Predicted IL-18/IL-18R Binding Improvement Through Protein Interface Modification with Computer-Aided Design
Abstract
1. Introduction
2. Materials and Methods
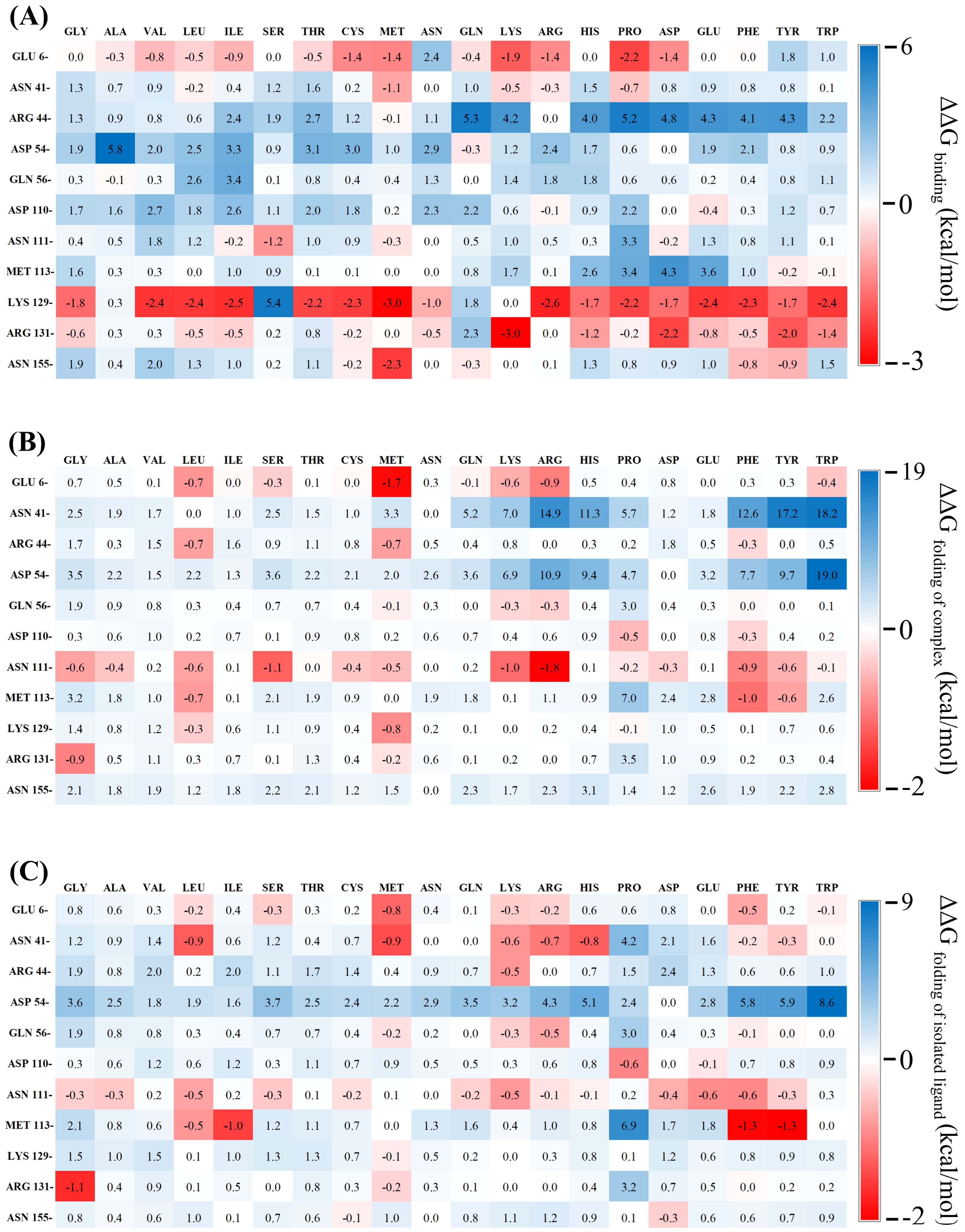
2.1. In Silico Mutagenesis and Energy Decomposition
2.2. Analysis of Relative Binding Free Energy Change upon Mutation
2.3. Molecular Dynamics (MD) Simulation
2.4. Trajectory Analysis
3. Results and Discussion
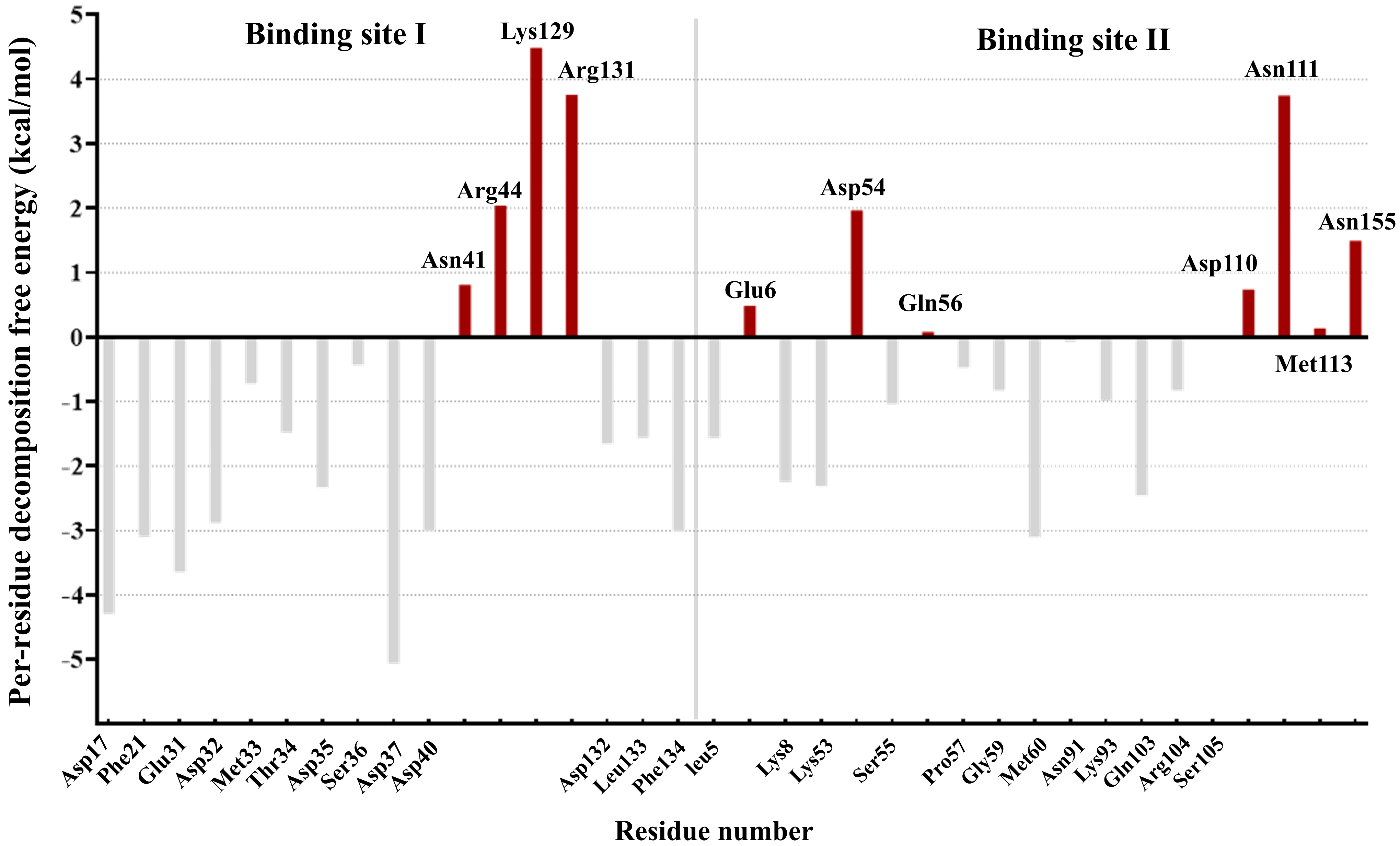
3.1. Per-Residue Decomposition Energy
3.2. Validation of the Computational Approach
- (1)
- “ΔΔGbinding”, which estimated the change in the binding affinity of the wild-type and the mutant variant toward its receptor;
- (2)
- “ΔΔGfolding of complex”, which determined the impact of mutations on the stability of the IL-18/IL-18R complex;
- (3)
- “ΔΔGfolding of free IL-18”, which investigated the effect of mutations on the stability of isolated IL-18.
ΔΔG folding of complex : Y = −0.4544x + 0.5580
ΔΔG folding of isolated ligand : Y = −0.2829x + 0.5701
3.3. In Silico Screening of Favorable Mutation
3.3.1. Single Point Mutation Study
3.3.2. Double Mutation Study
3.3.3. Multiple Mutation Study
3.3.4. Selection of Favorable Mutations
- -
- E6M+K129M+R131G+N111S;
- -
- E6M+N111S+R131G;
- -
- E6M+K129M+R131G.
