Identification and Validation of a PEX5-Dependent Signature for Prognostic Prediction in Glioma
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Acquisition and Processing
2.2. Construction of the PEX5-Based Prognostic Gene Signature
2.3. Survival Analysis
2.4. Independent Prognostic Role of the Risk Signature
2.5. Construction of the Nomogram
2.6. Differential Expression Gene (DEG) and Functional Enrichment Analysis
2.7. Plasmid Construction
2.8. Cell Culture and PEX5 Knockout Cell Establishment
2.9. qPCR
2.10. Western Blot
2.11. Immunofluorescence
2.12. CCK-8 Assays
2.13. Colony Formation Assay
2.14. Cell Cycle
2.15. Wound-Healing Assay
2.16. Transwell
2.17. Statistical Analysis
3. Results
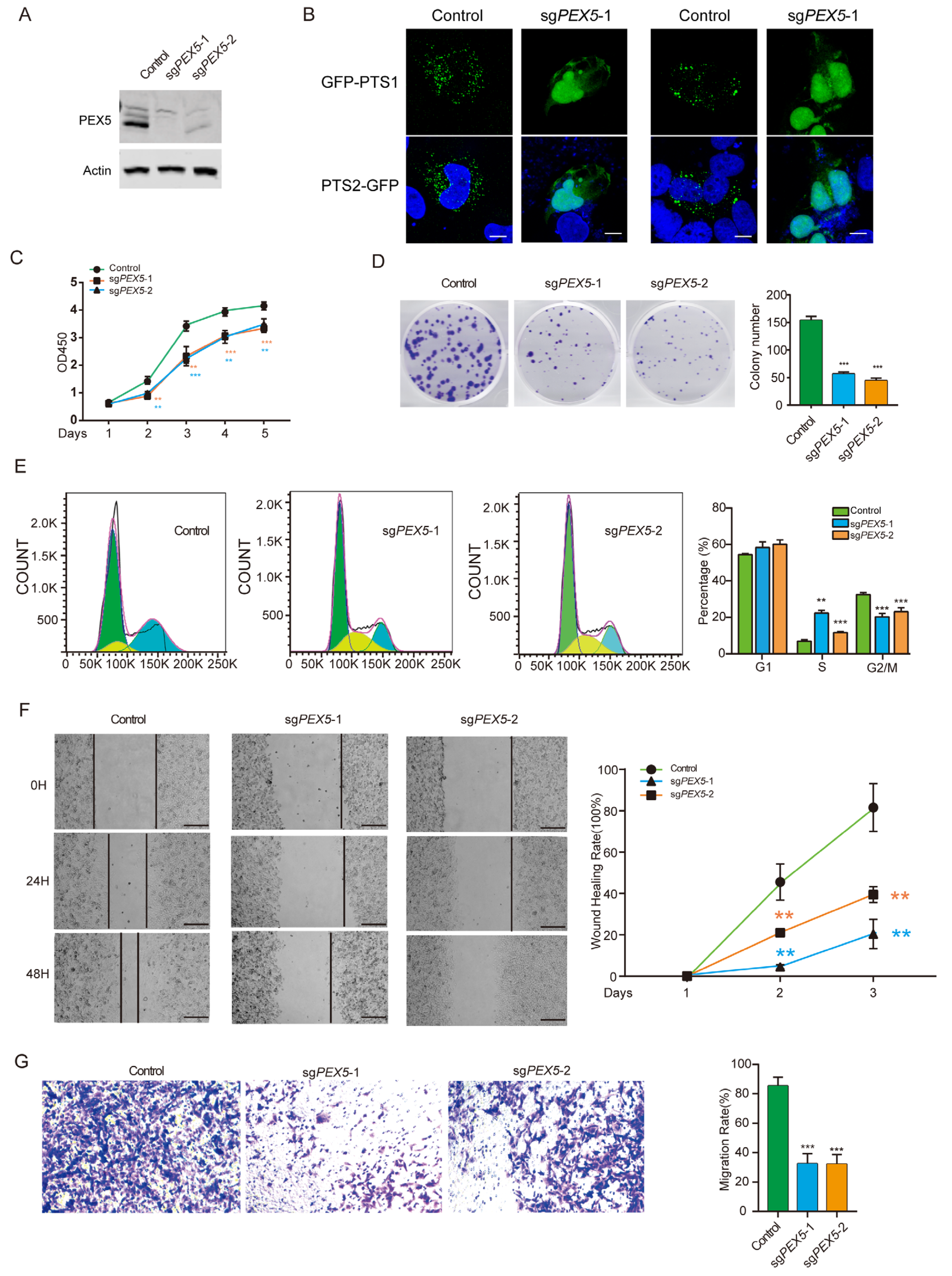
3.1. PEX5 Deletion Inhibits Cell Growth, Migration, and Invasion of Glioma Cells
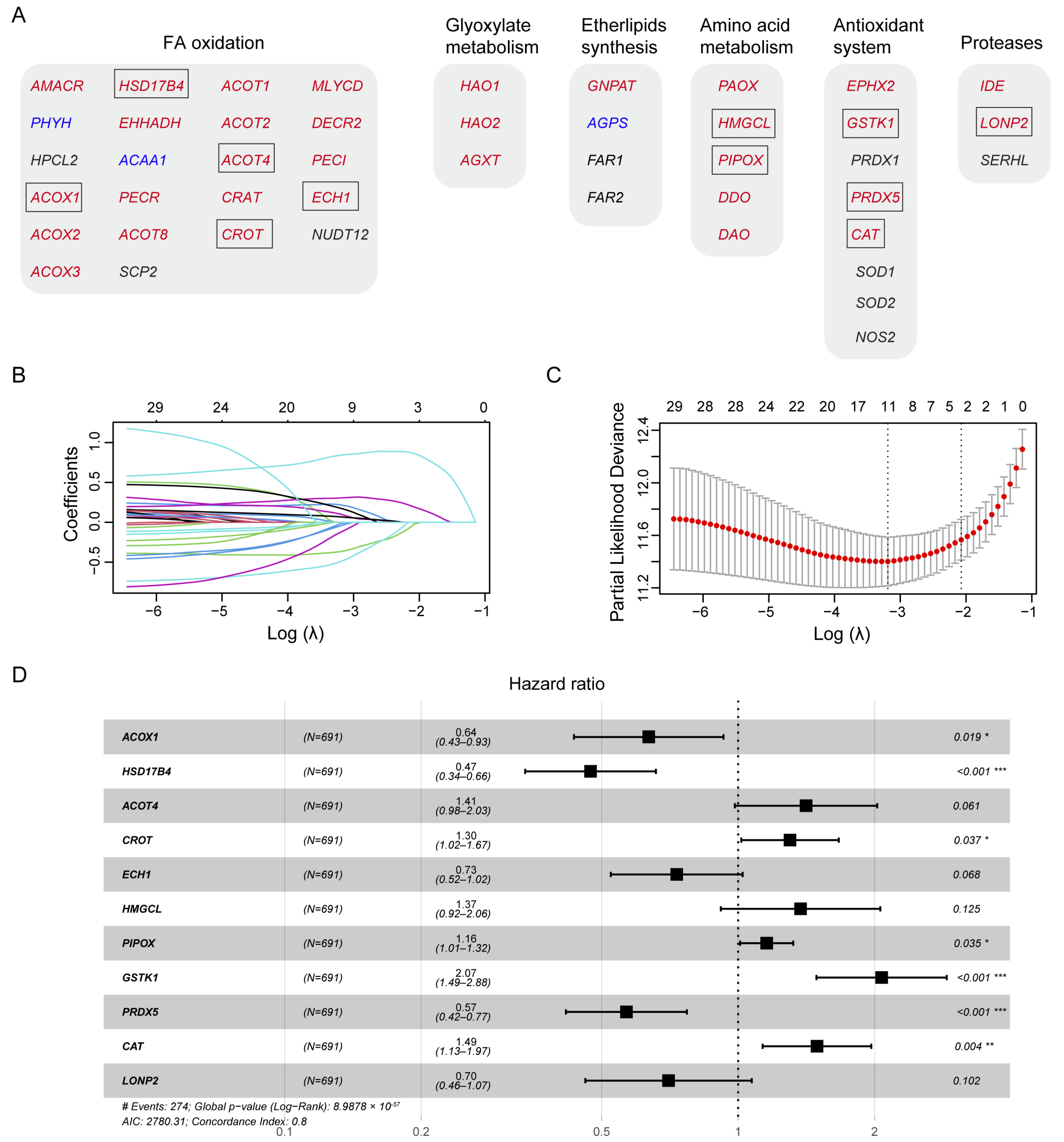
3.2. Construction of A Prognostic Gene Signature in the TCGA Training Cohort
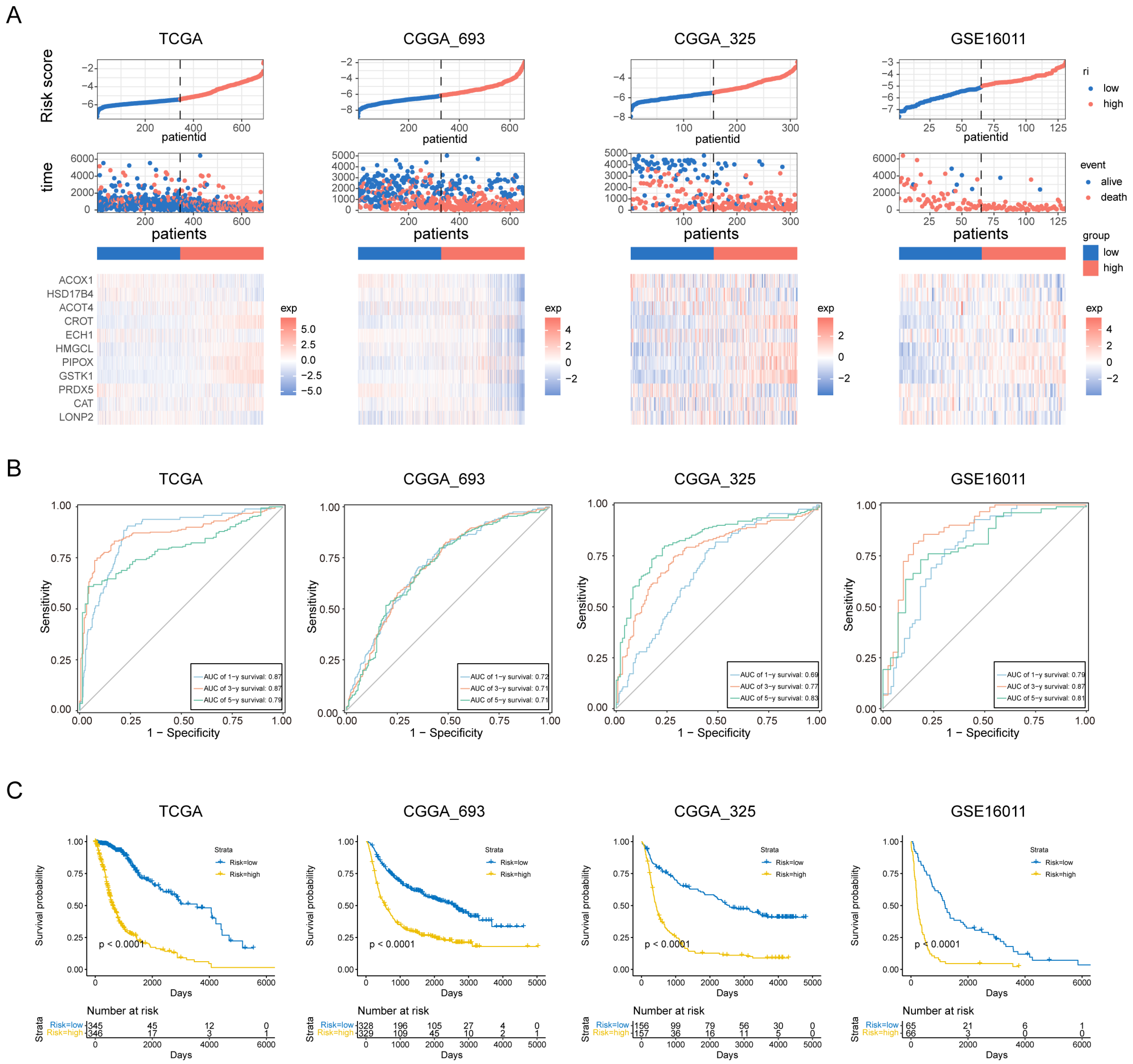
3.3. Evaluation of the Prognosis Model
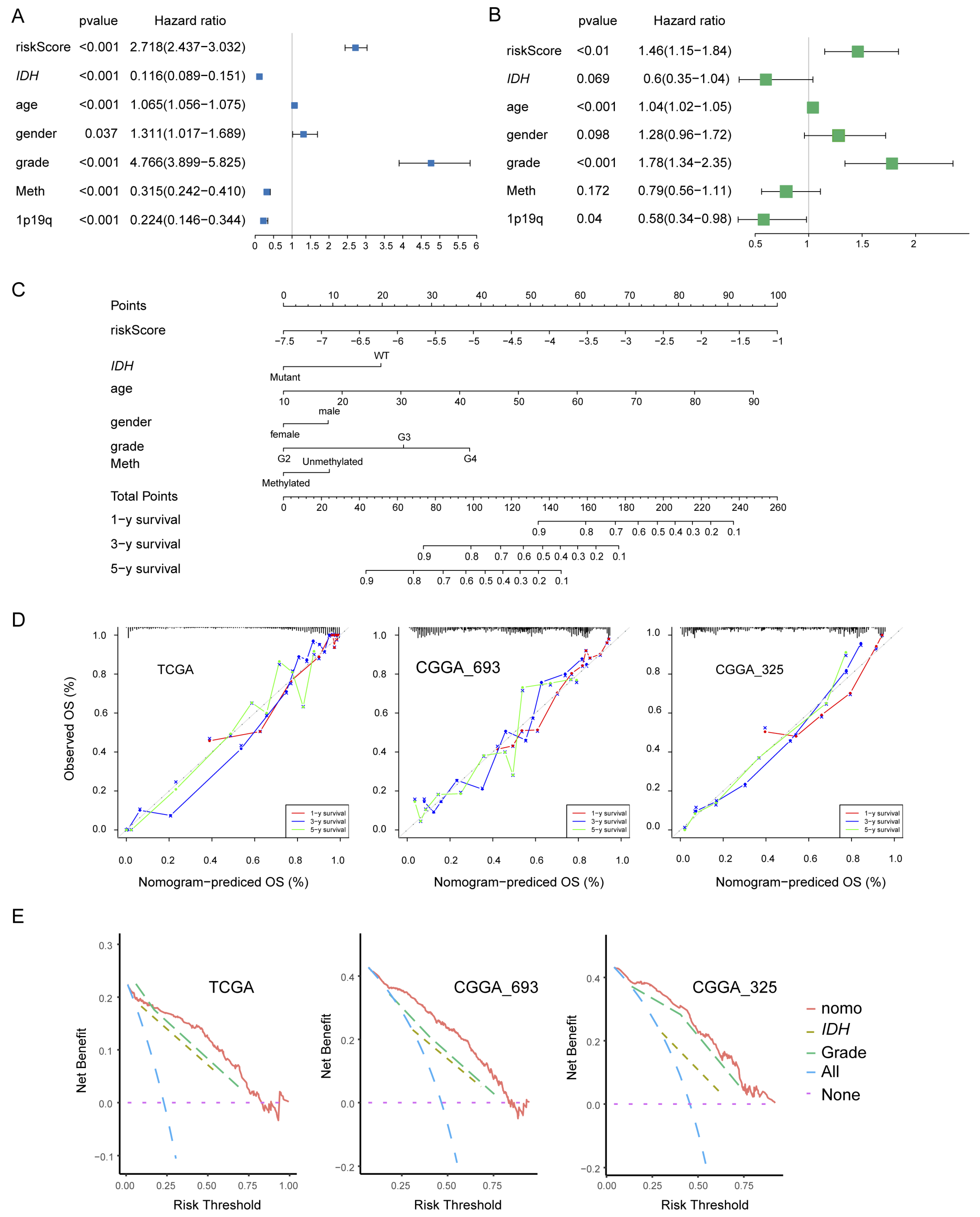
3.4. Construction and Validation of the Nomogram
3.5. High-Risk Group Had An Immunosuppressive Feature
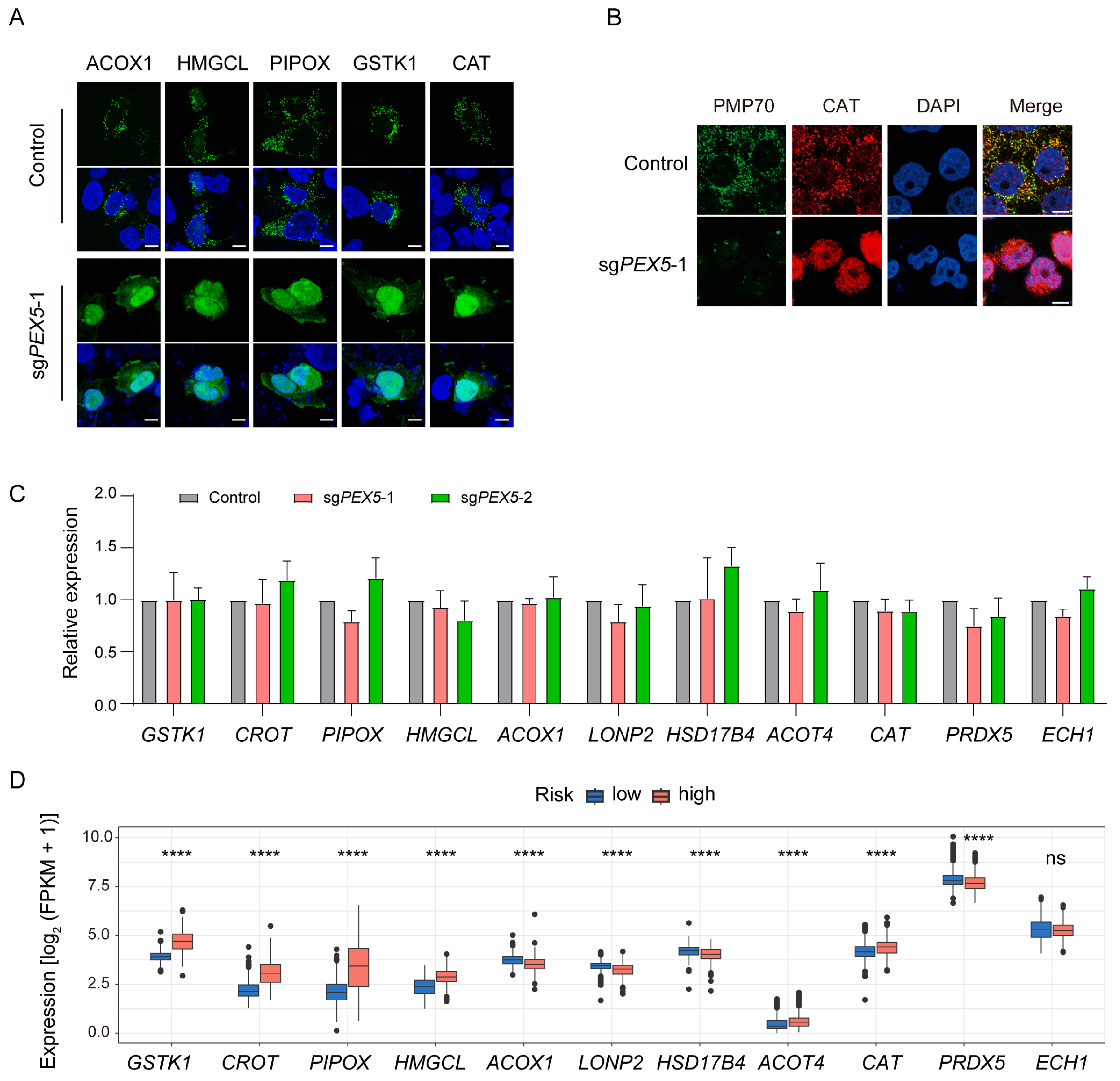
3.6. The Peroxisomal Localization of the Signature Genes Depends on PEX5
3.7. GSTK1 Inhibits the Growth, Migration, and Invasion of Glioma Cells
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Alexander, B.M.; Cloughesy, T.F. Adult Glioblastoma. J. Clin. Oncol. 2017, 35, 2402–2409. [Google Scholar] [CrossRef]
- Louis, D.N.; Perry, A.; Reifenberger, G.; von Deimling, A.; Figarella-Branger, D.; Cavenee, W.K.; Ohgaki, H.; Wiestler, O.D.; Kleihues, P.; Ellison, D.W. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: A summary. Acta Neuropathol. 2016, 131, 803–820. [Google Scholar] [CrossRef]
- Rodriguez-Camacho, A.; Flores-Vazquez, J.G.; Moscardini-Martelli, J.; Torres-Rios, J.A.; Olmos-Guzman, A.; Ortiz-Arce, C.S.; Cid-Sanchez, D.R.; Perez, S.R.; Macias-Gonzalez, M.D.S.; Hernandez-Sanchez, L.C.; et al. Glioblastoma Treatment: State-of-the-Art and Future Perspectives. Int. J. Mol. Sci. 2022, 23, 7207. [Google Scholar] [CrossRef]
