CrossRef]
- Gallisà-Suñé, N.; Sànchez-Fernàndez-de-Landa, P.; Zimmermann, F.; Serna, M.; Regué, L.; Paz, J.; Llorca, O.; Lüders, J.; Roig, J. BICD2 phosphorylation regulates dynein function and centrosome separation in G2 and M. Nat. Commun. 2023, 14, 2434. [Google Scholar] [CrossRef]
- Melixetian, M.; Klein, D.K.; Sørensen, C.S.; Helin, K. NEK11 regulates CDC25A degradation and the IR-induced G2/M checkpoint. Nat. Cell Biol. 2009, 11, 1247–1253. [Google Scholar] [CrossRef] [PubMed]
- Sørensen, C.S.; Melixetian, M.; Klein, D.K.; Helin, K. NEK11: Linking CHK1 and CDC25A in DNA damage checkpoint signaling. Cell Cycle 2010, 9, 450–455. [Google Scholar] [CrossRef] [PubMed]
- Fang, X.; Lin, H.; Wang, X.; Zuo, Q.; Qin, J.; Zhang, P. The NEK1 interactor, C21ORF2, is required for efficient DNA damage repair. Acta Biochim. Biophys. Sin. 2015, 47, 834–841. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Chen, C.-F.; Chiang, H.-C.; Pena, M.; Polci, R.; Wei, R.L.; Edwards, R.A.; Hansel, D.E.; Chen, P.-L.; Riley, D.J. Mutation of NIMA-related kinase 1 (NEK1) leads to chromosome instability. Mol. Cancer 2011, 10, 5. [Google Scholar] [CrossRef]
- Mann, J.R.; McKenna, E.D.; Mawrie, D.; Papakis, V.; Alessandrini, F.; Anderson, E.N.; Mayers, R.; Ball, H.E.; Kaspi, E.; Lubinski, K.; et al. Loss of function of the ALS-associated NEK1 kinase disrupts microtubule homeostasis and nuclear import. Sci. Adv. 2023, 9, 1–14. [Google Scholar] [CrossRef]
- Noh, M.-Y.; Oh, S.-i.; Kim, Y.-E.; Cha, S.J.; Sung, W.; Oh, K.-W.; Park, Y.; Mun, J.Y.; Ki, C.-S.; Nahm, M.; et al. Mutations in NEK1 cause ciliary dysfunction as a novel pathogenic mechanism in amyotrophic lateral sclerosis. Mol. Neurodegener. 2025, 20, 1–14. [Google Scholar] [CrossRef]
- Wang, H.; Qi, W.; Zou, C.; Xie, Z.; Zhang, M.; Naito, M.G.; Mifflin, L.; Liu, Z.; Najafov, A.; Pan, H.; et al. NEK1-mediated retromer trafficking promotes blood–brain barrier integrity by regulating glucose metabolism and RIPK1 activation. Nat. Commun. 2021, 12, 1–16. [Google Scholar] [CrossRef]
- Awuah, W.A.; Tan, J.K.; Shkodina, A.D.; Ferreira, T.; Adebusoye, F.T.; Mazzoleni, A.; Wellington, J.; David, L.; Chilcott, E.; Huang, H.; et al. Hereditary spastic paraplegia: Novel insights into the pathogenesis and management. SAGE Open Med. 2023, 12, 20503121231221941. [Google Scholar] [CrossRef] [PubMed]
- Martins, M.B.; Perez, A.M.; Bohr, V.A.; Wilson, D.M.; Kobarg, J. NEK1 deficiency affects mitochondrial functions and the transcriptome of key DNA repair pathways. Mutagenesis. 2021, 36, 223–236. [Google Scholar] [CrossRef]
- Thiel, C.; Kessler, K.; Giessl, A.; Dimmler, A.; Shalev, S.A.; von der Haar, S.; Zenker, M.; Zahnleiter, D.; Stöss, H.; Beinder, E.; et al. NEK1 mutations cause short-rib polydactyly syndrome type Majewski. Am. J. Hum. Genet. 2011, 88, 106–114. [Google Scholar] [CrossRef]
- He, Q.; Zhou, Y.; Jin, J.; Tian, Q.; Li, H.; Hou, B.; Xie, A. Association between NEK1 gene polymorphisms and the potential risk of sporadic Parkinson’s disease in the Chinese northern Han population: A case-control study. Neurosci. Lett. 2024, 837, 137913. [Google Scholar] [CrossRef]
- Shalom, O.; Shalva, N.; Altschuler, Y.; Motro, B. The mammalian Nek1 kinase is involved in primary cilium formation. FEBS Lett. 2008, 582, 1465–1470. [Google Scholar] [CrossRef]
- White, M.C.; Quarmby, L.M. The NIMA-family kinase, Nek1 affects the stability of centrosomes and ciliogenesis. BMC Cell Biol. 2008, 9, 29. [Google Scholar] [CrossRef]
- Chen, Y.; Chiang, H.C.; Litchfield, P.; Pena, M.; Juang, C.; Riley, D.J. Expression of Nek1 during kidney development and cyst formation in multiple nephron segments in the Nek1-deficient kat2J mouse model of polycystic kidney disease. J Biomed Sci. 2014, 21, 63. [Google Scholar] [CrossRef]
- Wang, W.; Wu, T.; Kirschner, M.W. The master cell cycle regulator APC-Cdc20 regulates ciliary length and disassembly of the primary cilium. eLife 2014, 3, e03083. [Google Scholar] [CrossRef]
- Zhu, J.; Cai, Y.; Liu, P.; Zhao, W. Frequent Nek1 overexpression in human gliomas. Biochem. Biophys. Res. Commun. 2016, 476, 522–527. [Google Scholar] [CrossRef] [PubMed]
