The Role of m6A Methylation in Tumor Immunity and Immune-Associated Disorder
Abstract
1. Introduction
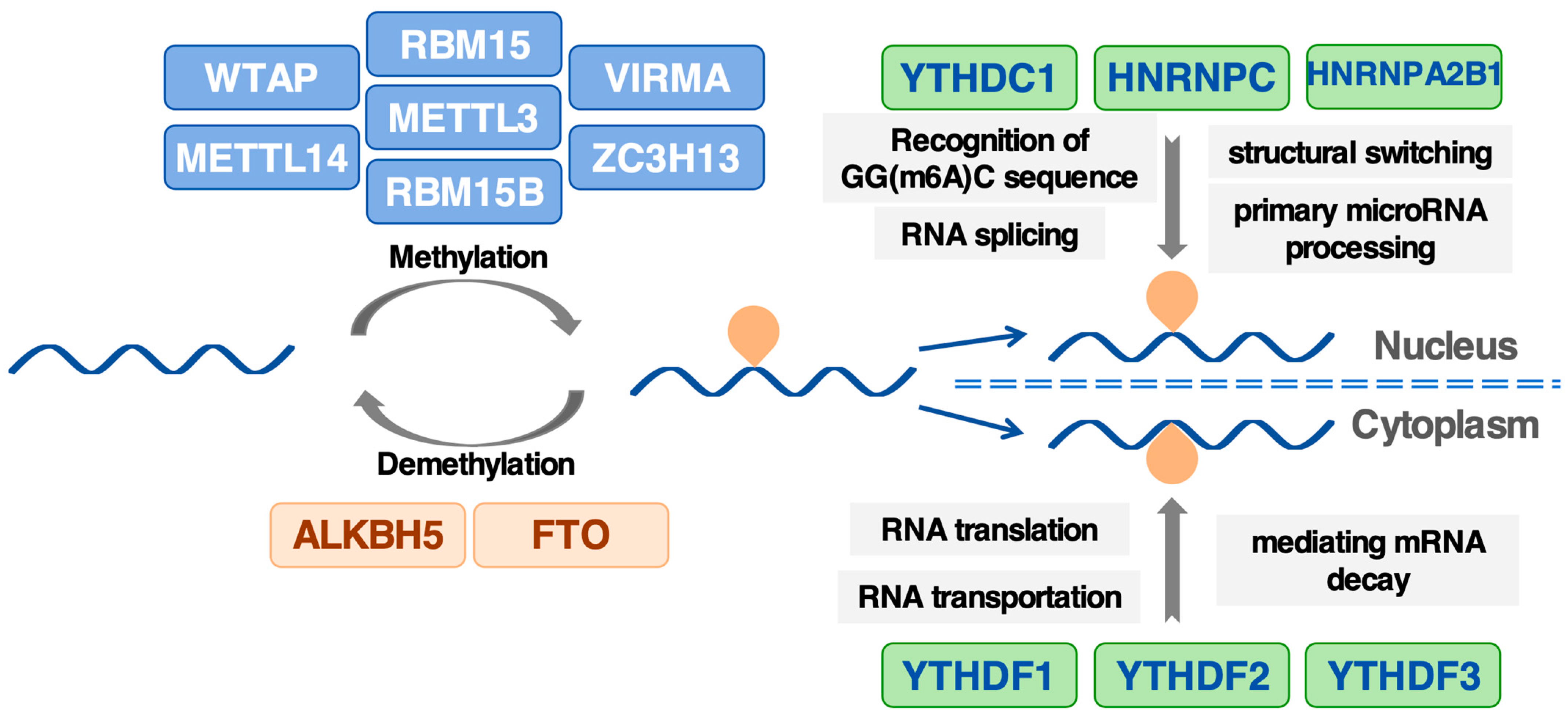
2. M6A Methylation-Related Proteins: From Writing to Interpreting
3. M6A Modification in Immune Cells
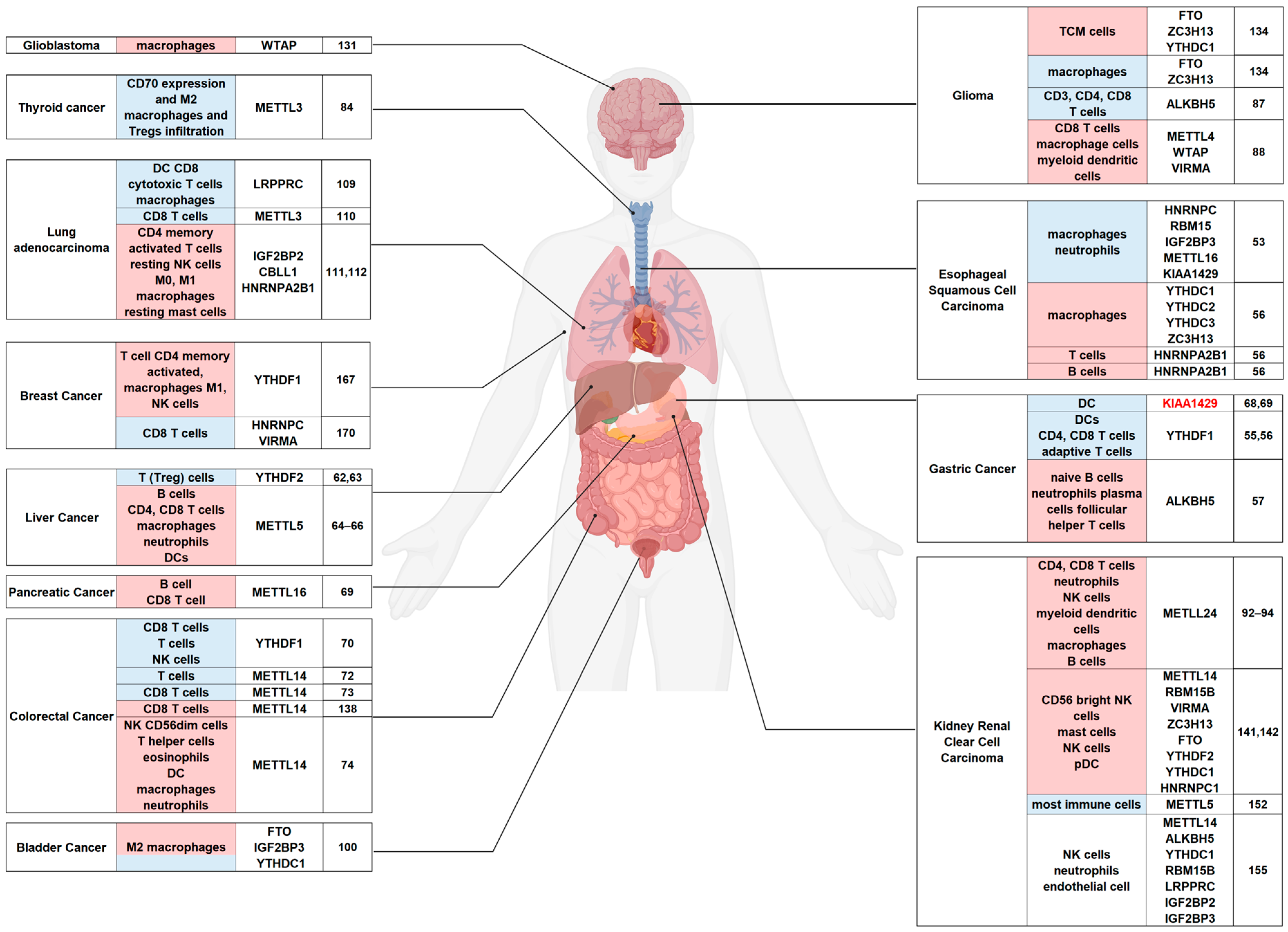
4. Aberrant m6A Modifications of Immune System Regulation in Human Cancer
4.1. Digestive System
4.1.1. Esophageal Cancer
4.1.2. Gastric Cancer
4.1.3. Liver Cancer
4.1.4. Pancreatic Cancer
4.1.5. Colorectal Cancer (CRC)
4.1.6. Gallbladder Cancer
4.2. Respiratory System
4.2.1. Lung Adenocarcinoma
4.2.2. Nasopharyngeal Carcinoma
4.3. Endocrine System
4.3.1. Thyroid Cancer
4.3.2. Pituitary Adenomas
4.4. Nervous System
4.5. Urinary System
4.5.1. Prostate Cancer
4.5.2. Renal Cell Carcinoma
4.5.3. Bladder Cancer
4.6. Reproductive System
4.6.1. Breast Cancer
4.6.2. Cervical Cancer
4.6.3. Endometrial Cancer
4.6.4. Ovarian Cancer
4.7. Musculoskeletal System
4.7.1. Melanoma
4.7.2. Osteosarcoma
4.8. Acute Myeloid Leukaemia
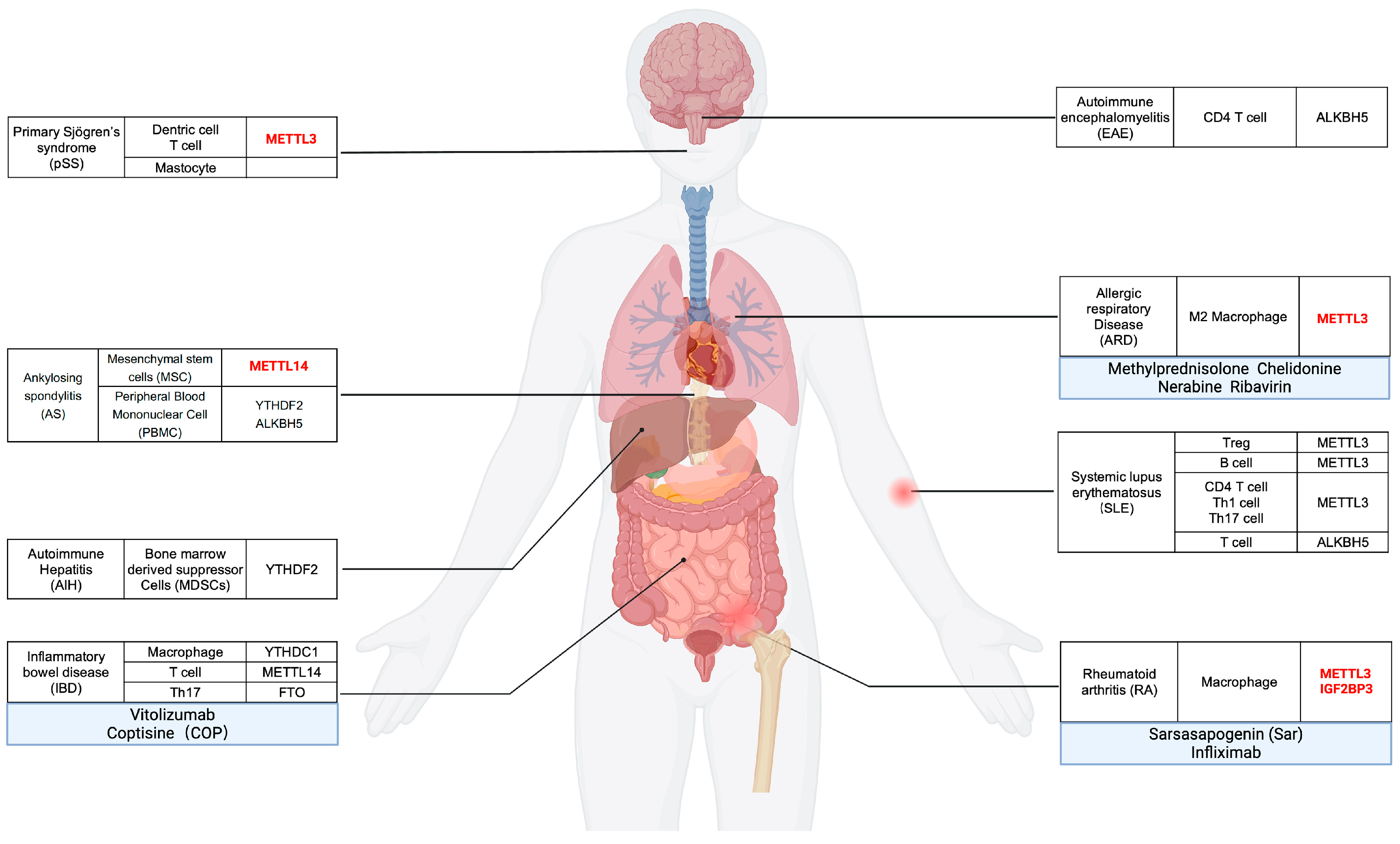
5. m6A Modification-Association with Immune Regulation and Implications on Autoimmune and Immune-Related Diseases
5.1. Rheumatoid Arthritis, RA
5.2. Systemic Lupus Erythematous, SLE
5.3. Inflammatory Bowel Disease, IBE
5.4. Primary Sjogren Syndrome, pSS
5.5. Ankylosing Spondylitis, AS
5.6. RNA Methylation and Other Autoimmune Diseases
5.7. M6A and Acquired Immunodeficiency Syndrome, AIDS
6. Conclusions and Prospects
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Huang, H.; Weng, H.; Chen, J. m(6)A Modification in Coding and Non-coding RNAs: Roles and Therapeutic Implications in Cancer. Cancer Cell 2020, 37, 270–288. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Lu, Z.; Gomez, A.; Hon, G.C.; Yue, Y.; Han, D.; Fu, Y.; Parisien, M.; Dai, Q.; Jia, G.; et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 2014, 505, 117–120. [Google Scholar] [CrossRef]
- Wang, X.; Zhao, B.S.; Roundtree, I.A.; Lu, Z.; Han, D.; Ma, H.; Weng, X.; Chen, K.; Shi, H.; He, C. N(6)-methyladenosine Modulates Messenger RNA Translation Efficiency. Cell 2015, 161, 1388–1399. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.; Ma, P.; Liu, Y.; Li, W.; Shu, Y. Multiple functions of m(6)A RNA methylation in cancer. J. Hematol. Oncol. 2018, 11, 48. [Google Scholar] [CrossRef]
- Wang, Y.; Li, L.; Li, J.; Zhao, B.; Huang, G.; Li, X.; Xie, Z.; Zhou, Z. The Emerging Role of m6A Modification in Regulating the Immune System and Autoimmune Diseases. Front. Cell Dev. Biol. 2021, 9, 755691. [Google Scholar] [CrossRef]
- Zhang, N.; Ding, C.; Zuo, Y.; Peng, Y.; Zuo, L. N6-methyladenosine and Neurological Diseases. Mol. Neurobiol. 2022, 59, 1925–1937. [Google Scholar] [CrossRef]
- Courtney, D.G.; Kennedy, E.M.; Dumm, R.E.; Bogerd, H.P.; Tsai, K.; Heaton, N.S.; Cullen, B.R. Epitranscriptomic Enhancement of Influenza A Virus Gene Expression and Replication. Cell Host Microbe 2017, 22, 377–386. [Google Scholar] [CrossRef] [PubMed]
- Perry, R.P.; Kelley, D.E.; Friderici, K.; Rottman, F. The methylated constituents of L cell messenger RNA: Evidence for an unusual cluster at the 5' terminus. Cell 1975, 4, 387–394. [Google Scholar] [CrossRef]
- Nichols, J.L. ‘Cap’ structures in maize poly(A)-containing RNA. Biochim. Biophys. Acta 1979, 563, 490–495. [Google Scholar] [CrossRef]
- Clancy, M.J.; Shambaugh, M.E.; Timpte, C.S.; Bokar, J.A. Induction of sporulation in Saccharomyces cerevisiae leads to the formation of N6-methyladenosine in mRNA: A potential mechanism for the activity of the IME4 gene. Nucleic Acids Res. 2002, 30, 4509–4518. [Google Scholar] [CrossRef]
- Levis, R.; Penman, S. 5′-terminal structures of poly(A)+ cytoplasmic messenger RNA and of poly(A)+ and poly(A)- heterogeneous nuclear RNA of cells of the dipteran Drosophila melanogaster. J. Mol. Biol. 1978, 120, 487–515. [Google Scholar] [CrossRef]
- Jia, G.; Fu, Y.; He, C. Reversible RNA adenosine methylation in biological regulation. Trends Genet. 2013, 29, 108–115. [Google Scholar] [CrossRef] [PubMed]
- Dominissini, D.; Moshitch-Moshkovitz, S.; Schwartz, S.; Salmon-Divon, M.; Ungar, L.; Osenberg, S.; Cesarkas, K.; Jacob-Hirsch, J.; Amariglio, N.; Kupiec, M.; et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 2012, 485, 201–206. [Google Scholar] [CrossRef]
- Linder, B.; Grozhik, A.V.; Olarerin-George, A.O.; Meydan, C.; Mason, C.E.; Jaffrey, S.R. Single-nucleotide-resolution mapping of m6A and m6Am throughout the transcriptome. Nat. Methods 2015, 12, 767–772. [Google Scholar] [CrossRef]
- Wang, H.; Hu, X.; Huang, M.; Liu, J.; Gu, Y.; Ma, L.; Zhou, Q.; Cao, X. Mettl3-mediated mRNA m(6)A methylation promotes dendritic cell activation. Nat. Commun. 2019, 10, 1898. [Google Scholar] [CrossRef] [PubMed]
- Lv, J.; Zhang, Y.; Gao, S.; Zhang, C.; Chen, Y.; Li, W.; Yang, Y.G.; Zhou, Q.; Liu, F. Endothelial-specific m(6)A modulates mouse hematopoietic stem and progenitor cell development via Notch signaling. Cell Res. 2018, 28, 249–252. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Song, J.; Yuan, W.; Zhang, W.; Sun, Z. Roles of RNA Methylation on Tumor Immunity and Clinical Implications. Front. Immunol. 2021, 12, 641507. [Google Scholar] [CrossRef]
- Meyer, K.D.; Saletore, Y.; Zumbo, P.; Elemento, O.; Mason, C.E.; Jaffrey, S.R. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell 2012, 149, 1635–1646. [Google Scholar] [CrossRef]
- Desrosiers, R.; Friderici, K.; Rottman, F. Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells. Proc. Natl. Acad. Sci. USA 1974, 71, 3971–3975. [Google Scholar] [CrossRef]
- Schöller, E.; Weichmann, F.; Treiber, T.; Ringle, S.; Treiber, N.; Flatley, A.; Feederle, R.; Bruckmann, A.; Meister, G. Interactions, localization, and phosphorylation of the m(6)A generating METTL3-METTL14-WTAP complex. Rna 2018, 24, 499–512. [Google Scholar] [CrossRef]
- Ping, X.L.; Sun, B.F.; Wang, L.; Xiao, W.; Yang, X.; Wang, W.J.; Adhikari, S.; Shi, Y.; Lv, Y.; Chen, Y.S.; et al. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res. 2014, 24, 177–189. [Google Scholar] [CrossRef] [PubMed]
- Patil, D.P.; Chen, C.K.; Pickering, B.F.; Chow, A.; Jackson, C.; Guttman, M.; Jaffrey, S.R. m(6)A RNA methylation promotes XIST-mediated transcriptional repression. Nature 2016, 537, 369–373. [Google Scholar] [CrossRef] [PubMed]
- Yue, Y.; Liu, J.; Cui, X.; Cao, J.; Luo, G.; Zhang, Z.; Cheng, T.; Gao, M.; Shu, X.; Ma, H.; et al. VIRMA mediates preferential m(6)A mRNA methylation in 3'UTR and near stop codon and associates with alternative polyadenylation. Cell Discov. 2018, 4, 10. [Google Scholar] [CrossRef] [PubMed]
- Knuckles, P.; Lence, T.; Haussmann, I.U.; Jacob, D.; Kreim, N.; Carl, S.H.; Masiello, I.; Hares, T.; Villaseñor, R.; Hess, D.; et al. Zc3h13/Flacc is required for adenosine methylation by bridging the mRNA-binding factor Rbm15/Spenito to the m(6)A machinery component Wtap/Fl(2)d. Genes Dev. 2018, 32, 415–429. [Google Scholar] [CrossRef] [PubMed]
- Alarcón, C.R.; Goodarzi, H.; Lee, H.; Liu, X.; Tavazoie, S.; Tavazoie, S.F. HNRNPA2B1 Is a Mediator of m(6)A-Dependent Nuclear RNA Processing Events. Cell 2015, 162, 1299–1308. [Google Scholar] [CrossRef]
- Xu, C.; Wang, X.; Liu, K.; Roundtree, I.A.; Tempel, W.; Li, Y.; Lu, Z.; He, C.; Min, J. Structural basis for selective binding of m6A RNA by the YTHDC1 YTH domain. Nat. Chem. Biol. 2014, 10, 927–929. [Google Scholar] [CrossRef]
- Yang, Y.; Hsu, P.J.; Chen, Y.S.; Yang, Y.G. Dynamic transcriptomic m(6)A decoration: Writers, erasers, readers and functions in RNA metabolism. Cell Res. 2018, 28, 616–624. [Google Scholar] [CrossRef]
- Li, A.; Chen, Y.S.; Ping, X.L.; Yang, X.; Xiao, W.; Yang, Y.; Sun, H.Y.; Zhu, Q.; Baidya, P.; Wang, X.; et al. Cytoplasmic m(6)A reader YTHDF3 promotes mRNA translation. Cell Res. 2017, 27, 444–447. [Google Scholar] [CrossRef]
- Shi, H.; Wang, X.; Lu, Z.; Zhao, B.S.; Ma, H.; Hsu, P.J.; Liu, C.; He, C. YTHDF3 facilitates translation and decay of N(6)-methyladenosine-modified RNA. Cell Res. 2017, 27, 315–328. [Google Scholar] [CrossRef]
- Jia, G.; Fu, Y.; Zhao, X.; Dai, Q.; Zheng, G.; Yang, Y.; Yi, C.; Lindahl, T.; Pan, T.; Yang, Y.G.; et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 2011, 7, 885–887. [Google Scholar] [CrossRef]
- Zheng, G.; Dahl, J.A.; Niu, Y.; Fedorcsak, P.; Huang, C.M.; Li, C.J.; Vågbø, C.B.; Shi, Y.; Wang, W.L.; Song, S.H.; et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol. Cell 2013, 49, 18–29. [Google Scholar] [CrossRef] [PubMed]
- Parkin, J.; Cohen, B. An overview of the immune system. Lancet 2001, 357, 1777–1789. [Google Scholar] [CrossRef] [PubMed]
- Teng, Y.; Yi, J.; Chen, J.; Yang, L. N6-Methyladenosine (m6A) Modification in Natural Immune Cell-Mediated Inflammatory Diseases. J. Innate Immun. 2023, 15, 804–821. [Google Scholar] [CrossRef]
- Hui, L.; Ziyue, Z.; Chao, L.; Bin, Y.; Aoyu, L.; Haijing, W. Epigenetic Regulations in Autoimmunity and Cancer: From Basic Science to Translational Medicine. Eur. J. Immunol. 2023, 53, e2048980. [Google Scholar] [CrossRef] [PubMed]
- Ma, Z.; Sugimura, R.; Lui, K.O. The role of m6A mRNA modification in normal and malignant hematopoiesis. J. Leukoc Biol. 2024, 115, 100–115. [Google Scholar] [CrossRef]
- Zhang, C.; Chen, Y.; Sun, B.; Wang, L.; Yang, Y.; Ma, D.; Lv, J.; Heng, J.; Ding, Y.; Xue, Y.; et al. m(6)A modulates haematopoietic stem and progenitor cell specification. Nature 2017, 549, 273–276. [Google Scholar] [CrossRef]
- Gao, Y.; Zimmer, J.T.; Vasic, R.; Liu, C.; Gbyli, R.; Zheng, S.J.; Patel, A.; Liu, W.; Qi, Z.; Li, Y.; et al. ALKBH5 modulates hematopoietic stem and progenitor cell energy metabolism through m(6)A modification-mediated RNA stability control. Cell Rep. 2023, 42, 113163. [Google Scholar] [CrossRef]
- Caux, C.; Ramos, R.N.; Prendergast, G.C.; Bendriss-Vermare, N.; Ménétrier-Caux, C. A Milestone Review on How Macrophages Affect Tumor Growth. Cancer Res. 2016, 76, 6439–6442. [Google Scholar] [CrossRef]
- Oishi, Y.; Manabe, I. Macrophages in inflammation, repair and regeneration. Int. Immunol. 2018, 30, 511–528. [Google Scholar] [CrossRef]
- Yunna, C.; Mengru, H.; Lei, W.; Weidong, C. Macrophage M1/M2 polarization. Eur. J. Pharmacol. 2020, 877, 173090. [Google Scholar] [CrossRef]
