IL6 and IL6R as Prognostic Biomarkers in Colorectal Cancer
Abstract
1. Introduction
2. Methods
2.1. Patient Cohorts
2.1.1. Cohort 1
2.1.2. Cohort 2
2.2. Immunohistochemical Staining
2.3. Histological Scoring
2.4. RNAScope®
2.5. TempOSeq RNA Sequencing
2.6. Statistical Analysis
3. Results
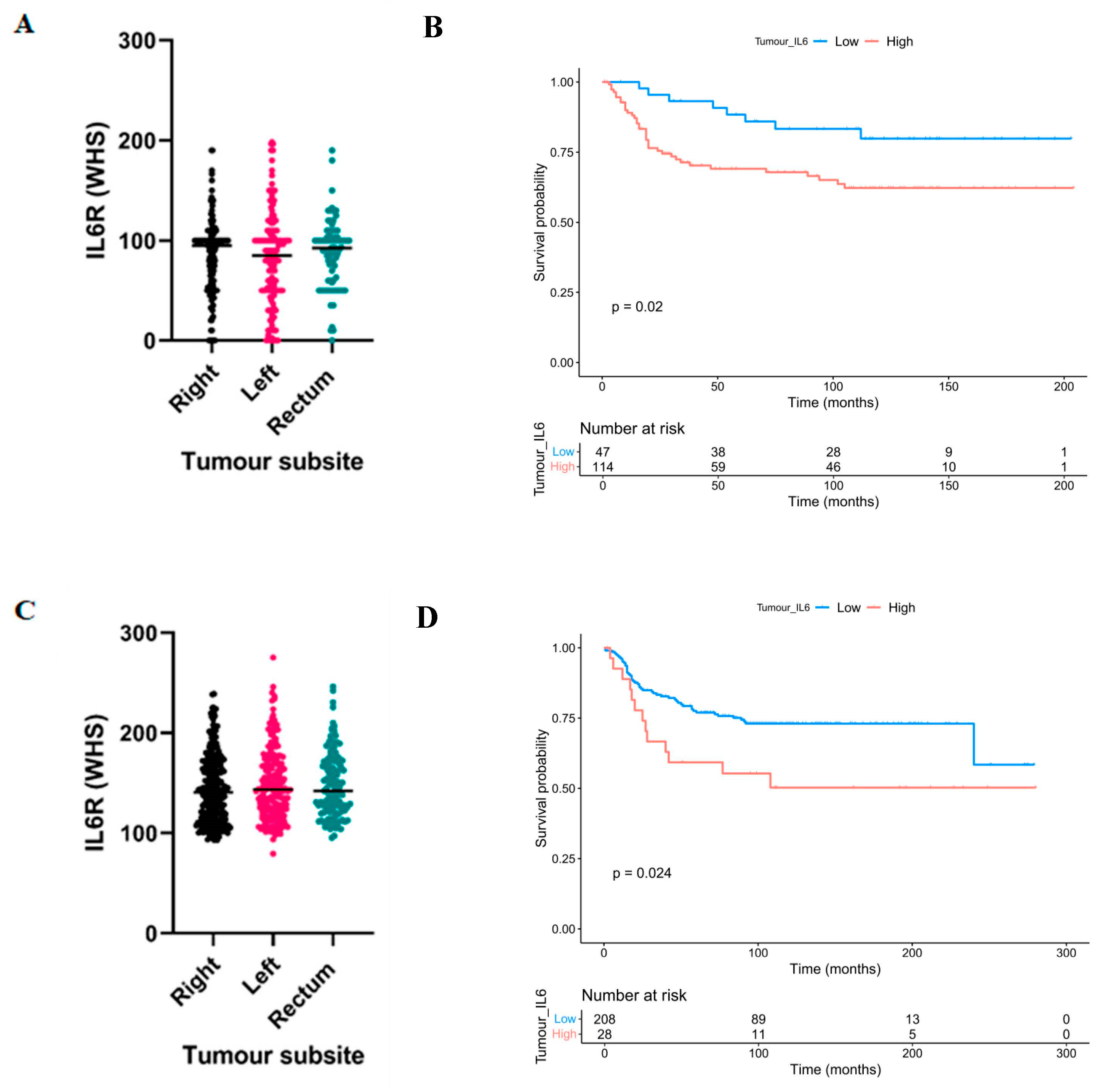
3.1. High Expression of Interleukin-6 Receptor Predicts Poor Clinical Outcomes in Right-Sided Colon Cancer
3.2. High Expression of Interleukin-6 Receptor Associates with Adverse Clinicopathological Characteristics in Right-Sided Colon Cancers
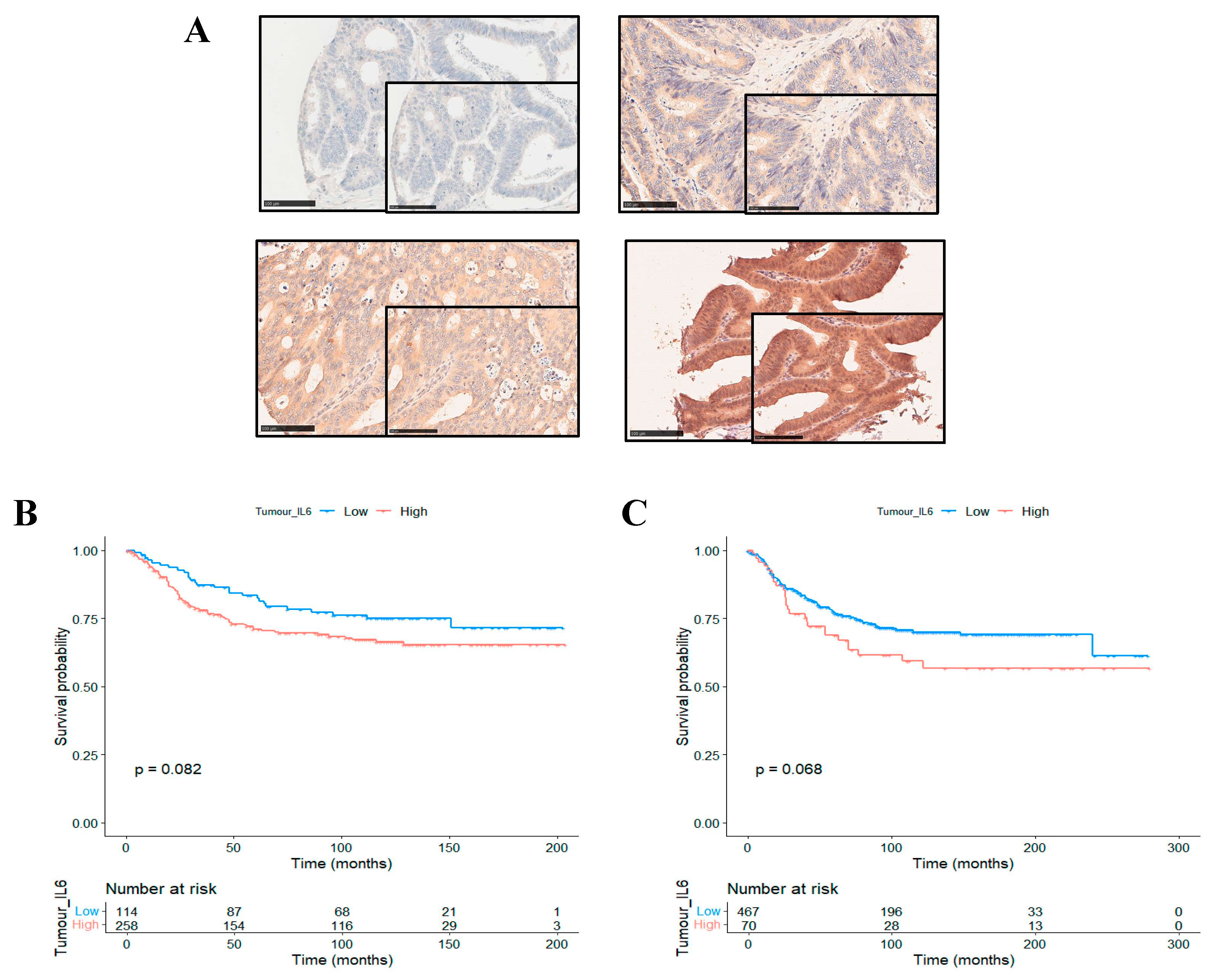
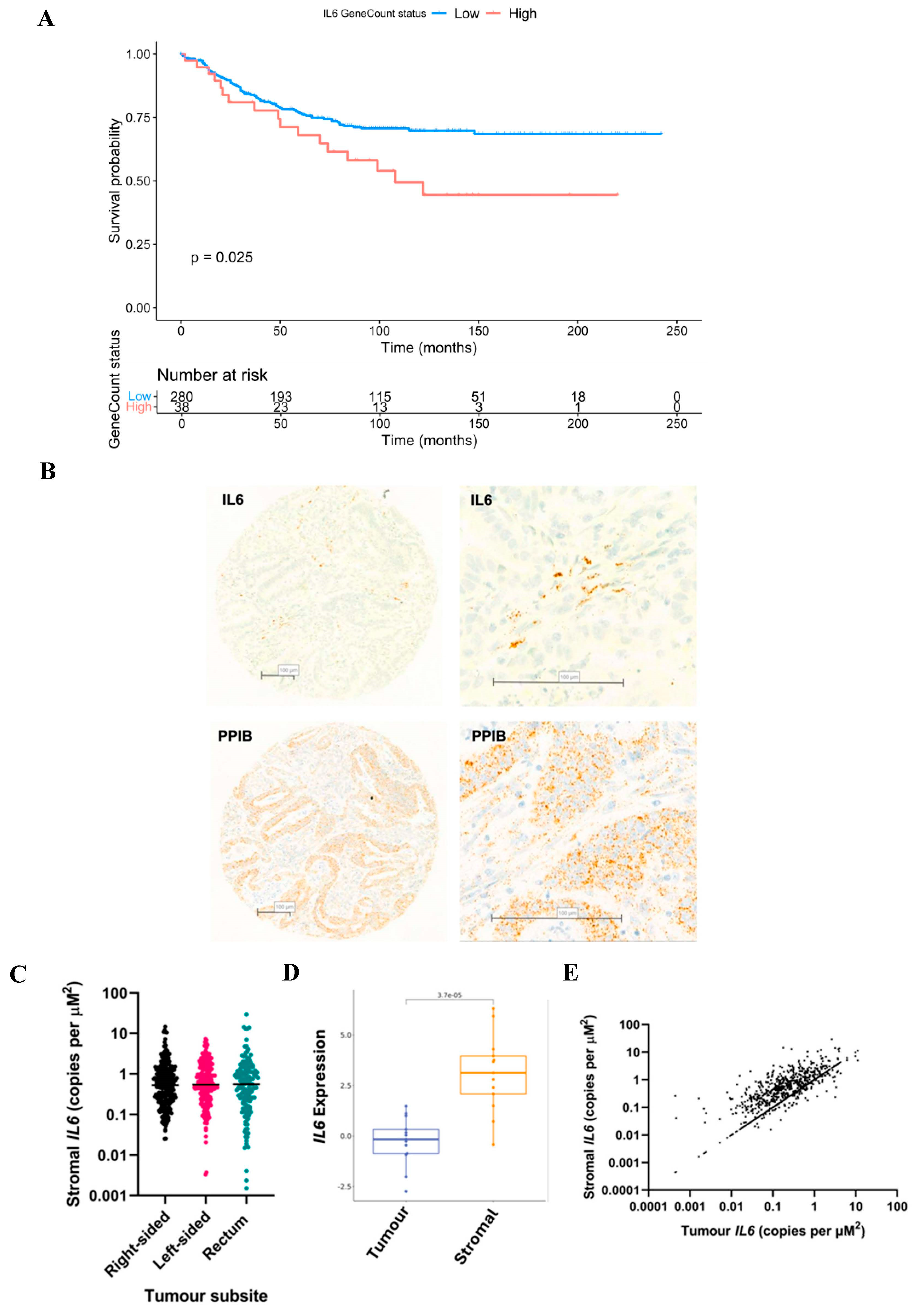
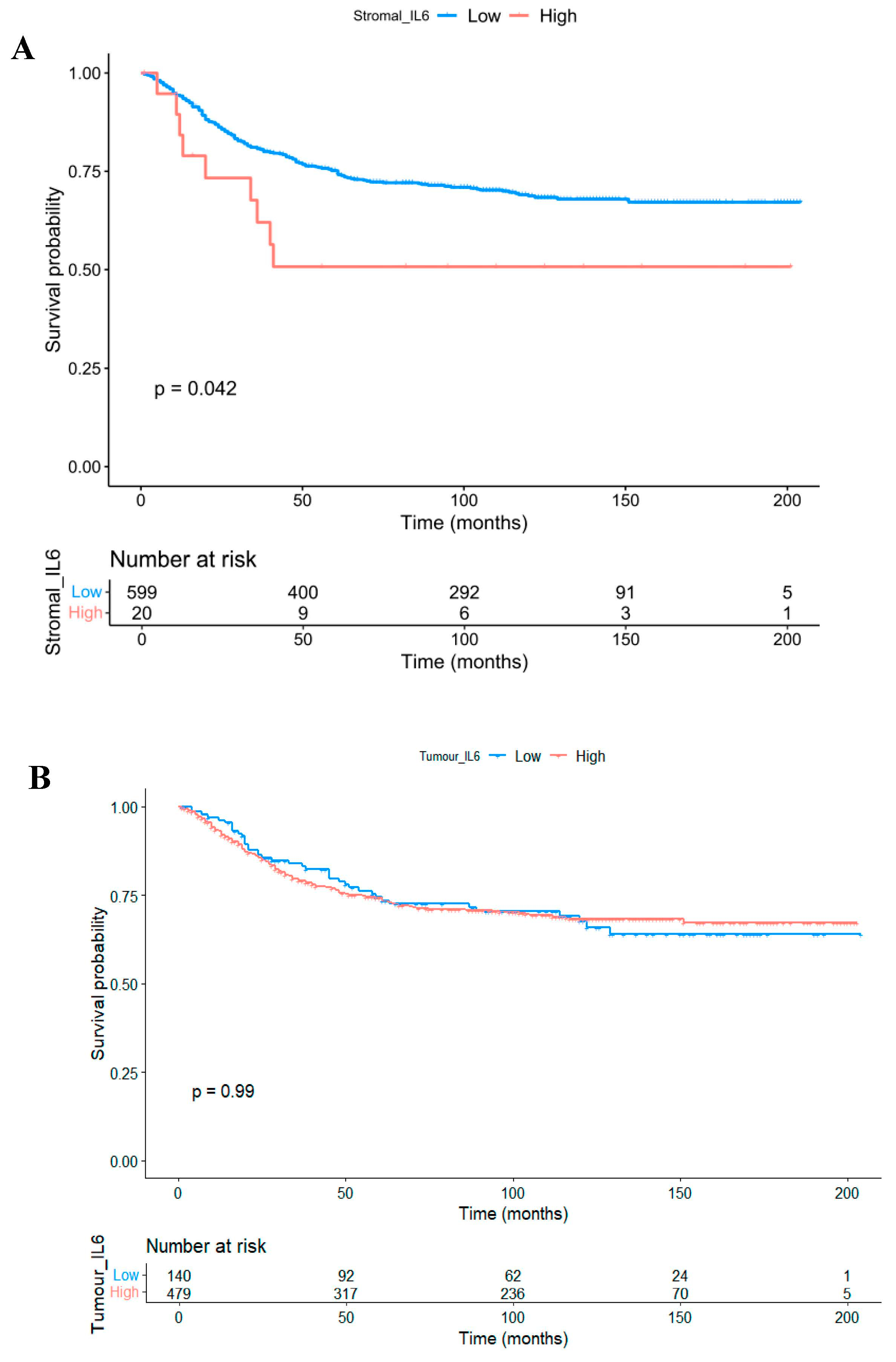
3.3. The Prognostic Value of Interleukin-6 Is Spatially Resolved
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Roxburgh, C.S.; McMillan, D.C. Cancer and systemic inflammation: Treat the tumour and treat the host. Br. J. Cancer 2014, 110, 1409–1412. [Google Scholar] [CrossRef] [PubMed]
- Guthrie, G.J.; Roxburgh, C.S.D.; Richards, C.H.; McMillan, D.C. Circulating IL-6 concentrations link tumour necrosis and systemic and local inflammatory responses in patients undergoing resection for colorectal cancer. Br. J. Cancer 2013, 109, 131–137. [Google Scholar] [CrossRef] [PubMed]
