Association of Altered Plasma Lipidome with Disease Severity in COVID-19 Patients
Abstract
1. Introduction
2. Materials and Methods
2.1. Cohort
2.2. Sample Collection
2.3. Sample Analysis
2.4. Statistical Analysis
3. Results
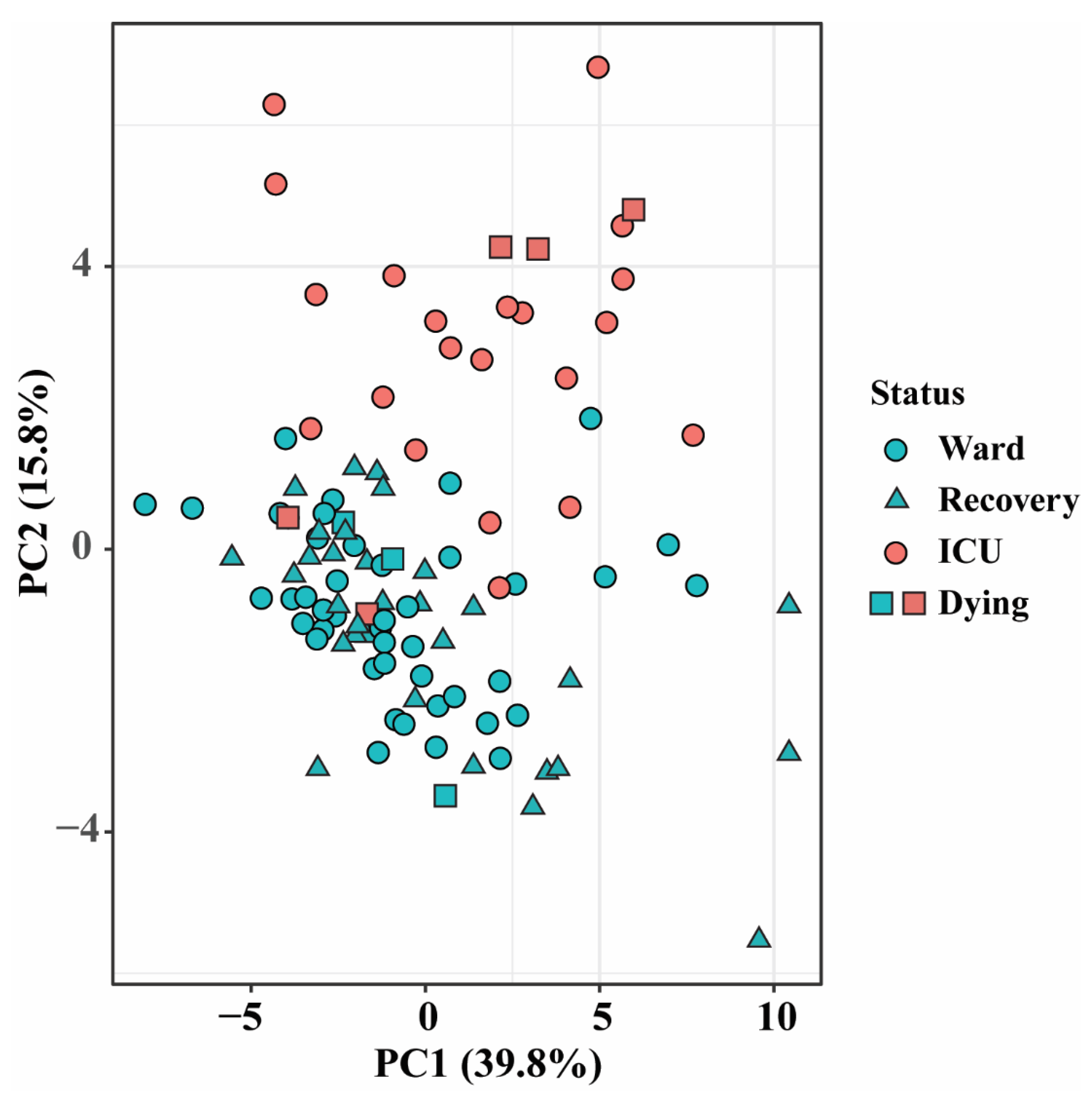
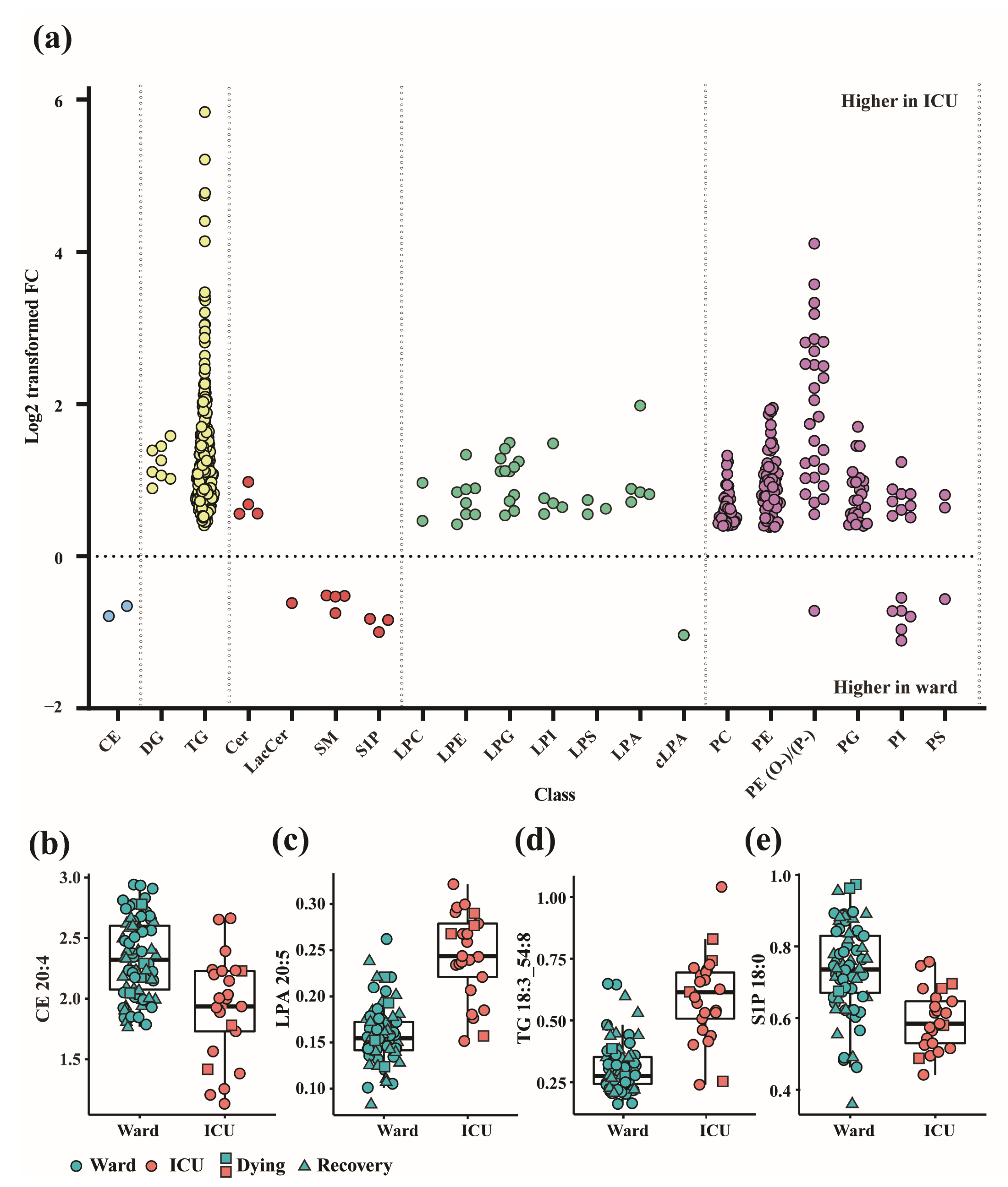
3.1. Altered Lipid Profiles in COVID-19 Patients in the Ward and ICU
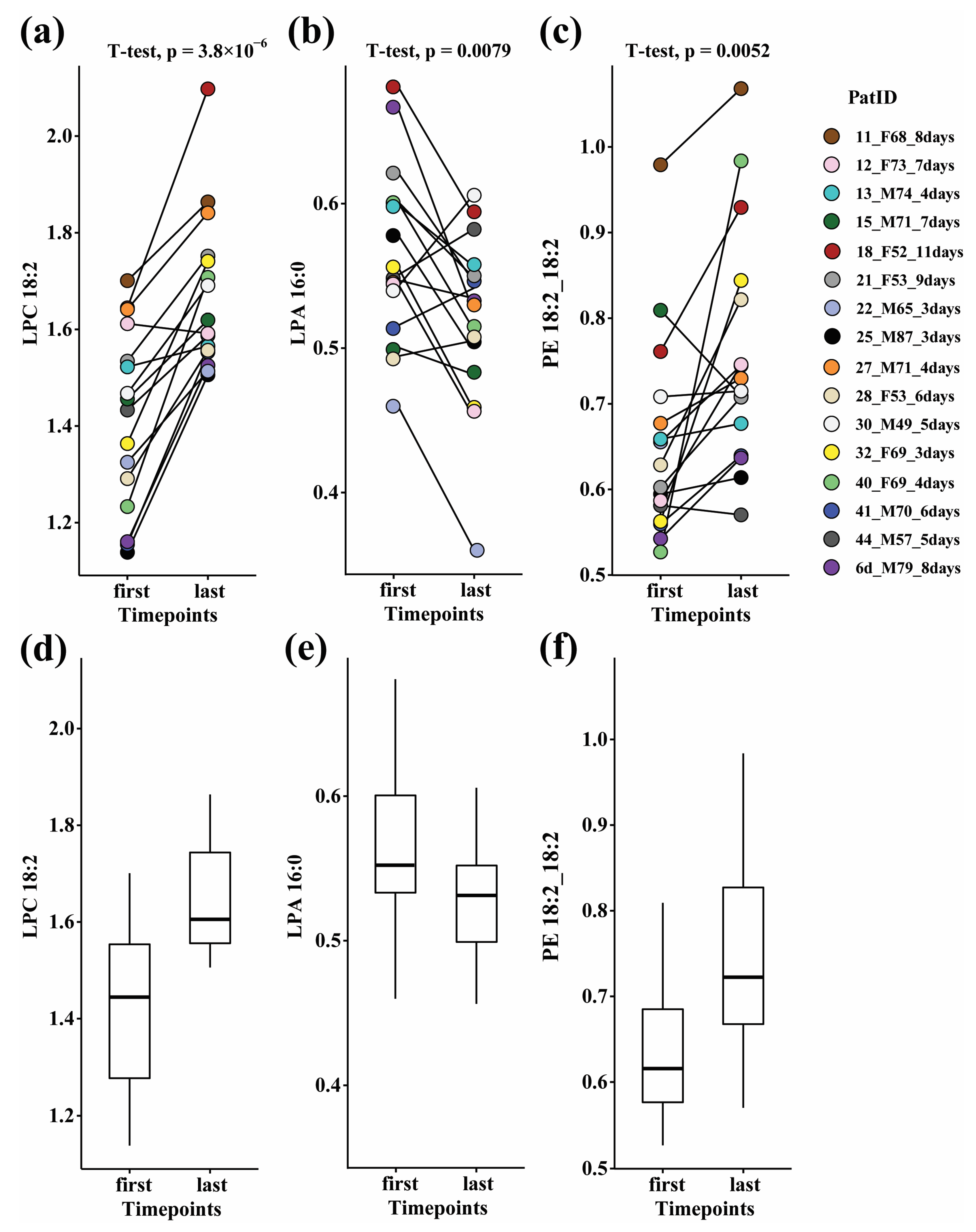
3.2. Paired Analysis in Ward Patients
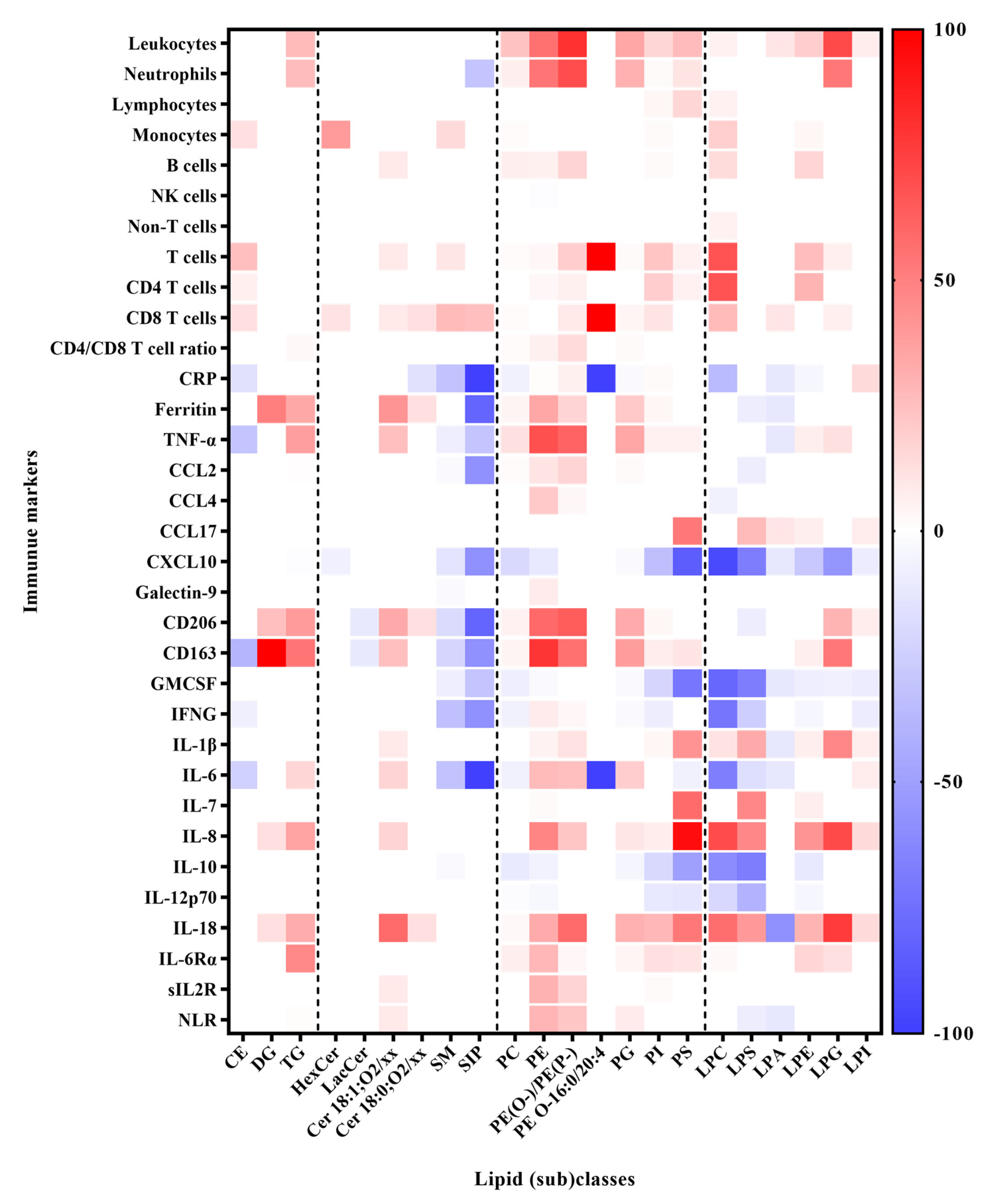
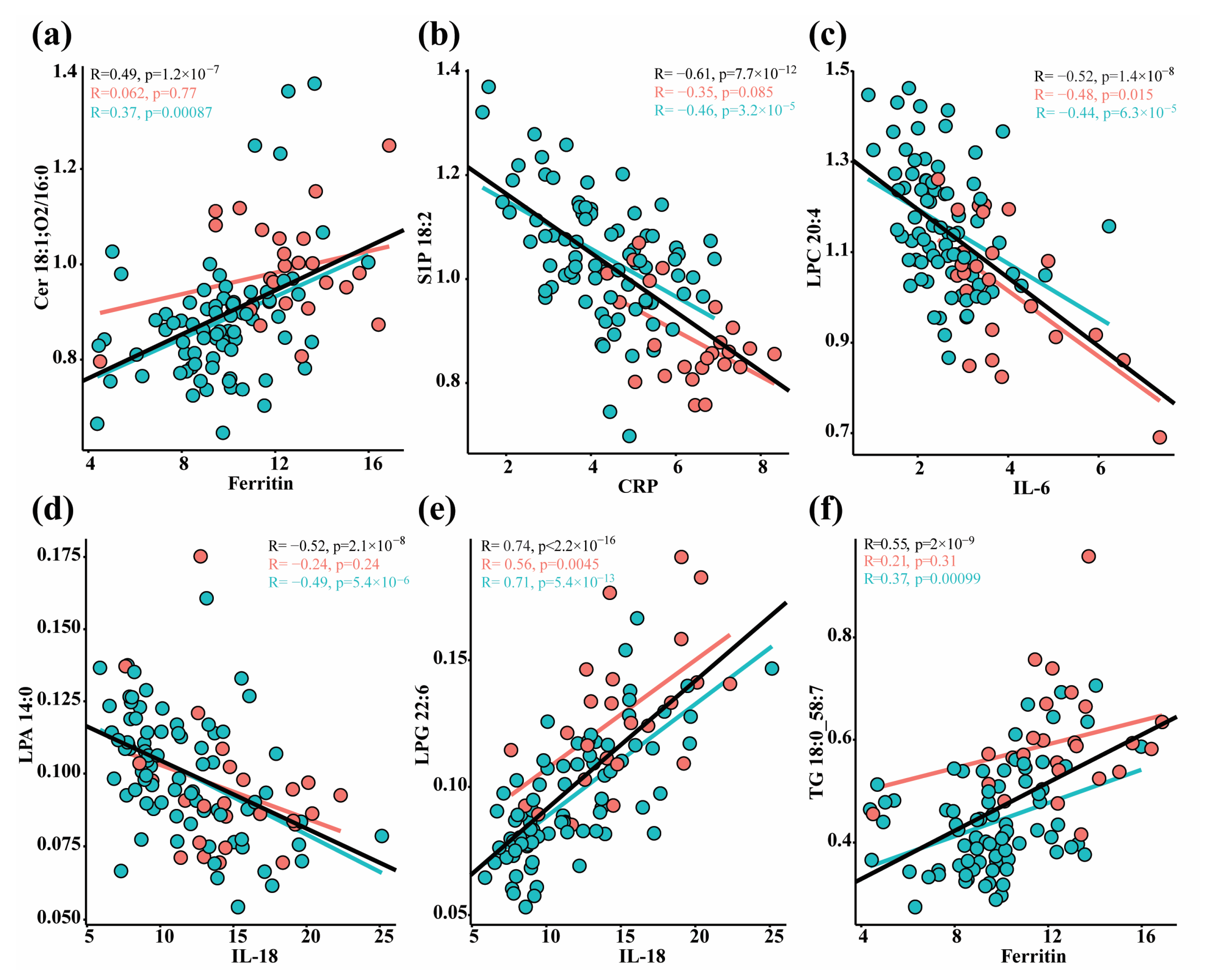
3.3. Correlation between Lipids and Immune Response Markers
4. Discussion
4.1. Sphingolipids Metabolism
4.2. Sphingoid Base 1-Phosphates
4.3. Glycerophospholipids
4.4. Glycerolipids and Other Neutral Lipids
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bruzzone, C.; Bizkarguenaga, M.; Gil-Redondo, R.; Diercks, T.; Arana, E.; de Vicuña, A.G.; Seco, M.; Bosch, A.; Palazón, A.; San Juan, I.; et al. SARS-CoV-2 Infection Dysregulates the Metabolomic and Lipidomic Profiles of Serum. iScience 2020, 23, 101645. [Google Scholar] [CrossRef]
- Hussein, M.A.; Ismail, N.E.M.; Mohamed, A.H.; Borik, R.M.; Ali, A.A.; Mosaad, Y.O. Plasma Phospholipids: A Promising Simple Biochemical Parameter to Evaluate COVID-19 Infection Severity. Bioinform. Biol. Insights 2021, 15, 117793222110558. [Google Scholar] [CrossRef] [PubMed]
- Ripa, I.; Andreu, S.; López-Guerrero, J.A.; Bello-Morales, R. Membrane Rafts: Portals for Viral Entry. Front. Microbiol. 2021, 12, 631274. [Google Scholar] [CrossRef] [PubMed]
- Lorizate, M.; Krausslich, H.-G. Role of Lipids in Virus Replication. Cold Spring Harb. Perspect. Biol. 2011, 3, a004820. [Google Scholar] [CrossRef] [PubMed]
- Kyle, J.E. How lipidomics can transform our understanding of virus infections. Expert Rev. Proteom. 2021, 18, 329–332. [Google Scholar] [CrossRef] [PubMed]
- Goodwin, C.M.; Xu, S.; Munger, J. Stealing the Keys to the Kitchen: Viral Manipulation of the Host Cell Metabolic Network. Trends Microbiol. 2015, 23, 789–798. [Google Scholar] [CrossRef] [PubMed]
- Bai, Y.; Huang, W.; Li, Y.; Lai, C.; Huang, S.; Wang, G.; He, Y.; Hu, L.; Chen, C. Lipidomic alteration of plasma in cured COVID-19 patients using ultra high-performance liquid chromatography with high-resolution mass spectrometry. Biosci. Rep. 2021, 41, BSR20204305. [Google Scholar] [CrossRef] [PubMed]
