Cloflucarban Illuminates Specificity and Context-Dependent Activation of the PINK1–Parkin Pathway by Mitochondrial Complex Inhibition
Abstract
1. Introduction
2. Results
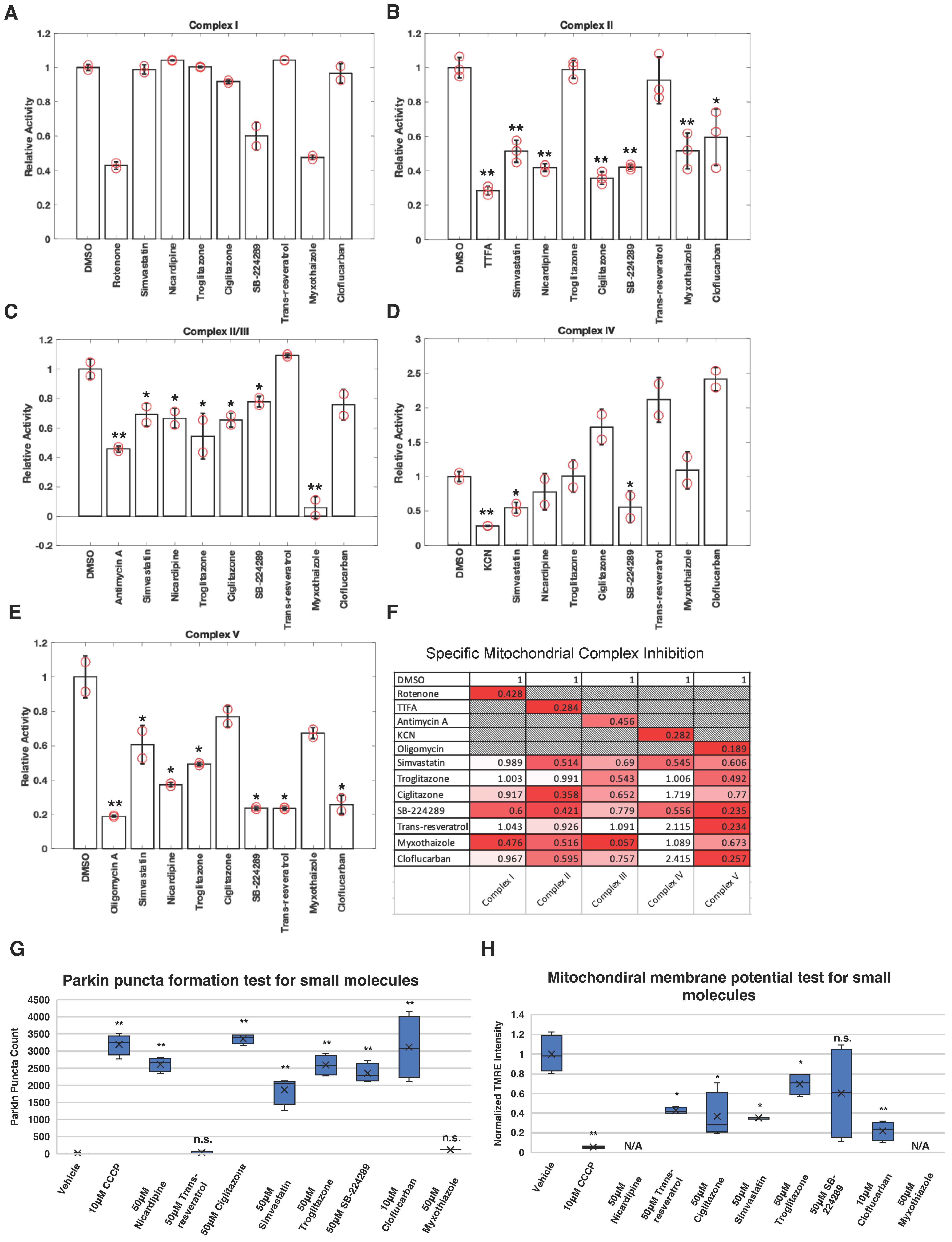
2.1. Profiling the Activity of Electron Transport Chain Inhibitors on Activation of the PINK1–Parkin Pathway
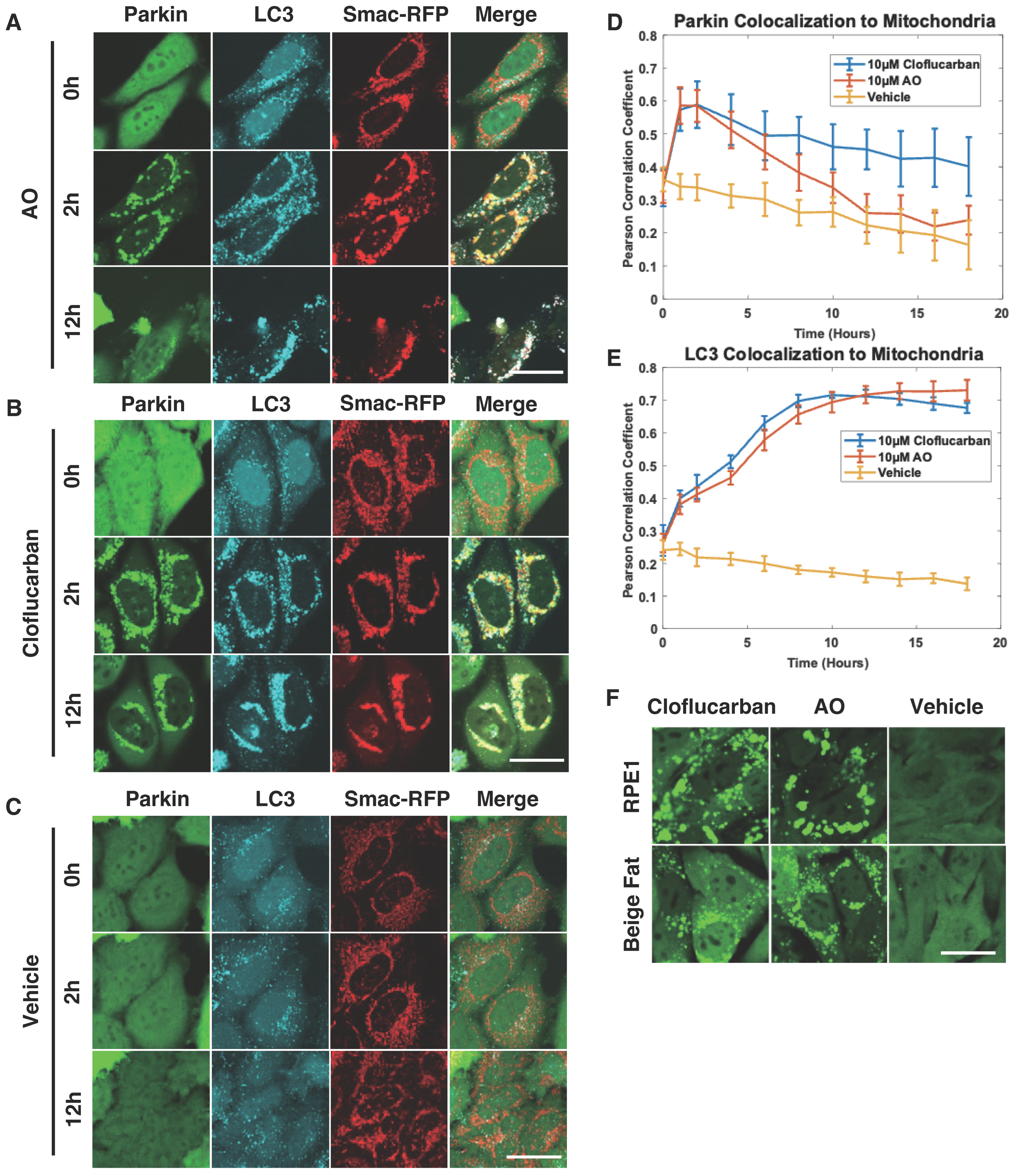
2.2. Cloflucarban Induces Mitophagy by Recruiting LC3
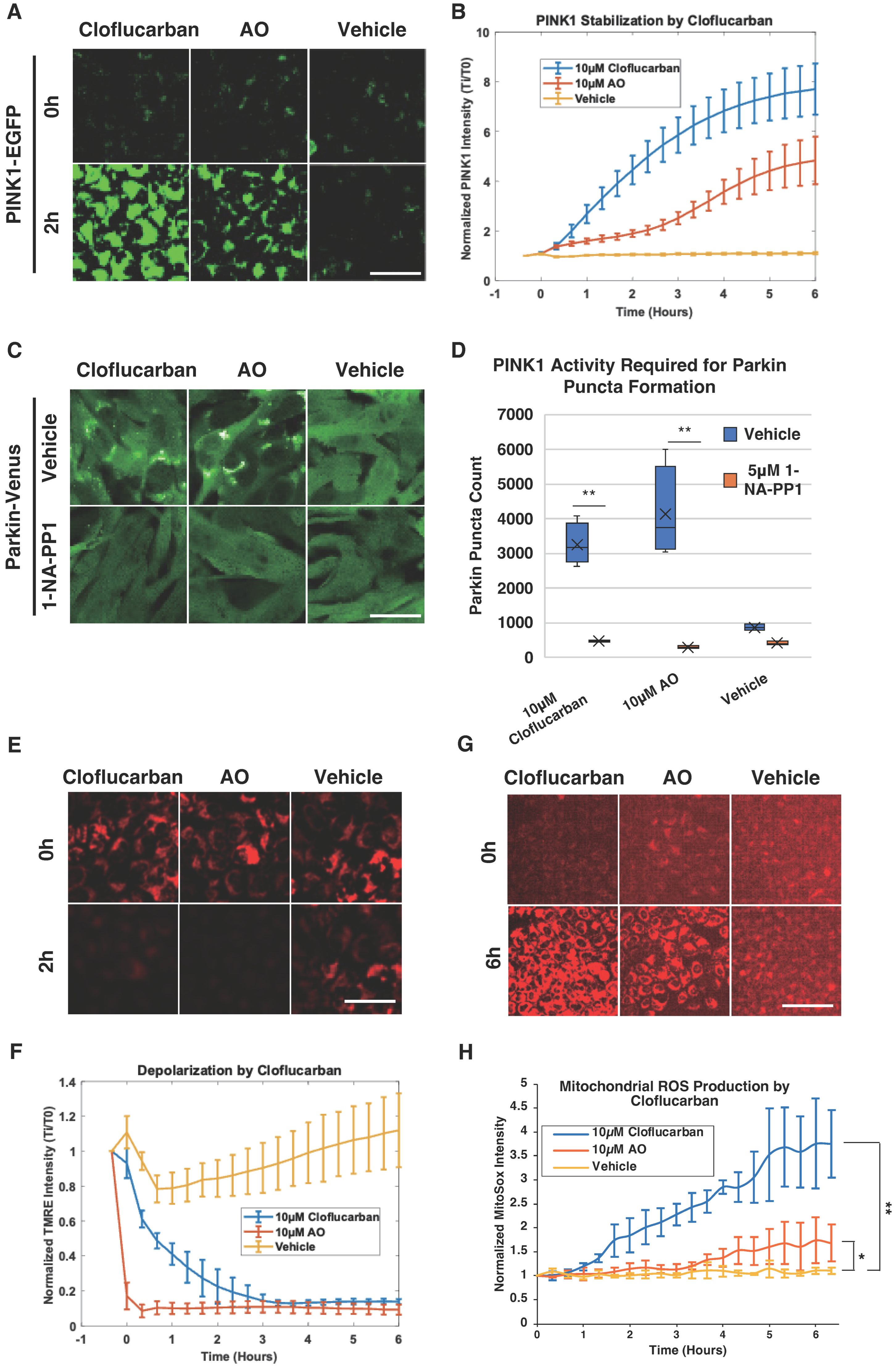
2.3. Cloflucarban-Induced Parkin Mitochondrial Translocation Is PINK1-Dependent
2.4. Complex III and V Knockdowns Confirm the Activation of PINK1–Parkin Pathway Occurs via Dual Inhibition
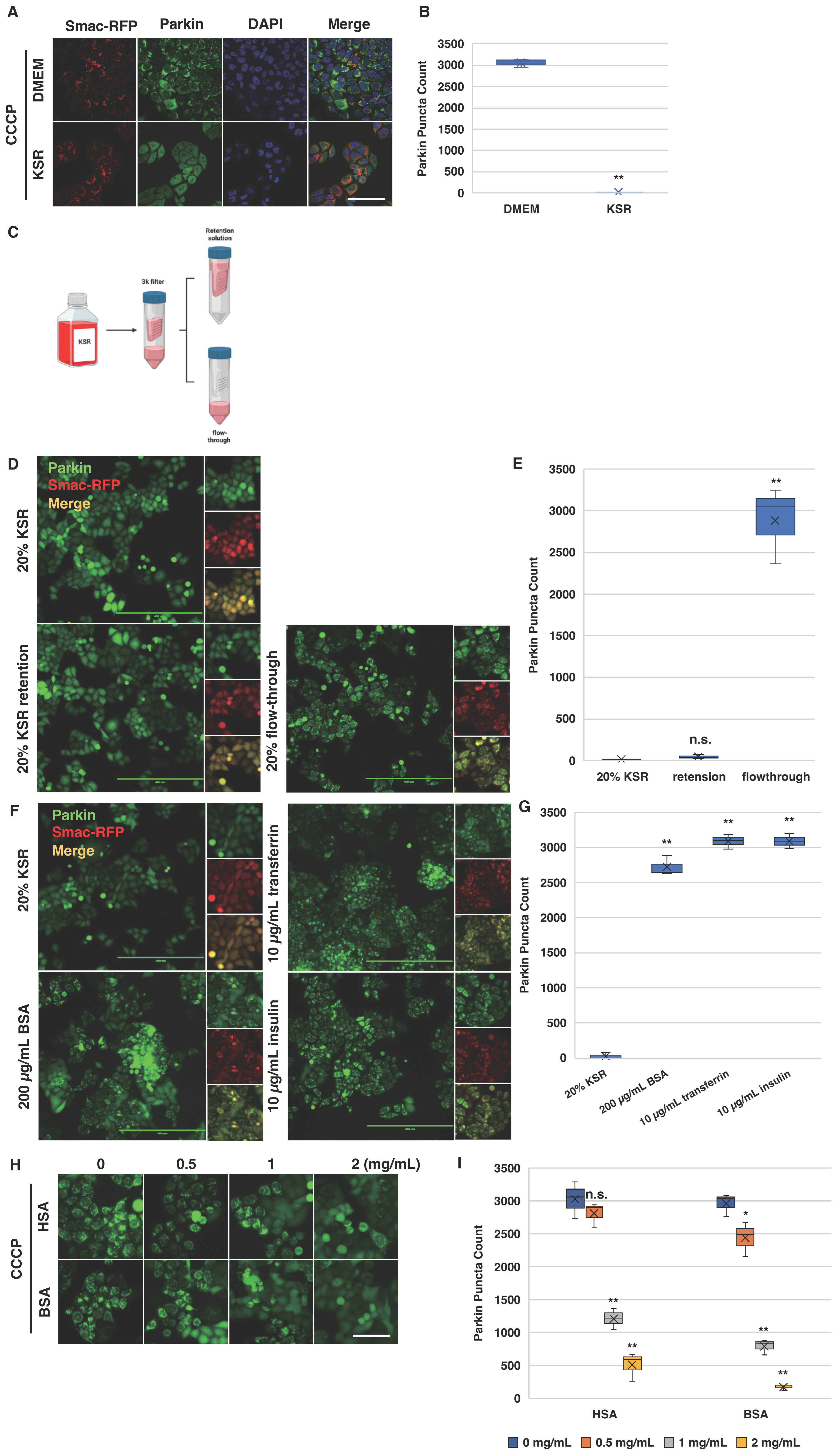
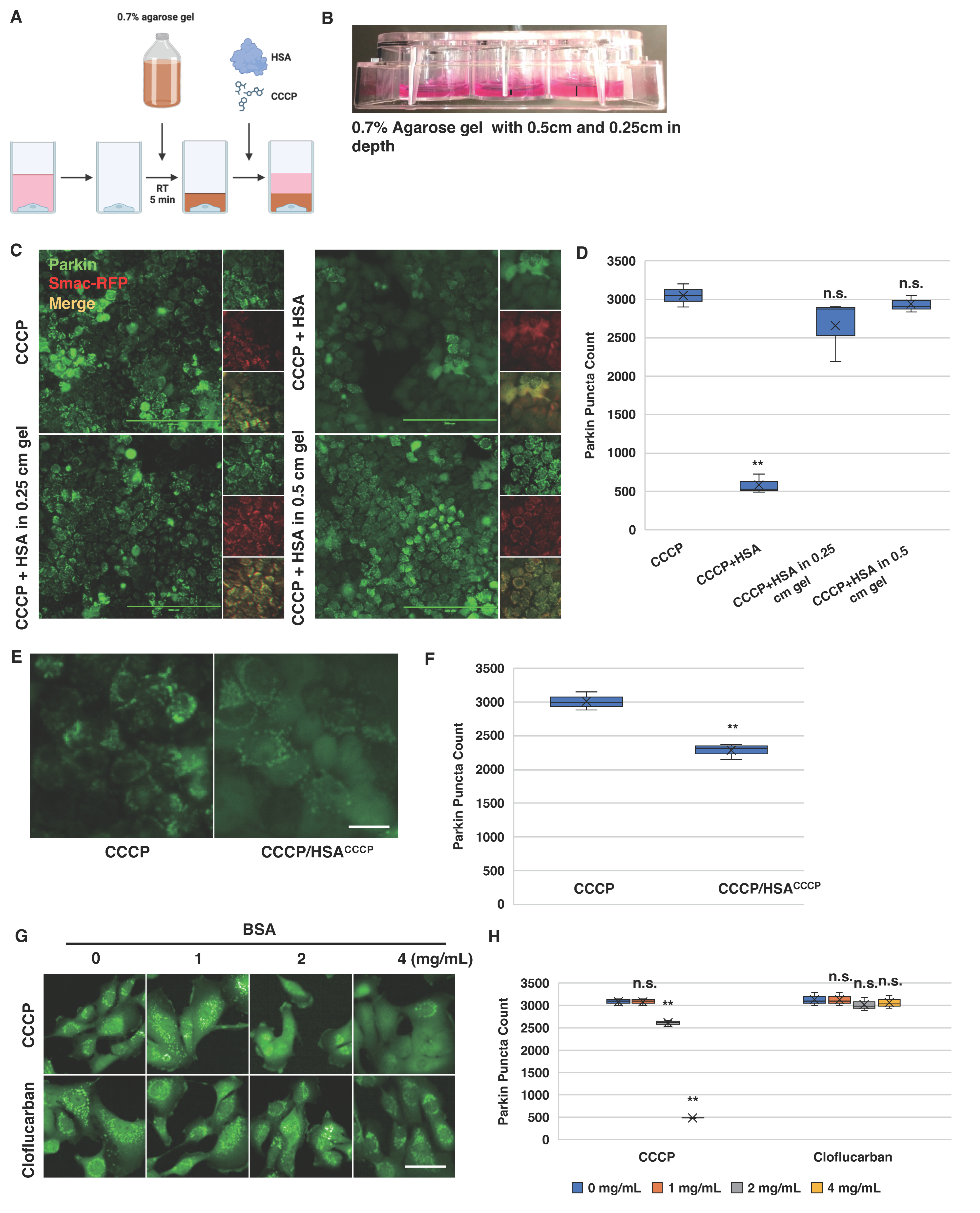
2.5. Bovine Serum Albumin Suppresses Parkin Mitochondrial Translocation Induced by CCCP
2.6. Albumin Modulates Cellular Response to CCCP-Induced Parkin Mitochondrial Translocation
2.7. Cloflucarban Induces Parkin Mitochondrial Translocation Independent of BSA
3. Discussion
4. Methods and Materials
4.1. Mitochondrial Complex Activity Assays
4.2. Cell Culture, Transfection, and Reagent Treatment
4.3. Live Cell Imaging and Fluorescence Microscopy
4.4. Mitochondrial Membrane Potential Assay and Quantification
4.5. Parkin Mitochondrial Recruitment Assays
4.6. Mitophagy Assays
4.7. PINK1 Stabilization Assay
4.8. Mitochondria Superoxide Production Assay
4.9. Parkin Mitochondrial Recruitment Assay with PINK1-M318A Activity Inhibition
4.10. shRNA Knockdown and Lentiviral Transduction
4.11. RNA Extraction and Quantitative Real-Time PCR
4.12. Agarose Diffusion Assay
4.13. HSA Saturation Assay
4.14. Cloflucarban Evaluation Assay
4.15. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Youle, R.J.; Narendra, D.P. Mechanisms of mitophagy. Nat. Rev. Mol. Cell Biol. 2011, 12, 9–14. [Google Scholar] [CrossRef]
- Pickrell, A.M.; Youle, R.J. The roles of PINK1, parkin, and mitochondrial fidelity in Parkinson’s disease. Neuron 2015, 85, 257–273. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, S.; Muqit, M.M.K. PTEN-induced kinase 1 (PINK1) and Parkin: Unlocking a mitochondrial quality control pathway linked to Parkinson’s disease. Curr. Opin. Neurobiol. 2022, 72, 111–119. [Google Scholar] [CrossRef] [PubMed]
- Jin, S.M.; Lazarou, M.; Wang, C.; Kane, L.A.; Narendra, D.P.; Youle, R.J. Mitochondrial membrane potential regulates PINK1 import and proteolytic destabilization by PARL. J. Cell Biol. 2010, 191, 933–942. [Google Scholar] [CrossRef]
- Deas, E.; Plun-Favreau, H.; Gandhi, S.; Desmond, H.; Kjaer, S.; Loh, S.H.; Renton, A.E.; Harvey, R.J.; Whitworth, A.J.; Martins, L.M.; et al. PINK1 cleavage at position A103 by the mitochondrial protease PARL. Hum. Mol. Genet. 2011, 20, 867–879. [Google Scholar] [CrossRef]