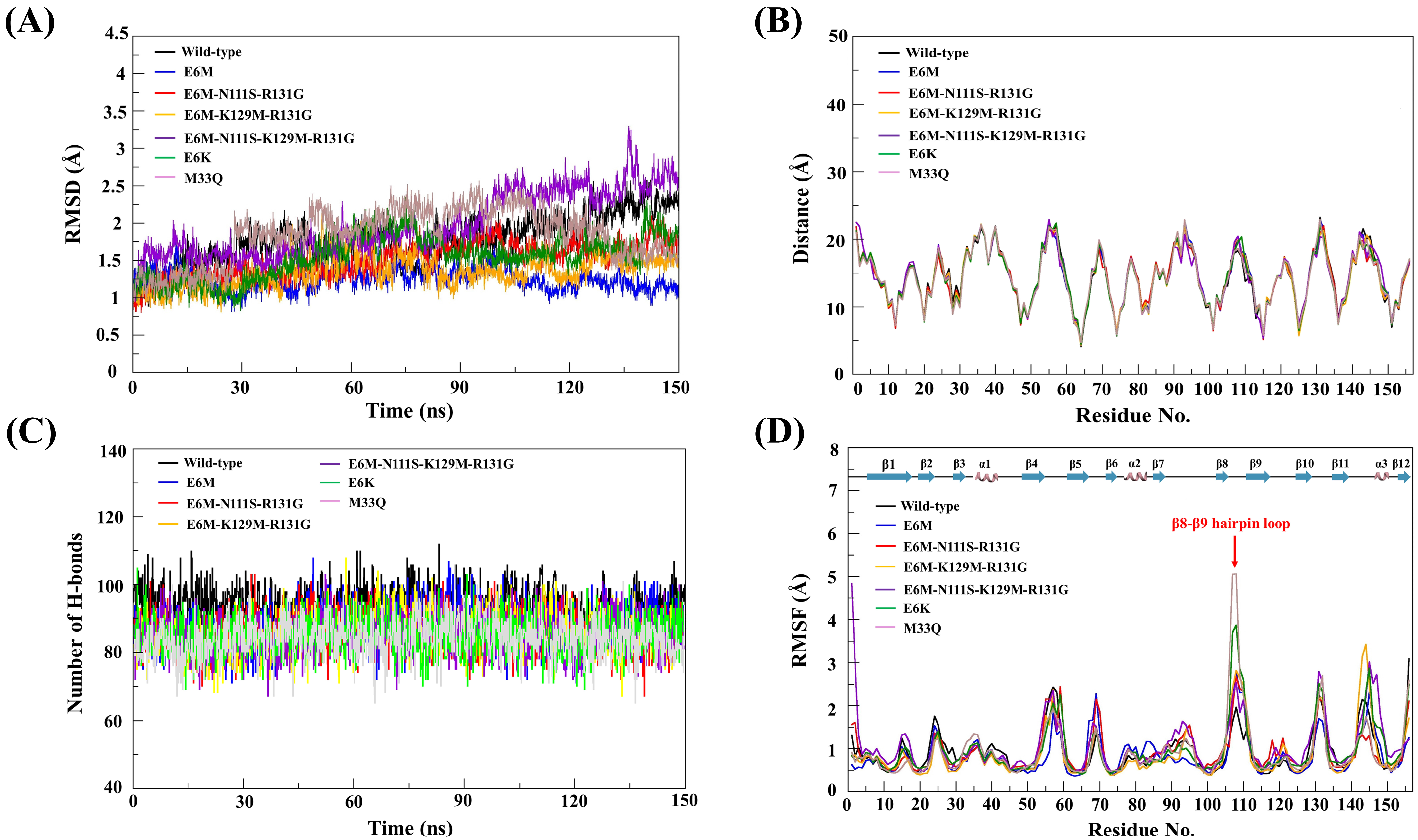
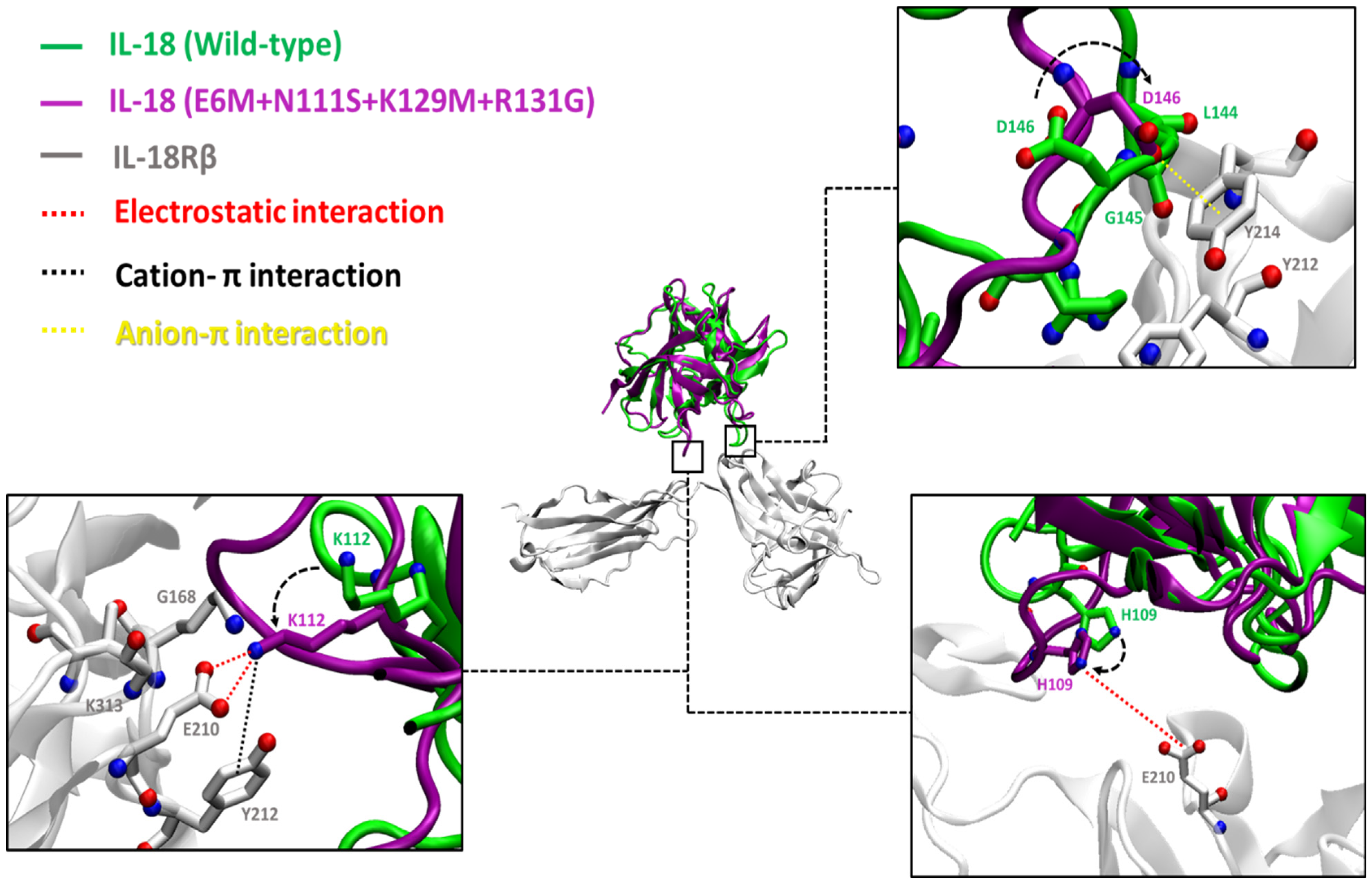
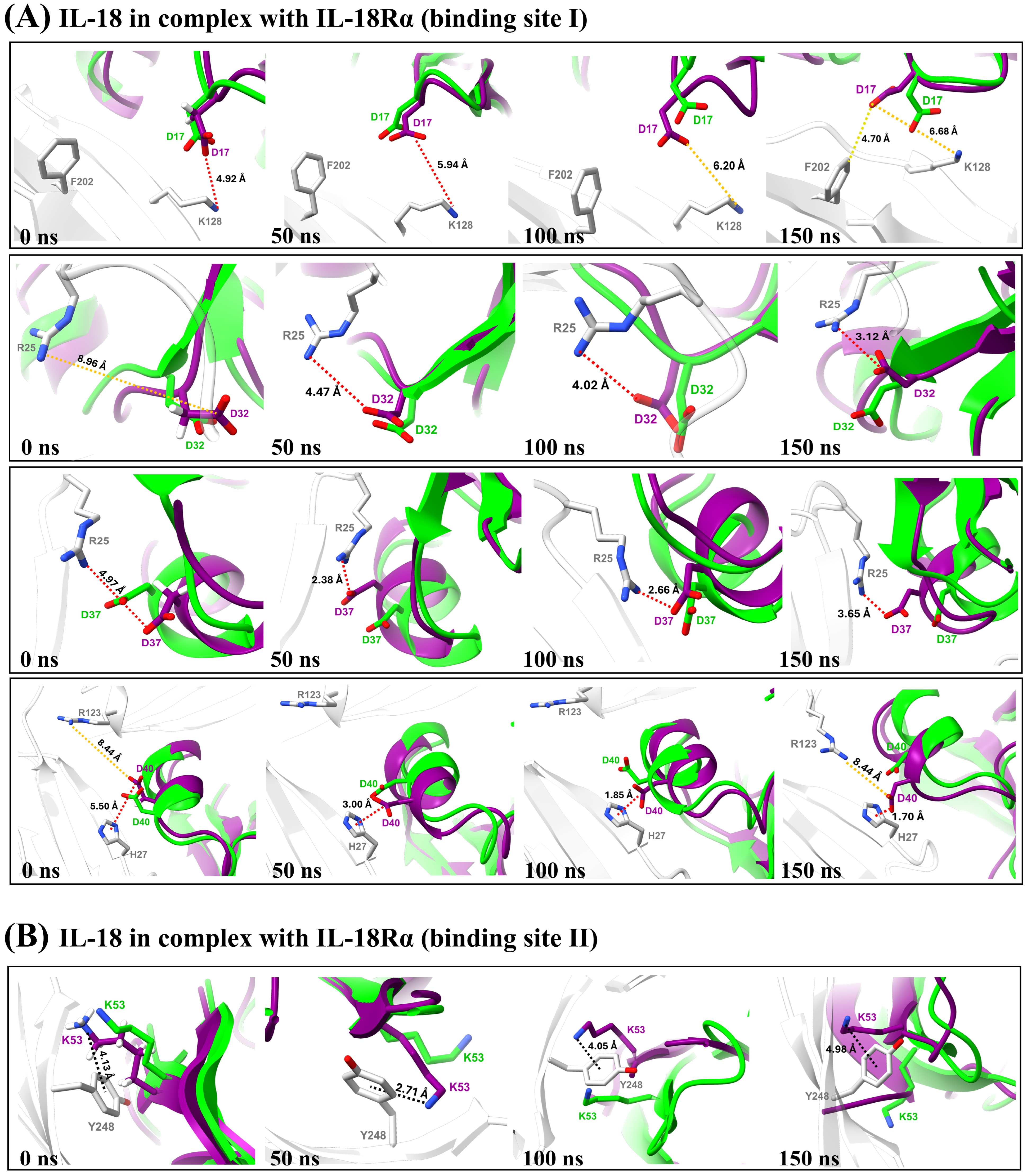
3.4. MD Simulation Analysis of Mutations
3.4.1. Conformational Stability Analysis
3.4.2. Structural Comparison
3.4.3. Hydrogen Bond Analysis
3.4.4. Structural Flexibility Analysis
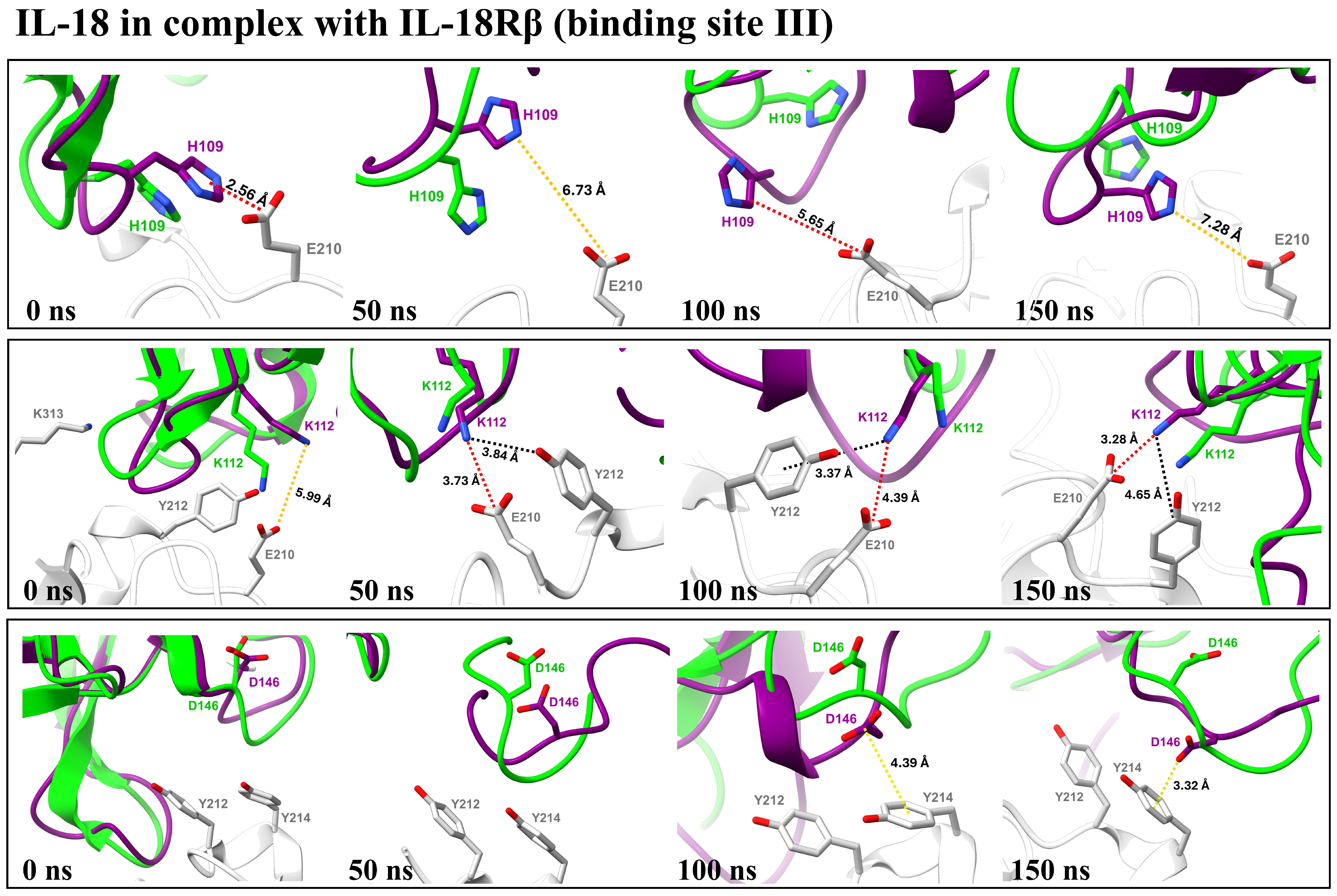
3.4.5. Binding Interaction Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed]
- Lee Ventola, C. Cancer Immunotherapy, Part 1: Current Strategies and Agents. Pharm. Ther. 2017, 42, 375–383. [Google Scholar]
- Koury, J.; Lucero, M.; Cato, C.; Chang, L.; Geiger, J.; Henry, D.; Hernandez, J.; Hung, F.; Kaur, P.; Teskey, G.; et al. Immunotherapies: Exploiting the Immune System for Cancer Treatment. J. Immunol. Res. 2018, 2018, 9585614. [Google Scholar] [CrossRef]
- Esfahani, K.; Roudaia, L.; Buhlaiga, N.; Del Rincon, S.V.; Papneja, N.; Miller, W.H. A Review of Cancer Immunotherapy: From the Past, to the Present, to the Future. Curr. Oncol. 2020, 27 (Suppl. 2), S87–S97. [Google Scholar] [CrossRef]
- Kaplanski, G. Interleukin-18: Biological Properties and Role in Disease Pathogenesis. Immunol. Rev. 2018, 281, 138–153. [Google Scholar] [CrossRef]
- Lee, S.; Margolin, K. Cytokines in Cancer Immunotherapy. Cancers 2011, 3, 3856. [Google Scholar] [CrossRef]
- Tsutsumi, N.; Kimura, T.; Arita, K.; Ariyoshi, M.; Ohnishi, H.; Yamamoto, T.; Zuo, X.; Maenaka, K.; Park, E.Y.; Kondo, N.; et al. The Structural Basis for Receptor Recognition of Human Interleukin-18. Nat. Commun. 2014, 5, 5340. [Google Scholar] [CrossRef]