- Khalil, M.I.; De Benedetti, A. Tousled-like kinase 1: A novel factor with multifaceted role in mCRPC progression and development of therapy resistance. Cancer Drug Resist. 2022, 5, 93–101. [Google Scholar] [CrossRef] [PubMed]
- Polci, R.; Peng, A.; Chen, P.L.; Riley, D.J.; Chen, Y. NIMA-related protein kinase 1 is involved early in the ionizing radiation-induced DNA damage response. Cancer Res. 2004, 64, 8800–8803. [Google Scholar] [CrossRef]
- Dyrskjøt, L.; Reinert, T.; Algaba, F.; Christensen, E.; Nieboer, D.; Hermann, G.G.; Mogensen, K.; Beukers, W.; Marquez, M.; Segersten, U.; et al. Prognostic impact of a 12-gene progression score in non-muscle-invasive bladder cancer: A prospective multicentre validation study. Eur. Urol. 2017, 72, 461–469. [Google Scholar] [CrossRef] [PubMed]
- Lindskrog, S.V.; Prip, F.; Lamy, P.; Taber, A.; Groeneveld, C.S.; Birkenkamp-Demtröder, K.; Jensen, J.B.; Strandgaard, T.; Nordentoft, I.; Christensen, E.; et al. An integrated multi-Omics analysis identifies prognostic molecular subtypes of non-muscle-invasive bladder cancer. Nat. Commun. 2021, 12, 2301. [Google Scholar] [CrossRef]
- Rapley, J.; Baxter, J.E.; Blot, J.; Wattam, S.L.; Casenghi, M.; Meraldi, P.; Nigg, E.A.; Fry, A.M. Co-ordinate regulation of the mother centriole component Nlp by Nek2 and Plk1 protein kinases. Mol. Cell. Biol. 2005, 25, 1309–1324. [Google Scholar] [CrossRef]
- Mardin, B.R.; Agircan, F.G.; Lange, C.; Schiebel, E. Plk1 controls the Nek2A-PP1γ antagonism in centrosome disjunction. Curr. Biol. 2011, 21, 1145–1151. [Google Scholar] [CrossRef] [PubMed]
- Mayor, T.; Hacker, U.; Stierhof, Y.D.; Nigg, E.A. The mechanism regulating the dissociation of the centrosomal protein C-Nap1 from mitotic spindle poles. J. Cell Sci. 2002, 115, 3275–3284. [Google Scholar] [CrossRef]
- Chen, Y.; Riley, D.J.; Zheng, L.; Chen, P.L.; Lee, W.H. Phosphorylation of the mitotic regulator protein Hec1 by Nek2 kinase is essential for faithful chromosome segregation. J. Biol. Chem. 2002, 277, 49408–49416. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.T.; Zhang, H. The Mitotic Checkpoint Complex (MCC): Looking Back and Forth after 15 Years. AIMS Mol. Sci. 2016, 3, 597–634. [Google Scholar] [CrossRef]
- DeLuca, J.G.; Gall, W.E.; Ciferri, C.; Cimini, D.; Musacchio, A.; Salmon, E.D. Kinetochore microtubule dynamics and attachment stability are regulated by Hec1. Cell. 2006, 127, 969–982. [Google Scholar] [CrossRef]
- Sundin, L.J.; Guimaraes, G.J.; DeLuca, J.G. The NDC80 complex proteins Nuf2 and Hec1 make distinct contributions to kinetochore-microtubule attachment in mitosis. Mol. Biol. Cell 2011, 22, 759–768. [Google Scholar] [CrossRef]
- Nguyen, K.; Boehling, J.; Tran, M.N.; Cheng, T.; Rivera, A.; Collins-Burow, B.M.; Lee, S.B.; Drewry, D.H.; Burow, M.E. NEK family review and correlations with patient survival outcomes in various cancer types. Cancers 2023, 15, 2067. [Google Scholar] [CrossRef]
- Faragher, A.J.; Fry, A.M. Nek2A kinase stimulates centrosome disjunction and is required for formation of bipolar mitotic spindles. Mol. Biol. Cell 2003, 14, 2876–2889. [Google Scholar] [CrossRef]
- Wu, J.; Luo, D.; Tou, L.; Xu, H.; Jiang, C.; Wu, D.; Que, H.; Zheng, J. NEK2 affects the ferroptosis sensitivity of gastric cancer cells by regulating the expression of HMOX1 through Keap1/Nrf 2. Mol. Cell. Biochem. 2025, 480, 425–437. [Google Scholar] [CrossRef]
- Franqui-Machin, R.; Hao, M.; Bai, H.; Gu, Z.; Zhan, X.; Habelhah, H.; Jethava, Y.; Qiu, L.; Frech, I.; Tricot, G.; et al. Destabilizing NEK2 overcomes resistance to proteasome inhibition in multiple myeloma. J. Clin. Investig. 2018, 128, 2877–2893. [Google Scholar] [CrossRef] [PubMed]
- Howes, S.C.; Alushin, G.M.; Shida, T.; Nachury, M.V.; Nogales, E. Effects of tubulin acetylation and tubulin acetyltransferase binding on microtubule structure. Mol. Biol. Cell. 2014, 25, 257–266. [Google Scholar] [CrossRef]
- Zhang, Y.; Chen, W.; Zeng, W.; Lu, Z.; Zhou, X. Biallelic loss of function NEK3 mutations deacetylate α-tubulin and downregulate NUP205 that predispose individuals to cilia-related abnormal cardiac left-right patterning. Cell Death Dis. 2020, 11, 1005. [Google Scholar] [CrossRef]