- Molinaro, A.M.; Taylor, J.W.; Wiencke, J.K.; Wrensch, M.R. Genetic and molecular epidemiology of adult diffuse glioma. Nat. Rev. Neurol. 2019, 15, 405–417. [Google Scholar] [CrossRef] [PubMed]
- Waterham, H.R.; Ferdinandusse, S.; Wanders, R.J. Human disorders of peroxisome metabolism and biogenesis. Biochim. Biophys. Acta 2016, 1863, 922–933. [Google Scholar] [CrossRef] [PubMed]
- Smith, J.J.; Aitchison, J.D. Peroxisomes take shape. Nat. Rev. Mol. Cell Biol. 2013, 14, 803–817. [Google Scholar] [CrossRef] [PubMed]
- Gould, S.J.; Keller, G.A.; Hosken, N.; Wilkinson, J.; Subramani, S. A conserved tripeptide sorts proteins to peroxisomes. J. Cell Biol. 1989, 108, 1657–1664. [Google Scholar] [CrossRef] [PubMed]
- Swinkels, B.W.; Gould, S.J.; Bodnar, A.G.; Rachubinski, R.A.; Subramani, S. A novel, cleavable peroxisomal targeting signal at the amino-terminus of the rat 3-ketoacyl-CoA thiolase. EMBO J. 1991, 10, 3255–3262. [Google Scholar] [CrossRef] [PubMed]
- Gatto, G.J., Jr.; Geisbrecht, B.V.; Gould, S.J.; Berg, J.M. Peroxisomal targeting signal-1 recognition by the TPR domains of human PEX5. Nat. Struct. Biol. 2000, 7, 1091–1095. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Subramani, S. Role of PEX5 ubiquitination in maintaining peroxisome dynamics and homeostasis. Cell Cycle 2017, 16, 2037–2045. [Google Scholar] [CrossRef] [PubMed]
- Braverman, N.; Dodt, G.; Gould, S.J.; Valle, D. An isoform of pex5p, the human PTS1 receptor, is required for the import of PTS2 proteins into peroxisomes. Hum. Mol. Genet. 1998, 7, 1195–1205. [Google Scholar] [CrossRef]
- Otera, H.; Harano, T.; Honsho, M.; Ghaedi, K.; Mukai, S.; Tanaka, A.; Kawai, A.; Shimizu, N.; Fujiki, Y. The mammalian peroxin Pex5pL, the longer isoform of the mobile peroxisome targeting signal (PTS) type 1 transporter, translocates the Pex7p.PTS2 protein complex into peroxisomes via its initial docking site, Pex14p. J. Biol. Chem. 2000, 275, 21703–21714. [Google Scholar] [CrossRef]
