Detecting the FLJ22447 lncRNA in Ovarian Cancer with Cyclopentane-Modified FIT-PNAs (cpFIT-PNAs)
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemical Synthesis of FIT-PNAs and cpFIT-PNAs
2.1.1. General Procedures and Materials
2.1.2. Solid-Phase Synthesis of FIT-PNA
Coupling of First Amino Acid (L-Lysine) onto NovaSyn TGA Resin
- -
- Fmoc Cleavage. A solution of DMF/piperidine (4:1, 1 mL) was added to the resin. After 2 min, the procedure was repeated. Finally, the resin was washed with DMF (3 × 1 mL) and DCM (3 × 1 mL).
- -
- Coupling of Fmoc-Bhoc-PNA-Monomers. An amount of 4 eq. of PNA monomer, 4 eq. HATU, 4 eq. HOBt, and 8 eq. of dry DIPEA in DMF (to 0.1 M PNA) were mixed in a glass vial equipped with a screw cap. After 3 min of pre-activation, the solution was transferred to the resin. After 60 min, the reaction mixture was discarded, and the resin was washed with DMF (2 × 1 mL) and DCM (2 × 1 mL).
- -
- Coupling of BisQ. An amount of 4 eq. of BisQ monomer, 4 eq. HATU, 4 eq. HOBt, and 8 eq. of dry DIPEA in DMF (to 0.1 M BisQ monomer) were mixed in a glass vial equipped with screw cap. Following 3 min of pre-activation, the solution was transferred to the resin. After 60 min, the procedure was repeated, and finally, the resin was washed with DMF (2 × 1 mL) and DCM (2 × 1 mL).
- -
- Cleavage of PNA from resin. An amount of 1 ml TFA was added to the dry resin. After 2 h, another portion of TFA was added. The combined TFA solutions were concentrated in vacuo.
- -
- PNA Purification. PNAs were precipitated from the concentrated TFA solution by addition of cold diethyl ether (10 mL). The precipitate was collected by centrifugation and decantation of the supernatant. The residue was dissolved in water and purified by semi-preparative HPLC. The purified PNAs were analyzed by ESI-MS. In some FIT-PNAs (e.g., K4 FIT-PNA, Table 1) a difference of 5 Daltons is found between observed and predicted masses, which is in the range of the MS accuracy (below 0.1%).
2.2. In Vitro Studies with Synthetic RNA
2.2.1. Tm Measurements
2.2.2. Fluorescence Measurements
2.3. Cell Lines and Culture Conditions
2.4. RT-qPCR
2.5. Confocal Microscopy Analysis
2.6. Flow Cytometry Analysis
2.7. MTT Assay
2.8. Statistical Analysis
3. Results
3.1. FIT-PNA and cpFIT-PNA Design and Synthesis
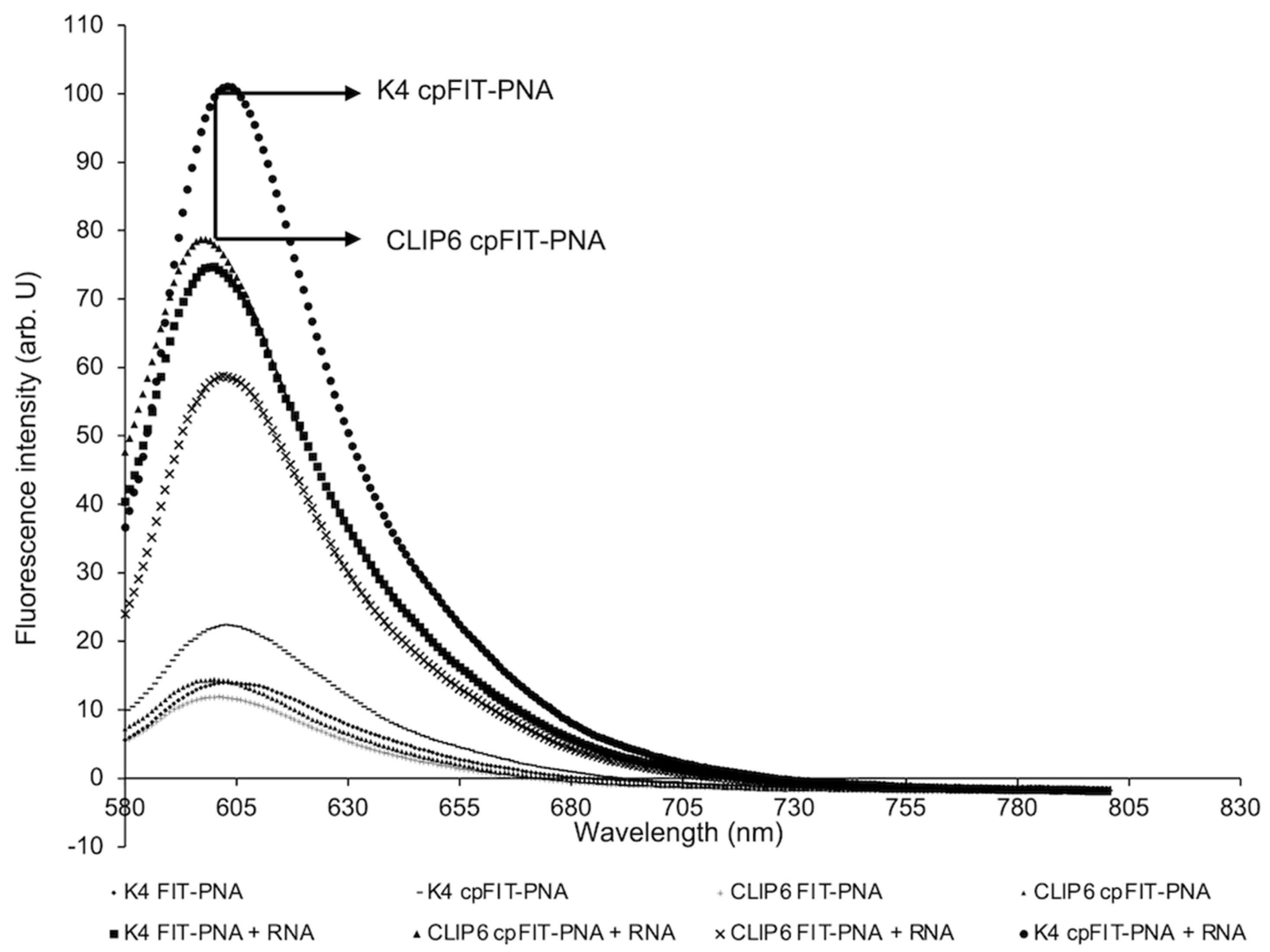
3.2. Flourescence Enhancements and Binding Affinities of FIT-PNAs and cpFIT-PNA with Synthetic RNA