- Soppina, V.; Herbstman, J.F.; Skiniotis, G.; Verhey, K.J. Luminal localization of α-tubulin K40 acetylation by cryo-EM analysis of fab-labeled microtubules. PLoS ONE 2012, 7, e48204. [Google Scholar] [CrossRef]
- Chen, Y.; Zhou, G.; Yu, M. Conformational dynamics of the nuclear pore complex central channel. Biochem. Soc. Trans. 2025, 53, 267–279. [Google Scholar] [CrossRef]
- Basei, F.L.; de Castro Ferezin, C.; Rodrigues de Oliveira, A.L.; Muñoz, J.P.; Zorzano, A.; Kobarg, J. Nek4 regulates mitochondrial respiration and morphology. FEBS J. 2022, 289, 3262–3279. [Google Scholar] [CrossRef] [PubMed]
- Ding, N.H.; Zhang, L.; Xiao, Z.; Rong, Z.X.; Li, Z.; He, J.; Chen, L.; Ou, D.M.; Liao, W.H.; Sun, L.Q. NEK4 kinase regulates EMT to promote lung cancer metastasis. J. Cell. Mol. Med. 2018, 22, 5877–5887. [Google Scholar] [CrossRef] [PubMed]
- Thiery, J.P.; Acloque, H.; Huang, R.Y.J.; Nieto, M.A. Epithelial-mesenchymal transitions in development and disease. Cell. 2009, 139, 871–890. [Google Scholar] [CrossRef] [PubMed]
- Park, S.J.; Jo, D.S.; Jo, S.-Y.; Shin, D.W.; Shim, S.; Jo, Y.K.; Shin, J.H.; Ha, Y.J.; Jeong, S.-Y.; Hwang, J.J.; et al. Inhibition of never in mitosis A (NIMA)-related kinase-4 reduces survivin expression and sensitizes cancer cells to TRAIL-induced cell death. Oncotarget. 2016, 7, 65957–65967. [Google Scholar] [CrossRef]
- Chandra, D.; Liu, J.W.; Tang, D.G. Early mitochondrial activation and cytochrome c up-regulation during apoptosis. J. Biol. Chem. 2002, 277, 50842–50854. [Google Scholar] [CrossRef]
- Wang, C.; Youle, R.J. The role of mitochondria in apoptosis. Annu. Rev. Genet. 2009, 43, 95–118. [Google Scholar] [CrossRef]
- Jiang, X.; Wang, X. Cytochrome C-mediated apoptosis. Annu Rev Biochem. 2004, 73, 87–106. [Google Scholar] [CrossRef] [PubMed]
- Ambrosini, G.; Adida, C.; Altieri, D.C. IAP-family protein survivin inhibits caspase activity and apoptosis induced by Fas (CD95), Bax, caspases, and anticancer drugs. Cancer Res. 1998, 58, 5315–5320. [Google Scholar] [PubMed]
- Doles, J.; Hemann, M.T. Nek4 status differentially alters sensitivity to distinct microtubule poisons. Cancer Res. 2010, 70, 1033–1041. [Google Scholar] [CrossRef]
- Abouzeid, H.A.; Kassem, L.; Liu, X.; Abuelhana, A. Paclitaxel resistance in breast cancer: Current challenges and recent advanced therapeutic strategies. Cancer Treat. Res. Commun. 2025, 43, 100918. [Google Scholar] [CrossRef] [PubMed]
- Xiao, H.; Verdier-Pinard, P.; Fernandez-Fuentes, N.; Burd, B.; Angeletti, R.; Fiser, A.; Horwitz, S.B.; Orr, G.A. Insights into the mechanism of microtubule stabilization by Taxol. Proc. Natl. Acad. Sci. USA 2006, 103, 10166–10173. [Google Scholar] [CrossRef]
- Prosser, S.L.; Sahota, N.K.; Pelletier, L.; Morrison, C.G.; Fry, A.M. Nek5 promotes centrosome integrity in interphase and loss of centrosome cohesion in mitosis. J. Cell Biol. 2015, 209, 339–348. [Google Scholar] [CrossRef]
- Sigal, Y.M.; Zhou, R.; Zhuang, X. Visualizing and discovering cellular structures with super-resolution microscopy. Science. 2018, 361, 880–887. [Google Scholar] [CrossRef]
- Matossian, M.D.; Wells, C.I.; Zuercher, W.J.; Collins-Burow, B.M.; Drewry, D.H.; Burow, M.E. NEK5 activity regulates the mesenchymal and migratory phenotype in breast cancer cells. Breast Cancer Res. Treatment. 2021, 188, 609–622. [Google Scholar] [CrossRef]
- Zhou, X.; Nie, H.; Wang, C.; Yu, X.; Yang, X.; He, X.; Ou, C. Prognostic value and therapeutic potential of NEK family in stomach adenocarcinoma. J. Cancer 2024, 15, 3154–3172. [Google Scholar] [CrossRef]
- Nusinow, D.P.; Szpyt, J.; Ghandi, M.; Rose, C.M.; McDonald, E.R.; Kalocsay, M.; Jané-Valbuena, J.; Gelfand, E.; Schweppe, D.K.; Jedrychowski, M.; et al. Quantitative proteomics of the cancer cell line encyclopedia. Cell. 2020, 180, 387–402.e16. [Google Scholar] [CrossRef]
- Li, T.; Fan, J.; Wang, B.; Traugh, N.; Chen, Q.; Liu, J.S.; Li, B.; Liu, X.S. TIMER: A web server for comprehensive analysis of tumor-infiltrating immune cells. Cancer Res. 2017, 77, e108–e110. [Google Scholar] [CrossRef]