- Liu, Y.; Liu, Z.; Tang, H.; Shen, Y.; Gong, Z.; Xie, N.; Zhang, X.; Wang, W.; Kong, W.; Zhou, Y.; et al. The N(6)-methyladenosine (m(6)A)-forming enzyme METTL3 facilitates M1 macrophage polarization through the methylation of STAT1 mRNA. Am. J. Physiol. Cell Physiol 2019, 317, C762–C775. [Google Scholar] [CrossRef] [PubMed]
- Yin, H.; Zhang, X.; Yang, P.; Zhang, X.; Peng, Y.; Li, D.; Yu, Y.; Wu, Y.; Wang, Y.; Zhang, J.; et al. RNA m6A methylation orchestrates cancer growth and metastasis via macrophage reprogramming. Nat. Commun. 2021, 12, 1394. [Google Scholar] [CrossRef] [PubMed]
- Dong, L.; Chen, C.; Zhang, Y.; Guo, P.; Wang, Z.; Li, J.; Liu, Y.; Liu, J.; Chang, R.; Li, Y.; et al. The loss of RNA N(6)-adenosine methyltransferase Mettl14 in tumor-associated macrophages promotes CD8(+) T cell dysfunction and tumor growth. Cancer Cell 2021, 39, 945–957.e10. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Q.; Hou, J.; Zhou, Y.; Li, Z.; Cao, X. The RNA helicase DDX46 inhibits innate immunity by entrapping m(6)A-demethylated antiviral transcripts in the nucleus. Nat. Immunol. 2017, 18, 1094–1103. [Google Scholar] [CrossRef]
- Han, D.; Liu, J.; Chen, C.; Dong, L.; Liu, Y.; Chang, R.; Huang, X.; Liu, Y.; Wang, J.; Dougherty, U.; et al. Anti-tumour immunity controlled through mRNA m(6)A methylation and YTHDF1 in dendritic cells. Nature 2019, 566, 270–274. [Google Scholar] [CrossRef]
- Takaba, H.; Takayanagi, H. The Mechanisms of T Cell Selection in the Thymus. Trends Immunol. 2017, 38, 805–816. [Google Scholar] [CrossRef]
- Germain, R.N. T-cell development and the CD4-CD8 lineage decision. Nat. Rev. Immunol. 2002, 2, 309–322. [Google Scholar] [CrossRef]
- Raza, I.G.A.; Clarke, A.J. B Cell Metabolism and Autophagy in Autoimmunity. Front. Immunol. 2021, 12, 681105. [Google Scholar] [CrossRef]
- Liu, X.S.; Zhou, L.M.; Yuan, L.L.; Gao, Y.; Kui, X.Y.; Liu, X.Y.; Pei, Z.J. NPM1 Is a Prognostic Biomarker Involved in Immune Infiltration of Lung Adenocarcinoma and Associated With m6A Modification and Glycolysis. Front. Immunol. 2021, 12, 724741. [Google Scholar] [CrossRef]
- Zhang, J.; Dong, Y.; Di, S.; Xie, S.; Fan, B.; Gong, T. Tumor associated macrophages in esophageal squamous carcinoma: Promising therapeutic implications. Biomed. Pharmacother. 2023, 167, 115610. [Google Scholar] [CrossRef]
- Guo, W.; Tan, F.; Huai, Q.; Wang, Z.; Shao, F.; Zhang, G.; Yang, Z.; Li, R.; Xue, Q.; Gao, S.; et al. Comprehensive Analysis of PD-L1 Expression, Immune Infiltrates, and m6A RNA Methylation Regulators in Esophageal Squamous Cell Carcinoma. Front. Immunol. 2021, 12, 669750. [Google Scholar] [CrossRef] [PubMed]
- Bian, Y.; Bi, G.; Shan, G.; Liang, J.; Yao, G.; Sui, Q.; Hu, Z.; Zhan, C.; Chen, Z.; Wang, Q. Identification of the relationship between single-cell N6-methyladenosine regulators and the infiltrating immune cells in esophageal carcinoma. Heliyon 2023, 9, e18132. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, S.; Mumbach, M.R.; Jovanovic, M.; Wang, T.; Maciag, K.; Bushkin, G.G.; Mertins, P.; Ter-Ovanesyan, D.; Habib, N.; Cacchiarelli, D.; et al. Perturbation of m6A writers reveals two distinct classes of mRNA methylation at internal and 5' sites. Cell Rep. 2014, 8, 284–296. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Wu, Q.; Li, B.; Wang, D.; Wang, L.; Zhou, Y.L. m(6)A regulator-mediated methylation modification patterns and tumor microenvironment infiltration characterization in gastric cancer. Mol. Cancer 2020, 19, 53. [Google Scholar] [CrossRef] [PubMed]
- Bai, X.; Wong, C.C.; Pan, Y.; Chen, H.; Liu, W.; Zhai, J.; Kang, W.; Shi, Y.; Yamamoto, M.; Tsukamoto, T.; et al. Loss of YTHDF1 in gastric tumors restores sensitivity to antitumor immunity by recruiting mature dendritic cells. J. Immunother. Cancer 2022, 10, e003663. [Google Scholar] [CrossRef]
- Liao, Q.; Xiong, J. YTHDF1 regulates immune cell infiltration in gastric cancer via interaction with p53. Exp. Ther. Med. 2024, 27, 255. [Google Scholar] [CrossRef]
- Ji, T.; Gao, X.; Li, D.; Huai, S.; Chi, Y.; An, X.; Ji, W.; Yang, S.; Li, J. Identification and validation of signature for prognosis and immune microenvironment in gastric cancer based on m6A demethylase ALKBH5. Front. Oncol. 2022, 12, 1079402. [Google Scholar] [CrossRef]
- Zhang, C.; Zhang, M.; Ge, S.; Huang, W.; Lin, X.; Gao, J.; Gong, J.; Shen, L. Reduced m6A modification predicts malignant phenotypes and augmented Wnt/PI3K-Akt signaling in gastric cancer. Cancer Med. 2019, 8, 4766–4781. [Google Scholar] [CrossRef]
- Yu, Y.; Meng, L.L.; Chen, X.Y.; Fan, H.N.; Chen, M.; Zhang, J.; Zhu, J.S. m(6)A reader YTHDF3 is associated with clinical prognosis, related RNA signatures and immunosuppression in gastric cancer. Cell. Signal. 2023, 108, 110699. [Google Scholar] [CrossRef]
- Chen, Y.; Zhao, Y.; Chen, J.; Peng, C.; Zhang, Y.; Tong, R.; Cheng, Q.; Yang, B.; Feng, X.; Lu, Y.; et al. ALKBH5 suppresses malignancy of hepatocellular carcinoma via m(6)A-guided epigenetic inhibition of LYPD1. Mol. Cancer 2020, 19, 123. [Google Scholar] [CrossRef] [PubMed]
- You, Y.; Wen, D.; Zeng, L.; Lu, J.; Xiao, X.; Chen, Y.; Song, H.; Liu, Z. ALKBH5/MAP3K8 axis regulates PD-L1+ macrophage infiltration and promotes hepatocellular carcinoma progression. Int. J. Biol. Sci. 2022, 18, 5001–5018. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Chen, Y.; Mao, Q.; Jiang, X.; Jiang, W.; Chen, J.; Xu, W.; Zhong, L.; Sun, X. Overexpression of YTHDF1 is associated with poor prognosis in patients with hepatocellular carcinoma. Cancer Biomark. 2018, 21, 859–868. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Li, J.; Feng, G.; Gao, S.; Wang, Y.; Zhang, S.; Liu, Y.; Ye, L.; Li, Y.; Zhang, X. MicroRNA-145 Modulates N(6)-Methyladenosine Levels by Targeting the 3'-Untranslated mRNA Region of the N(6)-Methyladenosine Binding YTH Domain Family 2 Protein. J. Biol. Chem. 2017, 292, 3614–3623. [Google Scholar] [CrossRef]
- Wang, L.; Peng, J.L. METTL5 serves as a diagnostic and prognostic biomarker in hepatocellular carcinoma by influencing the immune microenvironment. Sci. Rep. 2023, 13, 10755. [Google Scholar] [CrossRef]
- Xia, P.; Zhang, H.; Lu, H.; Xu, K.; Jiang, X.; Jiang, Y.; Gongye, X.; Chen, Z.; Liu, J.; Chen, X.; et al. METTL5 stabilizes c-Myc by facilitating USP5 translation to reprogram glucose metabolism and promote hepatocellular carcinoma progression. Cancer Commun. 2023, 43, 338–364. [Google Scholar] [CrossRef]
- Peng, H.; Chen, B.; Wei, W.; Guo, S.; Han, H.; Yang, C.; Ma, J.; Wang, L.; Peng, S.; Kuang, M.; et al. N(6)-methyladenosine (m(6)A) in 18S rRNA promotes fatty acid metabolism and oncogenic transformation. Nat. Metab. 2022, 4, 1041–1054. [Google Scholar] [CrossRef]
- Song, Z.; Wang, X.; Chen, F.; Chen, Q.; Liu, W.; Yang, X.; Zhu, X.; Liu, X.; Wang, P. LncRNA MALAT1 regulates METTL3-mediated PD-L1 expression and immune infiltrates in pancreatic cancer. Front. Oncol. 2022, 12, 1004212. [Google Scholar] [CrossRef]
- Li, L.; Wang, F.; Deng, Z.; Zhang, G.; Zhu, L.; Zhao, Z.; Liu, R. DCLRE1B promotes tumor progression and predicts immunotherapy response through METTL3-mediated m6A modification in pancreatic cancer. BMC Cancer 2023, 23, 1073. [Google Scholar] [CrossRef]
- Lu, L.; Zheng, D.; Qu, J.; Zhuang, Y.; Peng, J.; Lan, S.; Zhang, S.; Huang, F. METTL16 predicts a favorable outcome and primes antitumor immunity in pancreatic ductal adenocarcinoma. Front. Cell Dev. Biol. 2022, 10, 759020. [Google Scholar] [CrossRef]
- Li, T.; Tan, Y.T.; Chen, Y.X.; Zheng, X.J.; Wang, W.; Liao, K.; Mo, H.Y.; Lin, J.; Yang, W.; Piao, H.L.; et al. Methionine deficiency facilitates antitumour immunity by altering m(6)A methylation of immune checkpoint transcripts. Gut 2023, 72, 501–511. [Google Scholar] [CrossRef]
- Bao, Y.; Zhai, J.; Chen, H.; Wong, C.C.; Liang, C.; Ding, Y.; Huang, D.; Gou, H.; Chen, D.; Pan, Y.; et al. Targeting m(6)A reader YTHDF1 augments antitumour immunity and boosts anti-PD-1 efficacy in colorectal cancer. Gut 2023, 72, 1497–1509. [Google Scholar] [CrossRef]
- Wang, L.; Hui, H.; Agrawal, K.; Kang, Y.; Li, N.; Tang, R.; Yuan, J.; Rana, T.M. m(6) A RNA methyltransferases METTL3/14 regulate immune responses to anti-PD-1 therapy. EMBO J. 2020, 39, e104514. [Google Scholar] [CrossRef]
- Ge, J.; Liu, S.L.; Zheng, J.X.; Shi, Y.; Shao, Y.; Duan, Y.J.; Huang, R.; Yang, L.J.; Yang, T. RNA demethylase ALKBH5 suppresses tumorigenesis via inhibiting proliferation and invasion and promoting CD8(+) T cell infiltration in colorectal cancer. Transl. Oncol. 2023, 34, 101683. [Google Scholar] [CrossRef]
- Wu, T.; Zhang, X.; Xing, L.; Pan, D.; Liu, P.; Ding, R.; Yang, R.; Yang, X.; Li, Y. Abnormal Expression of N6-Methyladenosine RNA Methylation Regulator IGF2BP3 in Colon Cancer Predicts a Poor Prognosis. Dis. Markers 2022, 2022, 5883101. [Google Scholar] [CrossRef] [PubMed]
- Ji, J.; Liu, S.; Liang, Y.; Zheng, G. Comprehensive analysis of m6A regulators and relationship with tumor microenvironment, immunotherapy strategies in colorectal adenocarcinoma. BMC Genom. Data 2023, 24, 44. [Google Scholar] [CrossRef]
- Zhang, J.; Yang, K.; Bu, J.; Yan, J.; Hu, X.; Liu, K.; Gao, S.; Tang, S.; Gao, L.; Chen, W. IGF2BP3 promotes progression of gallbladder carcinoma by stabilizing KLK5 mRNA in N(6)-methyladenosine-dependent binding. Front. Oncol. 2022, 12, 1035871. [Google Scholar] [CrossRef]
- Ke, J.; Cui, J.; Yang, X.; Du, X.; Ma, B.; Yu, L. [Comprehensive Analysis of the Relationship between m6A Methylation Patterns and Immune Microenvironment in Lung Adenocarcinoma]. Zhongguo Fei Ai Za Zhi 2022, 25, 311–322. [Google Scholar] [CrossRef]
- Wu, L.; Cheng, D.; Yang, X.; Zhao, W.; Fang, C.; Chen, R.; Ji, M. M2-TAMs promote immunoresistance in lung adenocarcinoma by enhancing METTL3-mediated m6A methylation. Ann. Transl. Med. 2022, 10, 1380. [Google Scholar] [CrossRef]
- Kong, F.; Yang, X.; Lu, Z.; Liu, Z.; Yang, Y.; Wang, Z. A novel long noncoding RNA (lncRNA), LINC02657(LASTR), is a prognostic biomarker associated with immune infiltrates of lung adenocarcinoma based on unsupervised cluster analysis. PeerJ 2023, 11, e16167. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; Xu, Y.; Jin, L.; Wang, X.; Wu, S.; Wang, Y.; Zhao, J.; Zhou, F.; Ge, H. Effects of N6-Methyladenosine Regulators on LAG3 and Immune Infiltrates in Lung Adenocarcinoma. Dis. Markers 2022, 2022, 1829528. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; He, J.; Han, J.; Yang, J.; Liao, W.; Chen, N. m6A Regulators Mediated Methylation Modification Patterns and Tumor Microenvironment Infiltration Characterization In Nasopharyngeal Carcinoma. Front. Immunol. 2021, 12, 762243. [Google Scholar] [CrossRef] [PubMed]