- Abu-Farha, M.; Thanaraj, T.A.; Qaddoumi, M.G.; Hashem, A.; Abubaker, J.; Al-Mulla, F. The Role of Lipid Metabolism in COVID-19 Virus Infection and as a Drug Target. Int. J. Mol. Sci. 2020, 21, 3544. [Google Scholar] [CrossRef] [PubMed]
- Calder, P.C. Omega-3 Fatty Acids and Inflammatory Processes. Nutrients 2010, 2, 355–374. [Google Scholar] [CrossRef]
- Shen, B.; Yi, X.; Sun, Y.; Bi, X.; Du, J.; Zhang, C.; Quan, S.; Zhang, F.; Sun, R.; Qian, L.; et al. Proteomic and Metabolomic Characterization of COVID-19 Patient Sera. Cell 2020, 182, 59–72.e15. [Google Scholar] [CrossRef]
- Song, J.W.; Lam, S.M.; Fan, X.; Cao, W.J.; Wang, S.Y.; Tian, H.; Chua, G.H.; Zhang, C.; Meng, F.P.; Xu, Z.; et al. Omics-Driven Systems Interrogation of Metabolic Dysregulation in COVID-19 Pathogenesis. Cell Metab. 2020, 32, 188–202.e5. [Google Scholar] [CrossRef]
- Mohammedsaeed, W.; Alahamadey, Z.Z.; Khan, S.M. Alteration of lipid profile in COVID-19 saudi patients at al-madinah al-munawarah. Infection 2020, 14, 15. [Google Scholar]
- Osuna-Ramos, J.F.; Aguilar, H.R.; De Jesus-Gonzalez, L.A.; Ruiz, J.M.R.; Ortega, A.M.E.; Ramirez, L.A.O.; Utrilla, A.R.; Burgueno, E.R.; Soto-Almaral, A.; Rios-Tostado, J.J.; et al. Serum lipid profile changes and their clinical diagnostic significance in COVID-19 Mexican Patients. Infect. Dis. Except. HIV/AIDS 2020. [Google Scholar] [CrossRef]
- Farley, S.E.; Kyle, J.E.; Leier, H.C.; Bramer, L.M.; Weinstein, J.B.; Bates, T.A.; Lee, J.Y.; Metz, T.O.; Schultz, C.; Tafesse, F.G. A global lipid map reveals host dependency factors conserved across SARS-CoV-2 variants. Nat. Commun. 2022, 13, 3487. [Google Scholar] [CrossRef]
- Karu, N.; Kindt, A.; van Gammeren, A.J.; Ermens, A.A.; Harms, A.C.; Portengen, L.; Vermeulen, R.C.; Dik, W.A.; Langerak, A.W.; van der Velden, V.H.; et al. Severe COVID-19 Is Characterised by Perturbations in Plasma Amines Correlated with Immune Response Markers, and Linked to Inflammation and Oxidative Stress. Metabolites 2022, 12, 618. [Google Scholar] [CrossRef]
- Karu, N.; Kindt, A.; Lamont, L.; van Gammeren, A.J.; Ermens, A.A.; Harms, A.C.; Portengen, L.; Vermeulen, R.C.; Dik, W.A.; Langerak, A.W.; et al. Plasma Oxylipins and Their Precursors Are Strongly Associated with COVID-19 Severity and with Immune Response Markers. Metabolites 2022, 12, 619. [Google Scholar] [CrossRef]