- Yu, Y.; Shen, T.; Zhong, X.; Wang, L.-L.; Tai, W.; Zou, Y.; Qin, J.; Zhang, Z.; Zhang, C.-L. NEK6 is an injury-responsive kinase cooperating with STAT3 in regulation of reactive astrogliosis. Glia 2021, 70, 273–286. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; li, J.; Fu, M.; Wang, W. The JAK/STAT signaling pathway: From bench to clinic. Sig Transduct Target Ther. 2021, 6, 402. [Google Scholar] [CrossRef]
- Peron, M.; Dinarello, A.; Meneghetti, G.; Martorano, L.; Betto, R.M.; Facchinello, N.; Tesoriere, A.; Tiso, N.; Martello, G.; Argenton, F. Y705 and S727 are required for the mitochondrial import and transcriptional activities of STAT3, and for regulation of stem cell proliferation. Development 2021, 148, dev199477. [Google Scholar] [CrossRef]
- Zhu, M.; Sun, Y.; Xue, H.; Wu, G.; Wang, Z.; Shi, J.; Ma, J.; Gu, B.; Yan, X. NEK6 promotes the progression of osteosarcoma through activating STAT3 signaling pathway by down-regulation of miR-26a-5p. Int. J. Gen. Med. 2023, 16, 2831–2848. [Google Scholar] [CrossRef]
- Pavan, I.C.B.; Basei, F.L.; Brandemarte, M.; Rosa e Silva, I.; Issayama, L.K.; Mancini, M.C.S.; Goís, M.M.; da Silva, L.G.S.; Bezerra, R.M.N.; Simabuco, F.M.; et al. NEK6 Regulates Redox Balance and DNA Damage Response in DU-145 Prostate Cancer Cells. Cells. 2023, 12, 256. [Google Scholar] [CrossRef]
- De Donato, M.; Fanelli, M.; Mariani, M.; Raspaglio, G.; Pandya, D.; He, S.; Fiedler, P.; Petrillo, M.; Scambia, G.; Ferlini, C. Abstract 4327: Nek6 and Hif-1α cooperate with the cytoskeletal gateway of drug resistance to drive outcome in serous ovarian cancer. Cancer Res. 2015, 75 (Suppl. S15), 4327. [Google Scholar] [CrossRef]
- Carmeliet, P.; Dor, Y.; Herbert, J.-M.; Fukumura, D.; Brusselmans, K.; Dewerchin, M.; Neeman, M.; Bono, F.; Abramovitch, R.; Maxwell, P.; et al. Role of HIF-1α in hypoxia-mediated apoptosis, cell proliferation and tumour angiogenesis. Nature. 1998, 394, 485–490. [Google Scholar] [CrossRef]
- He, Y.; Zeng, M.Y.; Yang, D.; Motro, B.; Nunez, G. NEK7 is an essential mediator of NLRP3 activation downstream of potassium efflux. Nature 2016, 530, 354–357. [Google Scholar] [CrossRef] [PubMed]
- He, H.; Jiang, H.; Chen, Y.; Ye, J.; Wang, A.; Wang, C.; Liu, Q.; Liang, G.; Deng, X.; Jiang, W.; et al. Oridonin is a covalent NLRP3 inhibitor with strong anti-inflammasome activity. Nat. Commun. 2018, 9, 2550. [Google Scholar] [CrossRef] [PubMed]
- Kelley, N.; Jeltema, D.; Duan, Y.; He, Y. The NLRP3 inflammasome: An overview of mechanisms of activation and regulation. Int. J. Mol. Sci. 2019, 20, 3328. [Google Scholar] [CrossRef]
- Yan, Z.; Da, Q.; Li, Z.; Lin, Q.; Yi, J.; Su, Y.; Yu, G.; Ren, Q.; Liu, X.; Lin, Z.; et al. Inhibition of NEK7 suppressed hepatocellular carcinoma progression by mediating cancer cell pyroptosis. Front. Oncol. 2022, 12, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Dong, Y.; Liu, D.; Zou, Z.; Hao, G.; Gao, X.; Pan, P.; Liang, G. NEK7 coordinates rapid neuroinflammation after subarachnoid hemorrhage in mice. Front. Neurol. 2020, 11, 551. [Google Scholar] [CrossRef] [PubMed]
- Sharif, H.; Wang, L.; Wang, W.L.; Magupalli, V.G.; Andreeva, L.; Qiao, Q.; Hauenstein, A.V.; Wu, Z.; Núñez, G.; Mao, Y.; et al. Structural mechanism for NEK7-licensed activation of NLRP3 inflammasome. Nature. 2019, 570, 338–343. [Google Scholar] [CrossRef]
- Kim, S.K.; Choe, J.Y.; Park, K.Y. Anti-inflammatory effect of artemisinin on uric acid-induced NLRP3 inflammasome activation through blocking interaction between NLRP3 and NEK7. Biochem. Biophys. Res. Commun. 2019, 517, 338–345. [Google Scholar] [CrossRef]
- Chen, Y.; Meng, J.; Bi, F.; Li, H.; Chang, C.; Ji, C.; Liu, W. NEK7 regulates NLRP3 inflammasome activation and neuroinflammation post-traumatic brain injury. Front. Mol. Neurosci. 2019, 12, 202. [Google Scholar] [CrossRef]
- Li, D.; Wang, L.; Ou, J.; Wang, C.; Zhou, J.; Lu, L.; Wu, Y.; Gao, J. Reactive oxygen species induced by uric acid promote NRK-52E cell apoptosis through the NEK7-NLRP3 signaling pathway. Mol. Med. Rep. 2021, 24, 729. [Google Scholar] [CrossRef]