- Silva, E.M.; Mariano, V.A.; Pastrez, P.R.A.; Pinto, M.C.; Castro, A.G.; Syrjanen, K.J.; Longatto-Filho, A. High systemic IL-6 is associated with worse prognosis in patients with non-small cell lung cancer. PLoS ONE 2017, 12, e0181125. [Google Scholar] [CrossRef] [PubMed]
- Michalaki, V.; Syrigos, K.; Charles, P.; Waxman, J. Serum levels of IL-6 and TNF-alpha correlate with clinicopathological features and patient survival in patients with prostate cancer. Br. J. Cancer 2004, 90, 2312–2316. [Google Scholar] [CrossRef]
- Feng, S.; Li, Z.; Liu, M.; Ye, Q.; Xue, T.; Yan, B. Postoperative serum interleukin-6 levels correlate with survival in stage I-III colorectal cancer. BMC Gastroenterol. 2023, 23, 156. [Google Scholar] [CrossRef]
- Nagasaki, T.; Hara, M.; Nakanishi, H.; Takahashi, H.; Sato, M.; Takeyama, H. Interleukin-6 released by colon cancer-associated fibroblasts is critical for tumour angiogenesis: Anti-interleukin-6 receptor antibody suppressed angiogenesis and inhibited tumour-stroma interaction. Br. J. Cancer 2014, 110, 469–478. [Google Scholar] [CrossRef]
- Yin, Y.; Yao, S.; Hu, Y.; Feng, Y.; Li, M.; Bian, Z.; Zhang, J.; Qin, Y.; Qi, X.; Zhou, L.; et al. The Immune-microenvironment Confers Chemoresistance of Colorectal Cancer through Macrophage-Derived IL6. Clin. Cancer Res. 2017, 23, 7375–7387. [Google Scholar] [CrossRef]