- Schrijver, B.; Assmann, J.L.; van Gammeren, A.J.; Vermeulen, R.C.; Portengen, L.; Heukels, P.; Langerak, A.W.; Dik, W.A.; van der Velden, V.H.; Ermens, T.A. Extensive longitudinal immune profiling reveals sustained innate immune activaton in COVID-19 patients with unfavorable outcome. Eur. Cytokine Netw. 2020, 31, 154–167. [Google Scholar] [CrossRef]
- Schoeman, J.C.; Harms, A.C.; van Weeghel, M.; Berger, R.; Vreeken, R.J.; Hankemeier, T. Development and application of a UHPLC–MS/MS metabolomics based comprehensive systemic and tissue-specific screening method for inflammatory, oxidative and nitrosative stress. Anal. Bioanal. Chem. 2018, 410, 2551–2568. [Google Scholar] [CrossRef]
- Zhang, Z.; Singh, M.; Kindt, A.; Wegrzyn, A.B.; Pearson, M.J.; Ali, A.; Harms, A.C.; Baker, P. and Hankemeier, T. Development of a targeted hydrophilic interaction liquid chromatography-tandem mass spectrometry based lipidomics platform applied to a coronavirus disease severity study. J. Chromatogr. A 2023, 1708, 464342. [Google Scholar] [CrossRef]
- Chalfant, C.; Del Poeta, M. (Eds.) Sphingolipids as Signaling and Regulatory Molecules; Springer Science+Business Media: New York, NY, USA, 2010. [Google Scholar]
- Levental, I.; Levental, K.R.; Heberle, F.A. Lipid Rafts: Controversies Resolved, Mysteries Remain. Trends Cell Biol. 2020, 30, 341–353. [Google Scholar] [CrossRef]
- Sviridov, D.; Miller, Y.I.; Ballout, R.A.; Remaley, A.T.; Bukrinsky, M. Targeting Lipid Rafts—A Potential Therapy for COVID-19. Front. Immunol. 2020, 11, 574508. [Google Scholar] [CrossRef]
- Fecchi, K.; Anticoli, S.; Peruzzu, D.; Iessi, E.; Gagliardi, M.C.; Matarrese, P.; Ruggieri, A. Coronavirus Interplay With Lipid Rafts and Autophagy Unveils Promising Therapeutic Targets. Front. Microbiol. 2020, 11, 1821. [Google Scholar] [CrossRef]
- Marino, L.; Criniti, A.; Guida, S.; Bucci, T.; Ballesio, L.; Suppa, M.; Galardo, G.; Vacca, A.; Santulli, M.; Angeloni, A.; et al. Interleukin 18 and IL-18 BP response to Sars-CoV-2 virus infection. Clin. Exp. Med. 2022, 23, 1243–1250. [Google Scholar] [CrossRef]
- Schooling, C.M.; Li, M.; Au Yeung, S.L. Interleukin-18 and COVID-19. Epidemiol. Infect. 2022, 150, e14. [Google Scholar] [CrossRef]
- Cesta, M.C.; Zippoli, M.; Marsiglia, C.; Gavioli, E.M.; Mantelli, F.; Allegretti, M.; Balk, R.A. The Role of Interleukin-8 in Lung Inflammation and Injury: Implications for the Management of COVID-19 and Hyperinflammatory Acute Respiratory Distress Syndrome. Front. Pharmacol. 2022, 12, 808797. [Google Scholar] [CrossRef]