- Lu, M.; Zhang, Z.; Xue, M.; Zhao, B.S.; Harder, O.; Li, A.; Liang, X.; Gao, T.Z.; Xu, Y.; Zhou, J.; et al. N(6)-methyladenosine modification enables viral RNA to escape recognition by RNA sensor RIG-I. Nat. Microbiol. 2020, 5, 584–598. [Google Scholar] [CrossRef] [PubMed]
- Ning, J.; Hou, X.; Hao, J.; Zhang, W.; Shi, Y.; Huang, Y.; Ruan, X.; Zheng, X.; Gao, M. METTL3 inhibition induced by M2 macrophage-derived extracellular vesicles drives anti-PD-1 therapy resistance via M6A-CD70-mediated immune suppression in thyroid cancer. Cell Death Differ. 2023, 30, 2265–2279. [Google Scholar] [CrossRef]
- Yuan, F.; Cai, X.; Wang, Y.; Du, C.; Cong, Z.; Zeng, X.; Tang, C.; Ma, C. Comprehensive analysis of m(6)A subtype classification for immune microenvironment of pituitary adenomas. Int. Immunopharmacol. 2023, 124, 110784. [Google Scholar] [CrossRef]
- Lin, S.; Xu, H.; Zhang, A.; Ni, Y.; Xu, Y.; Meng, T.; Wang, M.; Lou, M. Prognosis Analysis and Validation of m(6)A Signature and Tumor Immune Microenvironment in Glioma. Front. Oncol. 2020, 10, 541401. [Google Scholar] [CrossRef]
- Tang, W.; Xu, N.; Zhou, J.; He, Z.; Lenahan, C.; Wang, C.; Ji, H.; Liu, B.; Zou, Y.; Zeng, H.; et al. ALKBH5 promotes PD-L1-mediated immune escape through m6A modification of ZDHHC3 in glioma. Cell Death Discov. 2022, 8, 497. [Google Scholar] [CrossRef]
- Wang, L.M.; Englander, Z.K.; Miller, M.L.; Bruce, J.N. Malignant Glioma. Adv. Exp. Med. Biol. 2023, 1405, 1–30. [Google Scholar] [CrossRef]
- Zhao, R.; Li, B.; Zhang, S.; He, Z.; Pan, Z.; Guo, Q.; Qiu, W.; Qi, Y.; Zhao, S.; Wang, S.; et al. The N(6)-Methyladenosine-Modified Pseudogene HSPA7 Correlates With the Tumor Microenvironment and Predicts the Response to Immune Checkpoint Therapy in Glioblastoma. Front. Immunol. 2021, 12, 653711. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Zhong, J.; Zeng, J.; Duan, X.; Lu, J.; Sun, X.; Liu, Q.; Liang, Y.; Lin, Z.; Zhong, W.; et al. Characterization of the m6A-Associated Tumor Immune Microenvironment in Prostate Cancer to Aid Immunotherapy. Front. Immunol. 2021, 12, 735170. [Google Scholar] [CrossRef]
- Cheng, Y.; Li, L.; Wei, X.; Xu, F.; Huang, X.; Qi, F.; Zhang, Y.; Li, X. HNRNPC suppresses tumor immune microenvironment by activating Treg cells promoting the progression of prostate cancer. Cancer Sci. 2023, 114, 1830–1845. [Google Scholar] [CrossRef]
- Jiang, Z.; Zhang, W.; Zeng, Z.; Tang, D.; Li, C.; Cai, W.; Chen, Y.; Li, Y.; Jin, Q.; Zhang, X.; et al. A comprehensive investigation discovered the novel methyltransferase METTL24 as one presumably prognostic gene for kidney renal clear cell carcinoma potentially modulating tumor immune microenvironment. Front. Immunol. 2022, 13, 926461. [Google Scholar] [CrossRef]
- Adelaja, A.; Taylor, B.; Sheu, K.M.; Liu, Y.; Luecke, S.; Hoffmann, A. Six distinct NFkappaB signaling codons convey discrete information to distinguish stimuli and enable appropriate macrophage responses. Immunity 2021, 54, 916–930.e7. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Xue, V.W.; Wang, Q.M.; Lian, G.Y.; Huang, X.R.; Lee, T.L.; To, K.F.; Tang, P.M.; Lan, H.Y. The Mincle/Syk/NF-kappaB Signaling Circuit Is Essential for Maintaining the Protumoral Activities of Tumor-Associated Macrophages. Cancer Immunol. Res. 2020, 8, 1004–1017. [Google Scholar] [CrossRef] [PubMed]
- Goto, Y.; Sakamoto, S.; Ichikawa, T. [Molecular biology and genetics of prostate cancer]. Nihon Rinsho 2016, 74 (Suppl. 3), 55–59. [Google Scholar]
- Sekhoacha, M.; Riet, K.; Motloung, P.; Gumenku, L.; Adegoke, A.; Mashele, S. Prostate Cancer Review: Genetics, Diagnosis, Treatment Options, and Alternative Approaches. Molecules 2022, 27, 5730. [Google Scholar] [CrossRef]
- Yu, L.; Sun, H.; Che, M.; Gai, W. Observation of Serological Index and Efficacy of Abiraterone Hydrochloride Tablets Combined with Endocrine Therapy in Patients with Metastatic Castration-Resistant Prostate Cancer. Arch. Esp. Urol. 2023, 76, 588–595. [Google Scholar] [CrossRef] [PubMed]
- Wei, Z.; Chen, Y.; Zeng, Z.; Peng, Y.; Li, L.; Hu, N.; Gao, X.; Cai, W.; Yin, L.; Xu, Y.; et al. The novel m6A writer METTL5 as prognostic biomarker probably associating with the regulation of immune microenvironment in kidney cancer. Heliyon 2022, 8, e12078. [Google Scholar] [CrossRef]
- Shao, Y.; Wu, B.; Yang, Z.; Liu, Z.; Ma, Y.; Huang, H.; Liu, Y.; Wang, Z.; Hu, W.; Wang, Y.; et al. ALDOB represents a potential prognostic biomarker for patients with clear cell renal cell carcinoma. Transl. Androl. Urol. 2023, 12, 549–571. [Google Scholar] [CrossRef]
- Zhao, J.; Huang, S.; Tan, D.; Yang, K.; Chen, M.; Jia, X.; Mao, X. PGM1 and ENO1 Promote the Malignant Progression of Bladder Cancer via Comprehensive Analysis of the m6A Signature and Tumor Immune Infiltration. J. Oncol. 2022, 2022, 8581805. [Google Scholar] [CrossRef]
- Ji, H.; Zhang, J.A.; Liu, H.; Li, K.; Wang, Z.W.; Zhu, X. Comprehensive characterization of tumor microenvironment and m6A RNA methylation regulators and its effects on PD-L1 and immune infiltrates in cervical cancer. Front. Immunol. 2022, 13, 976107. [Google Scholar] [CrossRef]
- Cai, X.; Liang, C.; Zhang, M.; Xu, Y.; Weng, Y.; Li, X.; Yu, W. N6-methyladenosine modification and metabolic reprogramming of digestive system malignancies. Cancer Lett. 2022, 544, 215815. [Google Scholar] [CrossRef] [PubMed]
- Weng, H.; Huang, F.; Yu, Z.; Chen, Z.; Prince, E.; Kang, Y.; Zhou, K.; Li, W.; Hu, J.; Fu, C.; et al. The m(6)A reader IGF2BP2 regulates glutamine metabolism and represents a therapeutic target in acute myeloid leukemia. Cancer Cell 2022, 40, 1566–1582.e10. [Google Scholar] [CrossRef] [PubMed]
- Ni, H.H.; Zhang, L.; Huang, H.; Dai, S.Q.; Li, J. Connecting METTL3 and intratumoural CD33(+) MDSCs in predicting clinical outcome in cervical cancer. J. Transl. Med. 2020, 18, 393. [Google Scholar] [CrossRef]
- Ma, J.; Yang, D.; Ma, X.X. Immune infiltration-related N6-methyladenosine RNA methylation regulators influence the malignancy and prognosis of endometrial cancer. Aging 2021, 13, 16287–16315. [Google Scholar] [CrossRef]
- Wang, B.; Mao, Z.; Ye, J.; Jiao, X.; Zhang, T.; Wang, Q.; Han, S.; Zhang, Y.; Wang, C.; Dong, T.; et al. Glycolysis Induced by METTL14 Is Essential for Macrophage Phagocytosis and Phenotype in Cervical Cancer. J. Immunol. 2024, 212, 723–736. [Google Scholar] [CrossRef]
- Li, Y.; Li, J.; Yu, Q.; Ji, L.; Peng, B. METTL14 regulates microglia/macrophage polarization and NLRP3 inflammasome activation after ischemic stroke by the KAT3B-STING axis. Neurobiol. Dis. 2023, 185, 106253. [Google Scholar] [CrossRef] [PubMed]
- Feng, Q.; Xu, D.; Zhou, M.; Wu, Z.; Wu, Z.; Wang, Z.; Bi, J.; Pei, W. CDC42EP3 promotes colorectal cancer through regulating cell proliferation, cell apoptosis and cell migration. Cancer Cell Int. 2021, 21, 169. [Google Scholar] [CrossRef]
- Maffei, R.; Bulgarelli, J.; Fiorcari, S.; Bertoncelli, L.; Martinelli, S.; Guarnotta, C.; Castelli, I.; Deaglio, S.; Debbia, G.; De Biasi, S.; et al. The monocytic population in chronic lymphocytic leukemia shows altered composition and deregulation of genes involved in phagocytosis and inflammation. Haematologica 2013, 98, 1115–1123. [Google Scholar] [CrossRef]
- Hu, Y.; Pan, Q.; Wang, M.; Ai, X.; Yan, Y.; Tian, Y.; Jing, Y.; Tang, P.; Jiang, J. m(6)A RNA Methylation Regulator YTHDF1 Correlated With Immune Microenvironment Predicts Clinical Outcomes and Therapeutic Efficacy in Breast Cancer. Front. Med. 2021, 8, 667543. [Google Scholar] [CrossRef]
- Lian, B.; Yan, S.; Li, J.; Bai, Z.; Li, J. HNRNPC promotes collagen fiber alignment and immune evasion in breast cancer via activation of the VIRMA-mediated TFAP2A/DDR1 axis. Mol. Med. 2023, 29, 103. [Google Scholar] [CrossRef]
- Yang, S.; Wei, J.; Cui, Y.H.; Park, G.; Shah, P.; Deng, Y.; Aplin, A.E.; Lu, Z.; Hwang, S.; He, C.; et al. m(6)A mRNA demethylase FTO regulates melanoma tumorigenicity and response to anti-PD-1 blockade. Nat. Commun. 2019, 10, 2782. [Google Scholar] [CrossRef]
- Wang, G.; Zeng, D.; Sweren, E.; Miao, Y.; Chen, R.; Chen, J.; Wang, J.; Liao, W.; Hu, Z.; Kang, S.; et al. N6-methyladenosine RNA Methylation Correlates with Immune Microenvironment and Immunotherapy Response of Melanoma. J. Investig. Dermatol. 2023, 143, 1579–1590.e5. [Google Scholar] [CrossRef]
- Fujimura, T.; Kambayashi, Y.; Aiba, S. Crosstalk between regulatory T cells (Tregs) and myeloid derived suppressor cells (MDSCs) during melanoma growth. Oncoimmunology 2012, 1, 1433–1434. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.; Skora, A.D.; Li, Z.; Liu, Q.; Tam, A.J.; Blosser, R.L.; Diaz, L.A., Jr.; Papadopoulos, N.; Kinzler, K.W.; Vogelstein, B.; et al. Eradication of metastatic mouse cancers resistant to immune checkpoint blockade by suppression of myeloid-derived cells. Proc. Natl. Acad. Sci. USA 2014, 111, 11774–11779. [Google Scholar] [CrossRef] [PubMed]
- Liao, X.; Chen, L.; Liu, J.; Hu, H.; Hou, D.; You, R.; Wang, X.; Huang, H. m(6)A RNA methylation regulators predict prognosis and indicate characteristics of tumour microenvironment infiltration in acute myeloid leukaemia. Epigenetics 2023, 18, 2160134. [Google Scholar] [CrossRef] [PubMed]
- Waters, J.K.; Reznik, S.I. Update on Management of Squamous Cell Esophageal Cancer. Curr. Oncol. Rep. 2022, 24, 375–385. [Google Scholar] [CrossRef]
- Huang, F.L.; Yu, S.J. Esophageal cancer: Risk factors, genetic association, and treatment. Asian J. Surg. 2018, 41, 210–215. [Google Scholar] [CrossRef]
- He, C.; Teng, X.; Wang, L.; Ni, M.; Zhu, L.; Liu, J.; Lv, W.; Hu, J. The implications of N6-methyladenosine (m6A) modification in esophageal carcinoma. Mol. Biol. Rep. 2023, 50, 8691–8703. [Google Scholar] [CrossRef]
- Jin, C.; Gao, J.; Zhu, J.; Ao, Y.; Shi, B.; Li, X. Exosomal NAT10 from esophageal squamous cell carcinoma cells modulates macrophage lipid metabolism and polarization through ac4C modification of FASN. Transl. Oncol. 2024, 45, 101934. [Google Scholar] [CrossRef]