- Seok, J.K.; Kang, H.C.; Cho, Y.Y.; Lee, H.S.; Lee, J.Y. Therapeutic regulation of the NLRP3 inflammasome in chronic inflammatory diseases. Arch. Pharm. Res. 2021, 44, 16–35. [Google Scholar] [CrossRef]
- Chen, Y.; Ye, X.; Escames, G.; Lei, W.; Zhang, X.; Li, M.; Jing, T.; Yao, Y.; Qiu, Z.; Wang, Z.; et al. The NLRP3 inflammasome: Contributions to inflammation-related diseases. Cell Mol. Biol. Lett. 2023, 28, 51. [Google Scholar] [CrossRef]
- Yao, J.; Sterling, K.; Wang, Z.; Zhang, Y.; Song, W. The role of inflammasomes in human diseases and their potential as therapeutic targets. Signal Transduct. Target Ther. 2024, 9, 10. [Google Scholar] [CrossRef] [PubMed]
- Lui, K.; Huang, Y. RanGTPase: A key regulator of nucleocytoplasmic trafficking. Mol Cell Pharmacol. 2009, 1, 148–156. [Google Scholar] [CrossRef]
- Kalab, P.; Heald, R. The RanGTP gradient—a GPS for the mitotic spindle. J Cell Sci. 2008, 121, 1577–1586. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Han, X.; Chen, J.; Xie, X.; Xu, J.; Zhao, Y.; Shen, J.; Hu, L.; Xu, P.; Song, H.; et al. Yes-associated protein (YAP) and transcriptional coactivator with PDZ-binding motif (TAZ) mediate cell density-dependent proinflammatory responses. J. Biol. Chem. 2018, 293, 18071–18085. [Google Scholar] [CrossRef] [PubMed]
- Choi, H.J.C.; Lin, J.-R.; Vannier, J.-B.; Slaats, G.G.; Kile, A.C.; Paulsen, R.D.; Manning, D.K.; Beier, D.R.; Giles, R.H.; Boulton, S.J.; et al. NEK8 links the ATR-regulated replication stress response and S phase CDK activity to renal ciliopathies. Mol. Cell. 2013, 51, 423–439. [Google Scholar] [CrossRef]
- Otto, E.A.; Trapp, M.L.; Schultheiss, U.T.; Helou, J.; Quarmby, L.M.; Hildebrandt, F. NEK8 mutations affect ciliary and centrosomal localization and may cause nephronophthisis. J. Am. Soc. Nephrol. 2008, 19, 587–592. [Google Scholar] [CrossRef]
- Shiba, D.; Manning, D.K.; Koga, H.; Beier, D.R.; Yokoyama, T. Inv acts as a molecular anchor for Nphp3 and Nek8 in the proximal segment of primary cilia. Cytoskeleton 2010, 67, 112–119. [Google Scholar] [CrossRef] [PubMed]
- Grampa, V.; Delous, M.; Zaidan, M.; Odye, G.; Thomas, S.; Elkhartoufi, N.; Filhol, E.; Niel, O.; Silbermann, F.; Lebreton, C.; et al. Novel NEK8 mutations cause severe syndromic renal cystic dysplasia through YAP dysregulation. PLoS Genet. 2016, 12, e1005894. [Google Scholar] [CrossRef]
- Abeyta, A.; Castella, M.; Jacquemont, C.; Taniguchi, T. NEK8 regulates DNA damage-induced RAD51 foci formation and replication fork protection. Cell Cycle. 2017, 16, 335–347. [Google Scholar] [CrossRef]
- Kang, E.; Kim, H.K.; Lee, H.B.; Han, W. Never in mitosis gene A-related kinase-8 promotes proliferation, migration, invasion, and stemness of breast cancer cells via β-catenin signalling activation. Sci. Rep. 2023, 13, 6829. [Google Scholar] [CrossRef]
- Bowers, A.J.; Boylan, J.F. Nek8, a NIMA family kinase member, is overexpressed in primary human breast tumors. Gene 2004, 328, 135–142. [Google Scholar] [CrossRef]
- Yamamoto, Y.; Chino, H.; Tsukamoto, S.; Ode, K.L.; Ueda, H.R.; Mizushima, N. NEK9 regulates primary cilia formation by acting as a selective autophagy adaptor for MYH9/Myosin IIA. Nat. Commun. 2021, 12, 3292. [Google Scholar] [CrossRef]
- Smith, S.C.; Petrova, A.V.; Madden, M.Z.; Wang, H.; Pan, Y.; Warren, M.D.; Hardy, C.W.; Liang, D.; Liu, E.A.; Robinson, M.H.; et al. A gemcitabine sensitivity screen identifies a role for NEK9 in the replication stress response. Nucleic Acids Res. 2014, 42, 11517–11527. [Google Scholar] [CrossRef]
- Tan, B.C.; Lee, S.C. Nek9, a novel FACT-associated protein, modulates interphase progression. J Biol Chem. 2004, 279, 9321–9330. [Google Scholar] [CrossRef]
- Roig, J.; Groen, A.; Caldwell, J.; Avruch, J. Active Nercc1 protein kinase concentrates at centrosomes early in mitosis and is necessary for proper spindle assembly. Mol. Biol. Cell 2005, 16, 4827–4840. [Google Scholar] [CrossRef] [PubMed]
- Kaneta, Y.; Ullrich, A. NEK9 depletion induces catastrophic mitosis by impairment of mitotic checkpoint control and spindle dynamics. Biochem. Biophys. Res. Commun. 2013, 442, 139–146. [Google Scholar] [CrossRef]
