Dclre1c-Mutation-Induced Immunocompromised Mice Are a Novel Model for Human Xenograft Research
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Materials and Reagents
2.3. Microinjection
2.4. Management of the Homozygous Dclre1c Knockout Mouse Colonies
2.5. Organ Index and Hematoxylin and Eosin Staining (H&E) Assay
2.6. Flow Cytometry
2.7. Cell Culture
2.8. Real Time qPCR
2.9. DNA Damage Assay
2.10. Human Xenograft Model
2.11. Multiplexed Immunofluorescence
2.12. Statistical Analysis
3. Results
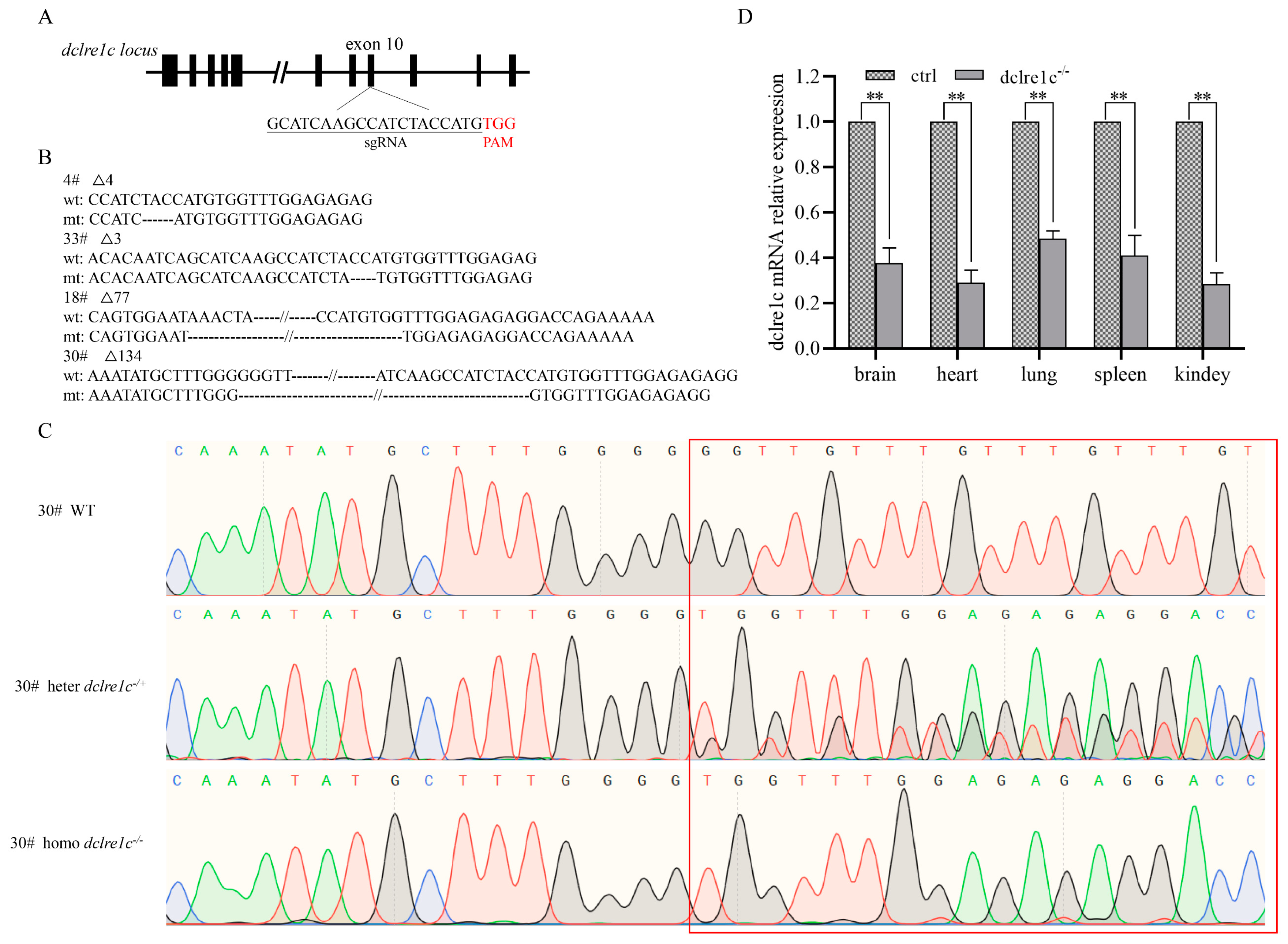
3.1. Generation of Homozygous Dclre1c Knockout Mouse Colonies
3.2. Dclre1c Knockout Aggravates DNA Damage
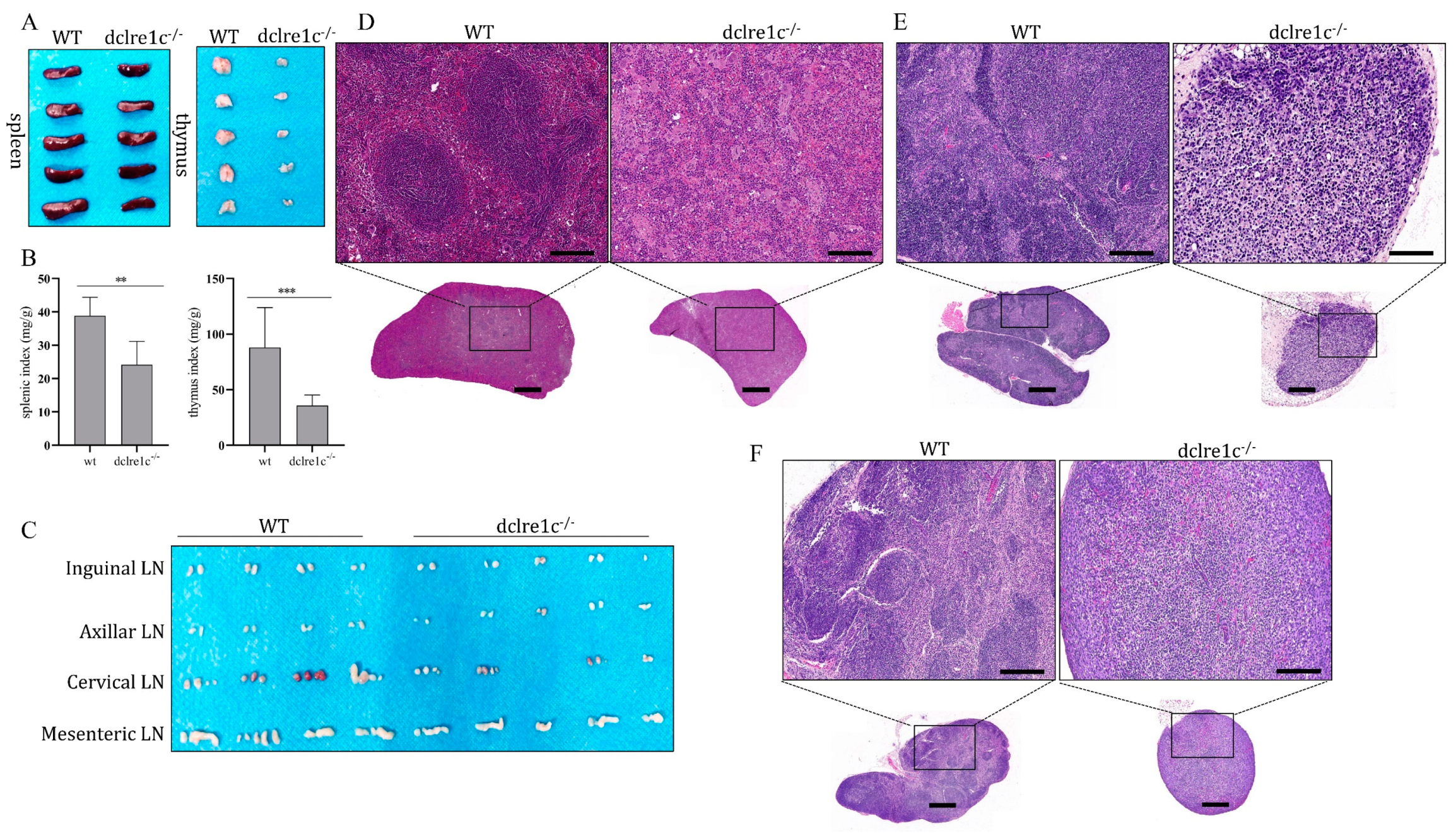
3.3. Dclre1c Knockout Obstructs Immune Organ Development
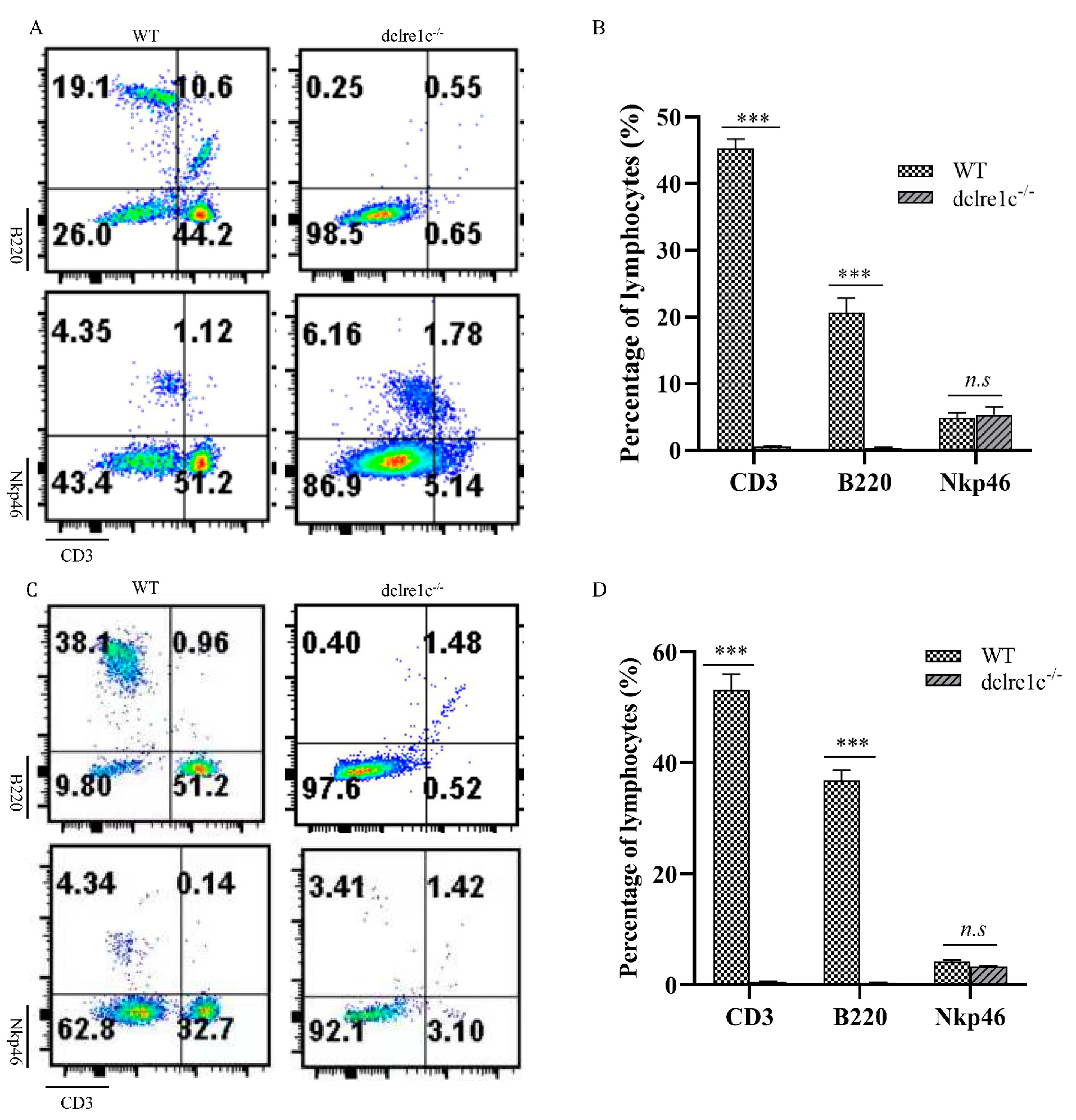
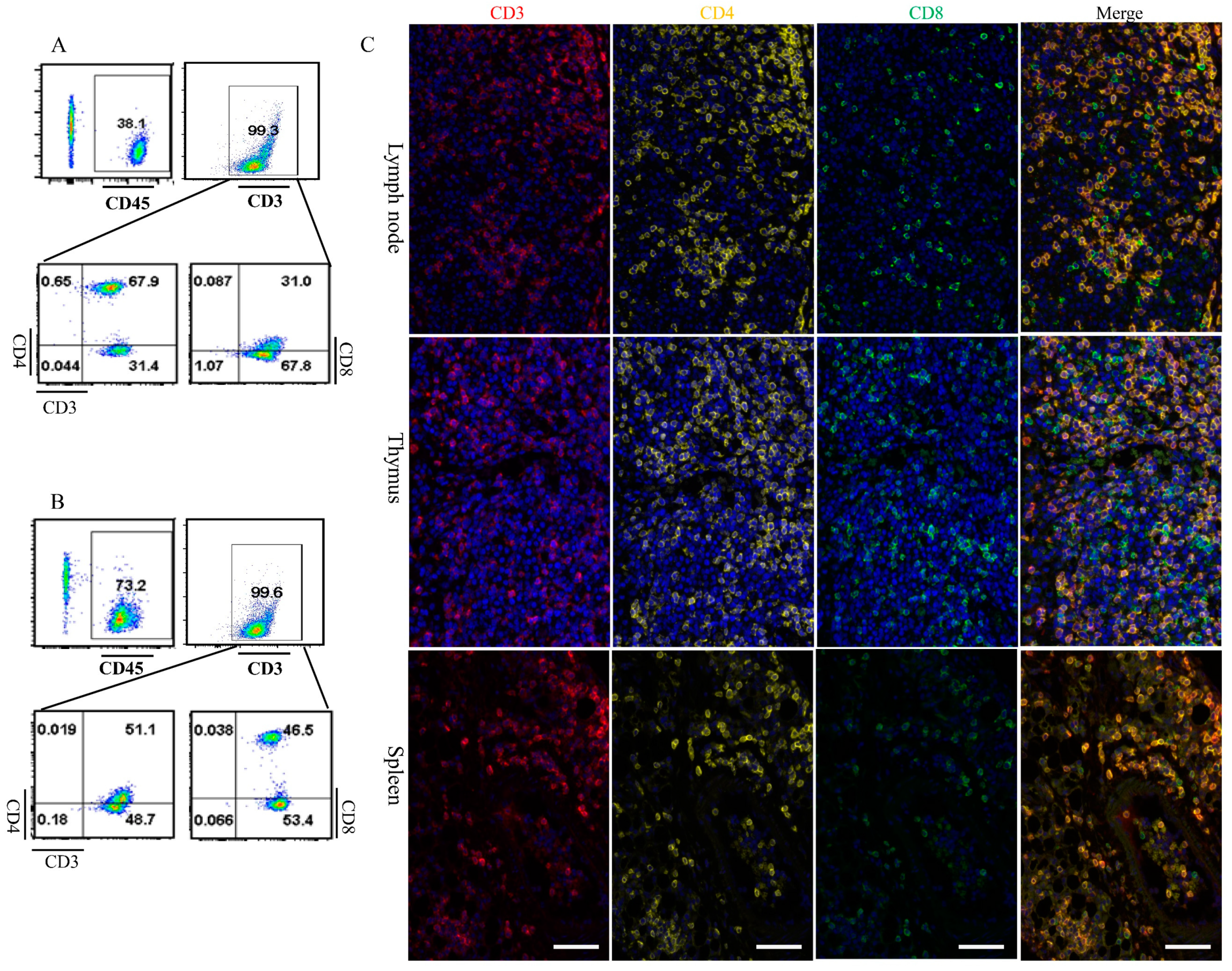
3.4. Dclre1c Knockout Contributes to Immune Cell Insufficiecy
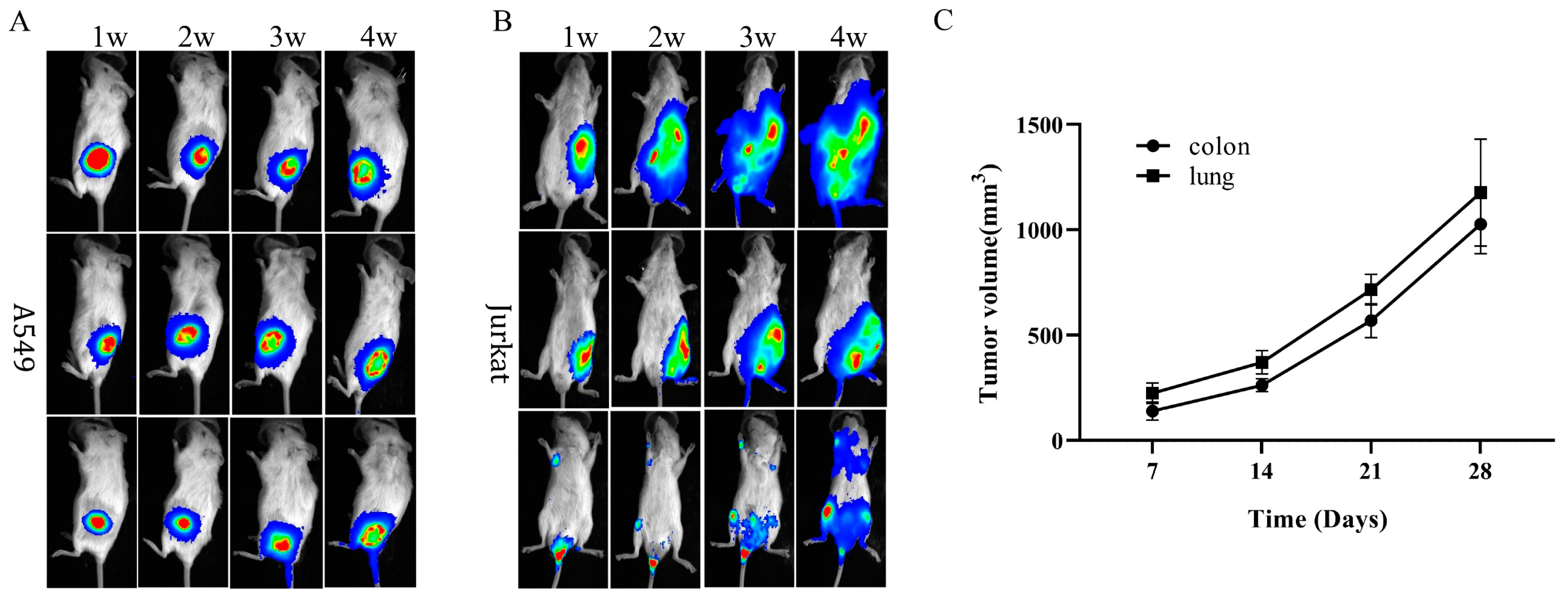
3.5. Immunodeficiency Caused by Dclre1c Knockout Is Suitable for Human Tumor Xenograft Models
3.6. Dclre1c Knockout Mice Are Ideal for Human Immune System Reconstitution
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rooney, S.; Sekiguchi, J.; Zhu, C.; Cheng, H.-L.; Manis, J.; Whitlow, S.; DeVido, J.; Foy, D.; Chaudhuri, J.; Lombard, D.; et al. Leaky Scid Phenotype Associated with Defective V(D)J Coding End Processing in Artemis-Deficient Mice. Mol. Cell 2002, 10, 1379–1390. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, G.; Lieber, M.R. Dynamics of the Artemis and DNA-PKcs Complex in the Repair of Double-Strand Breaks. J. Mol. Biol. 2022, 434, 167858. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, G.; Lieber, M.R.; Williams, D.R. Structural Analysis of the Basal State of the Artemis: DNA-PKcs Complex. Nucleic Acids Res. 2022, 50, 7697–7720. [Google Scholar] [CrossRef]