- Kato, Z.; Jee, J.G.; Shikano, H.; Mishima, M.; Ohki, I.; Ohnishi, H.; Li, A.; Hashimoto, K.; Matsukuma, E.; Omoya, K.; et al. The Structure and Binding Mode of Interleukin-18. Nat. Struct. Biol. 2003, 10, 966–971. [Google Scholar] [CrossRef] [PubMed]
- Saetang, J.; Puseenam, A.; Roongsawang, N.; Voravuthikunchai, S.P.; Sangkhathat, S.; Tipmanee, V. Immunologic Function and Molecular Insight of Recombinant Interleukin-18. PLoS ONE 2016, 11, e0160321. [Google Scholar] [CrossRef]
- Kim, S.H.M.; Azam, T.; Yoon, D.Y.; Reznikov, L.L.; Novick, D.; Rubinstein, M.; Dinarello, C.A. Site-Specific Mutations in the Mature Form of Human IL-18 with Enhanced Biological Activity and Decreased Neutralization by IL-18 Binding Protein. Proc. Natl. Acad. Sci. USA 2001, 98, 3304–3309. [Google Scholar] [CrossRef] [PubMed]
- Swencki-Underwood, B.; Cunningham, M.R.; Heavner, G.A.; Blasie, C.; McCarthy, S.G.; Dougherty, T.; Brigham-Burke, M.; Gunn, G.R.; Goletz, T.J.; Snyder, L.A. Engineering Human IL-18 with Increased Bioactivity and Bioavailability. Cytokine 2006, 34, 114–124. [Google Scholar] [CrossRef]
- Saetang, J.; Chonpathompikunlert, P.; Sretrirutchai, S.; Roongsawang, N.; Kayasut, K.; Voravuthikunchai, S.P.; Sukketsiri, W.; Tipmanee, V.; Sangkhathat, S. Anti-cancer effect of engineered recombinant interleukin 18. Adv. Clin. Exp. Med. 2020, 29, 1135. [Google Scholar] [CrossRef]
- Peeyatu, C.; Prompat, N.; Voravuthikunchai, S.P.; Roongsawang, N.; Sangkhathat, S.; Khongkow, P.; Saetang, J.; Tipmanee, V. Roles of Non-Binding T63 Alteration on IL-18 Binding. Int. J. Mol. Sci. 2024, 25, 12992. [Google Scholar] [CrossRef]