- Sexton, R.E.; Al Hallak, M.N.; Diab, M.; Azmi, A.S. Gastric cancer: A comprehensive review of current and future treatment strategies. Cancer Metastasis Rev. 2020, 39, 1179–1203. [Google Scholar] [CrossRef] [PubMed]
- Machlowska, J.; Baj, J.; Sitarz, M.; Maciejewski, R.; Sitarz, R. Gastric Cancer: Epidemiology, Risk Factors, Classification, Genomic Characteristics and Treatment Strategies. Int. J. Mol. Sci. 2020, 21, 4012. [Google Scholar] [CrossRef]
- Chen, X.Y.; Liang, R.; Yi, Y.C.; Fan, H.N.; Chen, M.; Zhang, J.; Zhu, J.S. The m(6)A Reader YTHDF1 Facilitates the Tumorigenesis and Metastasis of Gastric Cancer via USP14 Translation in an m(6)A-Dependent Manner. Front. Cell Dev. Biol. 2021, 9, 647702. [Google Scholar] [CrossRef] [PubMed]
- Ge, L.; Zhang, N.; Chen, Z.; Song, J.; Wu, Y.; Li, Z.; Chen, F.; Wu, J.; Li, D.; Li, J.; et al. Level of N6-Methyladenosine in Peripheral Blood RNA: A Novel Predictive Biomarker for Gastric Cancer. Clin. Chem. 2020, 66, 342–351. [Google Scholar] [CrossRef]
- Zhang, J.; Guo, S.; Piao, H.Y.; Wang, Y.; Wu, Y.; Meng, X.Y.; Yang, D.; Zheng, Z.C.; Zhao, Y. ALKBH5 promotes invasion and metastasis of gastric cancer by decreasing methylation of the lncRNA NEAT1. J. Physiol. Biochem. 2019, 75, 379–389. [Google Scholar] [CrossRef]
- Suo, D.; Gao, X.; Chen, Q.; Zeng, T.; Zhan, J.; Li, G.; Zheng, Y.; Zhu, S.; Yun, J.; Guan, X.Y.; et al. HSPA4 upregulation induces immune evasion via ALKBH5/CD58 axis in gastric cancer. J. Exp. Clin. Cancer Res. 2024, 43, 106. [Google Scholar] [CrossRef] [PubMed]
- Anwanwan, D.; Singh, S.K.; Singh, S.; Saikam, V.; Singh, R. Challenges in liver cancer and possible treatment approaches. Biochim. Biophys. Acta Rev. Cancer 2020, 1873, 188314. [Google Scholar] [CrossRef]
- Chen, M.; Wong, C.M. The emerging roles of N6-methyladenosine (m6A) deregulation in liver carcinogenesis. Mol. Cancer 2020, 19, 44. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.; Liu, B.; Nie, Z.; Duan, L.; Xiong, Q.; Jin, Z.; Yang, C.; Chen, Y. The role of m6A modification in the biological functions and diseases. Signal. Transduct. Target. Ther. 2021, 6, 74. [Google Scholar] [CrossRef]
- Yang, Q.; Liang, Y.; Shi, Y.; Shang, J.; Huang, X. The ALKBH5/SOX4 axis promotes liver cancer stem cell properties via activating the SHH signaling pathway. J. Cancer Res. Clin. Oncol. 2023, 149, 15499–15510. [Google Scholar] [CrossRef]
- Qiu, X.; Yang, S.; Wang, S.; Wu, J.; Zheng, B.; Wang, K.; Shen, S.; Jeong, S.; Li, Z.; Zhu, Y.; et al. M(6)A Demethylase ALKBH5 Regulates PD-L1 Expression and Tumor Immunoenvironment in Intrahepatic Cholangiocarcinoma. Cancer Res. 2021, 81, 4778–4793. [Google Scholar] [CrossRef]
- Zhang, L.; Dou, X.; Zheng, Z.; Ye, C.; Lu, T.X.; Liang, H.L.; Wang, L.; Weichselbaum, R.R.; He, C. YTHDF2/m(6) A/NF-kappaB axis controls anti-tumor immunity by regulating intratumoral Tregs. EMBO J. 2023, 42, e113126. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Liu, S.; Zhang, G.; Liu, J.; Cao, G. Knockdown of METTL5 inhibits the Myc pathway to downregulate PD-L1 expression and inhibits immune escape of hepatocellular carcinoma cells. J. Chemother. 2023, 35, 455–464. [Google Scholar] [CrossRef]
- Kolbeinsson, H.M.; Chandana, S.; Wright, G.P.; Chung, M. Pancreatic Cancer: A Review of Current Treatment and Novel Therapies. J. Investig. Surg. 2023, 36, 2129884. [Google Scholar] [CrossRef] [PubMed]
- Michot, J.M.; Bigenwald, C.; Champiat, S.; Collins, M.; Carbonnel, F.; Postel-Vinay, S.; Berdelou, A.; Varga, A.; Bahleda, R.; Hollebecque, A.; et al. Immune-related adverse events with immune checkpoint blockade: A comprehensive review. Eur. J. Cancer 2016, 54, 139–148. [Google Scholar] [CrossRef]
- Zou, T.; Shi, D.; Wang, W.; Chen, G.; Zhang, X.; Tian, Y.; Gong, P. Identification of a New m6A Regulator-Related Methylation Signature for Predicting the Prognosis and Immune Microenvironment of Patients with Pancreatic Cancer. Mediat. Inflamm. 2023, 2023, 5565054. [Google Scholar] [CrossRef]
- Baidoun, F.; Elshiwy, K.; Elkeraie, Y.; Merjaneh, Z.; Khoudari, G.; Sarmini, M.T.; Gad, M.; Al-Husseini, M.; Saad, A. Colorectal Cancer Epidemiology: Recent Trends and Impact on Outcomes. Curr. Drug Targets 2021, 22, 998–1009. [Google Scholar] [CrossRef]
- Xiong, J.; He, J.; Zhu, J.; Pan, J.; Liao, W.; Ye, H.; Wang, H.; Song, Y.; Du, Y.; Cui, B.; et al. Lactylation-driven METTL3-mediated RNA m(6)A modification promotes immunosuppression of tumor-infiltrating myeloid cells. Mol. Cell 2022, 82, 1660–1677.e10. [Google Scholar] [CrossRef]
- Li, N.; Kang, Y.; Wang, L.; Huff, S.; Tang, R.; Hui, H.; Agrawal, K.; Gonzalez, G.M.; Wang, Y.; Patel, S.P.; et al. ALKBH5 regulates anti-PD-1 therapy response by modulating lactate and suppressive immune cell accumulation in tumor microenvironment. Proc. Natl. Acad. Sci. USA 2020, 117, 20159–20170. [Google Scholar] [CrossRef]
- Belharazem, D.; Magdeburg, J.; Berton, A.K.; Beissbarth, L.; Sauer, C.; Sticht, C.; Marx, A.; Hofheinz, R.; Post, S.; Kienle, P.; et al. Carcinoma of the colon and rectum with deregulation of insulin-like growth factor 2 signaling: Clinical and molecular implications. J. Gastroenterol. 2016, 51, 971–984. [Google Scholar] [CrossRef] [PubMed]
- Lochhead, P.; Imamura, Y.; Morikawa, T.; Kuchiba, A.; Yamauchi, M.; Liao, X.; Qian, Z.R.; Nishihara, R.; Wu, K.; Meyerhardt, J.A.; et al. Insulin-like growth factor 2 messenger RNA binding protein 3 (IGF2BP3) is a marker of unfavourable prognosis in colorectal cancer. Eur. J. Cancer 2012, 48, 3405–3413. [Google Scholar] [CrossRef]
- Goetze, T.O. Gallbladder carcinoma: Prognostic factors and therapeutic options. World J. Gastroenterol. 2015, 21, 12211–12217. [Google Scholar] [CrossRef]
- Bai, X.; Chen, J.; Zhang, W.; Zhou, S.; Dong, L.; Huang, J.; He, X. YTHDF2 promotes gallbladder cancer progression and gemcitabine resistance via m6A-dependent DAPK3 degradation. Cancer Sci. 2023, 114, 4299–4313. [Google Scholar] [CrossRef]
- Qin, J.; Cui, Z.; Zhou, J.; Zhang, B.; Lu, R.; Ding, Y.; Hu, H.; Cai, J. IGF2BP3 drives gallbladder cancer progression by m6A-modified CLDN4 and inducing macrophage immunosuppressive polarization. Transl. Oncol. 2023, 37, 101764. [Google Scholar] [CrossRef] [PubMed]
- Hutchinson, B.D.; Shroff, G.S.; Truong, M.T.; Ko, J.P. Spectrum of Lung Adenocarcinoma. Semin. Ultrasound. CT MR 2019, 40, 255–264. [Google Scholar] [CrossRef]
- Saar, M.; Lavogina, D.; Lust, H.; Tamm, H.; Jaal, J. Immune checkpoint inhibitors modulate the cytotoxic effect of chemotherapy in lung adenocarcinoma cells. Oncol. Lett. 2023, 25, 152. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Liu, W.K.; Du, X.W.; Liu, X.L.; Li, G.; Yao, Y.; Han, T.; Li, W.Y.; Gu, J. Large-scale transcriptome analysis identified RNA methylation regulators as novel prognostic signatures for lung adenocarcinoma. Ann. Transl. Med. 2020, 8, 751. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Gu, X.; Xu, D.; Liu, B.; Qin, K.; Yuan, X. Comprehensive analysis of m6A modification patterns and m6A-related lncRNAs as potential biomarkers in lung adenocarcinoma. Env. Toxicol. 2024, 39, 2285–2303. [Google Scholar] [CrossRef]
- Hou, Q.; Zhong, Y.; Liu, L.; Wu, L.; Liu, J. Construction of a lung adenocarcinoma prognostic model based on N6-methyl-adenosine-related long noncoding RNA and screening of potential drugs based on this model. Anticancer Drugs 2022, 33, 371–383. [Google Scholar] [CrossRef]
- Shen, H.Y.; Zhang, J.; Xu, D.; Xu, Z.; Liang, M.X.; Chen, W.Q.; Tang, J.H.; Xia, W.J. Construction of an m6A-related lncRNA model for predicting prognosis and immunotherapy in patients with lung adenocarcinoma. Medicine 2023, 102, e33530. [Google Scholar] [CrossRef]
- Chang, E.T.; Ye, W.; Zeng, Y.X.; Adami, H.O. The Evolving Epidemiology of Nasopharyngeal Carcinoma. Cancer Epidemiol. Biomark. Prev. 2021, 30, 1035–1047. [Google Scholar] [CrossRef]
- Juarez-Vignon Whaley, J.J.; Afkhami, M.; Onyshchenko, M.; Massarelli, E.; Sampath, S.; Amini, A.; Bell, D.; Villaflor, V.M. Recurrent/Metastatic Nasopharyngeal Carcinoma Treatment from Present to Future: Where Are We and Where Are We Heading? Curr. Treat. Options Oncol. 2023, 24, 1138–1166. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Han, J.; Zhen, X.; Liu, Y.; Cui, Z.; Yue, Z.; Ding, L.; Xu, S. Analysis of Genetic Alteration Signatures and Prognostic Values of m6A Regulatory Genes in Head and Neck Squamous Cell Carcinoma. Front. Oncol. 2020, 10, 718. [Google Scholar] [CrossRef]
- Jin, S.; Li, M.; Chang, H.; Wang, R.; Zhang, Z.; Zhang, J.; He, Y.; Ma, H. The m6A demethylase ALKBH5 promotes tumor progression by inhibiting RIG-I expression and interferon alpha production through the IKKepsilon/TBK1/IRF3 pathway in head and neck squamous cell carcinoma. Mol. Cancer 2022, 21, 97. [Google Scholar] [CrossRef] [PubMed]
- Nabhan, F.; Dedhia, P.H.; Ringel, M.D. Thyroid cancer, recent advances in diagnosis and therapy. Int. J. Cancer 2021, 149, 984–992. [Google Scholar] [CrossRef]
- Zhou, X.; Chang, L.; Liang, Q.; Zhao, R.; Xiao, Y.; Xu, Z.; Yu, L. The m6A methyltransferase METTL3 drives thyroid cancer progression and lymph node metastasis by targeting LINC00894. Cancer Cell Int. 2024, 24, 47. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Chen, Y.; Chen, R.; Zhou, H.; Lin, Y.; Li, B.; Song, H.; Zhou, G.; Dong, M.; Xu, H. YTHDF3as a prognostic predictive biomarker of thyroid cancer and its correlation with immune infiltration. BMC Cancer 2023, 23, 882. [Google Scholar] [CrossRef]
- Tritos, N.A.; Miller, K.K. Diagnosis and Management of Pituitary Adenomas: A Review. JAMA 2023, 329, 1386–1398. [Google Scholar] [CrossRef]
- Chang, M.; Wang, Z.; Gao, J.; Yang, C.; Feng, M.; Niu, Y.; Tong, W.M.; Bao, X.; Wang, R. METTL3-mediated RNA m6A Hypermethylation Promotes Tumorigenesis and GH Secretion of Pituitary Somatotroph Adenomas. J. Clin. Endocrinol. Metab. 2022, 107, 136–149. [Google Scholar] [CrossRef]
- Davis, M.E. Glioblastoma: Overview of Disease and Treatment. Clin. J. Oncol. Nurs. 2016, 20, S2–S8. [Google Scholar] [CrossRef]
- Zhang, S.; Zhao, B.S.; Zhou, A.; Lin, K.; Zheng, S.; Lu, Z.; Chen, Y.; Sulman, E.P.; Xie, K.; Bogler, O.; et al. m(6)A Demethylase ALKBH5 Maintains Tumorigenicity of Glioblastoma Stem-like Cells by Sustaining FOXM1 Expression and Cell Proliferation Program. Cancer Cell 2017, 31, 591–606.e6. [Google Scholar] [CrossRef]