- O’Regan, L.; Barone, G.; Adib, R.; Woo, C.G.; Jeong, H.J.; Richardson, E.L.; Richards, M.W.; Muller, P.A.J.; Collis, S.J.; Fennell, D.A.; et al. EML4-ALK V3 oncogenic fusion proteins promote microtubule stabilization and accelerated migration through NEK9 and NEK7. J. Cell Sci. 2020, 133, jcs241505. [Google Scholar] [CrossRef]
- de Oliveira, A.P.; Basei, F.L.; Slepicka, P.F.; de Castro Ferezin, C.; Melo-Hanchuk, T.D.; de Souza, E.E.; Lima, T.I.; dos Santos, V.T.; Mendes, D.; Silveira, L.R.; et al. NEK10 interactome and depletion reveal new roles in mitochondria. Proteome Sci. 2020, 18, 1–15. [Google Scholar] [CrossRef]
- de Oliveira, A.P.; Navarro, C.D.C.; Dias, P.R.F.; Arguello, T.; Walker, B.R.; Bacman, S.R.; Sousa, L.M.; Castilho, R.F.; Consonni, S.R.; Moraes, C.T.; et al. NEK10 kinase ablation affects mitochondrial morphology, function and protein phosphorylation status. Proteome Sci. 2024, 22, 8. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.; Wang, Z.B.; Wang, H.H.; Zhang, T.; Qi, S.T.; Ouyang, Y.C.; Sun, Q.Y. Nek11 regulates asymmetric cell division during mouse oocyte meiotic maturation. Biochem. Biophys. Res. Commun. 2016, 474, 667–672. [Google Scholar] [CrossRef] [PubMed]
- Minoguchi, S.; Minoguchi, M.; Yoshimura, A. Differential control of the NIMA-related kinases, Nek6 and Nek7, by serum stimulation. Biochem. Biophys. Res. Commun. 2003, 301, 899–906. [Google Scholar] [CrossRef]
- Panchal, N.K.; Mohanty, S.; Prince, S.E. NIMA-related kinase-6 (NEK6) as an executable target in cancer. Clin. Transl. Oncol. 2023, 25, 66–77. [Google Scholar] [CrossRef]
- Branon, T.C.; Bosch, J.A.; Sanchez, A.D.; Udeshi, N.D.; Svinkina, T.; Carr, S.A.; Feldman, J.L.; Perrimon, N.; Ting, A.Y. Efficient proximity labeling in living cells and organisms with TurboID. Nat Biotechnol. 2018, 36, 880–887. [Google Scholar] [CrossRef] [PubMed]
- May, D.G.; Scott, K.L.; Campos, A.R.; Roux, K.J. Comparative application of BioID and TurboID for protein-proximity biotinylation. Cells. 2020, 9, 1070. [Google Scholar] [CrossRef]
- Newman, R.H.; Zhang, J.; Zhu, H. Toward a systems level view of dynamic phosphorylation networks. Front. Genet. 2014, 5, 263. [Google Scholar] [CrossRef]
- Zhu, H.; Cox, E.; Qian, J. Functional protein microarray as molecular decathlete: A versatile player in clinical proteomics. Proteom. Clin. Appl. 2012, 6, 548–562. [Google Scholar] [CrossRef]
- Newman, R.H.; Hu, J.; Rho, H.S.; Xie, Z.; Woodard, C.; Neiswinger, J.; Cooper, C.; Shirley, M.; Clark, H.M.; Hu, S.; et al. Construction of human activity-based phosphorylation networks. Mol. Syst. Biol. 2013, 9, 1–26. [Google Scholar] [CrossRef]
- Jonić, S. Cryo-electron Microscopy Analysis of Structurally Heterogeneous Macromolecular Complexes. Comput. Struct. Biotechnol. J. 2016, 14, 385–390. [Google Scholar] [CrossRef] [PubMed]
- Abramson, J.; Adler, J.; Dunger, J.; Evans, R.; Green, T.; Pritzel, A.; Ronneberger, O.; Willmore, L.; Ballard, A.J.; Bambrick, J.; et al. Accurate structure prediction of biomolecular interactions with AlphaFold 3. Nature. 2024, 630, 493–500. [Google Scholar] [CrossRef]
- Hu, C.-M.; Zhu, J.; Guo, X.E.; Chen, W.; Qiu, X.L.; Ngo, B.; Chien, R.; Wang, Y.V.; Tsai, C.Y.; Wu, G.; et al. Novel small molecules disrupting Hec1/Nek2 interaction ablate tumor progression by triggering Nek2 degradation through a death-trap mechanism. Oncogene. 2015, 34, 1220–1230. [Google Scholar] [CrossRef] [PubMed]
- Greenwald, E.C.; Mehta, S.; Zhang, J. Genetically Encoded Fluorescent Biosensors Illuminate the Spatiotemporal Regulation of Signaling Networks. Chem. Rev. 2018, 118, 11707–11794. [Google Scholar] [CrossRef]
- Newman, R.H.; Fosbrink, M.; Zhang, J. Fluorescent biosensors for tracking signaling dynamics in living cells. Chem. Rev. 2011, 111, 3614–3666. [Google Scholar] [CrossRef] [PubMed]
- Newman, R.H.; Zhang, J. Integrated strategies to gain a systems-level view of dynamic signaling networks. Methods Enzymol. 2017, 589, 133–170. [Google Scholar] [CrossRef] [PubMed]
- Nijenhuis, W.; van Grinsven, M.M.P.; Kapitein, L.C. An optimized toolbox for the optogenetic control of intracellular transport. J. Cell Biol. 2020, 219, e201907149. [Google Scholar] [CrossRef] [PubMed]
