The Function of H2A Histone Variants and Their Roles in Diseases
Abstract
1. Introduction
2. The Functions of H2A Histone Variants
2.1. Gene Transcription
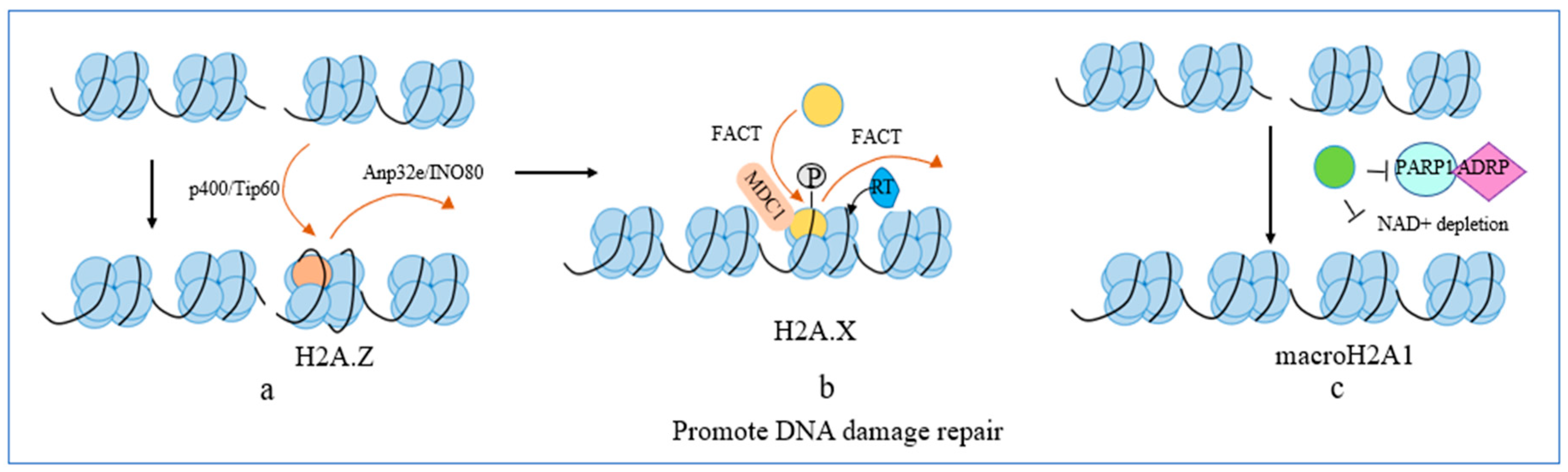
2.2. DNA Damage Repair
3. The Roles of Histone H2A Variants in Diseases
3.1. Cancer
| Histone Variants | Types of Cancer | Potential Mechanisms |
|---|---|---|
| H2A.Z | Lung cancer | Increases the sensitivity of lung cancer cells to radiotherapy [117]. |
| Breast cancer | Activates the expression of cyclin A1, thus promoting breast cancer development [37]. | |
| Malignant melanoma | H2A.Z2 enhances tumor sensitivity to drugs by gene transcriptional regulation for E2F1 in malignant melanoma [119]. | |
| Uterine leiomyoma | Deficient H2A.Z disposition in uterine leiomyoma cells with SRCAP complex mutations suggests a cancer inhibition role [118]. | |
| Hepatocellular carcinoma | H2A.Z1 promotes tumorigenesis, mainly through cell cycle signaling and the DNA damage pathway [113], and H2A.Z acetylation promotes downstream pro-oncogene transcription in liver cancer [110]. | |
| Bladder cancer | Promotes pro-oncogene expression in bladder cancer [120]. | |
| H2A.X | Gastrointestinal cancer | Inhibits EMT and promotes autophagy in colon cancer [123]. |
| Breast cancer | Increases Twist1 and Slug transcription factors, regulates EMT, and facilitates tumor cell migration and invasion after H2A.X deletion in breast cancer [125]. | |
| Prostate cancer | Inhibits the proliferation of prostate cancer cells [111]. | |
| Lung cancer | H2A.X deletion causes DNA breaks and cell cycle arrest in lung cancer [130]. | |
| Head and neck carcinoma | A marker of DNA damage repair in head and neck carcinoma [131]. | |
| Leukemia | Patients diagnosed with cancer usually occur in the absence of H2A.X in leukemia [133]. | |
| macroH2A | Malignant melanoma | Activates CDK8 to promote the development of malignant melanoma [134]. |
| Anal carcinoma | MacroH2A2 promotes the progression of anal carcinoma [135]. | |
| Glioblastoma | MacroH2A2 antagonizes tumor self-renewal and inhibits glioblastoma growth [136]. | |
| Solitary dormant disseminated cancer | nhibit cell cycle and pro-tumor-related signaling pathways in solitary dormant disseminated cancer [137]. | |
| Breast cancer | IMacroH2A1.1 acts as a suppressor of EMT in breast cells [140]. | |
| Colon cancer | Knockdown of macroH2A1.1 promotes tumor cell growth and proliferation in colon cancer [139]. | |
| Hematologic malignancies | Lacking macroH2A1.1 induces the development of hematologic malignancies [145]. | |
| Prostate cancers | MacroH2A1.2 is a tumor suppressor and inhibits osteoclast formation in prostate cancers [141]. | |
| H2A.B | Hodgkin’s lymphoma | Interacts with RNA polymerase II, increases ribosome biosynthesis in tumor cells, and promotes tumor development [112]. |
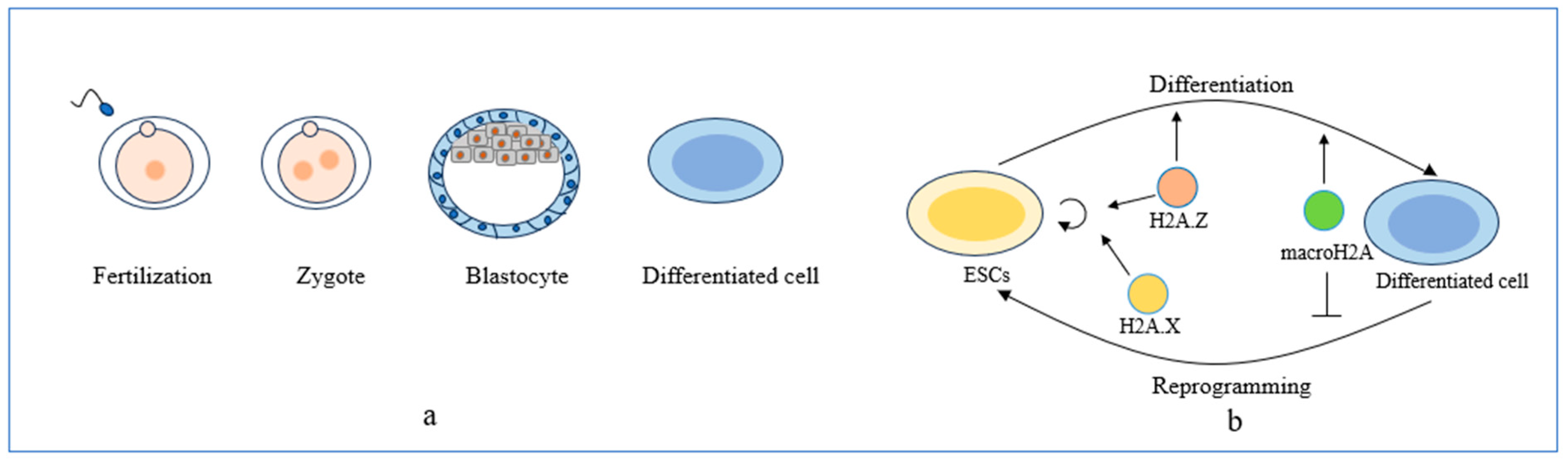
3.2. Embryonic Development Abnormalities
3.3. Other Diseases
4. Conclusions
| Histone H2A Variants | Distribution | Isoforms | Role in Gene Transcription | Role in DNA Damage Repair | Pathophysiological Processes Involved | The Roles in Disease |
|---|---|---|---|---|---|---|
| H2A.Z | Global | H2A.Z1 H2A.Z2 H2A.Z2.2 | Gene transcription activation or inhibition [32,33]. The mechanism is associated with the stability of nucleosomes [32], enhancer activity [25], RNA polymerase II pausing [29,31], and PTMs of H2A.Z nucleosomes [93,181,182]. | Promotes DNA repair [60,61,63]. Removing from nucleosomes recruiting repair factors to the break region and promoting DNA damage repair [61,63]. | Tumorigenesis [37,110,117] Embryonic development disorders [25,95,148] Neurological diseases [97,98] Heart diseases [105] | Promotes tumorigenesis [37,110,117]. Inhibits tumorigenesis in uterine leiomyoma cells with SRCAP complex mutations [118]. H2A.Z knockout mouse embryos die [148]. Regulates neurogenesis [97,98], affects learning and memory [175], and mediates the progression of schizophrenia [176]. Cardiac hypertrophy [105] and regulation of cardiac growth [36]. |
| H2A.X | Global | Transcription related [81]. The phosphorylation H2A.X axis mediates TGFB1-associated gene transcription activation, aggravating pulmonary fibrosis [81]. | Accelerates DNA damage repair [6,71] γ-H2A.X amplifies the signal and functions as a platform for assembling the DNA damage repair machinery [6,71]. | Tumorigenesis [93,125,126] Embryonic development disorders [82] Neurological diseases [177] Heart diseases [106,107] Metabolic diseases [177] | Inhibits tumorigenesis [93,125,126]. H2A.X knockout mice exhibit reduced fertility and a decreased number of lymphocytes, resulting in immune deficiencies [149]. Depletion of H2A.X causes neurological disorders [177,183]. Aberrant expression in heart diseases [106,107]. Mediating mitochondrial function [177] and oxidative stress processes [178]. | |
| macroH2A | Global | macroH2A1.1 macroH2A1.2 macroH2A2 | Mostly represses gene transcription [50]. The mechanism is associated with RNA polymerase II initiation [53], chromatin remodeling [54], histone acetylation [56], transcription factors activity [184], and chromatin condensation [50]. | Promotes DNA damage repair. Required for transcriptional repression near breaks following DNA damage [50,83,84,90] MacroH2A1.2 recruits DNA damage repair mediators [84]. MacroH2A1.1 impedes the activity of PARP1 [83,85]. | Tumorigenesis [94,135,138] Embryonic development disorder [150,166] Neurological diseases [179] Metabolic diseases [102,104] | Promotes tumorigenesis or inhibits tumorigenesis [94,135]. MacroH2A-deficient mice’s growth and development are restricted [150]. The absence of macroH2A1.2 in mice leads to the manifestation of ASD [179]. Promotes adipogenesis [104]. Reduces lipid accumulation in hepatocytes [102]. |
| H2A.B | Mainly in the testes and brain | Promotes gene transcription [19,20]. H2A.B nucleosomes are poorly stable, resulting in easy dissociation from chromatin, the formation of open chromatin structures, and gene transcription activation [19,20]. | Causes DNA damage [48] H2A.B overexpression in Hela cells induces DNA damage and subsequent apoptosis by activating the NF-κβ pathway [48]. | Tumorigenesis [112] Embryonic development disorders [42] Metabolic diseases [180] | Promotes tumor development [112]. The viability of embryos from H2A.B knockout male mice mated with female mice is reduced [172]. Aberrant expression in diabetic oocytes [180]. |