- Strubbe, S.; De Bruyne, M.; Pannicke, U.; Beyls, E.; Vandekerckhove, B.; Leclercq, G.; De Baere, E.; Bordon, V.; Vral, A.; Schwarz, K.; et al. A Novel Non-Coding Variant in DCLRE1C Results in Deregulated Splicing and Induces SCID through the Generation of a Truncated ARTEMIS Protein That Fails to Support V(D)J Recombination and DNA Damage Repair. Front. Immunol. 2021, 12, 674226. [Google Scholar] [CrossRef]
- Moshous, D.; Callebaut, I.; de Chasseval, R.; Corneo, B.; Cavazzana-Calvo, M.; Le Deist, F.; Tezcan, I.; Sanal, O.; Bertrand, Y.; Philippe, N.; et al. Artemis, a novel DNA Double-strand Break Repair/V(D)J Recombination Protein, is Mutated in Human Severe Combined Immune Deficiency. Cell 2001, 105, 177–186. [Google Scholar] [CrossRef]
- Wang, W.; Li, Y.; Lin, K.; Wang, X.; Tu, Y.; Zhuo, Z. Progress in Building Clinically Relevant Patient-derived Tumor Xenograft Models for Cancer Research. Anim. Model. Exp. Med. 2023, 6, 381–398. [Google Scholar] [CrossRef]
- Tracey, A.T.; Murray, K.S.; Coleman, J.A.; Kim, K. Patient-Derived Xenograft Models in Urological Malignancies: Urothelial Cell Carcinoma and Renal Cell Carcinoma. Cancers 2020, 12, 439. [Google Scholar] [CrossRef]
- Shultz, L.D.; Goodwin, N.; Ishikawa, F.; Hosur, V.; Lyons, B.L.; Greiner, D.L. Human Cancer Growth and Therapy in Immunodeficient Mouse Models. Cold Spring Harb. Protoc. 2014, 2014, 694–708. [Google Scholar] [CrossRef]
- Tanaka, T.; Nishie, R.; Ueda, S.; Miyamoto, S.; Hashida, S.; Konishi, H.; Terada, S.; Kogata, Y.; Sasaki, H.; Tsunetoh, S.; et al. Patient-Derived Xenograft Models in Cervical Cancer: A Systematic Review. Int. J. Mol. Sci. 2021, 22, 9369. [Google Scholar] [CrossRef]