- Meissner, C.; Lorenz, H.; Weihofen, A.; Selkoe, D.J.; Lemberg, M.K. The mitochondrial intramembrane protease PARL cleaves human Pink1 to regulate Pink1 trafficking. J. Neurochem. 2011, 117, 856–867. [Google Scholar] [CrossRef]
- Yamano, K.; Youle, R.J. PINK1 is degraded through the N-end rule pathway. Autophagy 2013, 9, 1758–1769. [Google Scholar] [CrossRef]
- Okatsu, K.; Oka, T.; Iguchi, M.; Imamura, K.; Kosako, H.; Tani, N.; Kimura, M.; Go, E.; Koyano, F.; Funayama, M.; et al. PINK1 autophosphorylation upon membrane potential dissipation is essential for Parkin recruitment to damaged mitochondria. Nat. Commun. 2012, 3, 1016. [Google Scholar] [CrossRef]
- Gladkova, C.; Maslen, S.L.; Skehel, J.M.; Komander, D. Mechanism of parkin activation by PINK1. Nature 2018, 559, 410–414. [Google Scholar] [CrossRef]
- Geisler, S.; Holmstrom, K.M.; Treis, A.; Skujat, D.; Weber, S.S.; Fiesel, F.C.; Kahle, P.J.; Springer, W. The PINK1/Parkin-mediated mitophagy is compromised by PD-associated mutations. Autophagy 2010, 6, 871–878. [Google Scholar] [CrossRef]
- Matsuda, N.; Sato, S.; Shiba, K.; Okatsu, K.; Saisho, K.; Gautier, C.A.; Sou, Y.S.; Saiki, S.; Kawajiri, S.; Sato, F.; et al. PINK1 stabilized by mitochondrial depolarization recruits Parkin to damaged mitochondria and activates latent Parkin for mitophagy. J. Cell Biol. 2010, 189, 211–221. [Google Scholar] [CrossRef]
- Narendra, D.P.; Jin, S.M.; Tanaka, A.; Suen, D.F.; Gautier, C.A.; Shen, J.; Cookson, M.R.; Youle, R.J. PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol. 2010, 8, e1000298. [Google Scholar] [CrossRef]
- Vives-Bauza, C.; Zhou, C.; Huang, Y.; Cui, M.; de Vries, R.L.; Kim, J.; May, J.; Tocilescu, M.A.; Liu, W.; Ko, H.S.; et al. PINK1-dependent recruitment of Parkin to mitochondria in mitophagy. Proc. Natl. Acad. Sci. USA 2010, 107, 378–383. [Google Scholar] [CrossRef]
- Kazlauskaite, A.; Kelly, V.; Johnson, C.; Baillie, C.; Hastie, C.J.; Peggie, M.; Macartney, T.; Woodroof, H.I.; Alessi, D.R.; Pedrioli, P.G.; et al. Phosphorylation of Parkin at Serine65 is essential for activation: Elaboration of a Miro1 substrate-based assay of Parkin E3 ligase activity. Open Biol. 2014, 4, 130213. [Google Scholar] [CrossRef]
- Kane, L.A.; Lazarou, M.; Fogel, A.I.; Li, Y.; Yamano, K.; Sarraf, S.A.; Banerjee, S.; Youle, R.J. PINK1 phosphorylates ubiquitin to activate Parkin E3 ubiquitin ligase activity. J. Cell Biol. 2014, 205, 143–153. [Google Scholar] [CrossRef]
- Koyano, F.; Okatsu, K.; Kosako, H.; Tamura, Y.; Go, E.; Kimura, M.; Kimura, Y.; Tsuchiya, H.; Yoshihara, H.; Hirokawa, T.; et al. Ubiquitin is phosphorylated by PINK1 to activate parkin. Nature 2014, 510, 162–166. [Google Scholar] [CrossRef]
- Zhang, C.; Lee, S.; Peng, Y.; Bunker, E.; Giaime, E.; Shen, J.; Zhou, Z.; Liu, X. PINK1 triggers autocatalytic activation of Parkin to specify cell fate decisions. Curr. Biol. 2014, 24, 1854–1865. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Dorn, G.W., 2nd. PINK1-phosphorylated mitofusin 2 is a Parkin receptor for culling damaged mitochondria. Science 2013, 340, 471–475. [Google Scholar] [CrossRef] [PubMed]