- Natsume, T.; Kanemaki, M.T. Conditional degrons for controlling protein expression at the protein level. Annu. Rev. Genet. 2017, 51, 83–102. [Google Scholar] [CrossRef]
- Han, X.; Sun, Y. PROTACs: A novel strategy for cancer drug discovery and development. MedComm 2023, 4, e290. [Google Scholar] [CrossRef]
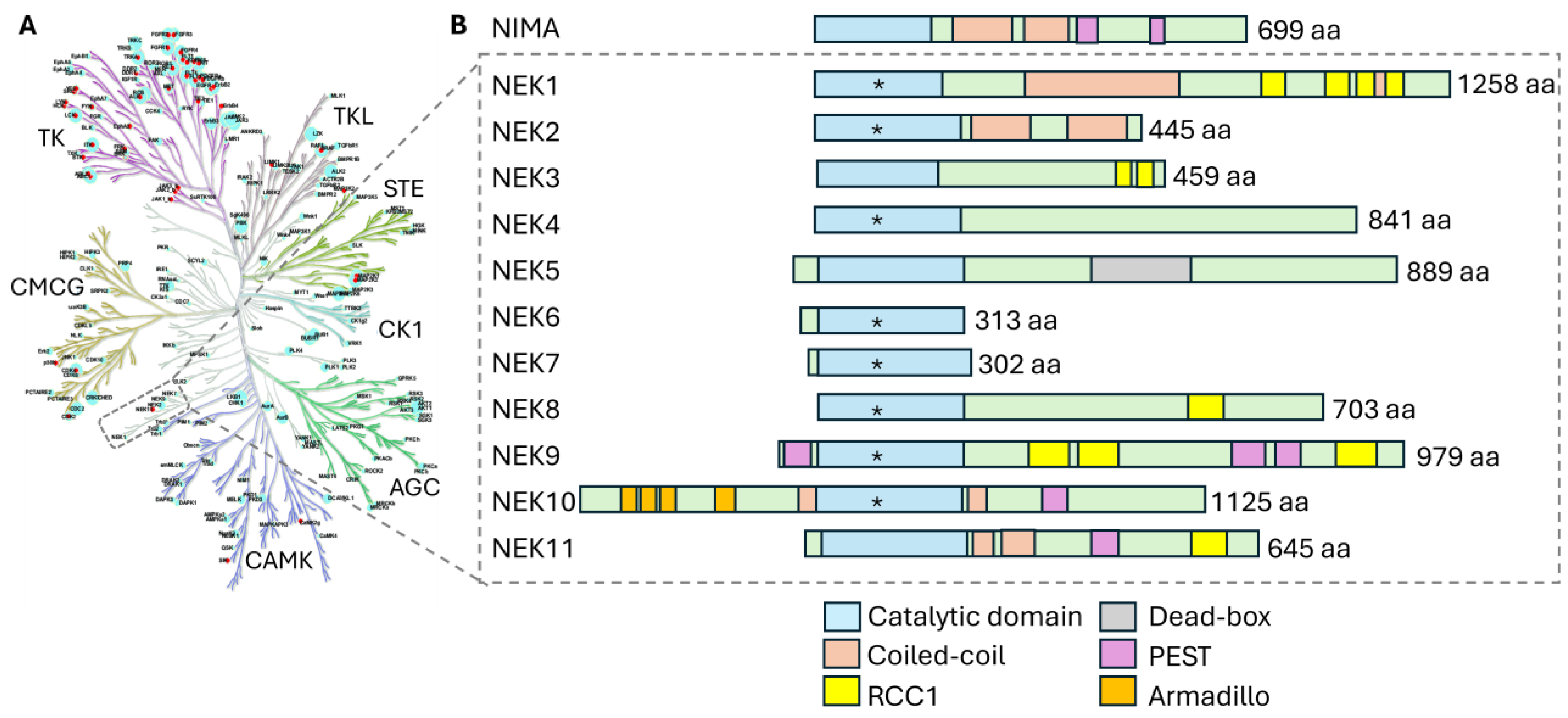

| Domain | NEK Family Members | Location | Functional Role |
|---|---|---|---|
| PEST sequence | NIMA; Fin1 (S. pombe); 5 of 11 human NEKs (e.g., NEK2A, 9, 10, 11) | Generally in the C-terminal non- catalytic region | Signals for rapid degradation by ubiquitin-proteosome system; ensures timely turn-over at mitotic exit to coordinate cell cycle progression |
| DEAD-box domain | NEK5 | C-terminal to the kinase domain | ATP-dependent RNA helicase activity; remodels RNA/RNPs; likely links NEK signaling to RNA processing, stress granule dynamics, or mitochondrial homeostasis |
| RCC1-like domain | NEK8, NEK9 | C-terminal to the kinase domain | Β-propeller scaffold for RAN-GTP interactions; enforces autoinhibition (NEK9), scaffolds NEK6/7 activation cascade; may target NEKs to chromatin or centrosomes |
| Coiled-coil domain | All human NEKs except NEK4, 6, 7; NIMA | C-terminal tail or flanking kinase domain | Drives homo- or hetero-oligomerization; enables trans-autophosphorylation (e.g., NEK2 dimerization); governs subcellular targeting (e.g., centrosome, nucleolus) |
| Armadillo Repeat domain | NEK10 | N-terminal region (4 tandem repeats) | Provides an elongated scaffold for protein–protein interactions; directs NEK10 to ciliary or stress-response complexes and mediates interactions (e.g., with HSPB1) |
| Kinase domain | All NEK family members | N-terminal 250–300 aa bilobed kinase fold | Binds ATP and catalyzes the transfer of phosphate to protein substrates; controlled by motifs (e.g., Gly-rich loop, VAIK, HRDLKPEN, DFG, and APE motifs) and regulatory features (“Tyr-down” autoinhibition, dimerization cues) |
| Function | NEK1 | NEK2 | NEK3 | NEK4 | NEK5 | NEK6 | NEK7 | NEK8 | NEK9 | NEK10 | NEK11 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| DNA Damage Response | |||||||||||
| Cell Cycle Regulation | |||||||||||
| Ciliary Function | |||||||||||
| Inflammation/ Immune Response | |||||||||||
| Microtubule Dynamics/ Intracellular Transport | |||||||||||
| Metabolism/ Mitochondrial Function | |||||||||||
| Cell Migration | |||||||||||
| Apoptosis | |||||||||||
| Hypoxia/ Redox Homeostasis | |||||||||||
| Unfolded Protein Response |
| NEK Member | Substrate | Phosphosite | Function of Phosphorylation | Ref |
|---|---|---|---|---|
| NEK1 | ||||
| ATRIP | T1989 | Interacts with ATR to maintain genomic stability in DNA damage repair pathways. | [59] | |
| Rad54 | S572 | Promotes the removal of Rad51 from chromatin during homologous recombination (HR) in the G2 phase of the cell cycle | [61] | |
| C21ORF2 | ND | Inhibits ubiquitylation, stabilizing C21ORF2 | [62,63] | |
| NEK2 | ||||
| c-NAP1 | S2131/T2132, S2128, S2229, S2234, S2322 | Separation of duplicated centrosomes at the onset of mitosis | [64] | |
| Rootletin | NH2- and COOH-term fragments | Triggers centrosome separation during the cell cycle | [65] | |
| HEC1 | S165 | Enhances HEC1 interaction with MAD1 at kinetochores | [66] | |
| NLP | ND | Triggers NLP’s removal from the centrosome, allowing reorganization of the microtubule network required for spindle formation. | [67] | |
| Centrobin | T35, S36, S41, S45 | Antagonizes centrobin’s microtubule-stabilizing activity | [68] | |
| β-catenin | S33, S37, T41 | Stabilizes β-catenin | [69] | |
| NEK3 | ||||
| VAV-2 | ND | Activates VAV2 during prolactin receptor signaling, leading to cytoskeletal reorganization and motility in breast cancer cells. | [70] | |
| NEK4 | ||||
| yH2AX | S139 (predicted) | Essential for the formation of γH2AX foci, a key marker of DSBs. | [71] | |
| NEK5 | ||||
| LonP1 | ND | Modulates LonP1 activity and affects mitochondrial function | [72] | |
| NEK6 | ||||
| EML4 | S144 | Reduces its affinity for microtubules, promoting chromosome congression during mitosis | [73] | |