- Saetang, J.; Roongsawang, N.; Sangkhathat, S.; Voravuthikunchai, S.P.; Sangkaew, N.; Prompat, N.; Srichana, T.; Tipmanee, V. Surface cysteine to serine substitutions in IL-18 reduce aggregation and enhance activity. PeerJ 2022, 10, e13626. [Google Scholar] [CrossRef] [PubMed]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The protein data bank. Nucleic Acids Res. 2000, 28, 235. [Google Scholar] [CrossRef]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33. [Google Scholar] [CrossRef] [PubMed]
- Delgado, J.; Radusky, L.G.; Cianferoni, D.; Serrano, L. FoldX 5.0: Working with RNA, Small Molecules and a New Graphical Interface. Bioinformatics 2019, 35, 4168–4169. [Google Scholar] [CrossRef] [PubMed]
- Romero-Durana, M.; Jiménez-García, B.; Fernández-Recio, J. PyDockEneRes: Per-Residue Decomposition of Protein-Protein Docking Energy. Bioinformatics 2020, 36, 2284–2285. [Google Scholar] [CrossRef]
- Case, D.A.; Betz, R.M.R.; Cheatham, T.E., III; Cerutti, D.S.; Darden, T.A.; Duke, R.E.; Giese, T.J.; Gohlke, H.; Goetz, A.W.; Homeyer, N.; et al. Amber 2016; University of California: San Francisco, CA, USA, 2016. [Google Scholar]
- Rostkowski, M.; Olsson, M.H.; Søndergaard, C.R.; Jensen, J.H. Graphical Analysis of PH-Dependent Properties of Proteins Predicted Using PROPKA. BMC Struct. Biol. 2011, 11, 6. [Google Scholar] [CrossRef] [PubMed]
- Mark, P.; Nilsson, L. Structure and dynamics of the TIP3P, SPC, and SPC/E water models at 298 K. J. Phys. Chem. A. 2001, 105, 9954–9960. [Google Scholar] [CrossRef]