- Jones, J.M.; Morrell, J.C.; Gould, S.J. PEX19 is a predominantly cytosolic chaperone and import receptor for class 1 peroxisomal membrane proteins. J. Cell Biol. 2004, 164, 57–67. [Google Scholar] [CrossRef]
- Bi, J.; Chowdhry, S.; Wu, S.; Zhang, W.; Masui, K.; Mischel, P.S. Altered cellular metabolism in gliomas—An emerging landscape of actionable co-dependency targets. Nat. Rev. Cancer 2020, 20, 57–70. [Google Scholar] [CrossRef] [PubMed]
- Masui, K.; Cavenee, W.K.; Mischel, P.S. Cancer metabolism as a central driving force of glioma pathogenesis. Brain Tumor Pathol. 2016, 33, 161–168. [Google Scholar] [CrossRef] [PubMed]
- Dahabieh, M.S.; Di Pietro, E.; Jangal, M.; Goncalves, C.; Witcher, M.; Braverman, N.E.; Del Rincon, S.V. Peroxisomes and cancer: The role of a metabolic specialist in a disease of aberrant metabolism. Biochim. Biophys. Acta Rev. Cancer 2018, 1870, 103–121. [Google Scholar] [CrossRef]
- Cohen, A.L.; Holmen, S.L.; Colman, H. IDH1 and IDH2 mutations in gliomas. Curr. Neurol. Neurosci. Rep. 2013, 13, 345. [Google Scholar] [CrossRef] [PubMed]
- Geisbrecht, B.V.; Gould, S.J. The human PICD gene encodes a cytoplasmic and peroxisomal NADP(+)-dependent isocitrate dehydrogenase. J. Biol. Chem. 1999, 274, 30527–30533. [Google Scholar] [CrossRef]
- Benedetti, E.; Galzio, R.; Laurenti, G.; D’Angelo, B.; Melchiorre, E.; Cifone, M.G.; Fanelli, F.; Muzi, P.; Coletti, G.; Alecci, M.; et al. Lipid metabolism impairment in human gliomas: Expression of peroxisomal proteins in human gliomas at different grades of malignancy. Int. J. Immunopathol. Pharmacol. 2010, 23, 235–246. [Google Scholar] [CrossRef]
- Chen, R.; Xue, Y.; Yang, F.; Wei, X.; Song, L.; Liu, X. Lentivirus-mediated silencing of HSDL2 suppresses cell proliferation in human gliomas. Tumour Biol. 2016, 37, 15065–15077. [Google Scholar] [CrossRef]