- Burr, M.L.; Sparbier, C.E.; Chan, Y.C.; Williamson, J.C.; Woods, K.; Beavis, P.A.; Lam, E.Y.N.; Henderson, M.A.; Bell, C.C.; Stolzenburg, S.; et al. CMTM6 maintains the expression of PD-L1 and regulates anti-tumour immunity. Nature 2017, 549, 101–105. [Google Scholar] [CrossRef] [PubMed]
- Demuynck, R.; Efimova, I.; Naessens, F.; Krysko, D.V. Immunogenic ferroptosis and where to find it? J. Immunother. Cancer 2021, 9, e003430. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Lu, Z.R. Molecular imaging of the tumor microenvironment. Adv. Drug Deliv. Rev. 2017, 113, 24–48. [Google Scholar] [CrossRef]
- Wu, H.; He, H.; Huang, J.; Wang, C.; Dong, Y.; Lin, R.; Cheng, Z.; Qiu, Q.; Hong, L. Identification and validation of transferrin receptor protein 1 for predicting prognosis and immune infiltration in lower grade glioma. Front. Mol. Neurosci. 2022, 15, 972308. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Zhao, D.; Spring, D.J.; DePinho, R.A. Genetics and biology of prostate cancer. Genes Dev. 2018, 32, 1105–1140. [Google Scholar] [CrossRef]
- Zhao, Y.; Sun, H.; Zheng, J.; Shao, C. Analysis of RNA m(6)A methylation regulators and tumour immune cell infiltration characterization in prostate cancer. Artif. Cells Nanomed. Biotechnol. 2021, 49, 407–435. [Google Scholar] [CrossRef]
- Liu, Y.; Qiu, S.; Sun, D.; Xiong, T.; Xiang, Q.; Li, Q. Construction of a Comprehensive Diagnostic Scoring Model for Prostate Cancer Based on a Novel Six-Gene Panel. Front. Genet. 2022, 13, 831162. [Google Scholar] [CrossRef]
- Wang, S.; Xu, G.; Chao, F.; Zhang, C.; Han, D.; Chen, G. HNRNPC Promotes Proliferation, Metastasis and Predicts Prognosis in Prostate Cancer. Cancer Manag. Res. 2021, 13, 7263–7276. [Google Scholar] [CrossRef]
- Bahadoram, S.; Davoodi, M.; Hassanzadeh, S.; Bahadoram, M.; Barahman, M.; Mafakher, L. Renal cell carcinoma: An overview of the epidemiology, diagnosis, and treatment. G Ital. Nefrol. 2022, 39, 2022-vol3. [Google Scholar]
- Zheng, B.; Cheng, F.; Yao, Z.; Zhang, Y.; Cong, Z.; Wang, J.; Niu, Z.; He, W. Identification of New m(6)A Methylation Modification Patterns and Tumor Microenvironment Infiltration Landscape that Predict Clinical Outcomes for Papillary Renal Cell Carcinoma Patients. Front. Cell Dev. Biol. 2022, 10, 818194. [Google Scholar] [CrossRef]
- Shao, Y.; Li, W.; Zhang, L.; Xue, B.; Chen, Y.; Zhang, Z.; Wang, D.; Wu, B. CDH13 is a prognostic biomarker and a potential therapeutic target for patients with clear cell renal cell carcinoma. Am. J. Cancer Res. 2022, 12, 4520–4544. [Google Scholar]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Fontugne, J.; Xylinas, E.; Krucker, C.; Dixon, V.; Groeneveld, C.S.; Pinar, U.; Califano, G.; Bucau, M.; Verine, J.; Desgrandchamps, F.; et al. Transcriptomic Profiling of Upper Tract Urothelial Carcinoma: Bladder Cancer Consensus Classification Relevance, Molecular Heterogeneity, and Differential Immune Signatures. Mod. Pathol. 2023, 36, 100300. [Google Scholar] [CrossRef] [PubMed]
- Schwarzova, L.; Varchulova Novakova, Z.; Danisovic, L.; Ziaran, S. Molecular classification of urothelial bladder carcinoma. Mol. Biol. Rep. 2023, 50, 7867–7877. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Beltran, A.; Blanca, A.; Cimadamore, A.; Gogna, R.; Montironi, R.; Cheng, L. Molecular Classification of Bladder Urothelial Carcinoma Using NanoString-Based Gene Expression Analysis. Cancers 2021, 13, 5500. [Google Scholar] [CrossRef] [PubMed]
- Lacy, S.; Lopez-Beltran, A.; MacLennan, G.T.; Foster, S.R.; Montironi, R.; Cheng, L. Molecular pathogenesis of urothelial carcinoma: The clinical utility of emerging new biomarkers and future molecular classification of bladder cancer. Anal. Quant. Cytol. Histol. 2009, 31, 5–16. [Google Scholar] [PubMed]
- Cappello, P.; Principe, M.; Bulfamante, S.; Novelli, F. Alpha-Enolase (ENO1), a potential target in novel immunotherapies. Front. Biosci. 2017, 22, 944–959. [Google Scholar]
- Xu, X.; Chen, B.; Zhu, S.; Zhang, J.; He, X.; Cao, G.; Chen, B. Hyperglycemia promotes Snail-induced epithelial-mesenchymal transition of gastric cancer via activating ENO1 expression. Cancer Cell Int. 2019, 19, 344. [Google Scholar] [CrossRef]
- Barzaman, K.; Karami, J.; Zarei, Z.; Hosseinzadeh, A.; Kazemi, M.H.; Moradi-Kalbolandi, S.; Safari, E.; Farahmand, L. Breast cancer: Biology, biomarkers, and treatments. Int. Immunopharmacol. 2020, 84, 106535. [Google Scholar] [CrossRef]
- Wang, Y.; Sun, Y.; Tan, M.; Lin, X.; Tai, P.; Huang, X.; Jin, Q.; Yuan, D.; Xu, T.; He, B. Association Between Polymorphisms in DNA Damage Repair Pathway Genes and Female Breast Cancer Risk. DNA Cell Biol. 2024, 43, 219–231. [Google Scholar] [CrossRef]
- Sun, Y.; Dong, D.; Xia, Y.; Hao, L.; Wang, W.; Zhao, C. YTHDF1 promotes breast cancer cell growth, DNA damage repair and chemoresistance. Cell Death Dis. 2022, 13, 230. [Google Scholar] [CrossRef]
- Belfiore, A.; Malaguarnera, R.; Nicolosi, M.L.; Lappano, R.; Ragusa, M.; Morrione, A.; Vella, V. A novel functional crosstalk between DDR1 and the IGF axis and its relevance for breast cancer. Cell Adh. Migr. 2018, 12, 305–314. [Google Scholar] [CrossRef]
- Han, Q.; Xiao, F.; Ma, L.; Zhou, J.; Wang, L.; Cheng, H.; Zhu, J.; Yao, F.; Lyu, J.; Du, L. DDR1 promotes migration and invasion of breast cancer by modulating the Src-FAK signaling. Neoplasma 2022, 69, 1154–1164. [Google Scholar] [CrossRef] [PubMed]
- Johnson, C.A.; James, D.; Marzan, A.; Armaos, M. Cervical Cancer: An Overview of Pathophysiology and Management. Semin. Oncol. Nurs. 2019, 35, 166–174. [Google Scholar] [CrossRef]
- Zhou, T.; Xiao, Z.; Lu, J.; Zhang, L.; Bo, L.; Wang, J. IGF2BP3-mediated regulation of GLS and GLUD1 gene expression promotes treg-induced immune escape in human cervical cancer. Am. J. Cancer Res. 2023, 13, 5289–5305. [Google Scholar] [PubMed]
- De Jesus, D.F.; Zhang, Z.; Brown, N.K.; Li, X.; Gaffrey, M.J.; Kahraman, S.; Wei, J.; Hu, J.; Basile, G.; Xiao, L.; et al. Redox Regulation of m(6) A Methyltransferase METTL3 in Human beta-cells Controls the Innate Immune Response in Type 1 Diabetes. bioRxiv 2023. [Google Scholar] [CrossRef]
- Yu, R.; Wei, Y.; He, C.; Zhou, P.; Yang, H.; Deng, C.; Liu, R.; Wu, P.; Gao, Q.; Cao, C. Integrative Analyses of m6A Regulators Identify that METTL3 is Associated with HPV Status and Immunosuppressive Microenvironment in HPV-related Cancers. Int. J. Biol. Sci. 2022, 18, 3874–3887. [Google Scholar] [CrossRef] [PubMed]
- Mao, Z.; Wang, B.; Zhang, T.; Cui, B. The roles of m6A methylation in cervical cancer: Functions, molecular mechanisms, and clinical applications. Cell Death Dis. 2023, 14, 734. [Google Scholar] [CrossRef]
- Brooks, R.A.; Fleming, G.F.; Lastra, R.R.; Lee, N.K.; Moroney, J.W.; Son, C.H.; Tatebe, K.; Veneris, J.L. Current recommendations and recent progress in endometrial cancer. CA Cancer J. Clin. 2019, 69, 258–279. [Google Scholar] [CrossRef]
- Yang, X.; Zhang, S.; He, C.; Xue, P.; Zhang, L.; He, Z.; Zang, L.; Feng, B.; Sun, J.; Zheng, M. METTL14 suppresses proliferation and metastasis of colorectal cancer by down-regulating oncogenic long non-coding RNA XIST. Mol. Cancer 2020, 19, 46. [Google Scholar] [CrossRef]
- Liu, J.; Eckert, M.A.; Harada, B.T.; Liu, S.M.; Lu, Z.; Yu, K.; Tienda, S.M.; Chryplewicz, A.; Zhu, A.C.; Yang, Y.; et al. m(6)A mRNA methylation regulates AKT activity to promote the proliferation and tumorigenicity of endometrial cancer. Nat. Cell Biol. 2018, 20, 1074–1083. [Google Scholar] [CrossRef] [PubMed]
- Park, Y.A.; Lee, J.W.; Choi, J.J.; Jeon, H.K.; Cho, Y.; Choi, C.; Kim, T.J.; Lee, N.W.; Kim, B.G.; Bae, D.S. The interactions between MicroRNA-200c and BRD7 in endometrial carcinoma. Gynecol. Oncol. 2012, 124, 125–133. [Google Scholar] [CrossRef] [PubMed]
- de Bree, E.; Michelakis, D. An overview and update of hyperthermic intraperitoneal chemotherapy in ovarian cancer. Expert. Opin. Pharmacother. 2020, 21, 1479–1492. [Google Scholar] [CrossRef] [PubMed]
- Penny, S.M. Ovarian Cancer: An Overview. Radiol. Technol. 2020, 91, 561–575. [Google Scholar]
- Alexandrova, E.; Pecoraro, G.; Sellitto, A.; Melone, V.; Ferravante, C.; Rocco, T.; Guacci, A.; Giurato, G.; Nassa, G.; Rizzo, F.; et al. An Overview of Candidate Therapeutic Target Genes in Ovarian Cancer. Cancers 2020, 12, 1470. [Google Scholar] [CrossRef]
- Wang, R.; Ye, H.; Yang, B.; Ao, M.; Yu, X.; Wu, Y.; Xi, M.; Hou, M. m6A-modified circNFIX promotes ovarian cancer progression and immune escape via activating IL-6R/JAK1/STAT3 signaling by sponging miR-647. Int. Immunopharmacol. 2023, 124, 110879. [Google Scholar] [CrossRef]
- Yan, Y.; Liang, Q.; Xu, Z.; Yi, Q. Integrative bioinformatics and experimental analysis revealed down-regulated CDC42EP3 as a novel prognostic target for ovarian cancer and its roles in immune infiltration. PeerJ 2021, 9, e12171. [Google Scholar] [CrossRef]
- Song, Y.; Qu, H. Identification and validation of a seven m6A-related lncRNAs signature predicting prognosis of ovarian cancer. BMC Cancer 2022, 22, 633. [Google Scholar] [CrossRef]
- Tan, W.; Liu, S.; Deng, Z.; Dai, F.; Yuan, M.; Hu, W.; Li, B.; Cheng, Y. Gene signature of m6A-related targets to predict prognosis and immunotherapy response in ovarian cancer. J. Cancer Res. Clin. Oncol. 2023, 149, 593–608. [Google Scholar] [CrossRef]
- Zhang, C.; Liu, J.; Guo, H.; Hong, D.; Ji, J.; Zhang, Q.; Guan, Q.; Ren, Q. m6A RNA methylation regulators were associated with the malignancy and prognosis of ovarian cancer. Bioengineered 2021, 12, 3159–3176. [Google Scholar] [CrossRef]
- Ahmed, B.; Qadir, M.I.; Ghafoor, S. Malignant Melanoma: Skin Cancer-Diagnosis, Prevention, and Treatment. Crit. Rev. Eukaryot. Gene Expr. 2020, 30, 291–297. [Google Scholar] [CrossRef]
- Chen, C.; Xie, L.; Ren, T.; Huang, Y.; Xu, J.; Guo, W. Immunotherapy for osteosarcoma: Fundamental mechanism, rationale, and recent breakthroughs. Cancer Lett. 2021, 500, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Tian, Y.; Zhao, F.; Chen, Z.; Su, P.; Li, Y.; Qian, A. Bone Microenvironment and Osteosarcoma Metastasis. Int. J. Mol. Sci. 2020, 21, 6985. [Google Scholar] [CrossRef]
- Wu, Z.Y.; Shi, Z.Y. The prognostic value and immune landscapes of m1A/m5C/m6A-associated lncRNA signature in osteosarcoma. Eur. Rev. Med. Pharmacol. Sci. 2022, 26, 5868–5883. [Google Scholar] [CrossRef] [PubMed]