3.3. FIT-PNAs and cpFIT-PNAs Detect FLJ22447 lncRNA in Living OVCAR8 Cells and CAFs
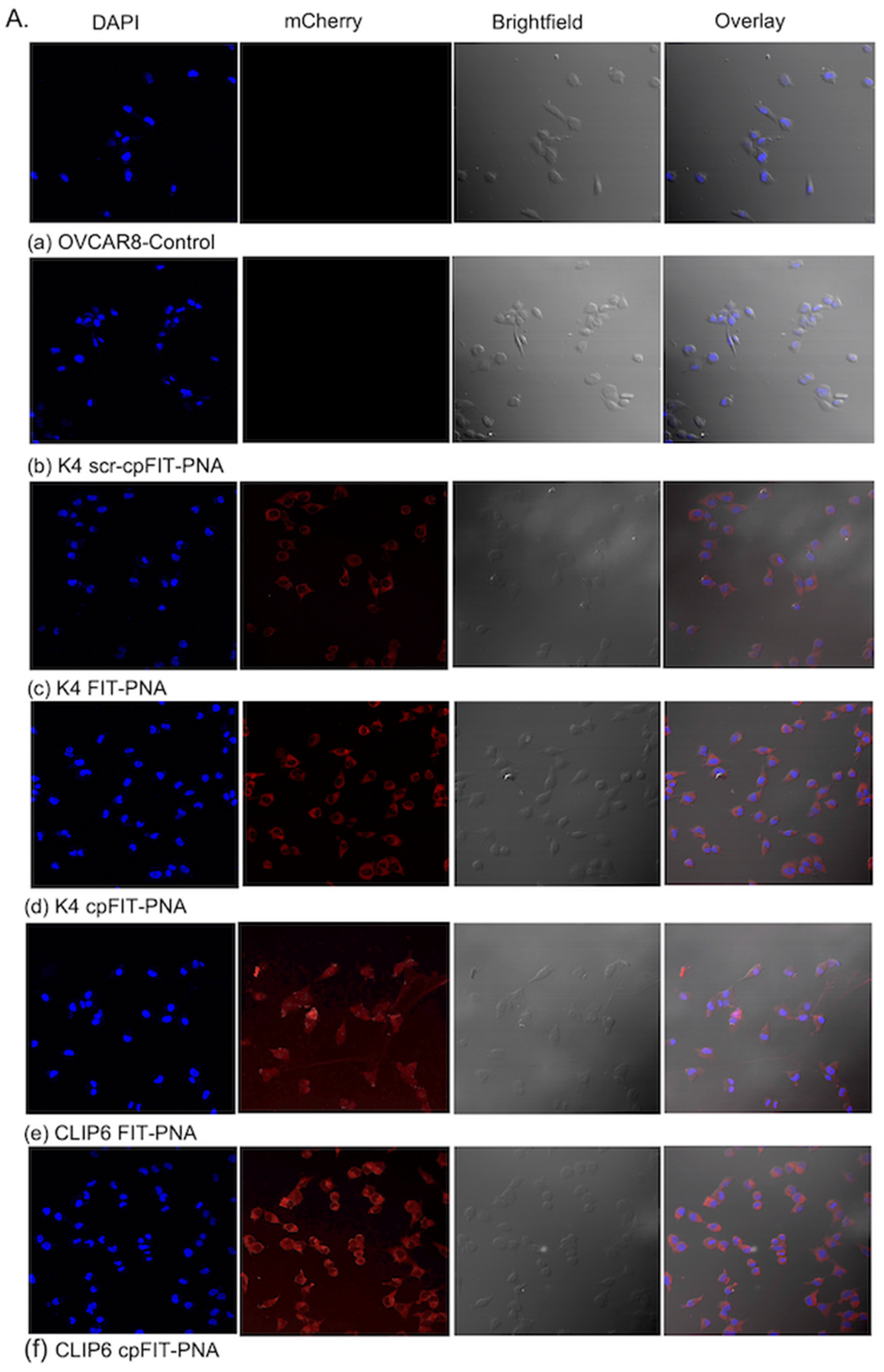
3.3.1. Cellular Uptake of FIT-PNAs and cpFIT-PNA in OVCAR8 Cells as Corroborated by Confocal Microscopy
3.3.2. Cellular Uptake of FIT-PNAs and cpFIT-PNA in OVCAR8 Cells and CAFs as Corroborated by FACS Analysis
3.3.3. FLJ22447 lncRNA Expression in OVCAR8 Cells and CAFs as Corroborated by RT-qPCR
3.3.4. cpFIT-PNAs Downregulate FLJ22447 lncRNA Expression
3.3.5. MTT Assay to Determine the Effect of cpFIT-PNAs on Cell Viability
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics. CA Cancer J. Clin. 2019, 69, 7–34. [Google Scholar] [CrossRef] [PubMed]
- Labiche, A.; Heutte, N.; Herlin, P.; Chasle, J.; Gauduchon, P.; Elie, N. Stromal compartment as a survival prognostic factor in advanced ovarian carcinoma. Int. J. Gynecol. Cancer 2010, 20, 28–33. [Google Scholar] [CrossRef]
- Aprile, M.; Costa, V.; Cimmino, A.; Calin, G.A. Emerging role of oncogenic long noncoding RNA as cancer biomarkers. Int. J. Cancer 2023, 152, 822–834. [Google Scholar] [CrossRef] [PubMed]
- Huarte, M. The emerging role of lncRNAs in cancer. Nat. Med. 2015, 21, 1253–1261. [Google Scholar] [CrossRef] [PubMed]
- Bahramian, S.; Sahebi, R.; Roohinejad, Z.; Delshad, E.; Javid, N.; Amini, A.; Razavi, A.E.; Shafiee, M.; Shamsabadi, F.T. Low expression of lncRNA-CAF attributed to the high expression of HIF1A in esophageal squamous cell carcinoma and gastric cancer patients. Mol. Biol. Rep. 2022, 49, 895–905. [Google Scholar] [CrossRef] [PubMed]
- Ding, L.; Ren, J.; Zhang, D.Y.; Li, Y.; Huang, X.F.; Hu, Q.G.; Wang, H.; Song, Y.X.; Ni, Y.H.; Hou, Y.Y. A novel stromal lncRNA signature reprograms fibroblasts to promote the growth of oral squamous cell carcinoma via lncRNA-CAF/interleukin-33. Carcinogenesis 2018, 39, 397–406. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, P.E.; Egholm, M.; Berg, R.H.; Buchardt, O. Sequence-selective recognition of DNA by strand displacement with a thymine-substituted polyamide. Science 1991, 254, 1497–1500. [Google Scholar] [CrossRef]
- Egholm, M.; Buchardt, O.; Christensen, L.; Behrens, C.; Freier, S.M.; Driver, D.A.; Berg, R.H.; Kim, S.K.; Norden, B.; Nielsen, P.E. PNA hybridizes to complementary oligonucleotides obeying the Watson-Crick hydrogen-bonding rules. Nature 1993, 365, 566–568. [Google Scholar] [CrossRef]