- Zhou, C.; Chen, Y.; Ji, Y.; He, X.; Xue, D. Increased Serum Levels of Hepcidin and Ferritin Are Associated with Severity of COVID-19. Med. Sci. Monit. 2020, 26, e926178-1. [Google Scholar] [CrossRef]
- Ottolenghi, S.; Zulueta, A.; Caretti, A. Iron and Sphingolipids as Common Players of (Mal)Adaptation to Hypoxia in Pulmonary Diseases. Int. J. Mol. Sci. 2020, 21, 307. [Google Scholar] [CrossRef]
- Torretta, E.; Garziano, M.; Poliseno, M.; Capitanio, D.; Biasin, M.; Santantonio, T.A.; Clerici, M.; Lo Caputo, S.; Trabattoni, D.; Gelfi, C. Severity of COVID-19 Patients Predicted by Serum Sphingolipids Signature. Int. J. Mol. Sci. 2021, 22, 10198. [Google Scholar] [CrossRef]
- Carpinteiro, A.; Edwards, M.J.; Hoffmann, M.; Kochs, G.; Gripp, B.; Weigang, S.; Adams, C.; Carpinteiro, E.; Gulbins, A.; Keitsch, S.; et al. Pharmacological Inhibition of Acid Sphingomyelinase Prevents Uptake of SARS-CoV-2 by Epithelial Cells. Cell Rep. Med. 2020, 1, 100142. [Google Scholar] [CrossRef]
- Törnquist, K.; Asghar, M.Y.; Srinivasan, V.; Korhonen, L.; Lindholm, D. Sphingolipids as Modulators of SARS-CoV-2 Infection. Front. Cell Dev. Biol. 2021, 9, 689854. [Google Scholar] [CrossRef]
- Schneider-Schaulies, J.; Schneider-Schaulies, S. Sphingolipids in viral infection. Biol. Chem. 2015, 396, 585–595. [Google Scholar] [CrossRef]
- Sakamoto, H.; Okamoto, K.; Aoki, M.; Kato, H.; Katsume, A.; Ohta, A.; Tsukuda, T.; Shimma, N.; Aoki, Y.; Arisawa, M.; et al. Host sphingolipid biosynthesis as a target for hepatitis C virus therapy. Nat. Chem. Biol. 2005, 1, 333–337. [Google Scholar] [CrossRef]
- Nguyen, A.; Guedán, A.; Mousnier, A.; Swieboda, D.; Zhang, Q.; Horkai, D.; Le Novere, N.; Solari, R.; Wakelam, M.J. Host lipidome analysis during rhinovirus replication in HBECs identifies potential therapeutic targets. J. Lipid Res. 2018, 59, 1671–1684. [Google Scholar] [CrossRef]
- Grassmé, H.; Riehle, A.; Wilker, B.; Gulbins, E. Rhinoviruses Infect Human Epithelial Cells via Ceramide-enriched Membrane Platforms. J. Biol. Chem. 2005, 280, 26256–26262. [Google Scholar] [CrossRef]
- Choi, R.H.; Tatum, S.M.; Symons, J.D.; Summers, S.A.; Holland, W.L. Ceramides and other sphingolipids as drivers of cardiovascular disease. Nat. Rev. Cardiol. 2021, 18, 701–711. [Google Scholar] [CrossRef]
- Wigger, D.; Schumacher, F.; Schneider-Schaulies, S.; Kleuser, B. Sphingosine 1-phosphate metabolism and insulin signaling. Cell Signal. 2021, 82, 109959. [Google Scholar] [CrossRef]