- Ryckaert, J.P.; Ciccotti, G.; Berendsen, H.J. Numerical integration of the cartesian equations of motion of a system with constraints: Molecular dynamics of n-alkanes. J. Comput. Phys. 1977, 23, 327–341. [Google Scholar] [CrossRef]
- Berendsen, H.J.C.; Postma, J.P.M.; van Gunsteren, W.F.; DiNola, A.; Haak, J.R. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 1984, 81, 3684–3690. [Google Scholar] [CrossRef]
- Maruyama, Y.; Igarashi, R.; Ushiku, Y.; Mitsutake, A. Analysis of Protein Folding Simulation with Moving Root Mean Square Deviation. J. Chem. Inf. Model. 2023, 63, 1529–1541. [Google Scholar] [CrossRef]
- Karplus, M.; McCammon, J.A. Molecular Dynamics Simulations of Biomolecules. Nat. Struct. Biol. 2002, 9, 646–652. [Google Scholar] [CrossRef] [PubMed]
- Farmer, J.; Kanwal, F.; Nikulsin, N.; Tsilimigras, M.C.B.; Jacobs, D.J. Statistical Measures to Quantify Similarity between Molecular Dynamics Simulation Trajectories. Entropy 2017, 19, 646. [Google Scholar] [CrossRef] [PubMed]
- Nutho, B.; Mahalapbutr, P.; Hengphasatporn, K.; Pattaranggoon, N.C.; Simanon, N.; Shigeta, Y.; Hannongbua, S.; Rungrotmongkol, T. Why Are Lopinavir and Ritonavir Effective against the Newly Emerged Coronavirus 2019? Atomistic Insights into the Inhibitory Mechanisms. Biochemistry 2020, 59, 1769–1779. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, Y.; Kato, Z.; Matsukuma, E.; Li, A.; Omoya, K.; Hashimoto, K.; Ohnishi, H.; Kondo, N. Generation of Highly Stable IL-18 Based on a Ligand-Receptor Complex Structure. Biochem. Biophys. Res. Commun. 2004, 317, 181–186. [Google Scholar] [CrossRef]
- Hibbert, E.G.; Dalby, P.A. Directed Evolution Strategies for Improved Enzymatic Performance. Microb. Cell Fact. 2005, 4, 29. [Google Scholar] [CrossRef]