- Gegg, M.E.; Cooper, J.M.; Chau, K.Y.; Rojo, M.; Schapira, A.H.; Taanman, J.W. Mitofusin 1 and mitofusin 2 are ubiquitinated in a PINK1/parkin-dependent manner upon induction of mitophagy. Hum. Mol. Genet. 2010, 19, 4861–4870. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, A.; Cleland, M.M.; Xu, S.; Narendra, D.P.; Suen, D.F.; Karbowski, M.; Youle, R.J. Proteasome and p97 mediate mitophagy and degradation of mitofusins induced by Parkin. J. Cell Biol. 2010, 191, 1367–1380. [Google Scholar] [CrossRef]
- Sarraf, S.A.; Raman, M.; Guarani-Pereira, V.; Sowa, M.E.; Huttlin, E.L.; Gygi, S.P.; Harper, J.W. Landscape of the PARKIN-dependent ubiquitylome in response to mitochondrial depolarization. Nature 2013, 496, 372–376. [Google Scholar] [CrossRef]
- Ordureau, A.; Sarraf, S.A.; Duda, D.M.; Heo, J.M.; Jedrychowski, M.P.; Sviderskiy, V.O.; Olszewski, J.L.; Koerber, J.T.; Xie, T.; Beausoleil, S.A.; et al. Quantitative proteomics reveal a feedforward mechanism for mitochondrial PARKIN translocation and ubiquitin chain synthesis. Mol. Cell 2014, 56, 360–375. [Google Scholar] [CrossRef]
- Ordureau, A.; Paulo, J.A.; Zhang, W.; Ahfeldt, T.; Zhang, J.; Cohn, E.F.; Hou, Z.; Heo, J.M.; Rubin, L.L.; Sidhu, S.S.; et al. Dynamics of PARKIN-Dependent Mitochondrial Ubiquitylation in Induced Neurons and Model Systems Revealed by Digital Snapshot Proteomics. Mol. Cell 2018, 70, 211–227.e218. [Google Scholar] [CrossRef]
- Lazarou, M.; Sliter, D.A.; Kane, L.A.; Sarraf, S.A.; Wang, C.; Burman, J.L.; Sideris, D.P.; Fogel, A.I.; Youle, R.J. The ubiquitin kinase PINK1 recruits autophagy receptors to induce mitophagy. Nature 2015, 524, 309–314. [Google Scholar] [CrossRef]
- Heo, J.M.; Ordureau, A.; Paulo, J.A.; Rinehart, J.; Harper, J.W. The PINK1-PARKIN Mitochondrial Ubiquitylation Pathway Drives a Program of OPTN/NDP52 Recruitment and TBK1 Activation to Promote Mitophagy. Mol. Cell 2015, 60, 7–20. [Google Scholar] [CrossRef]
- Richter, B.; Sliter, D.A.; Herhaus, L.; Stolz, A.; Wang, C.; Beli, P.; Zaffagnini, G.; Wild, P.; Martens, S.; Wagner, S.A.; et al. Phosphorylation of OPTN by TBK1 enhances its binding to Ub chains and promotes selective autophagy of damaged mitochondria. Proc. Natl. Acad. Sci. USA 2016, 113, 4039–4044. [Google Scholar] [CrossRef] [PubMed]
- Shimura, H.; Hattori, N.; Kubo, S.; Mizuno, Y.; Asakawa, S.; Minoshima, S.; Shimizu, N.; Iwai, K.; Chiba, T.; Tanaka, K.; et al. Familial Parkinson disease gene product, parkin, is a ubiquitin-protein ligase. Nat. Genet. 2000, 25, 302–305. [Google Scholar] [CrossRef] [PubMed]