- Saarbach, J.; Sabale, P.M.; Winssinger, N. Peptide nucleic acid (PNA) and its applications in chemical biology, diagnostics, and therapeutics. Curr. Opin. Chem. Biol. 2019, 52, 112–124. [Google Scholar] [CrossRef] [PubMed]
- Brodyagin, N.; Katkevics, M.; Kotikam, V.; Ryan, C.A.; Rozners, E. Chemical approaches to discover the full potential of peptide nucleic acids in biomedical applications. Beilstein J. Org. Chem. 2021, 17, 1641–1688. [Google Scholar]
- Perera, J.D.R.; Carufe, K.E.W.; Glazer, P.M. Peptide nucleic acids and their role in gene regulation and editing. Biopolymers 2021, 112, e23460. [Google Scholar] [CrossRef]
- Gupta, A.; Mishra, A.; Puri, N. Peptide nucleic acids: Advanced tools for biomedical applications. J. Biotechnol. 2017, 259, 148–159. [Google Scholar] [CrossRef] [PubMed]
- Pradeep, S.P.; Malik, S.; Slack, F.J.; Bahal, R. Unlocking the potential of chemically modified peptide nucleic acids for RNA-based therapeutics. RNA 2023, 29, 434–445. [Google Scholar] [CrossRef]
- Montazersaheb, S.; Hejazi, M.S.; Nozad Charoudeh, H. Potential of peptide nucleic acids in future therapeutic applications. Adv. Pharm. Bull. 2018, 8, 551–563. [Google Scholar] [CrossRef] [PubMed]
- MacLelland, V.; Kravitz, M.; Gupta, A. Therapeutic and diagnostic applications of antisense peptide nucleic acids. Mol. Ther. Nucleic Acids 2023, 35, 102086. [Google Scholar] [CrossRef]
- Vilaivan, T. Fluorogenic PNA probes. Beilstein. J. Org. Chem. 2018, 14, 253–281. [Google Scholar] [CrossRef]
- Koehler, O.; Jarikote, D.V.; Seitz, O. Forced intercalation probes (FIT Probes): Thiazole orange as a fluorescent base in peptide nucleic acids for homogeneous single-nucleotide-polymorphism detection. ChemBioChem 2005, 6, 69–77. [Google Scholar] [CrossRef]
- Socher, E.; Jarikote, D.V.; Knoll, A.; Roeglin, L.; Burmeister, J.; Seitz, O. FIT probes: Peptide nucleic acid probes with a fluorescent base surrogate enable real-time DNA quantification and single nucleotide polymorphism discovery. Anal. Biochem. 2008, 375, 318–330. [Google Scholar] [CrossRef]
- Karunakaran, V.; Pérez Lustres, J.L.; Zhao, L.; Ernsting, N.P.; Seitz, O. Large dynamic stokes shift of DNA intercalation dye thiazole orange has contribution from a high-frequency mode. J. Am. Chem. Soc. 2006, 128, 2954–2962. [Google Scholar] [CrossRef]
- Hashoul, D.; Shapira, R.; Falchenko, M.; Tepper, O.; Paviov, V.; Nissan, A.; Yavin, E. Red-emitting FIT-PNAs: “On site” detection of RNA biomarkers in fresh human cancer tissues. Biosens Bioelectron. 2019, 137, 271–278. [Google Scholar] [CrossRef]