- Zheng, D.; Yu, L.; Wei, Z.; Xia, K.; Guo, W. N6-Methyladenosine-Related lncRNAs Are Potential Prognostic Biomarkers and Correlated With Tumor Immune Microenvironment in Osteosarcoma. Front. Genet. 2021, 12, 805607. [Google Scholar] [CrossRef]
- Wu, Z.; Zhang, X.; Chen, D.; Li, Z.; Wu, X.; Wang, J.; Deng, Y. N6-Methyladenosine-Related LncRNAs Are Potential Remodeling Indicators in the Tumor Microenvironment and Prognostic Markers in Osteosarcoma. Front. Immunol. 2021, 12, 806189. [Google Scholar] [CrossRef]
- Menter, T.; Tzankov, A. Tumor Microenvironment in Acute Myeloid Leukemia: Adjusting Niches. Front. Immunol. 2022, 13, 811144. [Google Scholar] [CrossRef] [PubMed]
- Shimony, S.; Stahl, M.; Stone, R.M. Acute myeloid leukemia: 2023 update on diagnosis, risk-stratification, and management. Am. J. Hematol. 2023, 98, 502–526. [Google Scholar] [CrossRef]
- Zhong, F.; Yao, F.; Cheng, Y.; Liu, J.; Zhang, N.; Li, S.; Li, M.; Huang, B.; Wang, X. m6A-related lncRNAs predict prognosis and indicate immune microenvironment in acute myeloid leukemia. Sci. Rep. 2022, 12, 1759. [Google Scholar] [CrossRef]
- Li, D.; Liang, J.; Cheng, C.; Guo, W.; Li, S.; Song, W.; Song, Z.; Bai, Y.; Zhang, Y.; Wu, X.; et al. Identification of m6A-Related lncRNAs Associated With Prognoses and Immune Responses in Acute Myeloid Leukemia. Front. Cell Dev. Biol. 2021, 9, 770451. [Google Scholar] [CrossRef]
- Deng, X.; Su, R.; Weng, H.; Huang, H.; Li, Z.; Chen, J. RNA N(6)-methyladenosine modification in cancers: Current status and perspectives. Cell Res. 2018, 28, 507–517. [Google Scholar] [CrossRef]
- Wardowska, A. m6A RNA Methylation in Systemic Autoimmune Diseases-A New Target for Epigenetic-Based Therapy? Pharmaceuticals 2021, 14, 218. [Google Scholar] [CrossRef]
- Geng, Q.; Cao, X.; Fan, D.; Gu, X.; Zhang, Q.; Zhang, M.; Wang, Z.; Deng, T.; Xiao, C. Diagnostic gene signatures and aberrant pathway activation based on m6A methylation regulators in rheumatoid arthritis. Front. Immunol. 2022, 13, 1041284. [Google Scholar] [CrossRef]
- Wang, J.; Yan, S.; Lu, H.; Wang, S.; Xu, D. METTL3 Attenuates LPS-Induced Inflammatory Response in Macrophages via NF-κB Signaling Pathway. Mediat. Inflamm. 2019, 2019, 3120391. [Google Scholar] [CrossRef]
- Shi, H.; Liu, C.; Tan, H.; Li, Y.; Nguyen, T.M.; Dhungana, Y.; Guy, C.; Vogel, P.; Neale, G.; Rankin, S.; et al. Hippo Kinases Mst1 and Mst2 Sense and Amplify IL-2R-STAT5 Signaling in Regulatory T Cells to Establish Stable Regulatory Activity. Immunity 2018, 49, 899–914.e6. [Google Scholar] [CrossRef]
- Tong, J.; Cao, G.; Zhang, T.; Sefik, E.; Amezcua Vesely, M.C.; Broughton, J.P.; Zhu, S.; Li, H.; Li, B.; Chen, L.; et al. m(6)A mRNA methylation sustains Treg suppressive functions. Cell Res. 2018, 28, 253–256. [Google Scholar] [CrossRef]
- Ottens, K.; Hinman, R.M.; Barrios, E.; Skaug, B.; Davis, L.S.; Li, Q.Z.; Castrillon, D.H.; Satterthwaite, A.B. Foxo3 Promotes Apoptosis of B Cell Receptor-Stimulated Immature B Cells, Thus Limiting the Window for Receptor Editing. J. Immunol. 2018, 201, 940–949. [Google Scholar] [CrossRef]
- Lu, S.; Wei, X.; Zhu, H.; Hu, Z.; Zheng, M.; Wu, J.; Zhao, C.; Yang, S.; Feng, D.; Jia, S.; et al. m(6)A methyltransferase METTL3 programs CD4(+) T-cell activation and effector T-cell differentiation in systemic lupus erythematosus. Mol. Med. 2023, 29, 46. [Google Scholar] [CrossRef]
- Morales, E.; Trujillo, H.; Bada, T.; Alonso, M.; Gutiérrez, E.; Rodríguez, E.; Gutiérrez, E.; Galindo, M.; Praga, M. What is the value of repeat kidney biopsies in patients with lupus nephritis? Lupus 2021, 30, 25–34. [Google Scholar] [CrossRef]
- Ning, Y.; Chen, J.; Shi, Y.; Song, N.; Yu, X.; Fang, Y.; Ding, X. Genistein Ameliorates Renal Fibrosis Through Regulation Snail via m6A RNA Demethylase ALKBH5. Front. Pharmacol. 2020, 11, 579265. [Google Scholar] [CrossRef]
- Ge, X.; Xue, G.; Ding, Y.; Li, R.; Hu, K.; Xu, T.; Sun, M.; Liao, W.; Zhao, B.; Wen, C.; et al. The Loss of YTHDC1 in Gut Macrophages Exacerbates Inflammatory Bowel Disease. Adv. Sci. 2023, 10, e2205620. [Google Scholar] [CrossRef]
- Zhao, M.; Li, P.; Qiao, D.; Hua, S.; Yue, Q.; Dai, Y.; Huang, Y.; Jiang, J.; Yin, H.; Li, M.; et al. N6-methyladenosine modification of TSC1 mRNA contributes to macrophage polarization regulated by Coptisine in DSS-induced ulcerative colitis. Phytomedicine 2024, 122, 155153. [Google Scholar] [CrossRef]
- Ma, Y.; Zhang, X.; Xuan, B.; Li, D.; Yin, N.; Ning, L.; Zhou, Y.L.; Yan, Y.; Tong, T.; Zhu, X.; et al. Disruption of CerS6-mediated sphingolipid metabolism by FTO deficiency aggravates ulcerative colitis. Gut 2024, 73, 268–281. [Google Scholar] [CrossRef]
- Cheng, L.; Li, H.; Zhan, H.; Liu, Y.; Li, X.; Huang, Y.; Wang, L.; Zhang, F.; Li, Y. Alterations of m6A RNA methylation regulators contribute to autophagy and immune infiltration in primary Sjögren's syndrome. Front. Immunol. 2022, 13, 949206. [Google Scholar] [CrossRef]
- He, F.; Liu, H.; Yu, C. N(6)-Methyladenosine Regulator-Mediated RNA Methylation Is Involved in Primary Sjögren's Syndrome Immunoinfiltration. Dis. Markers 2022, 2022, 5242287. [Google Scholar] [CrossRef]
- Li, Z.; Wang, P.; Li, J.; Xie, Z.; Cen, S.; Li, M.; Liu, W.; Ye, G.; Zheng, G.; Ma, M.; et al. The N(6)-methyladenosine demethylase ALKBH5 negatively regulates the osteogenic differentiation of mesenchymal stem cells through PRMT6. Cell Death Dis. 2021, 12, 578. [Google Scholar] [CrossRef] [PubMed]
- Luo, Q.; Guo, Y.; Xiao, Q.; Fu, B.; Zhang, L.; Guo, Y.; Huang, Z.; Li, J. Expression and Clinical Significance of the m6A RNA-Binding Proteins YTHDF2 in Peripheral Blood Mononuclear Cells From New-Onset Ankylosing Spondylitis. Front. Med. 2022, 9, 922219. [Google Scholar] [CrossRef]
- Lyu, Z.; Huang, B.; Zhang, J.; Qian, Q.; Pu, X.; Cui, N.; Ou, Y.; Li, B.; You, Z.; Lian, M.; et al. Suppression of YTHDF2 attenuates autoimmune hepatitis by expansion of myeloid-derived suppressor cells. J. Autoimmun. 2023, 135, 102993. [Google Scholar] [CrossRef]
- Zhou, J.; Zhang, X.; Hu, J.; Qu, R.; Yu, Z.; Xu, H.; Chen, H.; Yan, L.; Ding, C.; Zou, Q.; et al. m(6)A demethylase ALKBH5 controls CD4(+) T cell pathogenicity and promotes autoimmunity. Sci. Adv. 2021, 7, eabg0470. [Google Scholar] [CrossRef]
- Han, X.; Liu, L.; Huang, S.; Xiao, W.; Gao, Y.; Zhou, W.; Zhang, C.; Zheng, H.; Yang, L.; Xie, X.; et al. RNA m(6)A methylation modulates airway inflammation in allergic asthma via PTX3-dependent macrophage homeostasis. Nat. Commun. 2023, 14, 7328. [Google Scholar] [CrossRef]
- Lichinchi, G.; Gao, S.; Saletore, Y.; Gonzalez, G.M.; Bansal, V.; Wang, Y.; Mason, C.E.; Rana, T.M. Dynamics of the human and viral m(6)A RNA methylomes during HIV-1 infection of T cells. Nat. Microbiol. 2016, 1, 16011. [Google Scholar] [CrossRef] [PubMed]
- Kennedy, E.M.; Bogerd, H.P.; Kornepati, A.V.; Kang, D.; Ghoshal, D.; Marshall, J.B.; Poling, B.C.; Tsai, K.; Gokhale, N.S.; Horner, S.M.; et al. Posttranscriptional m(6)A Editing of HIV-1 mRNAs Enhances Viral Gene Expression. Cell Host Microbe 2016, 19, 675–685. [Google Scholar] [CrossRef] [PubMed]
- Tirumuru, N.; Zhao, B.S.; Lu, W.; Lu, Z.; He, C.; Wu, L. N(6)-methyladenosine of HIV-1 RNA regulates viral infection and HIV-1 Gag protein expression. Elife 2016, 5, e15528. [Google Scholar] [CrossRef]
- McInnes, I.B.; Schett, G. The pathogenesis of rheumatoid arthritis. N. Engl. J. Med. 2011, 365, 2205–2219. [Google Scholar] [CrossRef]
- Li, X.F.; Sun, Y.Y.; Bao, J.; Chen, X.; Li, Y.H.; Yang, Y.; Zhang, L.; Huang, C.; Wu, B.M.; Meng, X.M.; et al. Functional role of PPAR-γ on the proliferation and migration of fibroblast-like synoviocytes in rheumatoid arthritis. Sci. Rep. 2017, 7, 12671. [Google Scholar] [CrossRef]
- Wan, L.; Liu, J.; Huang, C.; Zhu, Z.; Wang, K.; Sun, G.; Zhu, L.; Hu, Z. Comprehensive Analysis and Functional Characteristics of Differential Expression of N6-Methyladenosine Methylation Modification in the Whole Transcriptome of Rheumatoid Arthritis. Mediat. Inflamm. 2022, 2022, 4766992. [Google Scholar] [CrossRef]
- Aihaiti, Y.; Tuerhong, X.; Zheng, H.; Cai, Y.; Yang, M.; Xu, P. Peroxiredoxin 4 regulates tumor-cell-like characteristics of fibroblast-like synoviocytes in rheumatoid arthritis through PI3k/Akt signaling pathway. Clin. Immunol. 2022, 237, 108964. [Google Scholar] [CrossRef] [PubMed]
- Lin, X.; Tao, C.; Zhang, R.; Zhang, M.; Wang, Q.; Chen, J. N6-methyladenosine modification of TGM2 mRNA contributes to the inhibitory activity of sarsasapogenin in rheumatoid arthritis fibroblast-like synoviocytes. Phytomedicine 2022, 95, 153871. [Google Scholar] [CrossRef] [PubMed]
- Luo, Q.; Gao, Y.; Zhang, L.; Rao, J.; Guo, Y.; Huang, Z.; Li, J. Decreased ALKBH5, FTO, and YTHDF2 in Peripheral Blood Are as Risk Factors for Rheumatoid Arthritis. Biomed. Res. Int. 2020, 2020, 5735279. [Google Scholar] [CrossRef]
- Wu, S.; Li, X.F.; Wu, Y.Y.; Yin, S.Q.; Huang, C.; Li, J. N(6)-Methyladenosine and Rheumatoid Arthritis: A Comprehensive Review. Front. Immunol. 2021, 12, 731842. [Google Scholar] [CrossRef]
- Chen, Q.; Li, H.; Liu, Y.; Zhao, M. Epigenetic Regulation of Immune and Inflammatory Responses in Rheumatoid Arthritis. Front. Immunol. 2022, 13, 881191. [Google Scholar] [CrossRef] [PubMed]
- Song, S.; Zhao, R.; Qiao, J.; Liu, J.; Cheng, T.; Zhang, S.X.; Li, X.F. Predictive value of drug efficacy by M6A modification patterns in rheumatoid arthritis patients. Front. Immunol. 2022, 13, 940918. [Google Scholar] [CrossRef] [PubMed]
- Carter, E.E.; Barr, S.G.; Clarke, A.E. The global burden of SLE: Prevalence, health disparities and socioeconomic impact. Nat. Rev. Rheumatol. 2016, 12, 605–620. [Google Scholar] [CrossRef] [PubMed]
- Luo, Q.; Fu, B.; Zhang, L.; Guo, Y.; Huang, Z.; Li, J. Decreased Peripheral Blood ALKBH5 Correlates with Markers of Autoimmune Response in Systemic Lupus Erythematosus. Dis. Markers 2020, 2020, 8193895. [Google Scholar] [CrossRef]
- Luo, Q.; Rao, J.; Zhang, L.; Fu, B.; Guo, Y.; Huang, Z.; Li, J. The study of METTL14, ALKBH5, and YTHDF2 in peripheral blood mononuclear cells from systemic lupus erythematosus. Mol. Genet. Genom. Med. 2020, 8, e1298. [Google Scholar] [CrossRef]