- Valente, E.M.; Abou-Sleiman, P.M.; Caputo, V.; Muqit, M.M.; Harvey, K.; Gispert, S.; Ali, Z.; Del Turco, D.; Bentivoglio, A.R.; Healy, D.G.; et al. Hereditary early-onset Parkinson’s disease caused by mutations in PINK1. Science 2004, 304, 1158–1160. [Google Scholar] [CrossRef]
- Ge, P.; Dawson, V.L.; Dawson, T.M. PINK1 and Parkin mitochondrial quality control: A source of regional vulnerability in Parkinson’s disease. Mol. Neurodegener. 2020, 15, 20. [Google Scholar] [CrossRef]
- Narendra, D.; Tanaka, A.; Suen, D.F.; Youle, R.J. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J. Cell Biol. 2008, 183, 795–803. [Google Scholar] [CrossRef] [PubMed]
- Georgakopoulos, N.D.; Wells, G.; Campanella, M. The pharmacological regulation of cellular mitophagy. Nat. Chem. Biol. 2017, 13, 136–146. [Google Scholar] [CrossRef]
- Grenier, K.; McLelland, G.L.; Fon, E.A. Parkin- and PINK1-Dependent Mitophagy in Neurons: Will the Real Pathway Please Stand Up? Front. Neurol. 2013, 4, 100. [Google Scholar] [CrossRef]
- Kondapalli, C.; Kazlauskaite, A.; Zhang, N.; Woodroof, H.I.; Campbell, D.G.; Gourlay, R.; Burchell, L.; Walden, H.; Macartney, T.J.; Deak, M.; et al. PINK1 is activated by mitochondrial membrane potential depolarization and stimulates Parkin E3 ligase activity by phosphorylating Serine 65. Open Biol. 2012, 2, 120080. [Google Scholar] [CrossRef]
- Burman, J.L.; Pickles, S.; Wang, C.; Sekine, S.; Vargas, J.N.S.; Zhang, Z.; Youle, A.M.; Nezich, C.L.; Wu, X.; Hammer, J.A.; et al. Mitochondrial fission facilitates the selective mitophagy of protein aggregates. J. Cell Biol. 2017, 216, 3231–3247. [Google Scholar] [CrossRef] [PubMed]
- Jin, S.M.; Youle, R.J. The accumulation of misfolded proteins in the mitochondrial matrix is sensed by PINK1 to induce PARK2/Parkin-mediated mitophagy of polarized mitochondria. Autophagy 2013, 9, 1750–1757. [Google Scholar] [CrossRef] [PubMed]
- Fiesel, F.C.; James, E.D.; Hudec, R.; Springer, W. Mitochondrial targeted HSP90 inhibitor Gamitrinib-TPP (G-TPP) induces PINK1/Parkin-dependent mitophagy. Oncotarget 2017, 8, 106233–106248. [Google Scholar] [CrossRef] [PubMed]
- Munch, C.; Harper, J.W. Mitochondrial unfolded protein response controls matrix pre-RNA processing and translation. Nature 2016, 534, 710–713. [Google Scholar] [CrossRef]
- Spelbrink, J.N.; Li, F.Y.; Tiranti, V.; Nikali, K.; Yuan, Q.P.; Tariq, M.; Wanrooij, S.; Garrido, N.; Comi, G.; Morandi, L.; et al. Human mitochondrial DNA deletions associated with mutations in the gene encoding Twinkle, a phage T7 gene 4-like protein localized in mitochondria. Nat. Genet. 2001, 28, 223–231. [Google Scholar] [CrossRef] [PubMed]
- Sliter, D.A.; Martinez, J.; Hao, L.; Chen, X.; Sun, N.; Fischer, T.D.; Burman, J.L.; Li, Y.; Zhang, Z.; Narendra, D.P.; et al. Parkin and PINK1 mitigate STING-induced inflammation. Nature 2018, 561, 258–262. [Google Scholar] [CrossRef]