- Kam, Y.; Rubinstein, A.; Naik, S.; Djavsarov, I.; Halle, D.; Ariel, I.; Gure, A.O.; Stojadinovic, A.; Pan, H.; Tsivin, V.; et al. Detection of a long non-coding RNA (CCAT1) in living cells and human adenocarcinoma of colon tissues using FIT-PNA molecular beacons. Cancer Lett. 2014, 352, 90–96. [Google Scholar] [CrossRef] [PubMed]
- Loibl, N.; Arenz, C.; Seitz, O. Monitoring dicer-mediated miRNA-21 maturation and Ago2 loading by a dual-colour FIT PNA probe set. Chembiochem 2020, 21, 2527–2532. [Google Scholar] [CrossRef] [PubMed]
- Kolevzon, N.; Hashoul, D.; Naik, S.; Rubinstein, A.; Yavin, E. Single point mutation detection in living cancer cells by far-red emitting PNA–FIT probes. Chem. Commun. 2016, 52, 2405–2407. [Google Scholar] [CrossRef]
- Sonar, M.V.; Wampole, M.E.; Jin, Y.Y.; Chen, C.P.; Thakur, M.L.; Wickstrom, E. Fluorescence detection of KRAS2 mRNA hybridization in lung cancer cells with PNA-peptides containing an internal thiazole orange. Bioconjug. Chem. 2014, 25, 1697–1708. [Google Scholar] [CrossRef] [PubMed]
- Kummer, S.; Knoll, A.; Socher, E.; Bethge, L.; Herrmann, A.; Seitz, O. Fluorescence imaging of Influenza H1N1 mRNA in living infected cells using single-chromophore FIT-PNA. Angew. Chem. Int. Ed. 2011, 50, 1931–1934. [Google Scholar] [CrossRef] [PubMed]
- Kummer, S.; Knoll, A.; Socher, E.; Bethge, L.; Herrmann, A.; Seitz, O. PNA FIT-probes for the dual color imaging of two viral mRNA targets in Influenza H1N1 infected live cells. Bioconjug. Chem. 2012, 23, 2051–2060. [Google Scholar] [CrossRef]
- Sato, Y.; Miura, H.; Tanabe, T.; Okeke, C.U.; Kikuchi, A.; Nishizawa, S. Fluorescence sensing of the panhandle structure of the Influenza A virus RNA promoter by thiazole orange base surrogate-carrying peptide nucleic acid conjugated with small molecule. Anal. Chem. 2022, 94, 7814–7822. [Google Scholar] [CrossRef]
- Tepper, O.; Peled, I.; Fastman, Y.; Heinberg, A.; Mitesser, V.; Dzikowski, R.; Yavin, E. FIT-PNAs as RNA-sensing probes for drug-resistant Plasmodium falciparum. ACS Sens. 2022, 7, 50–59. [Google Scholar] [CrossRef] [PubMed]
- Lee, E.T.T.; Sato, Y.; Nishizawa, S. Small molecule–PNA oligomer conjugates for rRNA A-site at neutral pH for FID assays. Chem. Commun. 2020, 56, 14976–14979. [Google Scholar] [CrossRef]