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Luger, K.; Mäder, A.W.; Richmond, R.K.; Sargent, D.F.; Richmond, T.J. Crystal structure of the nucleosome core particle at 2.8 a resolution. Nature 1997, 389, 251–260. [Google Scholar] [CrossRef]
- Talbert, P.B.; Henikoff, S. Histone variants at a glance. J. Cell Sci. 2021, 134, jcs244749. [Google Scholar] [CrossRef]
- Talbert, P.B.; Henikoff, S. Histone variants on the move: Substrates for chromatin dynamics. Nat. Rev. Mol. Cell Biol. 2017, 18, 115–126. [Google Scholar] [CrossRef] [PubMed]
- Billon, P.; Côté, J. Precise deposition of histone H2A.Z in chromatin for genome expression and maintenance. Biochim. Biophys. Acta (BBA)—Gene Regul. Mech. 2012, 1819, 290–302. [Google Scholar] [CrossRef] [PubMed]
- Hammond, C.M.; Strømme, C.B.; Huang, H.; Patel, D.J.; Groth, A. Histone chaperone networks shaping chromatin function. Nat. Rev. Mol. Cell Biol. 2017, 18, 141–158. [Google Scholar] [CrossRef] [PubMed]
- Sokolova, V.; Sarkar, S.; Tan, D. Histone variants and chromatin structure, update of advances. Comp. Struct. Biotechnol. J. 2023, 21, 299–311. [Google Scholar] [CrossRef] [PubMed]
- Talbert, P.B.; Ahmad, K.; Almouzni, G.; Ausió, J.; Berger, F.; Bhalla, P.L.; Bonner, W.M.; Cande, W.Z.; Chadwick, B.P.; Chan, S.W.; et al. A unified phylogeny-based nomenclature for histone variants. Epigenetics Chromatin 2012, 5, 7. [Google Scholar] [CrossRef] [PubMed]
- Kurumizaka, H.; Kujirai, T.; Takizawa, Y. Contributions of histone variants in nucleosome structure and function. J. Mol. Biol. 2021, 433, 166678. [Google Scholar] [CrossRef] [PubMed]
- Martire, S.; Banaszynski, L.A. The roles of histone variants in fine-tuning chromatin organization and function. Nat. Rev. Mol. Cell Biol. 2020, 21, 522–541. [Google Scholar] [CrossRef]
- Feng, J.X.; Riddle, N.C. Epigenetics and genome stability. Mamm. Genome 2020, 31, 181–195.11. [Google Scholar] [CrossRef]
- Dunjić, M.; Jonas, F.; Yaakov, G.; More, R.; Mayshar, Y.; Rais, Y.; Orenbuch, A.H.; Cheng, S.; Barkai, N.; Stelzer, Y. Histone exchange sensors reveal variant specific dynamics in mouse embryonic stem cells. Nat. Commun. 2023, 14, 3791. [Google Scholar] [CrossRef] [PubMed]
- Torres-Arciga, K.; Flores-León, M.; Ruiz-Pérez, S.; Trujillo-Pineda, M.; González-Barrios, R.; Herrera, L.A. Histones and their chaperones: Adaptive remodelers of an ever-changing chromatinic landscape. Front. Genet. 2022, 13, 1057846. [Google Scholar] [CrossRef] [PubMed]
- Pardal, A.J.; Fernandes-Duarte, F.; Bowman, A.J. The histone chaperoning pathway: From ribosome to nucleosome. Essays Biochem. 2019, 63, 29–43. [Google Scholar]
- Burgess, R.J.; Zhang, Z. Histone chaperones in nucleosome assembly and human disease. Nat. Struct. Mol. Biol. 2013, 20, 14–22. [Google Scholar] [CrossRef] [PubMed]
- Oberdoerffer, P.; Miller, K.M. Histone H2A variants: Diversifying chromatin to ensure genome integrity. Semin. Cell Dev. Biol. 2023, 135, 59–72. [Google Scholar] [CrossRef] [PubMed]
- Herchenrother, A.; Wunderlich, T.M.; Lan, J.; Hake, S.B. Spotlight on histone H2A variants: From B to X to Z. Semin. Cell Dev. Biol. 2023, 135, 3–12. [Google Scholar] [CrossRef] [PubMed]
- Ausió, J.; Abbott, D.W. The many tales of a tail: Carboxyl-terminal tail heterogeneity specializes histone H2A variants for defined chromatin function. Biochemistry 2002, 41, 5945–5949. [Google Scholar] [CrossRef] [PubMed]
- Bönisch, C.; Hake, S.B. Histone H2A variants in nucleosomes and chromatin: More or less stable? Nucleic Acids Res. 2012, 40, 10719–10741. [Google Scholar] [CrossRef] [PubMed]
- Peng, J.; Yuan, C.; Hua, X.; Zhang, Z. Molecular mechanism of histone variant H2A.B on stability and assembly of nucleosome and chromatin structures. Epigenet. Chromatin 2020, 13, 28. [Google Scholar] [CrossRef]
- Zhou, M.; Dai, L.; Li, C.; Shi, L.; Huang, Y.; Guo, Z.; Wu, F.; Zhu, P.; Zhou, Z. Structural basis of nucleosome dynamics modulation by histone variants H2A.B and H2A.Z.2.2. EMBO J. 2021, 40, e105907. [Google Scholar] [CrossRef]
- Giaimo, B.D.; Ferrante, F.; Herchenröther, A.; Hake, S.B.; Borggrefe, T. The histone variant H2A.Z in gene regulation. Epigenet. Chromatin 2019, 12, 37. [Google Scholar] [CrossRef] [PubMed]
- Sales-Gil, R.; Kommer, D.C.; de Castro, I.J.; Amin, H.A.; Vinciotti, V.; Sisu, C.; Vagnarelli, P. Non-redundant functions of H2A.Z.1 and H2A.Z.2 in chromosome segregation and cell cycle progression. EMBO Rep. 2021, 22, e52061. [Google Scholar] [CrossRef] [PubMed]
- Bönisch, C.; Schneider, K.; Pünzeler, S.; Wiedemann, S.M.; Bielmeier, C.; Bocola, M.; Eberl, H.C.; Kuegel, W.; Neumann, J.; Kremmer, E.; et al. H2A.Z.2.2 is an alternatively spliced histone H2A.Z variant that causes severe nucleosome destabilization. Nucleic Acids Res. 2012, 40, 5951–5964. [Google Scholar] [CrossRef] [PubMed]
- Greenberg, R.S.; Long, H.K.; Swigut, T.; Wysocka, J. Single amino acid change underlies distinct roles of H2A.Z subtypes in human syndrome. Cell 2019, 178, 1421–1436. [Google Scholar] [CrossRef] [PubMed]
- Colino-Sanguino, Y.; Clark, S.J.; Valdes-Mora, F. The H2A.Z-nucleosome code in mammals: Emerging functions. Trends Genet. 2022, 38, 516. [Google Scholar] [CrossRef] [PubMed]
- Hardy, S.; Jacques, P.E.; Gévry, N.; Forest, A.; Fortin, M.E.; Laflamme, L.; Gaudreau, L.; Robert, F. The euchromatic and heterochromatic landscapes are shaped by antagonizing effects of transcription on H2A.Z deposition. PLoS Genet. 2009, 5, e1000687. [Google Scholar] [CrossRef]
- Moreno-Andrés, D.; Yokoyama, H.; Scheufen, A.; Holzer, G.; Lue, H.; Schellhaus, A.K.; Weberruss, M.; Takagi, M.; Antonin, W. Vps72/yl1-mediated H2A.Z deposition is required for nuclear reassembly after mitosis. Cells 2020, 9, 1702. [Google Scholar] [CrossRef]
- Obri, A.; Ouararhni, K.; Papin, C.; Diebold, M.L.; Padmanabhan, K.; Marek, M.; Stoll, I.; Roy, L.; Reilly, P.T.; Mak, T.W.; et al. Anp32e is a histone chaperone that removes H2A.Z from chromatin. Nature 2014, 505, 648–653. [Google Scholar] [CrossRef]
- Rudnizky, S.; Bavly, A.; Malik, O.; Pnueli, L.; Melamed, P.; Kaplan, A. H2A.Z controls the stability and mobility of nucleosomes to regulate expression of the LH genes. Nat. Commun. 2016, 7, 12958. [Google Scholar] [CrossRef]
- Li, S.; Wei, T.; Panchenko, A.R. Histone variant H2A.Z modulates nucleosome dynamics to promote DNA accessibility. Nat. Commun. 2023, 14, 769. [Google Scholar] [CrossRef]