- Jungermann, E.; Brown, J., Jr.; Yackovich, F.; Taber, D. Comparative evaluation of antibacterial soaps. J. Am. Oil Chem. Soc. 1967, 44, 232–234. [Google Scholar] [CrossRef]
- Lee, S.; Zhang, C.; Liu, X. Role of glucose metabolism and ATP in maintaining PINK1 levels during Parkin-mediated mitochondrial damage responses. J. Biol. Chem. 2015, 290, 904–917. [Google Scholar] [CrossRef]
- Nezich, C.L.; Wang, C.; Fogel, A.I.; Youle, R.J. MiT/TFE transcription factors are activated during mitophagy downstream of Parkin and Atg5. J. Cell Biol. 2015, 210, 435–450. [Google Scholar] [CrossRef] [PubMed]
- McLelland, G.L.; Soubannier, V.; Chen, C.X.; McBride, H.M.; Fon, E.A. Parkin and PINK1 function in a vesicular trafficking pathway regulating mitochondrial quality control. EMBO J. 2014, 33, 282–295. [Google Scholar] [CrossRef]
- Durhuus, J.A.; Hansson, S.; Morville, T.; Kuhlman, A.B.; Dohlmann, T.L.; Larsen, S.; Helge, J.W.; Angleys, M.; Muniesa-Vargas, A.; Bundgaard, J.R.; et al. Simvastatin improves mitochondrial respiration in peripheral blood cells. Sci. Rep. 2020, 10, 17012. [Google Scholar] [CrossRef] [PubMed]
- Takei, M.; Hiramatsu, M.; Mori, A. Inhibitory effects of calcium antagonists on mitochondrial swelling induced by lipid peroxidation or arachidonic acid in the rat brain in vitro. Neurochem. Res. 1994, 19, 1199–1206. [Google Scholar] [CrossRef] [PubMed]
- Rachek, L.I.; Yuzefovych, L.V.; Ledoux, S.P.; Julie, N.L.; Wilson, G.L. Troglitazone, but not rosiglitazone, damages mitochondrial DNA and induces mitochondrial dysfunction and cell death in human hepatocytes. Toxicol. Appl. Pharmacol. 2009, 240, 348–354. [Google Scholar] [CrossRef] [PubMed]
- Scatena, R.; Bottoni, P.; Martorana, G.E.; Vincenzoni, F.; Botta, G.; Pastore, P.; Giardina, B. Mitochondria, ciglitazone and liver: A neglected interaction in biochemical pharmacology. Eur. J. Pharmacol. 2007, 567, 50–58. [Google Scholar] [CrossRef]
- Lagouge, M.; Argmann, C.; Gerhart-Hines, Z.; Meziane, H.; Lerin, C.; Daussin, F.; Messadeq, N.; Milne, J.; Lambert, P.; Elliott, P.; et al. Resveratrol improves mitochondrial function and protects against metabolic disease by activating SIRT1 and PGC-1alpha. Cell 2006, 127, 1109–1122. [Google Scholar] [CrossRef]
- Davoudi, M.; Kallijarvi, J.; Marjavaara, S.; Kotarsky, H.; Hansson, E.; Leveen, P.; Fellman, V. A mouse model of mitochondrial complex III dysfunction induced by myxothiazol. Biochem. Biophys. Res. Commun. 2014, 446, 1079–1084. [Google Scholar] [CrossRef]
- Li, N.; Ragheb, K.; Lawler, G.; Sturgis, J.; Rajwa, B.; Melendez, J.A.; Robinson, J.P. Mitochondrial complex I inhibitor rotenone induces apoptosis through enhancing mitochondrial reactive oxygen species production. J. Biol. Chem. 2003, 278, 8516–8525. [Google Scholar] [CrossRef]
- Zhang, J.G.; Fariss, M.W. Thenoyltrifluoroacetone, a potent inhibitor of carboxylesterase activity. Biochem. Pharmacol. 2002, 63, 751–754. [Google Scholar] [CrossRef]