- Okeke, C.U.; Miura, H.; Sato, Y.; Nishizawa, S. Kinetic analysis of highly effective triplex formation between a small molecule–peptide nucleic acid conjugate probe and the influenza A virus RNA promoter region at neutral pH. Org. Biomol. Chem. 2023, 21, 3402–3410. [Google Scholar] [CrossRef]
- Zheng, H.; Botos, I.; Clausse, V.; Nikolayevskiy, H.; Rastede, E.E.; Fouz, M.F.; Mazur, S.J.; Appella, D.H. Conformational constraints of cyclopentane peptide nucleic acids facilitate tunable binding to DNA. Nucl. Acids Res. 2021, 49, 713–725. [Google Scholar] [CrossRef] [PubMed]
- Zheng, H.C.; Clausse, V.; Amarasekara, H.; Mazur, S.J.; Botos, I.; Appella, D.H. Variation of tetrahydrofurans in thyclotides enhances oligonucleotide binding and cellular uptake of peptide nucleic acids. JACS AU 2023, 3, 1952–1964. [Google Scholar] [CrossRef] [PubMed]
- Tepper, O.; Zheng, H.C.; Appella, D.H.; Yavin, E. Cyclopentane FIT-PNAs: Bright RNA sensors. Chem. Commun. 2021, 57, 540–543, Erratum in Chem. Commun. 2023, 59, 11593. [Google Scholar] [CrossRef] [PubMed]
- Tepper, O.; Appella, D.H.; Zheng, H.; Dzikowski, R.; Yavin, E. A biotinylated cpFIT-PNA platform for the facile detection of drug resistance to artemisinin in plasmodium falciparum. ACS Sens. 2024, 9, 1458–1464. [Google Scholar] [CrossRef] [PubMed]
- Tan, X.; Bruchez, M.P.; Armitage, B.A. Closing the loop: Constraining TAT peptide by γPNA hairpin for enhanced cellular delivery of biomolecules. Bioconjug. Chem. 2018, 29, 2892–2898. [Google Scholar] [CrossRef]
- Hakata, Y.; Ishikawa, S.; Ohtsuki, T.; Miyazawa, M.; Kitamatsu, M. Intracellular delivery of a peptide nucleic acid-based hybrid of an autophagy inducing peptide with a cell-penetrating peptide. Org. Biomol. Chem. 2020, 18, 1978–1986. [Google Scholar] [CrossRef] [PubMed]
- Kaplank, A.R.; Pham, H.; Liu, Y.; Oyaghire, S.; Bahal, R.; Engelman, D.; Glazer, P.M. Ku80-targeted pH-sensitive peptide-PNA conjugates are tumor selective and sensitize cancer cells to ionizing radiation. Mol. Cancer Res. 2020, 18, 873–882. [Google Scholar] [CrossRef] [PubMed]
- Fabbri, E.; Tamanini, A.; Jakova, T.; Gasparello, J.; Manicardi, A.; Corradini, R.; Finotti, A.; Borgatti, M.; Lampronti, I.; Munari, S.; et al. Treatment of human airway epithelial Calu-3 cells with a peptide-nucleic acid (PNA) targeting the microRNA miR-101-3p is associated with increased expression of the cystic fibrosis transmembrane conductance regulator gene. Eur. J. Med. Chem. 2021, 209, 112876. [Google Scholar] [CrossRef] [PubMed]