- Day, D.S.; Zhang, B.; Stevens, S.M.; Ferrari, F.; Larschan, E.N.; Park, P.J.; Pu, W.T. Comprehensive analysis of promoter-proximal RNA polymerase II pausing across mammalian cell types. Genome Biol. 2016, 17, 120. [Google Scholar] [CrossRef]
- Cole, L.; Kurscheid, S.; Nekrasov, M.; Domaschenz, R.; Vera, D.L.; Dennis, J.H.; Tremethick, D.J. Multiple roles of H2A.Z in regulating promoter chromatin architecture in human cells. Nat. Commun. 2021, 12, 2524. [Google Scholar] [CrossRef] [PubMed]
- Domaschenz, R.; Kurscheid, S.; Nekrasov, M.; Han, S.; Tremethick, D.J. The histone variant H2A.Z is a master regulator of the epithelial-mesenchymal transition. Cell Rep. 2017, 21, 943–952. [Google Scholar] [CrossRef]
- Lewis, T.S.; Sokolova, V.; Jung, H.; Ng, H.; Tan, D. Structural basis of chromatin regulation by histone variant H2A.Z. Nucleic Acids Res. 2021, 49, 11379–11391. [Google Scholar] [CrossRef]
- Suto, R.K.; Clarkson, M.J.; Tremethick, D.J.; Luger, K. Crystal structure of a nucleosome core particle containing the variant histone H2A.Z. Nat. Struct. Biol. 2000, 7, 1121–1124. [Google Scholar]
- Shin, H.; He, M.; Yang, Z.; Jeon, Y.H.; Pfleger, J.; Sayed, D.; Abdellatif, M. Transcriptional regulation mediated by H2A.Z via Anp32e-dependent inhibition of protein phosphatase 2A. Biochim. Biophys. Acta-Gene Regul. Mech. 2018, 1861, 481–496. [Google Scholar] [CrossRef]
- Tsai, C.H.; Chen, Y.J.; Yu, C.J.; Tzeng, S.R.; Wu, I.C.; Kuo, W.H.; Lin, M.C.; Chan, N.L.; Wu, K.J.; Teng, S.C. SMYD3-mediated H2A.Z.1 methylation promotes cell cycle and cancer proliferation. Cancer Res. 2016, 76, 6043–6053. [Google Scholar] [CrossRef]
- Li, X.; Mei, Q.; Yu, Q.; Wang, M.; He, F.; Xiao, D.; Liu, H.; Ge, F.; Yu, X.; Li, S. The TORC1 activates Rpd3l complex to deacetylate INO80 and H2A.Z and repress autophagy. Sci. Adv. 2023, 9, e8312. [Google Scholar] [CrossRef]
- Soboleva, T.A.; Parker, B.J.; Nekrasov, M.; Hart-Smith, G.; Tay, Y.J.; Tng, W.; Wilkins, M.; Ryan, D.; Tremethick, D.J. A new link between transcriptional initiation and pre-mRNA splicing: The RNA binding histone variant H2A.B. PLoS Genet. 2017, 13, e1006633. [Google Scholar] [CrossRef] [PubMed]
- Okuwaki, M.; Kato, K.; Shimahara, H.; Tate, S.; Nagata, K. Assembly and disassembly of nucleosome core particles containing histone variants by human nucleosome assembly protein I. Mol. Cell. Biol. 2005, 25, 10639–10651. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.; Soboleva, T.A.; Tremethick, D.J. Short histone H2A variants: Small in stature but not in function. Cells 2020, 9, 867. [Google Scholar] [CrossRef] [PubMed]
- Soboleva, T.A.; Nekrasov, M.; Pahwa, A.; Williams, R.; Huttley, G.A.; Tremethick, D.J. A unique H2A histone variant occupies the transcriptional start site of active genes. Nat. Struct. Mol. Biol. 2011, 19, 25–30. [Google Scholar] [CrossRef] [PubMed]
- Kohestani, H.; Wereszczynski, J. Effects of H2A.B incorporation on nucleosome structures and dynamics. Biophys. J. 2021, 120, 1498–1509. [Google Scholar] [CrossRef] [PubMed]
- Anuar, N.D.; Kurscheid, S.; Field, M.; Zhang, L.; Rebar, E.; Gregory, P.; Buchou, T.; Bowles, J.; Koopman, P.; Tremethick, D.J.; et al. Gene editing of the multi-copy H2A.B gene and its importance for fertility. Genome Biol. 2019, 20, 23. [Google Scholar] [CrossRef] [PubMed]
- Baralle, F.E.; Giudice, J. Alternative splicing as a regulator of development and tissue identity. Nat. Rev. Mol. Cell Biol. 2017, 18, 437–451. [Google Scholar] [CrossRef]
- Hirano, R.; Arimura, Y.; Kujirai, T.; Shibata, M.; Okuda, A.; Morishima, K.; Inoue, R.; Sugiyama, M.; Kurumizaka, H. Histone variant H2A.B-H2B dimers are spontaneously exchanged with canonical H2A-H2B in the nucleosome. Commun. Biol. 2021, 4, 191. [Google Scholar] [CrossRef] [PubMed]
- Arimura, Y.; Kimura, H.; Oda, T.; Sato, K.; Osakabe, A.; Tachiwana, H.; Sato, Y.; Kinugasa, Y.; Ikura, T.; Sugiyama, M.; et al. Structural basis of a nucleosome containing histone H2A.B/H2A.Bbd that transiently associates with reorganized chromatin. Sci. Rep. 2013, 3, 3510. [Google Scholar] [CrossRef] [PubMed]
- Goshima, T.; Shimada, M.; Sharif, J.; Matsuo, H.; Misaki, T.; Johmura, Y.; Murata, K.; Koseki, H.; Nakanishi, M. Mammal-specific H2A variant, H2A.Bbd, is involved in apoptotic induction via activation of NF-κB signaling pathway. J. Biol. Chem. 2014, 289, 11656–11666. [Google Scholar] [CrossRef] [PubMed]
- Rivera-Casas, C.; Gonzalez-Romero, R.; Cheema, M.S.; Ausió, J.; Eirín-López, J.M. The characterization of macroH2A beyond vertebrates supports an ancestral origin and conserved role for histone variants in chromatin. Epigenetics 2016, 11, 415–425. [Google Scholar] [CrossRef]
- Sun, Z.; Bernstein, E. Histone variant macroH2A: From chromatin deposition to molecular function. Essays Biochem. 2019, 63, 59–74. [Google Scholar]
- Mehrotra, P.V.; Ahel, D.; Ryan, D.P.; Weston, R.; Wiechens, N.; Kraehenbuehl, R.; Owen-Hughes, T.; Ahel, I. DNA repair factor APLF is a histone chaperone. Mol. Cell 2011, 41, 46–55. [Google Scholar] [CrossRef] [PubMed]
- Mandemaker, I.K.; Fessler, E.; Corujo, D.; Kotthoff, C.; Wegerer, A.; Rouillon, C.; Buschbeck, M.; Jae, L.T.; Mattiroli, F.; Ladurner, A.G. The histone chaperone Anp32B regulates chromatin incorporation of the atypical human histone variant macroH2A. Cell Rep. 2023, 42, 113300. [Google Scholar] [CrossRef] [PubMed]
- Doyen, C.M.; An, W.; Angelov, D.; Bondarenko, V.; Mietton, F.; Studitsky, V.M.; Hamiche, A.; Roeder, R.G.; Bouvet, P.; Dimitrov, S. Mechanism of polymerase II transcription repression by the histone variant macroH2A. Mol. Cell. Biol. 2006, 26, 1156–1164. [Google Scholar] [CrossRef]
- Chakravarthy, S.; Patel, A.; Bowman, G.D. The basic linker of macroH2A stabilizes DNA at the entry/exit site of the nucleosome. Nucleic Acids Res. 2012, 40, 8285–8295.55. [Google Scholar] [CrossRef] [PubMed]
- Douet, J.; Corujo, D.; Malinverni, R.; Renauld, J.; Sansoni, V.; Marjanović, M.P.; Cantari’O, N.; Valero, V.; Mongelard, F.; Bouvet, P.; et al. MacroH2A histone variants maintain nuclear organization and heterochromatin architecture. J. Cell Sci. 2017, 130, 1570–1582. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.M.; Heo, K.; Choi, J.; Kim, K.; An, W. The histone variant macroH2A regulates Ca(2+) influx through TRPC3 and TRPC6 channels. Oncogenesis 2013, 2, e77. [Google Scholar] [CrossRef]
- Chang, E.Y.; Ferreira, H.; Somers, J.; Nusinow, D.A.; Owen-Hughes, T.; Narlikar, G.J. MacroH2A allows ATP-dependent chromatin remodeling by SWI/SNF and ACF complexes but specifically reduces recruitment of SWI/SNF. Biochemistry 2008, 47, 13726–13732. [Google Scholar] [CrossRef]