- Cartier, A.; Hla, T. Sphingosine 1-phosphate: Lipid signaling in pathology and therapy. Science 2019, 366, eaar5551. [Google Scholar] [CrossRef]
- Strub, G.M.; Maceyka, M.; Hait, N.C.; Milstien, S.; Spiegel, S. Extracellular and Intracellular Actions of Sphingosine-1-Phosphate. In Sphingolipids as Signaling and Regulatory Molecules; Chalfant, C., Poeta, M.D., Eds.; Springer: New York, NY, USA, 2010; pp. 141–155. [Google Scholar]
- Marfia, G.; Navone, S.; Guarnaccia, L.; Campanella, R.; Mondoni, M.; Locatelli, M.; Barassi, A.; Fontana, L.; Palumbo, F.; Garzia, E.; et al. Decreased serum level of sphingosine-1-phosphate: A novel predictor of clinical severity in COVID-19. EMBO Mol. Med. 2021, 13, e13424. [Google Scholar] [CrossRef] [PubMed]
- Rosen, H.; Oldstone, M.B.A. The riddle of the Sphinx: Why sphingosine-1-phosphate may help define molecular mechanisms underlying risk stratification for serious COVID-19 infections. EMBO Mol. Med. 2021, 13, e13533. [Google Scholar] [CrossRef]
- Stepanovska, B.; Huwiler, A. Targeting the S1P receptor signaling pathways as a promising approach for treatment of autoimmune and inflammatory diseases. Pharmacol. Res. 2020, 154, 104170. [Google Scholar] [CrossRef]
- Czeloth, N.; Bernhardt, G.; Hofmann, F.; Genth, H.; Förster, R. Sphingosine-1-Phosphate Mediates Migration of Mature Dendritic Cells. J. Immunol. 2005, 175, 2960–2967. [Google Scholar] [CrossRef]
- Hannun, Y.A.; Obeid, L.M. Sphingolipids and their metabolism in physiology and disease. Nat. Rev. Mol. Cell Biol. 2018, 19, 175–191. [Google Scholar] [CrossRef]
- Mandala, S.; Hajdu, R.; Bergstrom, J.; Quackenbush, E.; Xie, J.; Milligan, J.; Thornton, R.; Shei, G.J.; Card, D.; Keohane, C.; et al. Alteration of Lymphocyte Trafficking by Sphingosine-1-Phosphate Receptor Agonists. Science 2002, 296, 346–349. [Google Scholar] [CrossRef]
- Aoki, M.; Aoki, H.; Ramanathan, R.; Hait, N.C.; Takabe, K. Sphingosine-1-Phosphate Signaling in Immune Cells and Inflammation: Roles and Therapeutic Potential. Mediat. Inflamm. 2016, 2016, 8606878. [Google Scholar]
- Joshi, J.C.; Joshi, B.; Rochford, I.; Rayees, S.; Akhter, M.Z.; Baweja, S.; Chava, K.R.; Tauseef, M.; Abdelkarim, H.; Natarajan, V.; et al. SPHK2-Generated S1P in CD11b+ Macrophages Blocks STING to Suppress the Inflammatory Function of Alveolar Macrophages. Cell Rep. 2020, 30, 4096–4109.e5. [Google Scholar] [CrossRef]