- Lin, Z.; Niu, Y.; Wan, A.; Chen, D.; Liang, H.; Chen, X.; Sun, L.; Zhan, S.; Chen, L.; Cheng, C.; et al. RNA m(6) A methylation regulates sorafenib resistance in liver cancer through FOXO3-mediated autophagy. Embo J. 2020, 39, e103181. [Google Scholar] [CrossRef]
- Moulton, V.R.; Tsokos, G.C. T cell signaling abnormalities contribute to aberrant immune cell function and autoimmunity. J. Clin. Investig. 2015, 125, 2220–2227. [Google Scholar] [CrossRef]
- Deng, L.J.; Fang, X.Y.; Wu, J.; Li, Q.R.; Mao, Y.M.; Leng, R.X.; Fan, Y.G.; Ye, D.Q. ALKBH5 Expression could Affect the Function of T Cells in Systemic Lupus Erythematosus Patients: A Case-control Study. Curr. Pharm. Des. 2022, 28, 2270–2278. [Google Scholar] [CrossRef]
- Tang, Y.; Xie, H.; Chen, J.; Geng, L.; Chen, H.; Li, X.; Hou, Y.; Lu, L.; Shi, S.; Zeng, X.; et al. Activated NF-κB in bone marrow mesenchymal stem cells from systemic lupus erythematosus patients inhibits osteogenic differentiation through downregulating Smad signaling. Stem Cells Dev. 2013, 22, 668–678. [Google Scholar] [CrossRef]
- Gao, L.; Liesveld, J.; Anolik, J.; McDavid, A.; Looney, R.J. IFNβ signaling inhibits osteogenesis in human SLE bone marrow. Lupus 2020, 29, 1040–1049. [Google Scholar] [CrossRef]
- Lv, X.; Liu, X.; Zhao, M.; Wu, H.; Zhang, W.; Lu, Q.; Chen, X. RNA Methylation in Systemic Lupus Erythematosus. Front. Cell Dev. Biol. 2021, 9, 696559. [Google Scholar] [CrossRef] [PubMed]
- Nie, K.; Yi, J.; Yang, Y.; Deng, M.; Yang, Y.; Wang, T.; Chen, X.; Zhang, Z.; Wang, X. A Broad m6A Modification Landscape in Inflammatory Bowel Disease. Front. Cell Dev. Biol. 2021, 9, 782636. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Ding, C.; Chen, H.; Zhao, J.; Chen, Z.; Chen, B.; Mao, K.; Hao, Y.; Roulis, M.; Xu, H.; et al. m(6)A mRNA modification maintains colonic epithelial cell homeostasis via NF-κB-mediated antiapoptotic pathway. Sci. Adv. 2022, 8, eabl5723. [Google Scholar] [CrossRef]
- Lu, T.X.; Zheng, Z.; Zhang, L.; Sun, H.L.; Bissonnette, M.; Huang, H.; He, C. A New Model of Spontaneous Colitis in Mice Induced by Deletion of an RNA m(6)A Methyltransferase Component METTL14 in T Cells. Cell. Mol. Gastroenterol. Hepatol. 2020, 10, 747–761. [Google Scholar] [CrossRef]
- Keubler, L.M.; Buettner, M.; Häger, C.; Bleich, A. A Multihit Model: Colitis Lessons from the Interleukin-10-deficient Mouse. Inflamm. Bowel Dis. 2015, 21, 1967–1975. [Google Scholar] [CrossRef]
- Zong, X.; Wang, H.; Xiao, X.; Zhang, Y.; Hu, Y.; Wang, F.; Wang, Y.; Lu, Z. Enterotoxigenic Escherichia coli infection promotes enteric defensin expression via FOXO6-METTL3-m(6)A-GPR161 signalling axis. RNA Biol. 2021, 18, 576–586. [Google Scholar] [CrossRef]
- Zong, X.; Zhao, J.; Wang, H.; Lu, Z.; Wang, F.; Du, H.; Wang, Y. Mettl3 Deficiency Sustains Long-Chain Fatty Acid Absorption through Suppressing Traf6-Dependent Inflammation Response. J. Immunol. 2019, 202, 567–578. [Google Scholar] [CrossRef]
- Sun, L.; Ma, L.; Zhang, H.; Cao, Y.; Wang, C.; Hou, N.; Huang, N.; von Deneen, K.M.; Zhao, C.; Shi, Y.; et al. Fto Deficiency Reduces Anxiety- and Depression-Like Behaviors in Mice via Alterations in Gut Microbiota. Theranostics 2019, 9, 721–733. [Google Scholar] [CrossRef] [PubMed]
- Koelink, P.J.; Bloemendaal, F.M.; Li, B.; Westera, L.; Vogels, E.W.M.; van Roest, M.; Gloudemans, A.K.; van ‘t Wout, A.B.; Korf, H.; Vermeire, S.; et al. Anti-TNF therapy in IBD exerts its therapeutic effect through macrophage IL-10 signalling. Gut 2020, 69, 1053–1063. [Google Scholar] [CrossRef]
- Chen, Y.; Lei, J.; He, S. m(6)A Modification Mediates Mucosal Immune Microenvironment and Therapeutic Response in Inflammatory Bowel Disease. Front. Cell Dev. Biol. 2021, 9, 692160. [Google Scholar] [CrossRef]
- Zuo, F.; Wei, H.; Peng, J.; Li, S.; Zhou, Y. Effects on the Cell Barrier Function of L-Met and DL-HMTBA Is Related to Metabolic Characteristics and m(6)A Modification. Front. Nutr. 2022, 9, 836069. [Google Scholar] [CrossRef] [PubMed]
- Nishizawa, Y.; Konno, M.; Asai, A.; Koseki, J.; Kawamoto, K.; Miyoshi, N.; Takahashi, H.; Nishida, N.; Haraguchi, N.; Sakai, D.; et al. Oncogene c-Myc promotes epitranscriptome m(6)A reader YTHDF1 expression in colorectal cancer. Oncotarget 2018, 9, 7476–7486. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.; Liu, X.Q.; Lin, X.; Gao, L.Y.; Zhang, S.; Huang, X. Targeting YTHDF1 effectively re-sensitizes cisplatin-resistant colon cancer cells by modulating GLS-mediated glutamine metabolism. Mol. Ther. Oncolytics 2021, 20, 228–239. [Google Scholar] [CrossRef]
- Beroukas, D.; Hiscock, J.; Jonsson, R.; Waterman, S.A.; Gordon, T.P. Subcellular distribution of aquaporin 5 in salivary glands in primary Sjögren's syndrome. Lancet 2001, 358, 1875–1876. [Google Scholar] [CrossRef]
- Xiao, Q.; Wu, X.; Deng, C.; Zhao, L.; Peng, L.; Zhou, J.; Zhang, W.; Zhao, Y.; Fei, Y. The potential role of RNA N6-methyladenosine in primary Sjögren's syndrome. Front. Med. 2022, 9, 959388. [Google Scholar] [CrossRef] [PubMed]
- Schett, G.; Lories, R.J.; D'Agostino, M.A.; Elewaut, D.; Kirkham, B.; Soriano, E.R.; McGonagle, D. Enthesitis: From pathophysiology to treatment. Nat. Rev. Rheumatol. 2017, 13, 731–741. [Google Scholar] [CrossRef]
- Xie, Z.; Yu, W.; Zheng, G.; Li, J.; Cen, S.; Ye, G.; Li, Z.; Liu, W.; Li, M.; Lin, J.; et al. TNF-α-mediated m(6)A modification of ELMO1 triggers directional migration of mesenchymal stem cell in ankylosing spondylitis. Nat. Commun. 2021, 12, 5373. [Google Scholar] [CrossRef]
- Sun, X.; Zhou, C.; Zhu, J.; Wu, S.; Liang, T.; Jiang, J.; Chen, J.; Chen, T.; Huang, S.S.; Chen, L.; et al. Identification of clinical heterogeneity and construction of a novel subtype predictive model in patients with ankylosing spondylitis: An unsupervised machine learning study. Int. Immunopharmacol. 2023, 117, 109879. [Google Scholar] [CrossRef]
- Luan, Z.; Wang, Y. Association between ankylosing spondylitis and m6A methylation. J. Orthop. Surg. Res. 2023, 18, 757. [Google Scholar] [CrossRef]
- Du, J.; Liao, W.; Liu, W.; Deb, D.K.; He, L.; Hsu, P.J.; Nguyen, T.; Zhang, L.; Bissonnette, M.; He, C.; et al. N(6)-Adenosine Methylation of Socs1 mRNA Is Required to Sustain the Negative Feedback Control of Macrophage Activation. Dev. Cell 2020, 55, 737–753.e7. [Google Scholar] [CrossRef]
- Shu, B.; Zhou, Y.X.; Li, H.; Zhang, R.Z.; He, C.; Yang, X. The METTL3/MALAT1/PTBP1/USP8/TAK1 axis promotes pyroptosis and M1 polarization of macrophages and contributes to liver fibrosis. Cell Death Discov. 2021, 7, 368. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Ji, Y.; Feng, P.; Liu, R.; Li, G.; Zheng, J.; Xue, Y.; Wei, Y.; Ji, C.; Chen, D.; et al. The m6A Reader IGF2BP2 Regulates Macrophage Phenotypic Activation and Inflammatory Diseases by Stabilizing TSC1 and PPARγ. Adv. Sci. 2021, 8, 2100209. [Google Scholar] [CrossRef]
- Wang, X.; Ma, R.; Zhang, X.; Cui, L.; Ding, Y.; Shi, W.; Guo, C.; Shi, Y. Crosstalk between N6-methyladenosine modification and circular RNAs: Current understanding and future directions. Mol. Cancer 2021, 20, 121. [Google Scholar] [CrossRef]
- Hu, Y.; Lei, L.; Jiang, L.; Zeng, H.; Zhang, Y.; Fu, C.; Guo, H.; Dong, Y.; Ouyang, Y.; Zhang, X.; et al. LncRNA UCA1 promotes keratinocyte-driven inflammation via suppressing METTL14 and activating the HIF-1α/NF-κB axis in psoriasis. Cell Death Dis. 2023, 14, 279. [Google Scholar] [CrossRef]
- Yang, L.; Fu, J.; Han, X.; Zhang, C.; Xia, L.; Zhu, R.; Huang, S.; Xiao, W.; Yu, H.; Gao, Y.; et al. Hsa_circ_0004287 inhibits macrophage-mediated inflammation in an N(6)-methyladenosine-dependent manner in atopic dermatitis and psoriasis. J. Allergy Clin. Immunol. 2022, 149, 2021–2033. [Google Scholar] [CrossRef]
- Yang, J.; Wang, H.; Zhang, W. Regulation of Virus Replication and T Cell Homeostasis by N(6)-Methyladenosine. Virol. Sin. 2019, 34, 22–29. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Ma, S.; Deng, Y.; Yi, P.; Yu, J. Targeting the RNA m(6)A modification for cancer immunotherapy. Mol. Cancer 2022, 21, 76. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Yang, Z.; Li, R.; Wu, Y.; Chi, M.; Gao, S.; Sun, X.; Meng, X.; Wang, B. Potential roles of N6-methyladenosine (m6A) in immune cells. J. Transl. Med. 2021, 19, 251. [Google Scholar] [CrossRef]
- Ma, Z.; Gao, X.; Shuai, Y.; Xing, X.; Ji, J. The m6A epitranscriptome opens a new charter in immune system logic. Epigenetics 2021, 16, 819–837. [Google Scholar] [CrossRef]
- Pan, J.; Huang, T.; Deng, Z.; Zou, C. Roles and therapeutic implications of m6A modification in cancer immunotherapy. Front. Immunol. 2023, 14, 1132601. [Google Scholar] [CrossRef]
- Casey, S.C.; Tong, L.; Li, Y.; Do, R.; Walz, S.; Fitzgerald, K.N.; Gouw, A.M.; Baylot, V.; Gütgemann, I.; Eilers, M.; et al. MYC regulates the antitumor immune response through CD47 and PD-L1. Science 2016, 352, 227–231. [Google Scholar] [CrossRef]
- Wu, H.; Li, F.; Zhu, R. miR-338-5p inhibits cell growth and migration via inhibition of the METTL3/m6A/c-Myc pathway in lung cancer. Acta Biochim. Biophys. Sin. 2021, 53, 304–316. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.D.; Chen, Z.H.; Yu, K.; Lu, J.H.; Wu, Q.N.; Wang, Y.; Ju, H.Q.; Xu, R.H.; Liu, Z.X.; Zeng, Z.L. METTL3 Promotes the Progression of Gastric Cancer via Targeting the MYC Pathway. Front. Oncol. 2020, 10, 115. [Google Scholar] [CrossRef] [PubMed]
- Fu, C.; Liang, X.; Cui, W.; Ober-Blöbaum, J.L.; Vazzana, J.; Shrikant, P.A.; Lee, K.P.; Clausen, B.E.; Mellman, I.; Jiang, A. β-Catenin in dendritic cells exerts opposite functions in cross-priming and maintenance of CD8+ T cells through regulation of IL-10. Proc. Natl. Acad. Sci. USA 2015, 112, 2823–2828. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Li, S.W.; Liu, L.; Yang, J.; Huang, G.; Sang, Y. TRIM11 facilitates chemoresistance in nasopharyngeal carcinoma by activating the β-catenin/ABCC9 axis via p62-selective autophagic degradation of Daple. Oncogenesis 2020, 9, 45. [Google Scholar] [CrossRef]
- Roignant, J.Y.; Soller, M. m(6)A in mRNA: An Ancient Mechanism for Fine-Tuning Gene Expression. Trends Genet. 2017, 33, 380–390. [Google Scholar] [CrossRef] [PubMed]
- Motorin, Y.; Helm, M. Methods for RNA Modification Mapping Using Deep Sequencing: Established and New Emerging Technologies. Genes 2019, 10, 35. [Google Scholar] [CrossRef]