- Laurenti, G.; Benedetti, E.; D’Angelo, B.; Cristiano, L.; Cinque, B.; Raysi, S.; Alecci, M.; Ceru, M.P.; Cifone, M.G.; Galzio, R.; et al. Hypoxia induces peroxisome proliferator-activated receptor alpha (PPARalpha) and lipid metabolism peroxisomal enzymes in human glioblastoma cells. J. Cell Biochem. 2011, 112, 3891–3901. [Google Scholar] [CrossRef] [PubMed]
- Linden, A. Measuring diagnostic and predictive accuracy in disease management: An introduction to receiver operating characteristic (ROC) analysis. J. Eval. Clin. Pract. 2006, 12, 132–139. [Google Scholar] [CrossRef] [PubMed]
- Fitzgerald, M.; Saville, B.R.; Lewis, R.J. Decision curve analysis. JAMA 2015, 313, 409–410. [Google Scholar] [CrossRef] [PubMed]
- Dong, X.; Noorbakhsh, A.; Hirshman, B.R.; Zhou, T.; Tang, J.A.; Chang, D.C.; Carter, B.S.; Chen, C.C. Survival trends of grade I, II, and III astrocytoma patients and associated clinical practice patterns between 1999 and 2010: A SEER-based analysis. Neurooncol. Pract. 2016, 3, 29–38. [Google Scholar] [CrossRef] [PubMed]
- Yan, H.; Parsons, D.W.; Jin, G.; McLendon, R.; Rasheed, B.A.; Yuan, W.; Kos, I.; Batinic-Haberle, I.; Jones, S.; Riggins, G.J.; et al. IDH1 and IDH2 mutations in gliomas. N. Engl. J. Med. 2009, 360, 765–773. [Google Scholar] [CrossRef]
- Zhang, X.; Zhao, L.; Zhang, H.; Zhang, Y.; Ju, H.; Wang, X.; Ren, H.; Zhu, X.; Dong, Y. The immunosuppressive microenvironment and immunotherapy in human glioblastoma. Front. Immunol. 2022, 13, 1003651. [Google Scholar] [CrossRef]
- Mahajan, S.; Schmidt, M.H.H.; Schumann, U. The Glioma Immune Landscape: A Double-Edged Sword for Treatment Regimens. Cancers 2023, 15, 2024. [Google Scholar] [CrossRef]
- Charoentong, P.; Finotello, F.; Angelova, M.; Mayer, C.; Efremova, M.; Rieder, D.; Hackl, H.; Trajanoski, Z. Pan-cancer Immunogenomic Analyses Reveal Genotype-Immunophenotype Relationships and Predictors of Response to Checkpoint Blockade. Cell Rep. 2017, 18, 248–262. [Google Scholar] [CrossRef]
- Thorsson, V.; Gibbs, D.L.; Brown, S.D.; Wolf, D.; Bortone, D.S.; Ou Yang, T.H.; Porta-Pardo, E.; Gao, G.F.; Plaisier, C.L.; Eddy, J.A.; et al. The Immune Landscape of Cancer. Immunity 2018, 48, 812–830.e14. [Google Scholar] [CrossRef]