- Hargreaves, I.P.; Duncan, A.J.; Wu, L.; Agrawal, A.; Land, J.M.; Heales, S.J. Inhibition of mitochondrial complex IV leads to secondary loss complex II-III activity: Implications for the pathogenesis and treatment of mitochondrial encephalomyopathies. Mitochondrion 2007, 7, 284–287. [Google Scholar] [CrossRef]
- Roberts, H.; Choo, W.M.; Murphy, M.; Marzuki, S.; Lukins, H.B.; Linnane, S.W. mit- Mutations in the oli2 region of mitochondrial DNA affecting the 20 000 dalton subunit of the mitochondrial ATPase in Saccharomyces cerevisiae. FEBS Lett. 1979, 108, 501–504. [Google Scholar] [CrossRef]
- Pham, N.A.; Robinson, B.H.; Hedley, D.W. Simultaneous detection of mitochondrial respiratory chain activity and reactive oxygen in digitonin-permeabilized cells using flow cytometry. Cytometry 2000, 41, 245–251. [Google Scholar] [CrossRef] [PubMed]
- Ehrenberg, B.; Montana, V.; Wei, M.D.; Wuskell, J.P.; Loew, L.M. Membrane potential can be determined in individual cells from the nernstian distribution of cationic dyes. Biophys. J. 1988, 53, 785–794. [Google Scholar] [CrossRef]
- Strappazzon, F.; Nazio, F.; Corrado, M.; Cianfanelli, V.; Romagnoli, A.; Fimia, G.M.; Campello, S.; Nardacci, R.; Piacentini, M.; Campanella, M.; et al. AMBRA1 is able to induce mitophagy via LC3 binding, regardless of PARKIN and p62/SQSTM1. Cell Death Differ. 2015, 22, 419–432. [Google Scholar] [CrossRef]
- Bishop, A.C.; Ubersax, J.A.; Petsch, D.T.; Matheos, D.P.; Gray, N.S.; Blethrow, J.; Shimizu, E.; Tsien, J.Z.; Schultz, P.G.; Rose, M.D.; et al. A chemical switch for inhibitor-sensitive alleles of any protein kinase. Nature 2000, 407, 395–401. [Google Scholar] [CrossRef] [PubMed]
- Suski, J.M.; Lebiedzinska, M.; Bonora, M.; Pinton, P.; Duszynski, J.; Wieckowski, M.R. Relation between mitochondrial membrane potential and ROS formation. Methods Mol. Biol. 2012, 810, 183–205. [Google Scholar] [CrossRef]
- Pennacchio, L.A.; Bergmann, A.; Fukushima, A.; Okubo, K.; Salemi, A.; Lennon, G.G. Structure, sequence and location of the UQCRFS1 gene for the human Rieske Fe-S protein. Gene 1995, 155, 207–211. [Google Scholar] [CrossRef]
- Nicholls, D.G.; Budd, S.L. Mitochondria and neuronal survival. Physiol. Rev. 2000, 80, 315–360. [Google Scholar] [CrossRef] [PubMed]
- Kandagal, P.B.; Ashoka, S.; Seetharamappa, J.; Shaikh, S.M.; Jadegoud, Y.; Ijare, O.B. Study of the interaction of an anticancer drug with human and bovine serum albumin: Spectroscopic approach. J. Pharm. Biomed. Anal. 2006, 41, 393–399. [Google Scholar] [CrossRef]
- Ascenzi, P.; di Masi, A.; De Sanctis, G.; Coletta, M.; Fasano, M. Ibuprofen modulates allosterically NO dissociation from ferrous nitrosylated human serum heme-albumin by binding to three sites. Biochem. Biophys. Res. Commun. 2009, 387, 83–86. [Google Scholar] [CrossRef]