- Heichler, C.; Scheibe, K.; Schmied, A.; Geppert, C.I.; Schmid, B.; Wirtz, S.; Thoma, O.-M.; Kramer, V.; Waldner, M.J.; Büttner, C.; et al. STAT3 activation through IL-6/IL-11 in cancer-associated fibroblasts promotes colorectal tumour development and correlates with poor prognosis. Gut 2020, 69, 1269–1282. [Google Scholar] [CrossRef]
- Morrow, E.; Pennel, K.; Hatthakarnkul, P.; Leslie, H.; Mallon, E.; Andersen, D.; Jamieson, N.; McMillan, D.; Roseweir, A.; Edwards, J. High expression of STAT3 within the tumour-associated stroma predicts poor outcome in breast cancer patients. Cancer Med. 2023, 12, 13225–13240. [Google Scholar] [CrossRef]
- Sun, G.; Wang, T.; Shi, M.; Zhou, H.; Wang, J.; Huang, Z.; Zhang, H.; Shi, J. Low expression of IL6R predicts poor prognosis for lung adenocarcinoma. Ann. Transl. Med. 2021, 9, 1057. [Google Scholar] [CrossRef]
- Olsen, J.; Kirkeby, L.T.; Olsen, J.; Eiholm, S.; Jess, P.; Gögenur, I.; Troelsen, J.T. High interleukin-6 mRNA expression is a predictor of relapse in colon cancer. Anticancer Res. 2015, 35, 2235–2240. [Google Scholar] [PubMed]
- Gulubova, M.; Chonov, D.; Aleksandrova, E.; Ivanova, K.; Ignatova, M.M.; Vlaykova, T. Interleukin-6-Positive Immune Cells as a Possible New Immunologic Marker Associated With the Colorectal Cancer Prognosis. Appl. Immunohistochem. Mol. Morphol. 2024, 32, 233–243. [Google Scholar] [CrossRef] [PubMed]
- Rosen, R.D.; Sapra, A. TNM Classification. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2024. [Google Scholar]