- Chen, D.S.; Mellman, I. Oncology meets immunology: The cancer-immunity cycle. Immunity 2013, 39, 1–10. [Google Scholar] [CrossRef]
- Dyck, L.; Mills, K.H.G. Immune checkpoints and their inhibition in cancer and infectious diseases. Eur. J. Immunol. 2017, 47, 765–779. [Google Scholar] [CrossRef]
- Fu, J.; Li, K.; Zhang, W.; Wan, C.; Zhang, J.; Jiang, P.; Liu, X.S. Large-scale public data reuse to model immunotherapy response and resistance. Genome Med. 2020, 12, 21. [Google Scholar] [CrossRef]
- Chen, X.F.; Tian, M.X.; Sun, R.Q.; Zhang, M.L.; Zhou, L.S.; Jin, L.; Chen, L.L.; Zhou, W.J.; Duan, K.L.; Chen, Y.J.; et al. SIRT5 inhibits peroxisomal ACOX1 to prevent oxidative damage and is downregulated in liver cancer. EMBO Rep. 2018, 19, e45124. [Google Scholar] [CrossRef]
- Omi, S.; Nakata, R.; Okamura-Ikeda, K.; Konishi, H.; Taniguchi, H. Contribution of peroxisome-specific isoform of Lon protease in sorting PTS1 proteins to peroxisomes. J. Biochem. 2008, 143, 649–660. [Google Scholar] [CrossRef]
- Costa, C.F.; Lismont, C.; Chornyi, S.; Li, H.; Hussein, M.A.F.; Waterham, H.R.; Fransen, M. Functional Analysis of GSTK1 in Peroxisomal Redox Homeostasis in HEK-293 Cells. Antioxidants 2023, 12, 1236. [Google Scholar] [CrossRef]
- Hsu, J.; Fatuzzo, N.; Weng, N.; Michno, W.; Dong, W.; Kienle, M.; Dai, Y.; Pasca, A.; Abu-Remaileh, M.; Rasgon, N.; et al. Carnitine octanoyltransferase is important for the assimilation of exogenous acetyl-L-carnitine into acetyl-CoA in mammalian cells. J. Biol. Chem. 2023, 299, 102848. [Google Scholar] [CrossRef]
- Reuber, B.E.; Karl, C.; Reimann, S.A.; Mihalik, S.J.; Dodt, G. Cloning and functional expression of a mammalian gene for a peroxisomal sarcosine oxidase. J. Biol. Chem. 1997, 272, 6766–6776. [Google Scholar] [CrossRef] [PubMed]