- Danlos, F.X.; Grajeda-Iglesias, C.; Durand, S.; Sauvat, A.; Roumier, M.; Cantin, D.; Colomba, E.; Rohmer, J.; Pommeret, F.; Baciarello, G.; et al. Metabolomic analyses of COVID-19 patients unravel stage-dependent and prognostic biomarkers. Cell Death Dis. 2021, 12, 258. [Google Scholar] [CrossRef]
- Tasat, D.R.; Yakisich, J.S. Rationale for the use of sphingosine analogues in COVID-19 patients. Clin. Med. 2021, 21, e84–e87. [Google Scholar] [CrossRef]
- Naz, F.; Arish, M. Battling COVID-19 Pandemic: Sphingosine-1-Phosphate Analogs as an Adjunctive Therapy? Front. Immunol. 2020, 11, 1102. [Google Scholar] [CrossRef]
- Zhang, Z.; He, G.; Filipowicz, N.A.; Randall, G.; Belov, G.A.; Kopek, B.G.; Wang, X. Host Lipids in Positive-Strand RNA Virus Genome Replication. Front. Microbiol. 2019, 10, 286. [Google Scholar] [CrossRef] [PubMed]
- Izquierdo-Useros, N.; Naranjo-Gómez, M.; Erkizia, I.; Puertas, M.C.; Borràs, F.E.; Blanco, J.; Martinez-Picado, J. HIV and Mature Dendritic Cells: Trojan Exosomes Riding the Trojan Horse? PLoS Pathog. 2010, 6, e1000740. [Google Scholar] [CrossRef] [PubMed]
- Lin, D.A.; Boyce, J.A. Lysophospholipids as Mediators of Immunity. In Advances in Immunology; Alt, F.W., Austen, K.F., Honjo, T., Melchers, F., Uhr, J.W., Unanue, E.R., Eds.; Academic Press: Cambridge, MA, USA, 2006; pp. 141–167. ISBN 9780120224906. [Google Scholar] [CrossRef]
- Baral, P.K.; Amin, M.T.; Rashid, M.M.O.; Hossain, M.S. Assessment of Polyunsaturated Fatty Acids on COVID-19-Associated Risk Reduction. Rev. Bras. Farmacogn. 2022, 31, 50–64. [Google Scholar] [CrossRef]
- Farooqui, A.A.; Farooqui, T.; Sun, G.Y.; Lin, T.-N.; Teh, D.B.L.; Ong, W.-Y. COVID-19, Blood Lipid Changes, and Thrombosis. Biomedicines 2023, 11, 1181. [Google Scholar] [CrossRef]
- Hostetler, K.Y.; Reasor, M.; Yazaki, P.J. Chloroquine-induced phospholipid fatty liver. Measurement of drug and lipid concentrations in rat liver lysosomes. J. Biol. Chem. 1985, 260, 215–219. [Google Scholar] [CrossRef] [PubMed]
- Cruickshank-Quinn, C.; Zheng, L.K.; Quinn, K.; Bowler, R.; Reisdorph, R.; Reisdorph, N. Impact of Blood Collection Tubes and Sample Handling Time on Serum and Plasma Metabolome and Lipidome. Metabolites 2018, 8, 88. [Google Scholar] [CrossRef] [PubMed]
- Stephenson, D.J.; Hoeferlin, L.A.; Chalfant, C.E. Lipidomics in translational research and the clinical significance of lipid-based biomarkers. Transl. Res. 2017, 189, 13–29. [Google Scholar] [CrossRef]