- Lavigne, M.D.; Vatsellas, G.; Polyzos, A.; Mantouvalou, E.; Sianidis, G.; Maraziotis, I.; Agelopoulos, M.; Thanos, D. Composite macroH2A/NRF-1 nucleosomes suppress noise and generate robustness in gene expression. Cell Rep. 2015, 11, 1090–1101. [Google Scholar] [CrossRef] [PubMed]
- Recoules, L.; Heurteau, A.; Raynal, F.; Karasu, N.; Moutahir, F.; Bejjani, F.; Jariel-Encontre, I.; Cuvier, O.; Sexton, T.; Lavigne, A.C.; et al. The histone variant macroH2A1.1 regulates RNA polymerase II-paused genes within defined chromatin interaction landscapes. J. Cell Sci. 2022, 135, jcs259456. [Google Scholar] [CrossRef]
- Xu, Y.; Ayrapetov, M.K.; Xu, C.; Gursoy-Yuzugullu, O.; Hu, Y.; Price, B.D. Histone H2A.Z controls a critical chromatin re-modeling step required for DNA double-strand break repair. Mol. Cell 2012, 48, 723–733. [Google Scholar] [CrossRef]
- Gursoy-Yuzugullu, O.; Ayrapetov, M.K.; Price, B.D. Histone chaperone Anp32e removes H2A.Z from DNA double-strand breaks and promotes nucleosome reorganization and DNA repair. Proc. Natl. Acad. Sci. USA 2015, 112, 7507–7512. [Google Scholar] [CrossRef]
- Bao, Y. Chromatin response to DNA double-strand break damage. Epigenomics 2011, 3, 307–321. [Google Scholar] [CrossRef]
- Alatwi, H.E.; Downs, J.A. Removal of H2A.Z by INO80 promotes homologous recombination. EMBO Rep. 2015, 16, 986–994. [Google Scholar] [CrossRef] [PubMed]
- Rona, G.; Roberti, D.; Yin, Y.; Pagan, J.K.; Homer, H.; Sassani, E.; Zeke, A.; Busino, L.; Rothenberg, E.; Pagano, M. PARP1-dependent recruitment of the FBXL10-RNF68-RNF2 ubiquitin ligase to sites of DNA damage controls H2A.Z loading. eLife 2018, 7, e38771. [Google Scholar] [CrossRef]
- Ui, A.; Chiba, N.; Yasui, A. Relationship among DNA double-strand break (DSB), DSB repair, and transcription prevents genome instability and cancer. Cancer Sci. 2020, 111, 1443–1451. [Google Scholar] [CrossRef]
- Milano, L.; Gautam, A.; Caldecott, K.W. DNA damage and transcription stress. Mol. Cell 2024, 84, 70–79. [Google Scholar] [CrossRef] [PubMed]
- Henikoff, S.; Smith, M.M. Histone variants and epigenetics. Cold Spring Harb. Perspect. Biol. 2015, 7, a19364. [Google Scholar] [CrossRef] [PubMed]
- Seo, J.; Kim, S.C.; Lee, H.S.; Kim, J.K.; Shon, H.J.; Salleh, N.L.; Desai, K.V.; Lee, J.H.; Kang, E.S.; Kim, J.S.; et al. Genome-wide profiles of H2AX and γ-H2AX differentiate endogenous and exogenous DNA damage hotspots in human cells. Nucleic Acids Res. 2012, 40, 5965–5974. [Google Scholar] [CrossRef] [PubMed]
- Piquet, S.; Le Parc, F.; Bai, S.K.; Chevallier, O.; Adam, S.; Polo, S.E. The histone chaperone FACT coordinates H2A.X-dependent signaling and repair of DNA damage. Mol. Cell 2018, 72, 888–901. [Google Scholar] [CrossRef]
- Heo, K.; Kim, H.; Choi, S.H.; Choi, J.; Kim, K.; Gu, J.; Lieber, M.R.; Yang, A.S.; An, W. FACT-mediated exchange of histone variant H2Ax regulated by phosphorylation of H2AX and ADP-ribosylation of SPT16. Mol. Cell 2008, 30, 86–97. [Google Scholar] [CrossRef]
- Li, M.; Fang, Y. Histone variants: The artists of eukaryotic chromatin. Sci. China Life Sci. 2015, 58, 232–239. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Xiao, A.; Li, H.; Shechter, D.; Ahn, S.H.; Fabrizio, L.A.; Erdjument-Bromage, H.; Ishibe-Murakami, S.; Wang, B.; Tempst, P.; Hofmann, K.; et al. WSTF regulates the H2A.X DNA damage response via a novel tyrosine kinase activity. Nature 2009, 457, 57–62. [Google Scholar] [CrossRef] [PubMed]
- Schwertman, P.; Bekker-Jensen, S.; Mailand, N. Regulation of DNA double-strand break repair by ubiquitin and ubiquitin-like modifiers. Nat. Rev. Mol. Cell Biol. 2016, 17, 379–394. [Google Scholar] [CrossRef]
- Qiu, L.; Xu, W.; Lu, X.; Chen, F.; Chen, Y.; Tian, Y.; Zhu, Q.; Liu, X.; Wang, Y.; Pei, X.H.; et al. The HDAC6-RNF168 axis regulates H2A/H2A.X ubiquitination to enable double-strand break repair. Nucleic Acids Res. 2023, 51, 9166–9182. [Google Scholar] [CrossRef] [PubMed]
- Pan, M.R.; Peng, G.; Hung, W.C.; Lin, S.Y. Monoubiquitination of H2AX protein regulates DNA damage response signaling. J. Biol. Chem. 2011, 286, 28599–28607. [Google Scholar] [CrossRef] [PubMed]
- Mattiroli, F.; Vissers, J.H.; van Dijk, W.J.; Ikpa, P.; Citterio, E.; Vermeulen, W.; Marteijn, J.A.; Sixma, T.K. RNF168 ubiquitinates k13-15 on H2A/H2AX to drive DNA damage signaling. Cell 2012, 150, 1182–1195. [Google Scholar] [CrossRef]
- Sharma, D.; De Falco, L.; Padavattan, S.; Rao, C.; Geifman-Shochat, S.; Liu, C.F.; Davey, C.A. PARP1 exhibits enhanced association and catalytic efficiency with gammaH2A.X-nucleosome. Nat. Commun. 2019, 10, 5751. [Google Scholar] [CrossRef]
- Novo, N.; Romero-Tamayo, S.; Marcuello, C.; Boneta, S.; Blasco-Machin, I.; Velázquez-Campoy, A.; Villanueva, R.; Moreno-Loshuertos, R.; Lostao, A.; Medina, M.; et al. Beyond a platform protein for the degradosome assembly: The apoptosis-inducing factor as an efficient nuclease involved in chromatinolysis. Proc. Natl. Acad. Sci. USA Nexus 2023, 2, c312. [Google Scholar] [CrossRef] [PubMed]
- Artus, C.; Boujrad, H.; Bouharrour, A.; Brunelle, M.N.; Hoos, S.; Yuste, V.J.; Lenormand, P.; Rousselle, J.C.; Namane, A.; England, P.; et al. AIF promotes chromatinolysis and caspase-independent programmed necrosis by interacting with histone H2Ax. EMBO J. 2010, 29, 1585–1599. [Google Scholar] [CrossRef]
- Bano, D.; Prehn, J. Apoptosis-inducing factor (AIF) in physiology and disease: The tale of a repented natural born killer. EBioMedicine 2018, 30, 29–37. [Google Scholar] [CrossRef]
- Dobersch, S.; Rubio, K.; Singh, I.; Günther, S.; Graumann, J.; Cordero, J.; Castillo-Negrete, R.; Huynh, M.B.; Mehta, A.; Braubach, P.; et al. Positioning of nucleosomes containing γ-H2AX precedes active DNA demethylation and transcription initiation. Nat. Commun. 2021, 12, 1072. [Google Scholar] [CrossRef] [PubMed]
- Orlando, L.; Tanasijevic, B.; Nakanishi, M.; Reid, J.C.; García-Rodríguez, J.L.; Chauhan, K.D.; Porras, D.P.; Aslostovar, L.; Lu, J.D.; Shapovalova, Z.; et al. Phosphorylation state of the histone variant H2A.X controls human stem and progenitor cell fate decisions. Cell Rep. 2021, 34, 108818. [Google Scholar] [CrossRef] [PubMed]
- Kozlowski, M.; Corujo, D.; Hothorn, M.; Guberovic, I.; Mandemaker, I.K.; Blessing, C.; Sporn, J.; Gutierrez-Triana, A.; Smith, R.; Portmann, T.; et al. MacroH2A histone variants limit chromatin plasticity through two distinct mechanisms. EMBO Rep. 2018, 19, e44445. [Google Scholar] [CrossRef] [PubMed]