- Ashmarina, L.I.; Rusnak, N.; Miziorko, H.M.; Mitchell, G.A. 3-Hydroxy-3-methylglutaryl-CoA lyase is present in mouse and human liver peroxisomes. J. Biol. Chem. 1994, 269, 31929–31932. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.A.; Lee, J.; Kim, K.; Moon, H.; Min, C.; Moon, B.; Kim, D.; Yang, S.; Park, H.; Lee, G.; et al. The Peroxisomal Localization of Hsd17b4 Is Regulated by Its Interaction with Phosphatidylserine. Mol. Cells 2021, 44, 214–222. [Google Scholar] [CrossRef]
- Westin, M.A.; Hunt, M.C.; Alexson, S.E. The identification of a succinyl-CoA thioesterase suggests a novel pathway for succinate production in peroxisomes. J. Biol. Chem. 2005, 280, 38125–38132. [Google Scholar] [CrossRef] [PubMed]
- Purdue, P.E.; Lazarow, P.B. Targeting of human catalase to peroxisomes is dependent upon a novel COOH-terminal peroxisomal targeting sequence. J. Cell Biol. 1996, 134, 849–862. [Google Scholar] [CrossRef] [PubMed]
- Seo, M.S.; Kang, S.W.; Kim, K.; Baines, I.C.; Lee, T.H.; Rhee, S.G. Identification of a new type of mammalian peroxiredoxin that forms an intramolecular disulfide as a reaction intermediate. J. Biol. Chem. 2000, 275, 20346–20354. [Google Scholar] [CrossRef] [PubMed]
- FitzPatrick, D.R.; Germain-Lee, E.; Valle, D. Isolation and characterization of rat and human cDNAs encoding a novel putative peroxisomal enoyl-CoA hydratase. Genomics 1995, 27, 457–466. [Google Scholar] [CrossRef] [PubMed]
- Khan, F.; Pang, L.; Dunterman, M.; Lesniak, M.S.; Heimberger, A.B.; Chen, P. Macrophages and microglia in glioblastoma: Heterogeneity, plasticity, and therapy. J. Clin. Investig. 2023, 133, e163446. [Google Scholar] [CrossRef]
- Fransen, M.; Lismont, C.; Walton, P. The Peroxisome-Mitochondria Connection: How and Why? Int. J. Mol. Sci. 2017, 18, 1126. [Google Scholar] [CrossRef]
- Jansen, G.A.; Wanders, R.J. Alpha-oxidation. Biochim. Biophys. Acta 2006, 1763, 1403–1412. [Google Scholar] [CrossRef]
- Sassa, T.; Kihara, A. Metabolism of very long-chain Fatty acids: Genes and pathophysiology. Biomol. Ther. 2014, 22, 83–92. [Google Scholar] [CrossRef]