- Benjelloun, F.; Garrigue, A.; Chappedelaine, C.D.-D.; Soulas-Sprauel, P.; Malassis-Séris, M.; Stockholm, D.; Hauer, J.; Blondeau, J.; Rivière, J.; Lim, A.; et al. Stable and functional lymphoid reconstitution in artemis-deficient mice following lentiviral artemis gene transfer into hematopoietic stem cells. Mol. Ther. 2008, 16, 1490–1499. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Z.; Dunn, E.; Singh, K.; Khan, I.S.; Yannone, S.M.; Cowan, M.J. A non-leaky Artemis-deficient mouse that accurately models the human severe combined immune deficiency phenotype, including resistance to hematopoietic stem cell transplantation. Biol. Blood Marrow Transpl. 2009, 15, 1. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Li, L.; Salido, E.; Zhou, Y.; Bhattacharyya, S.; Yannone, S.M.; Dunn, E.; Meneses, J.; Feeney, A.J.; Cowan, M.J. Targeted disruption of the Artemis murine counterpart results in SCID and defective V(D)J recombination that is partially corrected with bone marrow transplantation. J. Immunol. 2005, 174, 2420–2428. [Google Scholar] [CrossRef] [PubMed]
- Bhatia, S.; Pooja; Yadav, S.K. CRISPR-Cas for Genome Editing: Classification, Mechanism, Designing and Applications. Int. J. Biol. Macromol. 2023, 238, 124054. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Huang, Y. The CRIPSR/Cas Gene-editing System—An Immature but Useful Toolkit for Experimental and Clinical Medicine. Anim. Model. Exp. Med. 2019, 2, 5–8. [Google Scholar] [CrossRef]
- Chen, X.-H.; Chen, R.; Shi, M.-Y.; Tian, R.-F.; Zhang, H.; Xin, Z.-Q.; Chen, Z.-N.; Wang, K. Chimeric Antigen Receptor T Cells Targeting CD147 for Non-small Cell Lung Cancer Therapy. Transl. Oncol. 2022, 16, 101309. [Google Scholar] [CrossRef]