| Mutation | ΔΔGbinding | ΔΔGfolding of complex | ΔΔGfolding of ligand | Score |
|---|---|---|---|---|
| Control group | ||||
| Wild-type a | 0 | 0 | 0 | 0 |
| E6K a | −1.9 | −0.6 | 0.3 | 1.95 |
| T63A a | −0.8 | 0.8 | 0.8 | −0.44 |
| M60Q a | 4.6 | 2.2 | 0.8 | −5.35 |
| M33Q a | 4.6 | 2.9 | 2.7 | −7.02 |
| Candidate group | ||||
| E6M | −1.4 | −1.7 | −0.8 | 2.76 |
| K129M | −3.0 | −0.8 | −0.1 | 2.74 |
| R131K | −3.0 | 0.2 | 0.0 | 1.93 |
| N111S | −1.2 | −1.1 | −0.3 | 1.87 |
| R131G | −0.6 | −0.9 | −1.1 | 1.70 |
| Mutation | ΔΔGbinding | ΔΔGfolding of complex | ΔΔGfolding of ligand | Score |
|---|---|---|---|---|
| Control group | ||||
| Wild-type a | 0 | 0 | 0 | 0 |
| E6K a | −1.9 | −0.6 | 0.3 | 1.95 |
| T63A a | −0.8 | 0.8 | 0.8 | −0.44 |
| M60Q a | 4.6 | 2.2 | 0.8 | −5.35 |
| M33Q a | 4.6 | 2.9 | 2.7 | −7.02 |
| Candidate group | ||||
| K129M+R131G | −2.0 | −2.5 | −2.1 | 4.48 |
| E6M+R131G | −1.9 | −2.6 | −1.9 | 4.43 |
| E6M+K129M | −2.3 | −2.6 | −0.9 | 4.02 |
| E6M+N111S | −2.0 | −2.6 | −1.1 | 3.98 |
| E6M+R131K | −3.4 | −1.5 | −0.7 | 3.94 |
| N111S+R131G | −2.2 | −1.9 | −1.4 | 3.82 |
| R131K+N111S | −4.0 | −0.9 | −0.2 | 3.63 |
| K129M+N111S | −2.6 | −2.0 | −0.4 | 3.52 |
| K129M+R131K | −2.4 | −0.6 | 0.0 | 2.09 |
| Mutation | ΔΔGbinding | ΔΔGfolding of complex | ΔΔGfolding of ligand | Score |
|---|---|---|---|---|
| Control group | ||||
| Wild-type a | 0 | 0 | 0 | 0 |
| E6K a | −1.9 | −0.6 | 0.3 | 1.95 |
| T63A a | −0.8 | 0.8 | 0.8 | −0.44 |
| M60Q a | 4.6 | 2.2 | 0.8 | −5.35 |
| M33Q a | 4.6 | 2.9 | 2.7 | −7.02 |
| Candidate group | ||||