- Zurlo, M.; Romagnoli, R.; Oliva, P.; Gasparello, J.; Finotti, A.; Gambari, R. Synergistic effects of the combined treatment of U251 and T98G glioma cells with an anti-tubulin tetrahydrothieno[2,3-c]pyridine derivative and a peptide nucleic acid targeting miR-221-3p. Int. J. Oncol. 2021, 59, 61. [Google Scholar] [CrossRef]
- Gasparello, J.; Papi, C.; Zurlo, M.; Gambari, L.; Rozzi, A.; Manicardi, A.; Corradini, R.; Gambari, R.; Finotti, A. Treatment of human glioblastoma U251 cells with sulforaphane and a peptide nucleic acid (PNA) targeting miR-15b-5p: Synergistic effects on induction of apoptosis. Molecules 2022, 27, 1299. [Google Scholar] [CrossRef]
- Shehzadi, K.; Yu, M.; Liang, J. De novo potent peptide nucleic acid antisense oligomer inhibitors targeting SARS-CoV-2 RNA-dependent RNA polymerase via structure-guided drug design. Int. J. Mol. Sci. 2023, 24, 17473. [Google Scholar] [CrossRef]
- Polak, A.; Machnik, G.; Bułdak, Ł.; Ruczyński, J.; Prochera, K.; Bujak, O.; Mucha, P.; Rekowski, P.; Okopień, B. The application of peptide nucleic acids (PNA) in the inhibition of proprotein convertase subtilisin/kexin 9 (PCSK9) gene expression in a cell-free transcription/translation system. Int. J. Mol. Sci. 2024, 25, 1463. [Google Scholar] [CrossRef] [PubMed]
- George, J.T.; Srivatsan, S.G. Vinyluridine as a versatile chemoselective handle for the post-transcriptional chemical functionalization of RNA. Bioconjug. Chem. 2017, 28, 1529–1536. [Google Scholar] [CrossRef]
- Milani, R.; Brognara, E.; Fabbri, E.; Manicardi, A.; Corradini, R.; Finotti, A.; Gasparello, J.; Borgatti, M.; Cosenza, L.C.; Lampronti, I.; et al. Targeting miR-155-5p and miR-221-3p by peptide nucleic acids induces caspase-3 activation and apoptosis in temozolomide-resistant T98G glioma cells. Int. J. Oncol. 2019, 55, 59–68. [Google Scholar] [CrossRef] [PubMed]
- Ndeboko, B.; Ramamurthy, N.; Lemamy, G.J.; Jamard, C.; Nielsen, P.E.; Cova, L. Role of cell-penetrating peptides in intracellular delivery of peptide nucleic acids targeting hepadnaviral replication. Mol. Ther. Nucleic Acids 2017, 9, 162–169. [Google Scholar] [CrossRef]
- Özeş, A.R.; Wang, Y.; Zong, X.; Fang, F.; Pilrose, J.; Nephew, K.P. Therapeutic targeting using tumor specific peptides inhibits long non-coding RNA HOTAIR activity in ovarian and breast cancer. Sci. Rep. 2017, 7, 894. [Google Scholar] [CrossRef] [PubMed]
- Castillo, J.I.; Równicki, M.; Wojciechowska, M.; Trylska, J. Antimicrobial synergy between mRNA targeted peptide nucleic acid and antibiotics in E. coli. Bioorg. Med. Chem. Lett. 2018, 28, 3094–3098. [Google Scholar] [CrossRef]