- Cui, H.; Wei, W.; Qian, M.; Tian, R.; Fu, X.; Li, H.; Nan, G.; Yang, T.; Lin, P.; Chen, X.; et al. PDGFA-associated Protein 1 is a Novel Target of C-Myc and Contributes to Colorectal Cancer Initiation and Progression. Cancer Commun. 2022, 42, 750–767. [Google Scholar] [CrossRef]
- Zhao, Y.; Liu, P.; Xin, Z.; Shi, C.; Bai, Y.; Sun, X.; Zhao, Y.; Wang, X.; Liu, L.; Zhao, X.; et al. Biological Characteristics of Severe Combined Immunodeficient Mice Produced by CRISPR/Cas9-Mediated Rag2 and IL2rg Mutation. Front. Genet. 2019, 10, 401. [Google Scholar] [CrossRef]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 2008, 3, 1101. [Google Scholar] [CrossRef]
- Yue, X.; Petersen, F.; Shu, Y.; Kasper, B.; Magatsin, J.; Ahmadi, M.; Yin, J.; Wax, J.; Wang, X.; Heidecke, H.; et al. Transfer of PBMC From SSc Patients Induces Autoantibodies and Systemic Inflammation in Rag2-/-/IL2rg-/- Mice. Front. Immunol. 2021, 12, 677970. [Google Scholar] [CrossRef]
- Bétous, R.; de Rugy, T.G.; Pelegrini, A.L.; Queille, S.; de Villartay, J.-P.; Hoffmann, J.-S. DNA Replication Stress Triggers Rapid DNA Replication Fork Breakage by Artemis and XPF. PLOS Genet. 2018, 14, e1007541. [Google Scholar] [CrossRef]
- Gill, R.P.K.; Gantchev, J.; Villarreal, A.M.; Ramchatesingh, B.; Netchiporouk, E.; Akilov, O.E.; Ødum, N.; Gniadecki, R.; Koralov, S.B.; Litvinov, I.V. Understanding Cell Lines, Patient-Derived Xenograft and Genetically Engineered Mouse Models Used to Study Cutaneous T-Cell Lymphoma. Cells 2022, 11, 593. [Google Scholar] [CrossRef]
- Eiseman, J.; Lan, J.; Guo, J.; Joseph, E.; Vucenik, I. Pharmacokinetics and Tissue Distribution of Inositol Hexaphosphate in C.B17 SCID Mice Bearing Human Breast Cancer Xenografts. Metabolism 2011, 60, 1465–1474. [Google Scholar] [CrossRef] [PubMed]