- Bankhead, P.; Loughrey, M.B.; Fernández, J.A.; Dombrowski, Y.; McArt, D.G.; Dunne, P.D.; McQuaid, S.; Gray, R.T.; Murray, L.J.; Coleman, H.G.; et al. QuPath: Open source software for digital pathology image analysis. Sci. Rep. 2017, 7, 16878. [Google Scholar] [CrossRef] [PubMed]
- Al-Badran, S.S.; Grant, L.; Campo, M.V.; Inthagard, J.; Pennel, K.; Quinn, J.; Quinn, J.; Hayman, L.; Horgan, P.G.; McMillan, D.C.; et al. Relationship between immune checkpoint proteins, tumour microenvironment characteristics, and prognosis in primary operable colorectal cancer. J. Pathol. Clin. Res. 2021, 7, 121–134. [Google Scholar] [CrossRef]
- Pennel, K.A.F.; Hatthakarnkul, P.; Wood, C.S.; Lian, G.-Y.; Al-Badran, S.; Quinn, J.A.; Legrini, A.; Inthagard, J.; Alexander, P.G.; van Wyk, H.; et al. JAK/STAT3 represents a therapeutic target for colorectal cancer patients with stromal-rich tumors. J. Exp. Clin. Cancer Res. 2024, 43, 64. [Google Scholar] [CrossRef]
- Patel, M.; McSorley, S.T.; Park, J.H.; Roxburgh, C.S.D.; Edwards, J.; Horgan, P.G.; McMillan, D.C. The relationship between right-sided tumour location, tumour microenvironment, systemic inflammation, adjuvant therapy and survival in patients undergoing surgery for colon and rectal cancer. Br. J. Cancer 2018, 118, 705–712. [Google Scholar] [CrossRef]