- Sudlow, G.; Birkett, D.J.; Wade, D.N. The characterization of two specific drug binding sites on human serum albumin. Mol. Pharmacol. 1975, 11, 824–832. [Google Scholar]
- Ghuman, J.; Zunszain, P.A.; Petitpas, I.; Bhattacharya, A.A.; Otagiri, M.; Curry, S. Structural basis of the drug-binding specificity of human serum albumin. J. Mol. Biol. 2005, 353, 38–52. [Google Scholar] [CrossRef] [PubMed]
- Deffieu, M.; Bhatia-Kissova, I.; Salin, B.; Klionsky, D.J.; Pinson, B.; Manon, S.; Camougrand, N. Increased levels of reduced cytochrome b and mitophagy components are required to trigger nonspecific autophagy following induced mitochondrial dysfunction. J. Cell Sci. 2013, 126, 415–426. [Google Scholar] [CrossRef]
- Mailloux, R.J.; McBride, S.L.; Harper, M.E. Unearthing the secrets of mitochondrial ROS and glutathione in bioenergetics. Trends Biochem. Sci. 2013, 38, 592–602. [Google Scholar] [CrossRef]
- Demers-Lamarche, J.; Guillebaud, G.; Tlili, M.; Todkar, K.; Belanger, N.; Grondin, M.; Nguyen, A.P.; Michel, J.; Germain, M. Loss of Mitochondrial Function Impairs Lysosomes. J. Biol. Chem. 2016, 291, 10263–10276. [Google Scholar] [CrossRef]
- Turrens, J.F.; Alexandre, A.; Lehninger, A.L. Ubisemiquinone is the electron donor for superoxide formation by complex III of heart mitochondria. Arch. Biochem. Biophys. 1985, 237, 408–414. [Google Scholar] [CrossRef]
- Xiao, B.; Goh, J.Y.; Xiao, L.; Xian, H.; Lim, K.L.; Liou, Y.C. Reactive oxygen species trigger Parkin/PINK1 pathway-dependent mitophagy by inducing mitochondrial recruitment of Parkin. J. Biol. Chem. 2017, 292, 16697–16708. [Google Scholar] [CrossRef]
- Xiao, B.; Deng, X.; Lim, G.G.Y.; Xie, S.; Zhou, Z.D.; Lim, K.L.; Tan, E.K. Superoxide drives progression of Parkin/PINK1-dependent mitophagy following translocation of Parkin to mitochondria. Cell Death Dis. 2017, 8, e3097. [Google Scholar] [CrossRef]
- Evstafieva, A.G.; Garaeva, A.A.; Khutornenko, A.A.; Klepikova, A.V.; Logacheva, M.D.; Penin, A.A.; Novakovsky, G.E.; Kovaleva, I.E.; Chumakov, P.M. A sustained deficiency of mitochondrial respiratory complex III induces an apoptotic cell death through the p53-mediated inhibition of pro-survival activities of the activating transcription factor 4. Cell Death Dis. 2014, 5, e1511. [Google Scholar] [CrossRef][Green Version]
- Zhang, C.; Liu, Z.; Bunker, E.; Ramirez, A.; Lee, S.; Peng, Y.; Tan, A.C.; Eckhardt, S.G.; Chapnick, D.A.; Liu, X. Sorafenib targets the mitochondrial electron transport chain complexes and ATP synthase to activate the PINK1-Parkin pathway and modulate cellular drug response. J. Biol. Chem. 2017, 292, 15105–15120. [Google Scholar] [CrossRef] [PubMed]
- Soutar, M.P.M.; Kempthorne, L.; Annuario, E.; Luft, C.; Wray, S.; Ketteler, R.; Ludtmann, M.H.R.; Plun-Favreau, H. FBS/BSA media concentration determines CCCP’s ability to depolarize mitochondria and activate PINK1-PRKN mitophagy. Autophagy 2019, 15, 2002–2011. [Google Scholar] [CrossRef]