- Villodre, E.S.; Hu, X.; Eckhardt, B.L.; Larson, R.; Huo, L.; Yoon, E.C.; Gong, Y.; Song, J.; Liu, S.; Ueno, N.T.; et al. NDRG1 in Aggressive Breast Cancer Progression and Brain Metastasis. JNCI J. Natl. Cancer Inst. 2022, 114, 579–591. [Google Scholar] [CrossRef] [PubMed]
- Stripecke, R.; Münz, C.; Schuringa, J.J.; Bissig, K.; Soper, B.; Meeham, T.; Yao, L.; Di Santo, J.P.; Brehm, M.; Rodriguez, E.; et al. Innovations, Challenges, and Minimal Information for Standardization of Humanized Mice. EMBO Mol. Med. 2020, 12, e8662. [Google Scholar] [CrossRef]
- De Barros, S.C.; Zimmermann, V.S.; Taylor, N. Concise Review: Hematopoietic Stem Cell Transplantation: Targeting the Thymus. Stem Cells 2013, 31, 1245–1251. [Google Scholar] [CrossRef]
- Muljo, S.A.; Schlissel, M.S. Pre-B and Pre-T-cell Receptors: Conservation of Strategies in Regulating Early Lymphocyte Development. Immunol. Rev. 2000, 175, 80–93. [Google Scholar] [CrossRef]
- Lee, Y.-R.; Kang, G.-S.; Oh, T.; Jo, H.-J.; Park, H.-J.; Ahn, G.-O. DNA-Dependent Protein Kinase Catalytic Subunit (DNA-PKcs): Beyond the DNA Double-Strand Break Repair. Mol. Cells 2023, 46, 200–205. [Google Scholar] [CrossRef]
- Rivera-Munoz, P.; Abramowski, V.; Jacquot, S.; André, P.; Charrier, S.; Lipson-Ruffert, K.; Fischer, A.; Galy, A.; Cavazzana, M.; de Villartay, J.-P. Lymphopoiesis in Transgenic Mice Over-expressing Artemis. Gene Ther. 2016, 23, 176–186. [Google Scholar] [CrossRef]
- Lee, P.P.; Woodbine, L.; Gilmour, K.C.; Bibi, S.; Cale, C.M.; Amrolia, P.J.; Veys, P.A.; Davies, E.G.; Jeggo, P.A.; Jones, A. The Many Faces of Artemis-deficient Combined Immunodeficiency-Two Patients with DCLRE1C Mutations and a Systematic Literature Review of Genotype-phenotype Correlation. Clin. Immunol. 2013, 149, 464–474. [Google Scholar] [CrossRef]