- Bourakkadi Idrissi, M.; El Bouhaddouti, H.; Mouaqit, O.; Ousadden, A.; Ait Taleb, K.; Benjelloun, E.B. Left-Sided Colon Cancer and Right-Sided Colon Cancer: Are They the Same Cancer or Two Different Entities? Cureus 2023, 15, e37563. [Google Scholar] [CrossRef]
- Asghari-Jafarabadi, M.; Wilkins, S.; Plazzer, J.P.; Yap, R.; McMurrick, P.J. Prognostic factors and survival disparities in right-sided versus left-sided colon cancer. Sci. Rep. 2024, 14, 12306. [Google Scholar] [CrossRef]
- Isobe, A.; Sawada, K.; Kinose, Y.; Ohyagi-Hara, C.; Nakatsuka, E.; Makino, H.; Ogura, T.; Mizuno, T.; Suzuki, N.; Morii, E.; et al. Interleukin 6 receptor is an independent prognostic factor and a potential therapeutic target of ovarian cancer. PLoS ONE 2015, 10, e0118080. [Google Scholar] [CrossRef]
- Yamamoto, T.; Tsunedomi, R.; Nakajima, M.; Suzuki, N.; Yoshida, S.; Tomochika, S.; Xu, M.; Nakagami, Y.; Matsui, H.; Tokumitsu, Y.; et al. IL-6 Levels Correlate with Prognosis and Immunosuppressive Stromal Cells in Patients with Colorectal Cancer. Ann. Surg. Oncol. 2023, 30, 5267–5277. [Google Scholar] [CrossRef]
- Toyoshima, Y.; Kitamura, H.; Xiang, H.; Ohno, Y.; Homma, S.; Kawamura, H.; Takahashi, N.; Kamiyama, T.; Tanino, M.; Taketomi, A. IL6 Modulates the Immune Status of the Tumor Microenvironment to Facilitate Metastatic Colonization of Colorectal Cancer Cells. Cancer Immunol. Res. 2019, 7, 1944–1957. [Google Scholar] [CrossRef]