- Khurana, S.; Kruhlak, M.J.; Kim, J.; Tran, A.D.; Liu, J.; Nyswaner, K.; Shi, L.; Jailwala, P.; Sung, M.H.; Hakim, O.; et al. A macrohistone variant links dynamic chromatin compaction to BRCA1-dependent genome maintenance. Cell Rep. 2014, 8, 1049–1062. [Google Scholar] [CrossRef]
- Kim, D.; Challa, S.; Jones, A.; Kraus, W.L. PARPs and ADP-ribosylation in RNA biology: From RNA expression and processing to protein translation and proteostasis. Genes. Dev. 2020, 34, 302–320. [Google Scholar] [CrossRef] [PubMed]
- Ruiz, P.D.; Hamilton, G.A.; Park, J.W.; Gamble, M.J. MacroH2A1 regulation of poly(ADP-ribose) synthesis and stability prevents necrosis and promotes DNA repair. Mol. Cell. Biol. 2019, 40, e00230-19. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Ruiz, P.D.; Novikov, L.; Casill, A.D.; Park, J.W.; Gamble, M.J. MacroH2A1.1 and PARP-1 cooperate to regulate transcription by promoting CBP-mediated H2B acetylation. Nat. Struct. Mol. Biol. 2014, 21, 981–989. [Google Scholar] [CrossRef] [PubMed]
- Sanchez, A.; Buck-Koehntop, B.A.; Miller, K.M. Joining the party: PARP regulation of during DNA repair (and transcription?). Bioessays 2022, 44, e2200015. [Google Scholar] [CrossRef]
- Kim, J.; Shin, Y.; Lee, S.; Kim, M.; Punj, V.; Lu, J.F.; Shin, H.; Kim, K.; Ulmer, T.S.; Koh, J.; et al. Regulation of breast cancer-induced osteoclastogenesis by macroH2A1.2 involving EZH2-mediated H3K27me3. Cell Rep. 2018, 24, 224–237. [Google Scholar] [CrossRef]
- Kumbhar, R.; Sanchez, A.; Perren, J.; Gong, F.; Corujo, D.; Medina, F.; Devanathan, S.K.; Xhemalce, B.; Matouschek, A.; Buschbeck, M.; et al. Poly(ADP-ribose) binding and macroH2A mediate recruitment and functions of KDM5A at DNA lesions. J. Cell Biol. 2021, 220, e202006149. [Google Scholar] [CrossRef]
- Na, Y.J.; Kim, B.R.; Kim, J.L.; Kang, S.; Jeong, Y.A.; Park, S.H.; Jo, M.J.; Kim, J.Y.; Kim, H.J.; Oh, S.C.; et al. Deficiency of 15-LOX-1 induces radioresistance through downregulation of macroH2A2 in colorectal cancer. Cancers 2019, 11, 1776. [Google Scholar] [CrossRef] [PubMed]
- Karras, G.I.; Kustatscher, G.; Buhecha, H.R.; Allen, M.D.; Pugieux, C.; Sait, F.; Bycroft, M.; Ladurner, A.G. The macro domain is an ADP-ribose binding module. EMBO J. 2005, 24, 1911–1920. [Google Scholar] [CrossRef] [PubMed]
- Corujo, D.; Buschbeck, M. Post-translational modifications of H2A histone variants and their role in cancer. Cancers 2018, 10, 59. [Google Scholar] [CrossRef] [PubMed]
- Monteiro, F.L.; Baptista, T.; Amado, F.; Vitorino, R.; Jerónimo, C.; Helguero, L.A. Expression and functionality of histone H2A variants in cancer. Oncotarget 2014, 5, 3428–3443. [Google Scholar] [CrossRef] [PubMed]
- Dijkwel, Y.; Tremethick, D.J. The role of the histone variant H2A.Z in metazoan development. J. Dev. Biol. 2022, 10, 28. [Google Scholar] [CrossRef]
- Li, Z.; Gadue, P.; Chen, K.; Jiao, Y.; Tuteja, G.; Schug, J.; Li, W.; Kaestner, K.H. FOXA2 and H2A.Z mediate nucleosome depletion during embryonic stem cell differentiation. Cell 2012, 151, 1608–1616. [Google Scholar] [CrossRef] [PubMed]
- Shen, T.; Ji, F.; Wang, Y.; Lei, X.; Zhang, D.; Jiao, J. Brain-specific deletion of histone variant H2A.Z results in cortical neurogenesis defects and neurodevelopmental disorder. Nucleic Acids Res. 2018, 46, 2290–2307. [Google Scholar] [CrossRef]
- Rao, V.K.; Swarnaseetha, A.; Tham, G.H.; Lin, W.Q.; Han, B.B.; Benoukraf, T.; Xu, G.L.; Ong, C.T. Phosphorylation of TET3 by CDK5 is critical for robust activation of BRN2 during neuronal differentiation. Nucleic Acids Res. 2020, 48, 1225–1238. [Google Scholar] [CrossRef]
- Karthik, N.; Taneja, R. Histone variants in skeletal myogenesis. Epigenetics 2021, 16, 243–262. [Google Scholar] [CrossRef]
- Saul, D.; Kosinsky, R.L. Epigenetics of aging and aging-associated diseases. Int. J. Mol. Sci. 2021, 22, 401. [Google Scholar] [CrossRef]
- Stefanelli, G.; Azam, A.B.; Walters, B.J.; Brimble, M.A.; Gettens, C.P.; Bouchard-Cannon, P.; Cheng, H.M.; Davidoff, A.M.; Narkaj, K.; Day, J.J.; et al. Learning and age-related changes in genome-wide H2A.Z binding in the mouse hippocampus. Cell Rep. 2018, 22, 1124–1131. [Google Scholar] [CrossRef]
- Pazienza, V.; Borghesan, M.; Mazza, T.; Sheedfar, F.; Panebianco, C.; Williams, R.; Mazzoccoli, G.; Andriulli, A.; Nakanishi, T.; Vinciguerra, M. Sirt1-metabolite binding histone macroH2A1.1 protects hepatocytes against lipid accumulation. Aging 2014, 6, 35–47. [Google Scholar] [CrossRef] [PubMed]
- Wan, D.; Liu, C.; Sun, Y.; Wang, W.; Huang, K.; Zheng, L. MacroH2A1.1 cooperates with EZH2 to promote adipogenesis by regulating WNT signaling. J. Mol. Cell Biol. 2017, 9, 325–337. [Google Scholar] [CrossRef] [PubMed]
- Pazienza, V.; Panebianco, C.; Rappa, F.; Memoli, D.; Borghesan, M.; Cannito, S.; Oji, A.; Mazza, G.; Tamburrino, D.; Fusai, G.; et al. Histone macroH2A1.2 promotes metabolic health and leanness by inhibiting adipogenesis. Epigenet. Chromatin 2016, 9, 45. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Fan, W.; Xu, M.; Lin, X.; Zhao, W.; Yang, Z. Critical role of Znhit1 for postnatal heart function and vacuolar cardiomyopathy. JCI Insight 2022, 7, e148752. [Google Scholar] [CrossRef] [PubMed]
- Eleftheriadou, O.; Boguslavskyi, A.; Longman, M.R.; Cowan, J.; Francois, A.; Heads, R.J.; Wadzinski, B.E.; Ryan, A.; Shattock, M.J.; Snabaitis, A.K. Expression and regulation of type 2a protein phosphatases and alpha4 signalling in cardiac health and hypertrophy. Basic Res. Cardiol. 2017, 112, 37. [Google Scholar] [CrossRef]
- Yoon, S.R.; Song, J.; Lee, J.H.; Kim, O.Y. Phosphorylation of histone H2A.X in peripheral blood mononuclear cells may be a useful marker for monitoring cardiometabolic risk in nondiabetic individuals. Dis. Markers 2017, 2017, 1–9. [Google Scholar] [CrossRef]
- Bray, F.; Laversanne, M.; Sung, H.; Ferlay, J.; Siegel, R.L.; Soerjomataram, I.; Jemal, A. Global cancer statistics 2022: Global estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA-Cancer J. Clin. 2024, 74, 229–263. [Google Scholar] [CrossRef]
- Balon, K.; Sheriff, A.; Jacków, J.; Baczmański, A. Targeting cancer with CRISPR/cas9-based therapy. Int. J. Mol. Sci. 2022, 23, 573. [Google Scholar] [CrossRef]
- Yuan, Y.; Cao, W.; Zhou, H.; Qian, H.; Wang, H. H2A.Z acetylation by lincznf337-as1 via kat5 implicated in the transcriptional misregulation in cancer signaling pathway in hepatocellular carcinoma. Cell Death Dis. 2021, 12, 609. [Google Scholar] [CrossRef]