- Chen, J.; Liao, S.; Xiao, Z.; Pan, Q.; Wang, X.; Shen, K.; Wang, S.; Yang, L.; Guo, F.; Liu, H.-F.; et al. The development and improvement of immunodeficient mice and humanized immune system mouse models. Front. Immunol. 2022, 13, 1007579. [Google Scholar] [CrossRef] [PubMed]
- Jiang, W.; Estes, V.M.; Wang, X.S.; Shao, Z.; Lee, B.J.; Lin, X.; Crowe, J.L.; Zha, S. Phosphorylation at S2053 in Murine (S2056 in Human) DNA-PKcs Is Dispensable for Lymphocyte Development and Class Switch Recombination. J. Immunol. 2019, 203, 178–187. [Google Scholar] [CrossRef]
- Bañuelos, C.; Banáth, J.; MacPhail, S.; Zhao, J.; Eaves, C.; O’connor, M.D.; Lansdorp, P.; Olive, P. Mouse but not Human Embryonic Stem Cells are Deficient in Rejoining of Ionizing Radiation-induced DNA Double-strand Breaks. DNA Repair 2008, 7, 1471–1483. [Google Scholar] [CrossRef]
- Roch, B.; Abramowski, V.; Etienne, O.; Musilli, S.; David, P.; Charbonnier, J.B.; Callebaut, I.; Boussin, F.D.; de Villartay, J.P. An XRCC4 Mutant Mouse, a Model for Human X4 Syndrome, Reveals Interplays with Xlf, PAXX, and ATM in Lymphoid Development. eLife 2021, 10, e69353. [Google Scholar] [CrossRef] [PubMed]
- Morio, T. Recent Advances in the Study of Immunodeficiency and DNA Damage Response. Int. J. Hematol. 2017, 106, 357–365. [Google Scholar] [CrossRef] [PubMed]
- Kumrah, R.; Vignesh, P.; Patra, P.; Singh, A.; Anjani, G.; Saini, P.; Sharma, M.; Kaur, A.; Rawat, A. Genetics of Severe Combined Immunodeficiency. Genes Dis. 2019, 7, 52–61. [Google Scholar] [CrossRef] [PubMed]
- Akamatsu, Y.; Oettinger, M.A. Distinct Roles of RAG1 and RAG2 in Binding the V(D)J Recombination Signal Sequences. Mol. Cell. Biol. 1998, 18, 4670–4678. [Google Scholar] [CrossRef]