- Miska, J.; Chandel, N.S. Targeting fatty acid metabolism in glioblastoma. J. Clin. Investig. 2023, 133, e163448. [Google Scholar] [CrossRef]
- Wang, L.B.; Karpova, A.; Gritsenko, M.A.; Kyle, J.E.; Cao, S.; Li, Y.; Rykunov, D.; Colaprico, A.; Rothstein, J.H.; Hong, R.; et al. Proteogenomic and metabolomic characterization of human glioblastoma. Cancer Cell 2021, 39, 509–528.e20. [Google Scholar] [CrossRef] [PubMed]
- DeNardo, D.G.; Ruffell, B. Macrophages as regulators of tumour immunity and immunotherapy. Nat. Rev. Immunol. 2019, 19, 369–382. [Google Scholar] [CrossRef] [PubMed]
- Bowman, R.L.; Klemm, F.; Akkari, L.; Pyonteck, S.M.; Sevenich, L.; Quail, D.F.; Dhara, S.; Simpson, K.; Gardner, E.E.; Iacobuzio-Donahue, C.A.; et al. Macrophage Ontogeny Underlies Differences in Tumor-Specific Education in Brain Malignancies. Cell Rep. 2016, 17, 2445–2459. [Google Scholar] [CrossRef]
- Vidyarthi, A.; Agnihotri, T.; Khan, N.; Singh, S.; Tewari, M.K.; Radotra, B.D.; Chatterjee, D.; Agrewala, J.N. Predominance of M2 macrophages in gliomas leads to the suppression of local and systemic immunity. Cancer Immunol. Immunother. 2019, 68, 1995–2004. [Google Scholar] [CrossRef]
- Tomaszewski, W.; Sanchez-Perez, L.; Gajewski, T.F.; Sampson, J.H. Brain Tumor Microenvironment and Host State: Implications for Immunotherapy. Clin. Cancer Res. 2019, 25, 4202–4210. [Google Scholar] [CrossRef] [PubMed]
- Bender, S.; Reuter, A.; Eberle, F.; Einhorn, E.; Binder, M.; Bartenschlager, R. Activation of Type I and III Interferon Response by Mitochondrial and Peroxisomal MAVS and Inhibition by Hepatitis C Virus. PLoS Pathog. 2015, 11, e1005264. [Google Scholar] [CrossRef]
- Choi, Y.B.; Choi, Y.; Harhaj, E.W. Peroxisomes support human herpesvirus 8 latency by stabilizing the viral oncogenic protein vFLIP via the MAVS-TRAF complex. PLoS Pathog. 2018, 14, e1007058. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, A.R.; Marques, M.; Ribeiro, D. Peroxisomes and Innate Immunity: Antiviral Response and Beyond. Int. J. Mol. Sci. 2019, 20, 3795. [Google Scholar] [CrossRef]