- Li, X.; Zhu, R.; Yuan, Y.; Cai, Z.; Liang, S.; Bian, J.; Xu, G. Double-stranded RNA-specific adenosine deaminase-knockdown inhibits the proliferation and induces apoptosis of DU145 and PC3 cells by promoting the phosphorylation of H2A.X variant histone. Oncol. Lett. 2021, 22, 764. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.; Wen, J.; Paver, E.; Wu, Y.H.; Sun, G.; Bullman, A.; Dahlstrom, J.E.; Tremethick, D.J.; Soboleva, T.A. H2A.B is cancer/testis factor involved in the activation of ribosome biogenesis in Hodgkin lymphoma. EMBO Rep. 2021, 22, e52462. [Google Scholar] [CrossRef] [PubMed]
- Chew, G.L.; Bleakley, M.; Bradley, R.K.; Malik, H.S.; Henikoff, S.; Molaro, A.; Sarthy, J. Short H2A histone variants are expressed in cancer. Nat. Commun. 2021, 12, 490. [Google Scholar] [CrossRef] [PubMed]
- Lone, I.N.; Sengez, B.; Hamiche, A.; Dimitrov, S.; Alotaibi, H. The role of histone variants in the epithelial-to-mesenchymal transition. Cells 2020, 9, 2499. [Google Scholar] [CrossRef] [PubMed]
- Babaei, G.; Aziz, S.G.; Jaghi, N. Emt, cancer stem cells and autophagy; The three main axes of metastasis. Biomed. Pharmacother. 2021, 133, 110909. [Google Scholar] [CrossRef]
- Saitoh, M. Involvement of partial EMT in cancer progression. J. Biochem. 2018, 164, 257–264. [Google Scholar] [CrossRef]
- Zheng, Y.; Han, X.; Wang, T. Role of H2A.Z.1 in epithelial-mesenchymal transition and radiation resistance of lung adenocarcinoma in vitro. Biochem. Biophys. Res. Commun. 2022, 611, 118–125. [Google Scholar] [CrossRef]
- Berta, D.G.; Kuisma, H.; Välimäki, N.; Räisänen, M.; Jäntti, M.; Pasanen, A.; Karhu, A.; Kaukomaa, J.; Taira, A.; Cajuso, T.; et al. Deficient H2A.Z deposition is associated with genesis of uterine leiomyoma. Nature 2021, 596, 398–403. [Google Scholar] [CrossRef]
- Vardabasso, C.; Gaspar-Maia, A.; Hasson, D.; Pünzeler, S.; Valle-Garcia, D.; Straub, T.; Keilhauer, E.C.; Strub, T.; Dong, J.; Panda, T.; et al. Histone variant H2A.Z.2 mediates proliferation and drug sensitivity of malignant melanoma. Mol. Cell 2015, 59, 75–88. [Google Scholar] [CrossRef]
- Kim, K.; Punj, V.; Choi, J.; Heo, K.; Kim, J.M.; Laird, P.W.; An, W. Gene dysregulation by histone variant H2A.Z in bladder cancer. Epigenetics Chromatin 2013, 6, 34. [Google Scholar] [CrossRef]
- Tang, S.; Huang, X.; Wang, X.; Zhou, X.; Huang, H.; Qin, L.; Tao, H.; Wang, Q.; Tao, Y. Vital and distinct roles of H2A.Z isoforms in hepatocellular carcinoma. OncoTargets Ther. 2020, 13, 4319–4337. [Google Scholar] [CrossRef]
- Dong, M.; Chen, J.; Deng, Y.; Zhang, D.; Dong, L.; Sun, D. H2AFZ is a prognostic biomarker correlated to TP53 mutation and immune infiltration in hepatocellular carcinoma. Front. Oncol. 2021, 11, 701736. [Google Scholar] [CrossRef]
- Weyemi, U.; Redon, C.E.; Choudhuri, R.; Aziz, T.; Maeda, D.; Boufraqech, M.; Parekh, P.R.; Sethi, T.K.; Kasoji, M.; Abrams, N.; et al. The histone variant H2A.X is a regulator of the epithelial–mesenchymal transition. Nat. Commun. 2016, 7, 10711. [Google Scholar] [CrossRef]
- Ge, Y.; Liu, B.; Cui, J.; Li, S. Livin regulates H2A.X y142 phosphorylation and promotes autophagy in colon cancer cells via a novel kinase activity. Front. Oncol. 2019, 9, 1233. [Google Scholar] [CrossRef]
- Weyemi, U.; Redon, C.E.; Sethi, T.K.; Burrell, A.S.; Jailwala, P.; Kasoji, M.; Abrams, N.; Merchant, A.; Bonner, W.M. Twist1 and slug mediate H2AX-regulated epithelial-mesenchymal transition in breast cells. Cell Cycle 2016, 15, 2398–2404. [Google Scholar] [CrossRef][Green Version]
- Guo, Z.F.; Kong, F.L. Akt regulates rsk2 to alter phosphorylation level of H2A.X in breast cancer. Oncol. Lett. 2021, 21, 187. [Google Scholar] [CrossRef]
- Ribeiro, I.P.; Caramelo, F.; Esteves, L.; Menoita, J.; Marques, F.; Barroso, L.; Miguéis, J.; Melo, J.B.; Carreira, I.M. Genomic predictive model for recurrence and metastasis development in head and neck squamous cell carcinoma patients. Sci. Rep. 2017, 7, 13897. [Google Scholar] [CrossRef]
- Knittel, G.; Liedgens, P.; Reinhardt, H.C. Targeting ATM-deficient CLL through interference with DNA repair pathways. Front. Genet. 2015, 6, 207. [Google Scholar] [CrossRef]
- Lord, C.J.; Ashworth, A. The DNA damage response and cancer therapy. Nature 2012, 481, 287–294. [Google Scholar] [CrossRef]
- Yin, H.; Jiang, Z.; Wang, S.; Zhang, P. Actinomycin D-activated RNase l promotes H2A.X/H2B-mediated DNA damage and apoptosis in lung cancer cells. Front. Oncol. 2019, 9, 1086. [Google Scholar] [CrossRef]
- Miyake, K.; Takano, N.; Kazama, H.; Kikuchi, H.; Hiramoto, M.; Tsukahara, K.; Miyazawa, K. Ricolinostat enhances adavosertib-induced mitotic catastrophe in TP53-mutated head and neck squamous cell carcinoma cells. Int. J. Oncol. 2022, 60, 54. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Chang, D.W.; Gong, Y.; Eng, C.; Wu, X. Measurement of DNA damage in peripheral blood by the γ-H2Ax assay as predictor of colorectal cancer risk. DNA Repair. 2017, 53, 24–30. [Google Scholar] [CrossRef]
- Fernández, M.I.; Gong, Y.; Ye, Y.; Lin, J.; Chang, D.W.; Kamat, A.M.; Wu, X. gamma-H2AX level in peripheral blood lymphocytes as a risk predictor for bladder cancer. Carcinogenesis 2013, 34, 2543–2547. [Google Scholar] [CrossRef] [PubMed]
- Kapoor, A.; Goldberg, M.S.; Cumberland, L.K.; Ratnakumar, K.; Segura, M.F.; Emanuel, P.O.; Menendez, S.; Vardabasso, C.; Leroy, G.; Vidal, C.I.; et al. The histone variant macroH2A suppresses melanoma progression through regulation of cdk8. Nature 2010, 468, 1105–1109. [Google Scholar] [CrossRef]
- Hu, W.; Miyai, K.; Sporn, J.C.; Luo, L.; Wang, J.Y.J.; Cosman, B.; Ramamoorthy, S. Loss of histone variant macroH2A2 expression associates with progression of anal neoplasm. J. Clin. Pathol. 2016, 69, 627–631. [Google Scholar] [CrossRef]
- Nikolic, A.; Maule, F.; Bobyn, A.; Ellestad, K.; Paik, S.; Marhon, S.A.; Mehdipour, P.; Lun, X.; Chen, H.M.; Mallard, C.; et al. MacroH2A2 antagonizes epigenetic programs of stemness in glioblastoma. Nat. Commun. 2023, 14, 3062. [Google Scholar] [CrossRef]
- Sun, D.; Singh, D.K.; Carcamo, S.; Filipescu, D.; Khalil, B.; Huang, X.; Miles, B.A.; Westra, W.; Sproll, K.C.; Hasson, D.; et al. MacroH2A impedes metastatic growth by enforcing a discrete dormancy program in disseminated cancer cells. Sci. Adv. 2022, 8, o876. [Google Scholar] [CrossRef]