- Allam, A.; Kabelitz, D. TCR trans-Rearrangements: Biological Significance in Antigen Recognition vs the Role as Lymphoma Biomarker. J. Immunol. 2006, 176, 5707–5712. [Google Scholar] [CrossRef]
- Bassing, C.H.; Swat, W.; Alt, F.W. The Mechanism and Regulation of Chromosomal V(D)J Recombination. Cell 2002, 109, S45–S55. [Google Scholar] [CrossRef]
- Chen, X.; Xu, X.; Chen, Y.; Cheung, J.C.; Wang, H.; Jiang, J.; de Val, N.; Fox, T.; Gellert, M.; Yang, W. Structure of an activated DNA-PK and its implications for NHEJ. Mol. Cell 2020, 81, 801–810.e3. [Google Scholar] [CrossRef]
- Niewolik, D.; Schwarz, K. Physical ARTEMIS:DNA-PKcs interaction is necessary for V(D)J recombination. Nucleic Acids Res. 2022, 50, 2096–2110. [Google Scholar] [CrossRef]
- Shultz, L.D.; Ishikawa, F.; Greiner, D.L. Humanized Mice in Translational Biomedical Research. Nat. Rev. Immunol. 2007, 7, 118–130. [Google Scholar] [CrossRef] [PubMed]
- Kanaji, N.; Tadokoro, A.; Susaki, K.; Yokokura, S.; Ohmichi, K.; Haba, R.; Watanabe, N.; Bandoh, S.; Ishii, T.; Dobashi, H.; et al. Higher Susceptibility of NOD/LtSz-scid Il2rg(−/−) NSG Mice to Xenotransplanted Lung Cancer Cell Lines. Cancer Manag. Res. 2014, 6, 431–436. [Google Scholar] [CrossRef] [PubMed]
- Ito, R.; Takahashi, T.; Katano, I.; Ito, M. Current Advances in Humanized Mouse Models. Cell. Mol. Immunol. 2012, 9, 208–214. [Google Scholar] [CrossRef] [PubMed]
- Aryee, K.E.; Burzenski, L.M.; Yao, L.C.; Keck, J.G.; Greiner, D.L.; Shultz, L.D.; Brehm, M.A. Enhanced Development of Functional Human NK cells in NOD-scid-IL2rg(null) Mice Expressing Human IL15. FASEB J. 2022, 36, e22476. [Google Scholar] [CrossRef]