| Eg5/KIF11 | S1033 | Regulation of spindle dynamics | [74] | |
| Histone H1 | ND | Regulation of chromatin condensation during mitosis | [75] | |
| Histone H3 | ND | Regulation of chromatin condensation during mitosis | [75] | |
| HSP72/HSPA1A | T66 | Regulation of spindle organization | [76] | |
| β-tubulin | T166 | Promotes the depolymerization of cortical microtubules (CMTs) | [77] | |
| p70S6K | T412 | Activation of enzymatic activity | [78] | |
| STAT3 | S727 | Promotes tumor growth | [79] | |
| NEK7 | ||||
| EML4 | S146 | Modulates microtubule stability | [73] | |
| TRF1 | S114 | Stabilizes the shelterin complex and limits ROS-induced telomeric attrition and DNA damage signaling. | [80] | |
| NLRP3 | S803 | NEK7 binds to and activates NLRP3 post-K+ efflux (non-catalytic); phosphorylation-independent interaction | [81] | |
| p70S6K | T412 | Activation of enzymatic activity | [78] | |
| Eg5/KIF11 | S146, S1033 | Regulation of spindle dynamics | [82] | |
| NEK8 | ||||
| ANKS6 | ND | Localization to the ciliary inversin compartment (IC) | [83] | |
| MYC | S405 | Inhibits ubiquitylation, stabilizing c-MYC | [84] | |
| BICD2 | ND | Potential role in modulating microtubule morphology | [85] | |
| NEK8 (Autophos) | ND | NEK8 functionally depends on CEP164 for localization and interaction; may act downstream | [83,84] | |
| NEK9 | ||||
| NEK6 | S206 | Activates NEK6, which is crucial for proper spindle formation | [74] | |
| NEK7 | S195 | Activates NEK7, which regulates centrosome separation | [86] | |
| NEDD1 | S377 | NEDD1 recruitment to centrosomes and proper γ-tubulin localization in mitotic cells | [87] | |
| TRIM28 | S473 | Enhances the transcriptional activity, promoting downstream expression of STAT3, NF-κB p100, and cortactin (CTTN), driving cytoskeletal reorganization and metastatic behavior in gastric cancer cells | [88] | |
| CTTN | S417 | Facilitates actin cytoskeletal remodeling and cell motility in the context of gastric cancer metastasis | [88] | |
| ARHGEF2 | ND | Activates ARHGEF2, promoting RhoA activation and cell motility | [89] | |
| BICD2 | ND | Regulates dynein–dynactin motor activity during mitosis | [90] | |
| NEK10 | ||||
| p53 | Y327 | Modulates p53’s stability and activity | [49] | |
| β-catenin (CTNNB1) | Y30 | Promotes degradation of CTNNB1 | [54] | |
| NEK11 | ||||
| CDC25A | S79, S82, S88 | Promotes CDC25A’s degradation in response to DNA damage | [91,92] | |
| CHK1 | S273 | Part of the DDR pathway, specifically regulating the G2/M checkpoint | [92] |
| Disease | NEK1 | NEK2 | NEK3 | NEK4 | NEK5 | NEK6 | NEK7 | NEK8 | NEK9 | NEK10 | NEK11 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Cancer | |||||||||||
| Ciliopathies (e.g., PKD) | |||||||||||
| Neurodegenerative Disorders | |||||||||||
| Inflammatory Disorders |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Baker, B.M.; Boehling, J.R.; Knopf, S.; Held, S.; Matossian, M.; Belgodere, J.A.; Hoang, V.T.; Collins-Burow, B.M.; Martin, E.C.; Lee, S.B.; et al. NEK Family Kinases: Structure, Function, and Role in Disease. Biomolecules 2025, 15, 1406. https://doi.org/10.3390/biom15101406
Baker BM, Boehling JR, Knopf S, Held S, Matossian M, Belgodere JA, Hoang VT, Collins-Burow BM, Martin EC, Lee SB, et al. NEK Family Kinases: Structure, Function, and Role in Disease. Biomolecules. 2025; 15(10):1406. https://doi.org/10.3390/biom15101406
Chicago/Turabian StyleBaker, Brandon M., Julia R. Boehling, Sarah Knopf, Stephanie Held, Margarite Matossian, Jorge A. Belgodere, Van T. Hoang, Bridgette M. Collins-Burow, Elizabeth C. Martin, Sean B. Lee, and et al. 2025. "NEK Family Kinases: Structure, Function, and Role in Disease" Biomolecules 15, no. 10: 1406. https://doi.org/10.3390/biom15101406
APA StyleBaker, B. M., Boehling, J. R., Knopf, S., Held, S., Matossian, M., Belgodere, J. A., Hoang, V. T., Collins-Burow, B. M., Martin, E. C., Lee, S. B., Burow, M. E., Drewry, D. H., & Newman, R. H. (2025). NEK Family Kinases: Structure, Function, and Role in Disease. Biomolecules, 15(10), 1406. https://doi.org/10.3390/biom15101406