- Durrington, P. Blood lipids after COVID-19 infection. Lancet Diabetes Endocrinol. 2023, 11, 68–69. [Google Scholar] [CrossRef]
- Masana, L.; Correig, E.; Ibarretxe, D.; Anoro, E.; Arroyo, J.A.; Jericó, C.; Guerrero, C.; Miret, M.L.; Näf, S.; Pardo, A.; et al. Low HDL and high triglycerides predict COVID-19 severity. Sci. Rep. 2021, 11, 7217. [Google Scholar] [CrossRef]
- Kovacevic, M.P.; Dube, K.M.; Lupi, K.E.; Szumita, P.M.; DeGrado, J.R. Evaluation of Hypertriglyceridemia in Critically Ill Patients With Coronavirus Disease 2019 Receiving Propofol. Crit. Care Explor. 2021, 3, e0330. [Google Scholar] [CrossRef]
- Zhang, C.; Shi, L.; Wang, F.-S. Liver injury in COVID-19: Management and challenges. Lancet Gastroenterol. Hepatol. 2020, 5, 428–430. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.; Shu, T.; Yang, X.; Song, J.X.; Zhang, M.; Yao, C.; Liu, W.; Huang, M.; Yu, Y.; Yang, Q.; et al. Plasma metabolomic and lipidomic alterations associated with COVID-19. Natl. Sci. Rev. 2020, 7, 1157–1168. [Google Scholar] [CrossRef]
- Baratta, F.; Pastori, D.; Ferro, D.; Carluccio, G.; Tozzi, G.; Angelico, F.; Violi, F.; Del Ben, M. Reduced lysosomal acid lipase activity: A new marker of liver disease severity across the clinical continuum of non-alcoholic fatty liver disease? World J. Gastroenterol. 2019, 25, 4172–4180. [Google Scholar] [PubMed]
- Trinder, M.; Boyd, J.H.; Brunham, L.R. Molecular regulation of plasma lipid levels during systemic inflammation and sepsis. Curr. Opin. Lipidol. 2019, 30, 108–116. [Google Scholar] [CrossRef]
- Barcia, A.M.; Harris, H.W. Triglyceride-Rich Lipoproteins as Agents of Innate Immunity. Clin. Infect. Dis. 2005, 41, S498–S503. [Google Scholar] [CrossRef]
| Patients (n = 44) | Samples (n = 103) | |
|---|---|---|
| Age (years) | 73 [49–87] | 71 [49–87] |
| Male (%) | 30 (68%) | 65 (63%) |
| BMI | 27 [19–42] | 27 [19–42] |
| Diabetes mellitus (DM) | 9 (20%) | |
| Chronic kidney disease (CKD) | 3 (7%) | |
| Cardiovascular disease (CVD) | 7 (16%) | |
| Chronic obstructive pulmonary disease (COPD) | 8 (18%) | |
| Past smoker | 17 (39%) | |
| Pre-admission beta-blockers, ACE inhibitors | 14 (32%) | |
| Pre-admission corticosteroids | 8 (18%) | |
| Pre-admission chloroquine | 27 (61%) | |
| Days with symptoms till hospitalization | 8 [1–19] | |