- Di Cara, F.; Sheshachalam, A.; Braverman, N.E.; Rachubinski, R.A.; Simmonds, A.J. Peroxisome-Mediated Metabolism is Required for Immune Response to Microbial Infection. Immunity 2018, 48, 832–833. [Google Scholar] [CrossRef] [PubMed]
- Facciotti, F.; Ramanjaneyulu, G.S.; Lepore, M.; Sansano, S.; Cavallari, M.; Kistowska, M.; Forss-Petter, S.; Ni, G.; Colone, A.; Singhal, A.; et al. Peroxisome-derived lipids are self antigens that stimulate invariant natural killer T cells in the thymus. Nat. Immunol. 2012, 13, 474–480. [Google Scholar] [CrossRef] [PubMed]
- Vijayan, V.; Srinu, T.; Karnati, S.; Garikapati, V.; Linke, M.; Kamalyan, L.; Mali, S.R.; Sudan, K.; Kollas, A.; Schmid, T.; et al. A New Immunomodulatory Role for Peroxisomes in Macrophages Activated by the TLR4 Ligand Lipopolysaccharide. J. Immunol. 2017, 198, 2414–2425. [Google Scholar] [CrossRef]
- Di Cara, F.; Andreoletti, P.; Trompier, D.; Vejux, A.; Bulow, M.H.; Sellin, J.; Lizard, G.; Cherkaoui-Malki, M.; Savary, S. Peroxisomes in Immune Response and Inflammation. Int. J. Mol. Sci. 2019, 20, 3877. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qin, X.; Wang, B.; Lu, X.; Song, Y.; Wang, W. Identification and Validation of a PEX5-Dependent Signature for Prognostic Prediction in Glioma. Biomolecules 2024, 14, 314. https://doi.org/10.3390/biom14030314
Qin X, Wang B, Lu X, Song Y, Wang W. Identification and Validation of a PEX5-Dependent Signature for Prognostic Prediction in Glioma. Biomolecules. 2024; 14(3):314. https://doi.org/10.3390/biom14030314
Chicago/Turabian StyleQin, Xuhui, Bing Wang, Xia Lu, Yanyang Song, and Wei Wang. 2024. "Identification and Validation of a PEX5-Dependent Signature for Prognostic Prediction in Glioma" Biomolecules 14, no. 3: 314. https://doi.org/10.3390/biom14030314
APA StyleQin, X., Wang, B., Lu, X., Song, Y., & Wang, W. (2024). Identification and Validation of a PEX5-Dependent Signature for Prognostic Prediction in Glioma. Biomolecules, 14(3), 314. https://doi.org/10.3390/biom14030314