- Dardenne, E.; Pierredon, S.; Driouch, K.; Gratadou, L.; Lacroix-Triki, M.; Espinoza, M.P.; Zonta, E.; Germann, S.; Mortada, H.; Villemin, J.P.; et al. Splicing switch of an epigenetic regulator by RNA helicases promotes tumor-cell invasiveness. Nat. Struct. Mol. Biol. 2012, 19, 1139–1146. [Google Scholar] [CrossRef]
- Sporn, J.C.; Jung, B. Differential regulation and predictive potential of macroH2A1 isoforms in colon cancer. Am. J. Pathol. 2012, 180, 2516–2526. [Google Scholar] [CrossRef]
- Hodge, D.Q.; Cui, J.; Gamble, M.J.; Guo, W. Histone variant macroH2A1 plays an isoform-specific role in suppressing epithelial-mesenchymal transition. Sci. Rep. 2018, 8, 841. [Google Scholar] [CrossRef]
- Kim, J.; Shin, Y.; Lee, S.; Kim, M.Y.; Punj, V.; Shin, H.; Kim, K.; Koh, J.; Jeong, D.; An, W. MacroH2A1.2 inhibits prostate cancer-induced osteoclastogenesis through cooperation with hp1α and h1.2. Oncogene 2018, 37, 5749–5765. [Google Scholar] [CrossRef] [PubMed]
- Giallongo, C.; Dulcamare, I.; Giallongo, S.; Duminuco, A.; Pieragostino, D.; Cufaro, M.C.; Amorini, A.M.; Lazzarino, G.; Romano, A.; Parrinello, N.; et al. MacroH2A1.1 as a crossroad between epigenetics, inflammation and metabolism of mesenchymal stromal cells in myelodysplastic syndromes. Cell Death Dis. 2023, 14, 686. [Google Scholar] [CrossRef] [PubMed]
- Novikov, L.; Park, J.W.; Chen, H.; Klerman, H.; Jalloh, A.S.; Gamble, M.J. QKI-mediated alternative splicing of the histone variant macroH2A1 regulates cancer cell proliferation. Mol. Cell. Biol. 2011, 31, 4244–4255. [Google Scholar] [CrossRef] [PubMed]
- Vieira-Silva, T.S.; Monteiro-Reis, S.; Barros-Silva, D.; Ramalho-Carvalho, J.; Graça, I.; Carneiro, I.; Martins, A.T.; Oliveira, J.; Antunes, L.; Hurtado-Bagès, S.; et al. Histone variant macroH2A1 is downregulated in prostate cancer and influences malignant cell phenotype. Cancer Cell Int. 2019, 19, 112. [Google Scholar] [CrossRef] [PubMed]
- Bereshchenko, O.; Lo Re, O.; Nikulenkov, F.; Flamini, S.; Kotaskova, J.; Mazza, T.; Le Pannérer, M.; Buschbeck, M.; Giallongo, C.; Palumbo, G.; et al. Deficiency and haploinsufficiency of histone macroH2A1.1 in mice recapitulate hematopoietic defects of human myelodysplastic syndrome. Clin. Epigenetics 2019, 11, 121. [Google Scholar] [CrossRef]
- Kang, L.; Cao, G.; Jing, W.; Liu, J.; Liu, M. Global, regional, and national incidence and mortality of congenital birth defects from 1990 to 2019. Eur. J. Pediatr. 2023, 182, 1781–1792. [Google Scholar] [CrossRef] [PubMed]
- Banaszynski, L.A.; Allis, C.D.; Lewis, P.W. Histone variants in metazoan development. Dev. Cell 2010, 19, 662–674. [Google Scholar] [CrossRef]
- Faast, R.; Thonglairoam, V.; Schulz, T.C.; Beall, J.; Wells, J.R.; Taylor, H.; Matthaei, K.; Rathjen, P.D.; Tremethick, D.J.; Lyons, I. Histone variant H2A.Z is required for early mammalian development. Curr. Biol. 2001, 11, 1183–1187. [Google Scholar] [CrossRef]
- Celeste, A.; Petersen, S.; Romanienko, P.J.; Fernandez-Capetillo, O.; Chen, H.T.; Sedelnikova, O.A.; Reina-San-Martin, B.; Coppola, V.; Meffre, E.; Difilippantonio, M.J.; et al. Genomic instability in mice lacking histone H2AX. Science 2002, 296, 922–927. [Google Scholar] [CrossRef]
- Pehrson, J.R.; Changolkar, L.N.; Costanzi, C.; Leu, N.A. Mice without macroH2A histone variants. Mol. Cell. Biol. 2014, 34, 4523–4533. [Google Scholar] [CrossRef]
- Law, C.; Cheung, P. Expression of non-acetylatable H2A.Z in myoblast cells blocks myoblast differentiation through disruption of myod expression. J. Biol. Chem. 2015, 290, 13234–13249. [Google Scholar] [CrossRef] [PubMed]
- Raja, D.A.; Subramaniam, Y.; Aggarwal, A.; Gotherwal, V.; Babu, A.; Tanwar, J.; Motiani, R.K.; Sivasubbu, S.; Gokhale, R.S.; Natarajan, V.T. Histone variant dictates fate biasing of neural crest cells to melanocyte lineage. Development 2020, 147, dev182576. [Google Scholar] [CrossRef]
- Zhao, B.; Chen, Y.; Jiang, N.; Yang, L.; Sun, S.; Zhang, Y.; Wen, Z.; Ray, L.; Liu, H.; Hou, G.; et al. Znhit1 controls intestinal stem cell maintenance by regulating H2A.Z incorporation. Nat. Commun. 2019, 10, 1071. [Google Scholar] [CrossRef] [PubMed]
- Hu, G.; Cui, K.; Northrup, D.; Liu, C.; Wang, C.; Tang, Q.; Ge, K.; Levens, D.; Crane-Robinson, C.; Zhao, K. H2A.Z facilitates access of active and repressive complexes to chromatin in embryonic stem cell self-renewal and differentiation. Cell Stem Cell 2013, 12, 180–192. [Google Scholar] [CrossRef] [PubMed]
- Jukam, D.; Shariati, S.; Skotheim, J.M. Zygotic genome activation in vertebrates. Dev. Cell 2017, 42, 316–332. [Google Scholar] [CrossRef] [PubMed]
- Deng, M.; Chen, B.; Liu, Z.; Cai, Y.; Wan, Y.; Zhou, J.; Wang, F. Exchanges of histone methylation and variants during mouse zygotic genome activation. Zygote 2020, 28, 51–58. [Google Scholar] [CrossRef] [PubMed]
- Ibarra-Morales, D.; Rauer, M.; Quarato, P.; Rabbani, L.; Zenk, F.; Schulte-Sasse, M.; Cardamone, F.; Gomez-Auli, A.; Cecere, G.; Iovino, N. Histone variant H2A.Z regulates zygotic genome activation. Nat. Commun. 2021, 12, 7002. [Google Scholar] [CrossRef]
- Yamada, S.; Kugou, K.; Ding, D.Q.; Fujita, Y.; Hiraoka, Y.; Murakami, H.; Ohta, K.; Yamada, T. The conserved histone variant H2A.Z illuminates meiotic recombination initiation. Curr. Genet. 2018, 64, 1015–1019. [Google Scholar] [CrossRef]
- Zhuo, M.L.P.W. Anp32e, a higher eukaryotic histone chaperone directs preferential recognition for H2A.Z. Cell Res. 2014, 24, 389–399. [Google Scholar]
- Pei, D.; Shu, X.; Gassama-Diagne, A.; Thiery, J.P. Mesenchymal-epithelial transition in development and reprogramming. Nat. Cell Biol. 2019, 21, 44–53. [Google Scholar] [CrossRef] [PubMed]
- Kafer, G.R.; Lehnert, S.A.; Pantaleon, M.; Kaye, P.L.; Moser, R.J. Expression of genes coding for histone variants and histone-associated proteins in pluripotent stem cells and mouse preimplantation embryos. Gene Expr. Patterns 2010, 10, 299–305. [Google Scholar] [CrossRef] [PubMed]
- Zha, S.; Sekiguchi, J.; Brush, J.W.; Bassing, C.H.; Alt, F.W. Complementary functions of atm and H2AX in development and suppression of genomic instability. Proc. Natl. Acad. Sci. USA 2008, 105, 9302–9306. [Google Scholar] [CrossRef]
- Turinetto, V.; Orlando, L.; Sanchez-Ripoll, Y.; Kumpfmueller, B.; Storm, M.P.; Porcedda, P.; Minieri, V.; Saviozzi, S.; Accomasso, L.; Cibrario, R.E.; et al. High basal γH2AX levels sustain self-renewal of mouse embryonic and induced pluripotent stem cells. Stem Cells 2012, 30, 1414–1423. [Google Scholar] [CrossRef] [PubMed]