| Clinical Characteristic | Low IL6R | High IL6R | p |
|---|---|---|---|
| Age | 0.324 | ||
| <65 | 15 (31.3) | 30 (26.3) | |
| >65 | 33 (68.8) | 84 (73.7) | |
| Sex | 0.061 | ||
| Female | 29 (60.4) | 52 (45.6) | |
| Male | 19 (39.6) | 62 (54.4) | |
| T stage | 0.602 | ||
| 1 | 1 (2.1) | 2 (1.8) | |
| 2 | 5 (10.4) | 9 (7.9) | |
| 3 | 31 (64.6) | 65 (57.0) | |
| 4 | 11 (22.9) | 38 (33.3) | |
| N stage | 0.298 | ||
| 0 | 34 (70.8) | 67 (58.8) | |
| 1 | 10 (20.8) | 30 (26.3) | |
| 2 | 4 (8.3) | 17 (14.9) | |
| mGPS | 0.059 | ||
| 0 | 20 (60.6) | 34 (38.6) | |
| 1 | 8 (24.2) | 25 (28.4) | |
| 2 | 5 (15.2) | 29 (33.0) | |
| MMR status | 0.335 | ||
| pMMR | 33 (68.8) | 83 (73.5) | |
| dMMR | 15 (31.3) | 30 (26.5) | |
| Marginal Involvement | 0.038 | ||
| Absent | 48 (100.0) | 105 (92.1) | |
| Present | 0 (0) | 9 (7.9) | |
| Peterson Index | 0.010 | ||
| Low | 43 (89.6) | 82 (71.9) | |
| High | 5 (10.4) | 32 (28.1) | |
| Vascular Invasion | 0.055 | ||
| Absent | 36 (75.0) | 69 (60.5) | |
| Present | 12 (25.0) | 45 (39.5) | |
| Peritoneal Involvement | 0.179 | ||
| Absent | 37 (77.1) | 78 (68.4) | |
| Present | 11 (22.9) | 36 (31.6) | |
| Tumour perforation | 0.204 | ||
| Low | 47 (97.9) | 104 (91.2) | |
| Moderate | 0 (0) | 5 (4.4) | |
| High | 1 (2.1) | 5 (4.4) | |
| GMS | 0.745 | ||