- Ren, J.; Yu, D.; Fu, R.; An, P.; Sun, R.; Wang, Z.; Guo, R.; Li, H.; Zhang, Y.; Li, Z.; et al. IL2RG-deficient Minipigs Generated via CRISPR/Cas9 Technology Support the Growth of Human Melanoma-derived Tumours. Cell Prolif. 2020, 53, e12863. [Google Scholar] [CrossRef]

| Name | Forward Primer | Reverse Primer |
|---|---|---|
| Dclre1c-seq | AAAACCTCATCTGCAATTGCTTTTA | CTTGGGCTTGTGCTGATGTG |
| Dclre1c-Qpcr | TGAGGCTTCGGGTGAGAAGG | AACTGATCCTGGGCAGTGAC |
| Bax | AAACTGGTGCTCAAGGCCC | GGTCCCGAAGTAGGAGAGGA |
| Xrcc1 | TCCGTCCGTCTGTTTGTCTG | GCTTCCTGGGAACCTGTTGT |
| Ogg1 | GAGACGACAGCCAGGCCTTT | GGAGGTTTGGGAAGCCATGAT |
| GAPDH | ACCCTTAAGAGGGATGCTGC | CCCAATACGGCCAAATCCGT |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bin, Y.; Wei, S.; Chen, R.; Zhang, H.; Ren, J.; Liu, P.; Xin, Z.; Zhang, T.; Yang, H.; Wang, K.; et al. Dclre1c-Mutation-Induced Immunocompromised Mice Are a Novel Model for Human Xenograft Research. Biomolecules 2024, 14, 180. https://doi.org/10.3390/biom14020180
Bin Y, Wei S, Chen R, Zhang H, Ren J, Liu P, Xin Z, Zhang T, Yang H, Wang K, et al. Dclre1c-Mutation-Induced Immunocompromised Mice Are a Novel Model for Human Xenograft Research. Biomolecules. 2024; 14(2):180. https://doi.org/10.3390/biom14020180
Chicago/Turabian StyleBin, Yixiao, Sanhua Wei, Ruo Chen, Haowei Zhang, Jing Ren, Peijuan Liu, Zhiqian Xin, Tianjiao Zhang, Haijiao Yang, Ke Wang, and et al. 2024. "Dclre1c-Mutation-Induced Immunocompromised Mice Are a Novel Model for Human Xenograft Research" Biomolecules 14, no. 2: 180. https://doi.org/10.3390/biom14020180
APA StyleBin, Y., Wei, S., Chen, R., Zhang, H., Ren, J., Liu, P., Xin, Z., Zhang, T., Yang, H., Wang, K., Feng, Z., Sun, X., Chen, Z., & Zhang, H. (2024). Dclre1c-Mutation-Induced Immunocompromised Mice Are a Novel Model for Human Xenograft Research. Biomolecules, 14(2), 180. https://doi.org/10.3390/biom14020180