- Ghosh, S.; Saini, S.; Saraogi, I. Peptide nucleic acid mediated inhibition of the bacterial signal recognition particle. Chem. Commun. 2018, 54, 8257–8260. [Google Scholar] [CrossRef]
- Lee, H.T.; Kim, S.K.; Lee, J.B.; Yoon, J.W. A novel peptide nucleic acid against the cytidine monophosphate kinase of S. aureus inhibits staphylococcal infection in vivo. Mol. Ther. Nucleic Acids 2019, 18, 245–252. [Google Scholar] [CrossRef]
- Barkowsky, G.; Lemster, A.L.; Pappesch, R.; Jacob, A.; Krüger, S.; Schröder, A.; Kreikemeyer, B.; Patenge, N. Influence of different cell-penetrating peptides on the antimicrobial efficiency of PNAs in streptococcus pyogenes. Mol. Ther. Nucleic Acids 2019, 18, 444–454. [Google Scholar] [CrossRef]
- Ghavami, M.; Shiraishi, T.; Nielsen, P.E. Cooperative cellular uptake and activity of octaarginine antisense peptide nucleic acid (PNA) conjugates. Biomolecules 2019, 9, 554. [Google Scholar] [CrossRef] [PubMed]
- Popella, L.; Jung, J.; Do, P.T.; Hayward, R.J.; Barquist, L.; Vogel, J. Comprehensive analysis of PNA-based antisense antibiotics targeting various essential genes in uropathogenic Escherichia coli. Nucleic Acids Res. 2022, 50, 6435–6452. [Google Scholar] [CrossRef] [PubMed]
- Abt, C.; Gerlach, L.M.; Bull, J.; Jacob, A.; Kreikemeyer, B.; Patenge, N. Pyrenebutyrate enhances the antibacterial effect of peptide-coupled antisense peptide nucleic acids in Streptococcus pyogenes. Microorganisms 2023, 11, 2131. [Google Scholar] [CrossRef] [PubMed]
- Pal, R.; Seleem, M.N. Antisense inhibition of RNA polymerase α subunit of Clostridioides difficile. Microbiol. Spectr. 2023, 11, e0175523. [Google Scholar] [CrossRef] [PubMed]
- Tsylents, U.; Siekierska, I.; Trylska, J. Peptide nucleic acid conjugates and their antimicrobial applications—A mini-review. Eur. Biophys. J. 2023, 52, 533–544. [Google Scholar] [CrossRef] [PubMed]
- Soudah, T.; Mogilevsky, M.; Karni, R.; Yavin, E. CLIP6-PNA-peptide conjugates: Non-endosomal delivery of splice switching oligonucleotides. Bioconjug. Chem. 2017, 28, 3036–3042. [Google Scholar] [CrossRef]
- Medina, S.H.; Miller, S.E.; Keim, A.I.; Gorka, A.P.; Schnermann, M.J.; Schneider, J.P. An intrinsically disordered peptide facilitates non-endosomal cell entry. Angew. Chem. Int. Ed. 2016, 55, 3369–3372. [Google Scholar] [CrossRef] [PubMed]