| E6M+K129M+R131G+N111S | −5.4 | −5.6 | −3.2 | 9.78 |
| E6M+N111S+R131G | −4.8 | −4.0 | −2.2 | 7.66 |
| E6M+K129M+R131G | −3.7 | −4.3 | −2.9 | 7.45 |
| K129M+N111S+R131G | −4.7 | −3.8 | −2.3 | 7.45 |
| E6M+K129M+R131K+N111S | −4.8 | −3.1 | −1.1 | 6.35 |
| E6M+R131K+N111S | −4.2 | −2.3 | −1.0 | 5.27 |
| E6M+K129M+N111S | −2.3 | −3.5 | −1.2 | 4.88 |
| E6M+K129M+R131K | −3.5 | −2.3 | −0.8 | 4.65 |
| K129M+R131K+N111S | −3.9 | −1.7 | −0.3 | 4.13 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Prompat, N.; Peeyatu, C.; Saetang, J.; Roongsawang, N.; Sangkhathat, S.; Tipmanee, V. Predicted IL-18/IL-18R Binding Improvement Through Protein Interface Modification with Computer-Aided Design. Biomolecules 2025, 15, 1360. https://doi.org/10.3390/biom15101360
Prompat N, Peeyatu C, Saetang J, Roongsawang N, Sangkhathat S, Tipmanee V. Predicted IL-18/IL-18R Binding Improvement Through Protein Interface Modification with Computer-Aided Design. Biomolecules. 2025; 15(10):1360. https://doi.org/10.3390/biom15101360
Chicago/Turabian StylePrompat, Napat, Chariya Peeyatu, Jirakrit Saetang, Niran Roongsawang, Surasak Sangkhathat, and Varomyalin Tipmanee. 2025. "Predicted IL-18/IL-18R Binding Improvement Through Protein Interface Modification with Computer-Aided Design" Biomolecules 15, no. 10: 1360. https://doi.org/10.3390/biom15101360
APA StylePrompat, N., Peeyatu, C., Saetang, J., Roongsawang, N., Sangkhathat, S., & Tipmanee, V. (2025). Predicted IL-18/IL-18R Binding Improvement Through Protein Interface Modification with Computer-Aided Design. Biomolecules, 15(10), 1360. https://doi.org/10.3390/biom15101360