- Mahalaxmi, G.; Ashok, S.; Arun, G.; Srinivas, G. Albumin binds to uncoupler CCCP to diminish depolarization of mitochondria. Toxicol. In Vitro 2022, 80, 105325. [Google Scholar] [CrossRef]
- Padman, B.S.; Bach, M.; Lucarelli, G.; Prescott, M.; Ramm, G. The protonophore CCCP interferes with lysosomal degradation of autophagic cargo in yeast and mammalian cells. Autophagy 2013, 9, 1862–1875. [Google Scholar] [CrossRef]
- Weinert, S.; Jabs, S.; Supanchart, C.; Schweizer, M.; Gimber, N.; Richter, M.; Rademann, J.; Stauber, T.; Kornak, U.; Jentsch, T.J. Lysosomal pathology and osteopetrosis upon loss of H+-driven lysosomal Cl- accumulation. Science 2010, 328, 1401–1403. [Google Scholar] [CrossRef]
- Burkhardt, J.K.; Hester, S.; Argon, Y. The glycoprotein of VSV accumulates in a distal Golgi compartment in the presence of CCCP. J. Cell Sci. 1989, 92 Pt 4, 643–654. [Google Scholar] [CrossRef]
- Llopis, J.; McCaffery, J.M.; Miyawaki, A.; Farquhar, M.G.; Tsien, R.Y. Measurement of cytosolic, mitochondrial, and Golgi pH in single living cells with green fluorescent proteins. Proc. Natl. Acad. Sci. USA 1998, 95, 6803–6808. [Google Scholar] [CrossRef]
- Maro, B.; Marty, M.C.; Bornens, M. In vivo and in vitro effects of the mitochondrial uncoupler FCCP on microtubules. EMBO J. 1982, 1, 1347–1352. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Lee, S.; Peng, Y.; Bunker, E.; Shen, C.; Giaime, E.; Shen, J.; Shen, J.; Zhou, Z.; Liu, X. A chemical genetic approach to probe the function of PINK1 in regulating mitochondrial dynamics. Cell Res. 2015, 25, 394–397. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ramirez, A.T.; Liu, Z.; Xu, Q.; Nowosadtko, S.; Liu, X. Cloflucarban Illuminates Specificity and Context-Dependent Activation of the PINK1–Parkin Pathway by Mitochondrial Complex Inhibition. Biomolecules 2024, 14, 248. https://doi.org/10.3390/biom14030248
Ramirez AT, Liu Z, Xu Q, Nowosadtko S, Liu X. Cloflucarban Illuminates Specificity and Context-Dependent Activation of the PINK1–Parkin Pathway by Mitochondrial Complex Inhibition. Biomolecules. 2024; 14(3):248. https://doi.org/10.3390/biom14030248
Chicago/Turabian StyleRamirez, Adrian T., Zeyu Liu, Quanbin Xu, Sarah Nowosadtko, and Xuedong Liu. 2024. "Cloflucarban Illuminates Specificity and Context-Dependent Activation of the PINK1–Parkin Pathway by Mitochondrial Complex Inhibition" Biomolecules 14, no. 3: 248. https://doi.org/10.3390/biom14030248
APA StyleRamirez, A. T., Liu, Z., Xu, Q., Nowosadtko, S., & Liu, X. (2024). Cloflucarban Illuminates Specificity and Context-Dependent Activation of the PINK1–Parkin Pathway by Mitochondrial Complex Inhibition. Biomolecules, 14(3), 248. https://doi.org/10.3390/biom14030248