| Total hospitalization days | 7 [2–62] | |
| Admitted to ward | 37 (84%) | 78 (76%) |
| Admitted to ICU | 7 (16%) | 25 (24%) |
| Organ failure | 7 (16%) | |
| Deceased | 9 (20%) | |
| Unfavorable outcome (ICU or death) | 12 (27%) | 36 (35%) |
| Invasive breathing support (intubated) | 6 (14%) | |
| Post-admission chloroquine | 35 (80%) | |
| Post-admission corticosteroids | 2 (5%) | |
| Post-admission antibiotics | 38 (86%) | |
| CRP, mg/L (normal < 10) | 104.5 [3–577] | |
| IL6, pg/mL (normal < 8) | 19.3 [1–397] | |
| Ferritin, ng/mL (normal 10–400) | 1035 [84–4807] | |
| Leukocytes, 109/L (normal 4.5–11) | 8 [4–20.5] | |
| Lymphocytes, 109/L (normal 0.8–5.0) | 0.95 [0.26–3.15] | |
| Neutrophils, 109/L (normal 1.7–6.5) | 6.36 [2.3–17.5] | |
| Age (years) | 73 [49–87] | 71 [49–87] |
| Male (%) | 30 (68%) | 65 (63%) |
| BMI | 27 [19–42] | 27 [19–42] |
| Diabetes mellitus (DM) | 9 (20%) | |
| Chronic kidney disease (CKD) | 3 (7%) | |
| Cardiovascular disease (CVD) | 7 (16%) | |
| Chronic obstructive pulmonary disease (COPD) | 8 (18%) | |
| Past smoker | 17 (39%) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Z.; Karu, N.; Kindt, A.; Singh, M.; Lamont, L.; van Gammeren, A.J.; Ermens, A.A.M.; Harms, A.C.; Portengen, L.; Vermeulen, R.C.H.; et al. Association of Altered Plasma Lipidome with Disease Severity in COVID-19 Patients. Biomolecules 2024, 14, 296. https://doi.org/10.3390/biom14030296
Zhang Z, Karu N, Kindt A, Singh M, Lamont L, van Gammeren AJ, Ermens AAM, Harms AC, Portengen L, Vermeulen RCH, et al. Association of Altered Plasma Lipidome with Disease Severity in COVID-19 Patients. Biomolecules. 2024; 14(3):296. https://doi.org/10.3390/biom14030296
Chicago/Turabian StyleZhang, Zhengzheng, Naama Karu, Alida Kindt, Madhulika Singh, Lieke Lamont, Adriaan J. van Gammeren, Anton A. M. Ermens, Amy C. Harms, Lutzen Portengen, Roel C. H. Vermeulen, and et al. 2024. "Association of Altered Plasma Lipidome with Disease Severity in COVID-19 Patients" Biomolecules 14, no. 3: 296. https://doi.org/10.3390/biom14030296
APA StyleZhang, Z., Karu, N., Kindt, A., Singh, M., Lamont, L., van Gammeren, A. J., Ermens, A. A. M., Harms, A. C., Portengen, L., Vermeulen, R. C. H., Dik, W. A., Langerak, A. W., van der Velden, V. H. J., & Hankemeier, T. (2024). Association of Altered Plasma Lipidome with Disease Severity in COVID-19 Patients. Biomolecules, 14(3), 296. https://doi.org/10.3390/biom14030296