| 0 | 13 (27.7) | 34 (30.1) | |
| 1 | 27 (57.4) | 55 (48.7) | |
| 2 | 7 (14.9) | 24 (21.2) |
| Clinical Characteristic | Low IL6R | High IL6R | p |
|---|---|---|---|
| Age | 0.778 | ||
| <65 | 63 (30.1) | 8 (28.6) | |
| >65 | 146 (69.9) | 20 (71.4) | |
| Sex | 0.460 | ||
| Female | 96 (45.9) | 12 (42.9) | |
| Male | 113 (54.1) | 16 (57.1) | |
| T stage | 0.294 | ||
| 1 | 5 (2.4) | 0 (0) | |
| 2 | 17 (8.1) | 0 (0) | |
| 3 | 115 (55.0) | 18 (64.3) | |
| 4 | 72 (34.4) | 10 (35.7) | |
| N stage | 0.163 | ||
| 0 | 127 (60.8) | 11 (39.3) | |
| 1 | 56 (26.8) | 14 (50.0) | |
| 2 | 26 (12.4) | 3 (10.7) | |
| mGPS | 0.499 | ||
| 0 | 112 (53.6) | 12 (42.9) | |
| 1 | 46 (22.0) | 9 (32.1) | |
| 2 | 51 (24.4) | 7 (25.0) | |
| MMR status | 0.093 | ||
| dMMR | 37 (32.3) | 10 (40.0) | |
| pMMR | 129 (77.7) | 15 (60.0) | |
| Marginal Involvement | 0.254 | ||
| Absent | 197 (94.3) | 25 (89.3) | |
| Present | 12 (5.7) | 3 (10.7) | |
| Peterson Index | 0.521 | ||
| Low | 153 (73.2) | 21 (75.0) | |
| High | 56 (26.8) | 7 (25.0) | |
| Vascular Invasion | <0.001 | ||
| Absent | 80 (38.3) | 20 (71.4) | |
| Present | 129 (61.7) | 8 (28.6) | |
| Peritoneal Involvement | 0.591 | ||
| Absent | 149 (71.3) | 20 (71.4) | |
| Present | 60 (28.7) | 8 (28.6) | |
| Tumour perforation | 0.335 | ||
| No | 201 (96.2) | 26 (92.9) | |
| Yes | 8 (3.8) | 2 (7.1) | |
| GMS | 0.120 | ||
| 0 | 27 (13.2) | 2 (7.4) | |