- Gokhan, G.; Kirit, H.A.; Kamatar, A.; Baghdasaryan, O.; Hamsici, S.; Acar, H. The effects of size and shape of the ovarian cancer spheroids on the drug resistance and migration. Gynecol. Oncol. 2020, 159, 563–572. [Google Scholar] [CrossRef]
- Fang, Z.; Xu, J.; Zhang, B.; Wang, W.; Liu, J.; Liang, C.; Hua, J.; Meng, Q.; Yu, X.; Shi, S. The promising role of noncoding RNAs in cancer-associated fibroblasts: An overview of current status and future perspectives. J. Hematol. Oncol 2020, 13, 154. [Google Scholar] [CrossRef]
- Wang, M.; Jiang, M.; Xie, A.; Zhang, N.; Xu, Y. Identification of CAF-related lncRNAs at the pan-cancer level represents a potential carcinogenic risk. Hum. Mol. Genet. 2024, ddae042. [Google Scholar] [CrossRef]
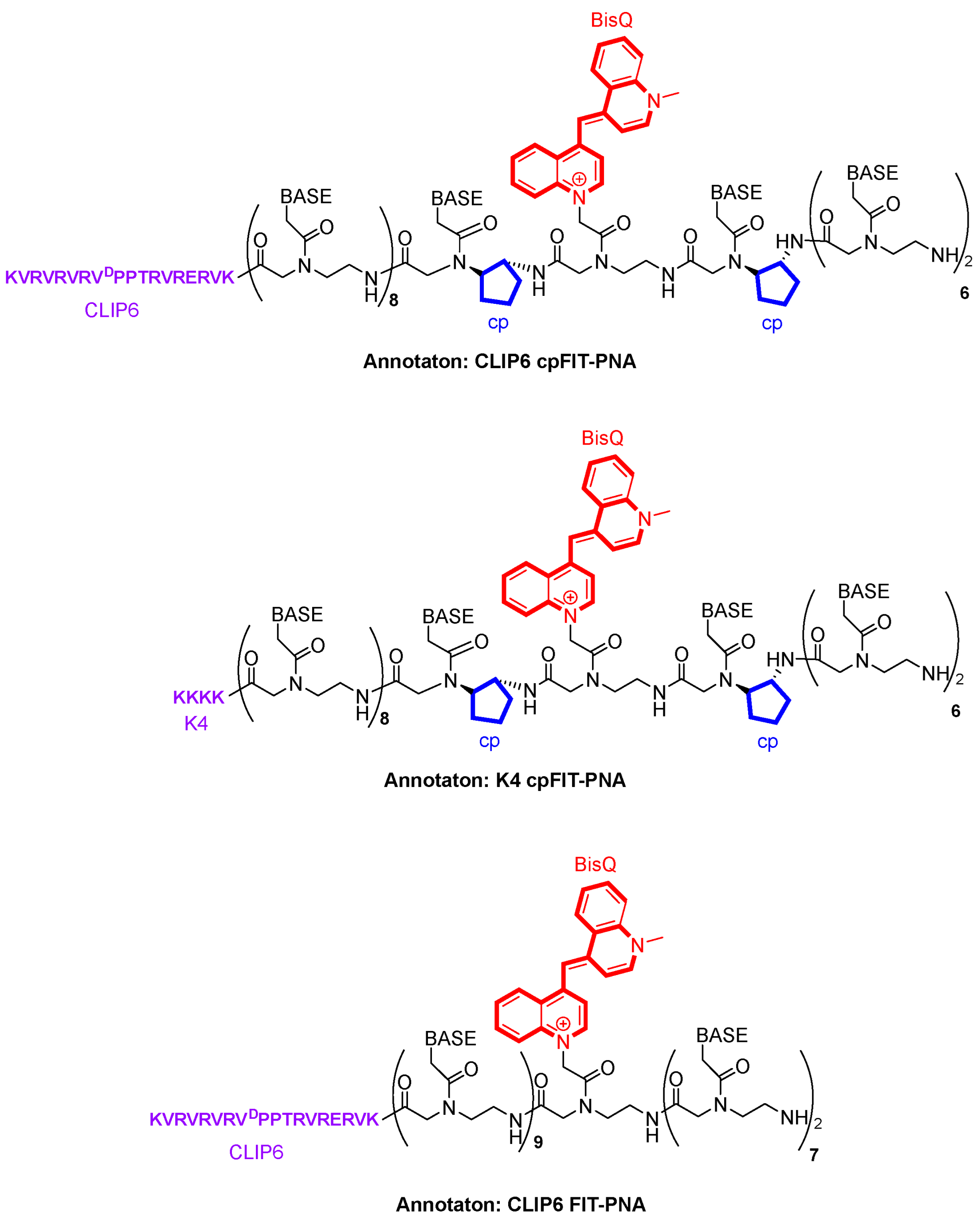






| Sequence | Construct | MW (Found) | MW (calc.) | Tm (°C) | ΔTm (°C) |
|---|---|---|---|---|---|
| K4 FIT-PNA | 3′ K4-TTCTAGATT-BisQ-ACAGTTT 5′ | 5271.3 | 5276.8 | 50.7 | |
| K4 cpFIT-PNA | 3′ K4-TTCTAGAT-cpT-BisQ-cpA-CAGTTT 5′ | 5351.3 | 5356.3 | 54.0 | 3.3 |
| K4 scr-cpFIT-PNA | 3′ K4-TTACGGAT-cpT-BisQ-CTTATAT 5′ | 5309.3 | 5309.3 | ||
| CLIP6 FIT-PNA | 3′ CLIP6-TTCTAGATT-BisQ-ACAGTTT 5′ | 6970.3 | 6975.84 | 59.3 | |
| CLIP6 cpFIT-PNA | 3′ CLIP6-TTCTAGAT-cpT-BisQ-cpA-CAGTTT 5′ | 7050.4 | 7053.96 | 62.3 | 3.0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mannully, S.T.; Mahajna, R.; Nazzal, H.; Maree, S.; Zheng, H.; Appella, D.H.; Reich, R.; Yavin, E. Detecting the FLJ22447 lncRNA in Ovarian Cancer with Cyclopentane-Modified FIT-PNAs (cpFIT-PNAs). Biomolecules 2024, 14, 609. https://doi.org/10.3390/biom14060609
Mannully ST, Mahajna R, Nazzal H, Maree S, Zheng H, Appella DH, Reich R, Yavin E. Detecting the FLJ22447 lncRNA in Ovarian Cancer with Cyclopentane-Modified FIT-PNAs (cpFIT-PNAs). Biomolecules. 2024; 14(6):609. https://doi.org/10.3390/biom14060609
Chicago/Turabian StyleMannully, Sheethal Thomas, Rawan Mahajna, Huda Nazzal, Salam Maree, Hongchao Zheng, Daniel H. Appella, Reuven Reich, and Eylon Yavin. 2024. "Detecting the FLJ22447 lncRNA in Ovarian Cancer with Cyclopentane-Modified FIT-PNAs (cpFIT-PNAs)" Biomolecules 14, no. 6: 609. https://doi.org/10.3390/biom14060609
APA StyleMannully, S. T., Mahajna, R., Nazzal, H., Maree, S., Zheng, H., Appella, D. H., Reich, R., & Yavin, E. (2024). Detecting the FLJ22447 lncRNA in Ovarian Cancer with Cyclopentane-Modified FIT-PNAs (cpFIT-PNAs). Biomolecules, 14(6), 609. https://doi.org/10.3390/biom14060609