- Eleuteri, B.; Aranda, S.; ERNFors, P. NORC recruitment by H2A.X deposition at rRNA gene promoter limits embryonic stem cell proliferation. Cell Rep. 2018, 23, 1853–1866. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.; Liu, Y.; Wen, D.; Tseng, Z.; Tahmasian, M.; Zhong, M.; RafII, S.; Stadtfeld, M.; Hochedlinger, K.; Xiao, A. Histone variant H2A.X deposition pattern serves as a functional epigenetic mark for distinguishing the developmental potentials of iPSCs. Cell Stem Cell 2014, 15, 281–294. [Google Scholar] [CrossRef]
- Creppe, C.; Janich, P.; Cantariño, N.; Noguera, M.; Valero, V.; Musulén, E.; Douet, J.; Posavec, M.; Martín-Caballero, J.; Sumoy, L.; et al. MacroH2A1 regulates the balance between self-renewal and differentiation commitment in embryonic and adult stem cells. Mol. Cell. Biol. 2012, 32, 1442–1452. [Google Scholar] [CrossRef] [PubMed]
- Pasque, V.; Radzisheuskaya, A.; Gillich, A.; Halley-Stott, R.P.; Panamarova, M.; Zernicka-Goetz, M.; Surani, M.A.; Silva, J.C. Histone variant macroH2A marks embryonic differentiation in vivo and acts as an epigenetic barrier to induced pluripotency. J. Cell Sci. 2012, 125, 6094–6104. [Google Scholar] [CrossRef] [PubMed]
- Barrero, M.J.; Sese, B.; Martí, M.; Izpisua, B.J. Macro histone variants are critical for the differentiation of human pluripotent cells. J. Biol. Chem. 2013, 288, 16110–16116. [Google Scholar] [CrossRef]
- Gonzalez-Munoz, E.; Arboleda-Estudillo, Y.; Chanumolu, S.K.; Otu, H.H.; Cibelli, J.B. Zebrafish macroH2A variants have distinct embryo localization and function. Sci. Rep. 2019, 9, 8632. [Google Scholar] [CrossRef]
- Costanzi, C.; Stein, P.; Worrad, D.M.; Schultz, R.M.; Pehrson, J.R. Histone macroH2A1 is concentrated in the inactive X chromosome of female preimplantation mouse embryos. Development 2000, 127, 2283–2289. [Google Scholar] [CrossRef]
- Chen, Y.; Chen, Q.; Mceachin, R.C.; Cavalcoli, J.D.; Yu, X. H2A.B facilitates transcription elongation at methylated CPG loci. Genome Res. 2014, 24, 570–579. [Google Scholar] [CrossRef] [PubMed]
- Molaro, A.; Wood, A.J.; Janssens, D.; Kindelay, S.M.; Eickbush, M.T.; Wu, S.; Singh, P.; Muller, C.H.; Henikoff, S.; Malik, H.S. Biparental contributions of the H2A.B histone variant control embryonic development in mice. PLoS. Biol. 2020, 18, e3001001. [Google Scholar] [CrossRef] [PubMed]
- Steinmetz, J.D.; Seeher, K.M.; Schiess, N.; Nichols, E.; Cao, B.; Servili, C.; Cavallera, V.; Cousin, E.; Hagins, H.; Moberg, M.E.; et al. Global, regional, and national burden of disorders affecting the nervous system, 1990–2021: A systematic analysis for the global burden of disease study 2021. Lancet Neurol. 2024, 23, 344–381. [Google Scholar] [CrossRef] [PubMed]
- Zovkic, I.B.; Paulukaitis, B.S.; Day, J.J.; Etikala, D.M.; Sweatt, J.D. Histone H2A.Z subunit exchange controls consolidation of recent and remote memory. Nature 2014, 515, 582–586. [Google Scholar] [CrossRef]
- Stefanelli, G.; Makowski, C.E.; Brimble, M.A.; Hall, M.; Reda, A.; Creighton, S.D.; Leonetti, A.M.; Mclean, T.; Zakaria, J.M.; Baumbach, J.; et al. The histone chaperone Anp32e regulates memory formation, transcription, and dendritic morphology by regulating steady-state H2A.Z binding in neurons. Cell Rep. 2021, 36, 109551. [Google Scholar] [CrossRef] [PubMed]
- Farrelly, L.A.; Zheng, S.; Schrode, N.; Topol, A.; Bhanu, N.V.; Bastle, R.M.; Ramakrishnan, A.; Chan, J.C.; Cetin, B.; Flaherty, E.; et al. Chromatin profiling in human neurons reveals aberrant roles for histone acetylation and bet family proteins in schizophrenia. Nat. Commun. 2022, 13, 2195. [Google Scholar] [CrossRef] [PubMed]
- Weyemi, U.; Paul, B.D.; Bhattacharya, D.; Malla, A.P.; Boufraqech, M.; Harraz, M.M.; Bonner, W.M.; Snyder, S.H. Histone H2AX promotes neuronal health by controlling mitochondrial homeostasis. Proc. Natl. Acad. Sci. USA 2019, 116, 7471–7476. [Google Scholar] [CrossRef] [PubMed]
- Weyemi, U.; Paul, B.D.; Snowman, A.M.; Jailwala, P.; Nussenzweig, A.; Bonner, W.M.; Snyder, S.H. Histone H2AX deficiency causes neurobehavioral deficits and impaired redox homeostasis. Nat. Commun. 2018, 9, 1526. [Google Scholar] [CrossRef]
- Ma, H.; Su, L.; Xia, W.; Wang, W.; Tan, G.; Jiao, J. MacroH2A1.2 deficiency leads to neural stem cell differentiation defects and autism-like behaviors. EMBO Rep. 2021, 22, e52150. [Google Scholar] [CrossRef]
- Berger, N.A.; Besson, V.C.; Boulares, A.H.; Bürkle, A.; Chiarugi, A.; Clark, R.S.; Curtin, N.J.; Cuzzocrea, S.; Dawson, T.M.; Dawson, V.L.; et al. Opportunities for the repurposing of PARP inhibitors for the therapy of non-oncological diseases. Br. J. Pharmacol. 2018, 175, 192–222. [Google Scholar] [CrossRef]
- Jiang, G.; Zhang, G.; An, T.; He, Z.; Kang, L.; Yang, X.; Gu, Y.; Zhang, D.; Wang, Y.; Gao, S. Effect of type I diabetes on the proteome of mouse oocytes. Cell Physiol. Biochem. 2016, 39, 2320–2330. [Google Scholar] [CrossRef] [PubMed]
- Ishibashi, T.; Dryhurst, D.; Rose, K.L.; Shabanowitz, J.; Hunt, D.F.; Ausió, J. Acetylation of vertebrate H2A.Z and its effect on the structure of the nucleosome. Biochemistry 2009, 48, 5007–5017. [Google Scholar] [CrossRef] [PubMed]
- Millar, C.B.; Xu, F.; Zhang, K.; Grunstein, M. Acetylation of H2AZ lys 14 is associated with genome-wide gene activity in yeast. Genes. Dev. 2006, 20, 711–722. [Google Scholar] [CrossRef] [PubMed]
- Perche, P.Y.; Vourc’H, C.; Konecny, L.; Souchier, C.; Robert-Nicoud, M.; Dimitrov, S.; Khochbin, S. Higher concentrations of histone macroH2A in the Barr body are correlated with higher nucleosome density. Curr. Biol. 2000, 10, 1531–1534. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yin, X.; Zeng, D.; Liao, Y.; Tang, C.; Li, Y. The Function of H2A Histone Variants and Their Roles in Diseases. Biomolecules 2024, 14, 993. https://doi.org/10.3390/biom14080993
Yin X, Zeng D, Liao Y, Tang C, Li Y. The Function of H2A Histone Variants and Their Roles in Diseases. Biomolecules. 2024; 14(8):993. https://doi.org/10.3390/biom14080993
Chicago/Turabian StyleYin, Xuemin, Dong Zeng, Yingjun Liao, Chengyuan Tang, and Ying Li. 2024. "The Function of H2A Histone Variants and Their Roles in Diseases" Biomolecules 14, no. 8: 993. https://doi.org/10.3390/biom14080993
APA StyleYin, X., Zeng, D., Liao, Y., Tang, C., & Li, Y. (2024). The Function of H2A Histone Variants and Their Roles in Diseases. Biomolecules, 14(8), 993. https://doi.org/10.3390/biom14080993