- Esteller, M.; Pandolfi, P.P. The Epitranscriptome of Noncoding RNAs in Cancer. Cancer Discov. 2017, 7, 359–368. [Google Scholar] [CrossRef]
- Shen, S.; Zhang, L.S. The regulation of antiviral innate immunity through non-m(6)A RNA modifications. Front. Immunol. 2023, 14, 1286820. [Google Scholar] [CrossRef]
| System | Cancer Type | m6A Regulator | Immune Cell Type | Target Genes | Mechanism | Immunity Expression | Methods of Treatment | Refs. |
|---|---|---|---|---|---|---|---|---|
| Digestive system | Esophageal squamous cell carcinoma (ESCC) | HNRNPC RBM15 IGF2BP3 METTL16 KIAA1429 | macrophages, neutrophils | negative | [50] | |||
| YTHDF2 METL14 KIAA1429 | negative | Immune-checkpointinhibition, PD-L1 | [51] | |||||
| YTHDC1 YTHDC2 YTHDC3 ZC3H13 | macrophages | HNRNPA2B1——p53 mutation | positive | [52] | ||||
| HNRNPA2B1 | T cell | positive | [52] | |||||
| HNRNPC | B cell | positive | Immune-checkpointinhibition, PD-L1 | [52] | ||||
| Gastric cancer (GC) | KIAA1429 (recruiting METTL3, METTL14, and WTAP) | DC | C-type lectin receptors, NOD-like receptors, T-cell receptors, toll-like receptors,NF-κB signaling pathways | negative | Immune-checkpointinhibition,PD1/L1 | [53,54] | ||
| YTHDF1 | recruitment of DCs, CD4+ and CD8+ T cells, adaptive T-cell | FNGR1 and JAK1/2-STAT1 pathway | negative | [55,56] | ||||
| ALKBH5 | naïve B cells, neutrophils, plasma cells, and follicular helper T cells | SLC7A2, CGB3 | positive | [57] | ||||
| YTHDF3 | PI3K/AKT pathway | positive | Immune-checkpointinhibition, PD-L1,CXCL1 | [58,59] | ||||
| Liver cancer | ALKBH5 | myeloid-derived suppressor-like cells | negative | [60] | ||||
| PD-L1+macrophage | MAP3K8 | JNK and ERK pathways | positive | Immune-checkpointinhibition,PD-L1 | [61] | |||
| YTHDF2 | T (Treg) cells | NF-κB signaling | negative | [62,63] | ||||
| METTL5 | B cells, CD8+ T cells, CD4+ T cells, macrophages, neutrophils and DCs | ribosomal and oxidative phosphorylation, mismatch repair, spliceosome and other signaling pathways | positive | PD1 (PDCD1),CTLA4 | [64,65,66] | |||
| Pancreatic cancer | METTL3 | lncRNA MALAT1 | positive | Immune-checkpointinhibition,PD-L1 | [67,68] | |||
| METTL16 | B and CD8+ T cell | positive | Immune-checkpointinhibition | [69] | ||||
| Colorectal cancer | YTHDF1 | CD8+Tcell, T cells and NK cells | IFN-γ-related gene | negative | Immune-checkpointinhibition,PD-1 | [70] | ||
| granulocytic myeloid-derived suppressor cells (G-MDSCs), neutrophils | YTHDF1-p65-CXCL1/CXCR2 axis | positive | [71] | |||||
| METTL14 | dysfunctional T cell | negative | ||||||
| METTL3 METTL14 | CD8+ T cells | Stat1 | IFN-γ-Stat1-Irf1 signaling pathway | negative | [72] | |||
| ALKBH5 | CD8+ T cells | NF-κB-CCL5 | positive | [73] | ||||
| IGF2BP3 | NK CD56dim cells, T helper cells, eosinophils, DC, and macrophage neutrophils | positive | Immune-checkpointinhibition,PD-1, CTLA-4 | [74,75] | ||||
| Gallbladder cancer | YTHDF2 | DAPK3 | negative | Chemotherapy, Gemcitabine | [76] | |||
| Respiratory system | lung adenocarcinoma | LRPPRC | DC, CD8+ cytotoxic T cells, macrophage | negative | Immune-checkpointinhibition,PD-1 | [77] | ||
| METTL3 | CD8 T cells | activation of chemokine signaling pathways, T cell receptor signaling pathways, and B cell receptor signaling pathway | negative | Immune-checkpointinhibition,PD-1 (Nivoumab) | [78] | |||
| IGF2BP2 CBLL1 HNRNPA2B1 | CD4 memory activated T cells, resting NK cells, M0 macrophages, M1 macrophages, and resting mast cells | positive | Immune-checkpointinhibition,LAG3 | [79,80] | ||||
| YTHDF2 YTHDF3 FTO | negative | Immune-checkpointinhibition, LAG3 | [80] | |||||
| Nasopharyngeal Carcinoma | LRPPRC | Th2 cell | negative | Immune-checkpointinhibition,PD-L1 | [81] | |||
| ALKBH5 | NK cells, T cells (CD8+ T and CD4+ T), DCs, M1 macrophages | RIG-I | ALKBH5/RIG-I/IFNα | negative | [82] | |||
| Endocrine System | Thyroid cancer | METTL3 | CD70 expression and M2 macrophages and Tregs infiltration | negative | Targeted therapy,cusatuzumab | [83] | ||
| Pituitary adenomas | YTHDF2 | M2 macrophages | positive | Immune-checkpointinhibition,PD-L1 | [84] | |||
| Nervous System | Glioblastoma | FTO ZC3H13 YTHDC1 | TCM cell | positive | [85] | |||
| FTO ZC3H13 | macrophages | negative | [85] | |||||
| ALKBH5 | CD3, CD4, CD8 T cells | negative | Immune-checkpointinhibition,PD-L1 | [86] | ||||
| METTL4 WTAP VIRMA | CD8 + T cells, macrophage cells, and myeloid dendritic cells | TFRC | positive | Immune-checkpointinhibition,PD-L1(cd274),CTLA4 | [87] | |||
| WTAP | macrophages | HSPA7 | YAP1-LOX | positive | Immune-checkpointinhibition,PD-1 | [88] | ||
| Urinary System | Prostate cancer | METTL14 ZC3H13 | Th1 cells, Th17 cells, stromal fraction and TGF-beta response | positive | Immune-checkpointinhibition,PD-1 | [39,89] | ||
| HNRNPC | Treg cells | positive | Immune-checkpointinhibition,CTLA-4 | [90] | ||||
| effector CD8 T cell | negative | [90] | ||||||
| Kidney renal clear cell carcinoma | METTL14 | CD4+ T cells, CD8+ T cells, neutrophils, natural killer (NK) cells, myeloid dendritic cells, macrophages, and B cells | NF-κB pathway | positive | Immune-checkpointinhibition, CD274(PD-L1),HAVCR2,PDCD1LG2(PD-L2),SIGLEC15,TIGIT | [91,92,93] | ||
| METTL14 RBM15B VIRMA ZC3H13 FTO YTHDF2 YTHDC1 HNRNPC1 | CD56 bright NK cells, mast cells, NK cells, pDC | CDH13 | positive | Immune-checkpointinhibition,PDCD1,CD274,PDCD1LG2,CTLA4,LAG3 | [94,95,96] | |||
| METTL5 | NK T cell, naïve CD8+T cell, effector memory CD8+ T cell, naïve CD4+ T cell, central memory CD4+ T cell, T helper 1 (Th1) CD4+ T cell, plasma cytoid dendritic cell, myeloid dendritic cell, mast cell, macrophage M2, macrophage M1, macrophage, eosinophil, class-switched memory B cell, plasma B cell, naïve B cell, memory B cell and B cell | negative | [97] | |||||
| METTL14 ALKBH5 YTHDC1 RBM15B LRPPRC IGF2BP2 IGF2BP3 | NK cells, neutrophils, endothelial cell | ALDOB | positive or negative correlation is unknown | [98] | ||||
| Bladder cancer | FTO IGF2BP3 | M2 macrophage | PGM1, ENO1 | positive | [99] | |||
| YTHDC1 | M2 macrophage | PGM1, ENO1 | negative | [99] | ||||
| Reproductive System | Cervical cancer | METTL16 YTHDF1 ZC3H13 | negative | Immune-checkpointinhibition,PD-L1 | [100] | |||
| IGF2BP3 | Treg cells | GLS, GLUD1 | positive | [101,102] | ||||
| METTL3 | CD33+MDSCs | positive | [103] | |||||
| Endometrial cancer | METTL14 ZC3H13 YTHDC1 | CD4+ Tell | positive | Immune-checkpointinhibition,PD-L1 | [104] | |||
| macrophage | negative | [105,106] | ||||||
| Ovarian cancer | NK, T central memory cells (Tcm) and T gamma delta cells | CDC42EP3 | positive | [107,108] | ||||
| Breast Cancer | YTHDF1 | T cells CD4 memory activated, macrophages M1, NK cells | WNT pathway | positive | Immune-checkpointinhibition | [109] | ||
| HNRNPC VIRMA | CD8 T cells | TFAP2A/DDR1 | negative | [110] | ||||
| Musculoskeletal System | Melanoma | FTO | Immune-checkpointinhibition,PD-1, CXCR4, SOX10 | [111,112] | ||||
| ALKBH5 | Tregs, PMN-MDSCs | negative | [113,114] | |||||
| DCs | positive | Immune-checkpointinhibition,PD-1 | [113,114] | |||||
| Acute myeloid leukaemia | METTL14 | most immune cells | negative | Immune-checkpointinhibition,CTAL4 (negative) | [115] | |||
| ZC3H13 | most immune cells | positive | Immune-checkpointinhibition,CTAL4 (negative) | [115] | ||||
| YTHDC2 | positive | Immune-checkpointinhibition,PD-L1 | [115] | |||||
| RBM15 | negative | Immune-checkpointinhibition,PD-L1 | [115] |
| Autoimmune Disease | m6A Regulator | Immune Cell Type | Immunity Expression | Target Gene | Mechanism | Treatment | Refs. |
|---|---|---|---|---|---|---|---|
| Rheumatoid arthritis (RA) | METTL3 | Macrophage polarization (M1) | positive | STAT1 | STAT1 methylation | Sarsasapogenin (Sar) | [41] |
| IGF2BP3 | Macrophage polarization (M1) | positive | Fibroblast-like synovial cells (FLS) | G2/M transition | Infliximab | [213] | |
| METTL3 | Macrophage | negative | NF-κB signaling pathway | [214] | |||
| Systemic lupus erythematosus (SLE) | METTL3 | Treg | negative | SOCS | il-2-stat5 signaling pathway | [215,216] | |
| METTL3 | B cell | positive | FOXO3 | [217] | |||
| METTL3 | CD4+ T cell, Th1 cell, Th17 cell | positive | [218] | ||||
| ALKBH5 | T cell | positive | [219,220] | ||||
| Inflammatory bowel disease (IBD) | YTHDC1 | Macrophage | positive | Fine-tuned NME nucleoside diphosphokinase 1 (NME1) | Vitolizumab | [221] | |
| METTL14 | T cell | positive | Naïve T cells induce inflammation | Coptisine (COP) | [222] | ||
| FTO | Th17 | positive | [223] | ||||
| Primary Sjögren’s syndrome (pSS) | METTL3 | Dentric cell and T cell | positive | [224] | |||
| Mastocyte | positive | FMR1 | [225] | ||||
| ankylosing spondylitis (AS) | METTL14 | Mesenchymal stem cells (MSC) migration | The ALKBH5-PRMT6 axis controls the activation of AKT signaling pathway | [226] | |||
| YTHDF2, ALKBH5 | Peripheral blood mononuclear cell (PBMC) | negative | [227] | ||||
| Autoimmune Hepatitis (AIH) | YTHDF2 | Bone marrow-derived suppressor cells (MDSCs) | positive | [228] | |||
| Autoimmune encephalomyelitis (EAE) | ALKBH5 | CD4+ T cell | negative | IFNG, CXCL2 | IL-17 signaling pathway | [229] | |
| Allergic respiratory Disease (ARD) | METTL3 | M2 macrophage | negative | PI3K/AKT and JAK/STAT6 signaling pathway | Methylprednisolone, Chelidonine, Nerabine, Ribavirin | [230] | |
| HIV | YTHDF3 | CD4+ T cell | negative | Increasing HIV-1 reverse transcription | Protease inhibitors (indinavir, amprenavir, etc.) | [231,232,233] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mu, S.; Zhao, K.; Zhong, S.; Wang, Y. The Role of m6A Methylation in Tumor Immunity and Immune-Associated Disorder. Biomolecules 2024, 14, 1042. https://doi.org/10.3390/biom14081042
Mu S, Zhao K, Zhong S, Wang Y. The Role of m6A Methylation in Tumor Immunity and Immune-Associated Disorder. Biomolecules. 2024; 14(8):1042. https://doi.org/10.3390/biom14081042
Chicago/Turabian StyleMu, Siyu, Kaiyue Zhao, Shanshan Zhong, and Yanli Wang. 2024. "The Role of m6A Methylation in Tumor Immunity and Immune-Associated Disorder" Biomolecules 14, no. 8: 1042. https://doi.org/10.3390/biom14081042
APA StyleMu, S., Zhao, K., Zhong, S., & Wang, Y. (2024). The Role of m6A Methylation in Tumor Immunity and Immune-Associated Disorder. Biomolecules, 14(8), 1042. https://doi.org/10.3390/biom14081042