| 1 | 141 (69.1) | 17 (63.0) | |
| 2 | 36 (17.6) | 8 (29.6) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pennel, K.A.F.; Kurniawan, A.; Samir Foad Al-Badran, S.; Schubert Santana, L.; Quinn, J.; Nixon, C.; Hatthakarnkul, P.; Maka, N.; Roxburgh, C.; McMillan, D.; et al. IL6 and IL6R as Prognostic Biomarkers in Colorectal Cancer. Biomolecules 2024, 14, 1629. https://doi.org/10.3390/biom14121629
Pennel KAF, Kurniawan A, Samir Foad Al-Badran S, Schubert Santana L, Quinn J, Nixon C, Hatthakarnkul P, Maka N, Roxburgh C, McMillan D, et al. IL6 and IL6R as Prognostic Biomarkers in Colorectal Cancer. Biomolecules. 2024; 14(12):1629. https://doi.org/10.3390/biom14121629
Chicago/Turabian StylePennel, Kathryn A. F., Ahmad Kurniawan, Sara Samir Foad Al-Badran, Leonor Schubert Santana, Jean Quinn, Colin Nixon, Phimmada Hatthakarnkul, Noori Maka, Campbell Roxburgh, Donald McMillan, and et al. 2024. "IL6 and IL6R as Prognostic Biomarkers in Colorectal Cancer" Biomolecules 14, no. 12: 1629. https://doi.org/10.3390/biom14121629
APA StylePennel, K. A. F., Kurniawan, A., Samir Foad Al-Badran, S., Schubert Santana, L., Quinn, J., Nixon, C., Hatthakarnkul, P., Maka, N., Roxburgh, C., McMillan, D., & Edwards, J. (2024). IL6 and IL6R as Prognostic Biomarkers in Colorectal Cancer. Biomolecules, 14(12), 1629. https://doi.org/10.3390/biom14121629






