Polymorphisms of KCNJ6 Gene and Their Correlation with Immune Indicators in Yaks (Bos grunniens)
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Approval
2.2. Sample Collection of Test Animals
2.3. Detection of Immune Indicators
2.4. Extraction of Blood Genomic DNA
2.5. Primer Design
2.6. PCR Amplification and Sequencing
2.7. Statistical Analysis
3. Results
3.1. Analysis of KCNJ6 Genotyping Results and Genetic Parameters in Yaks
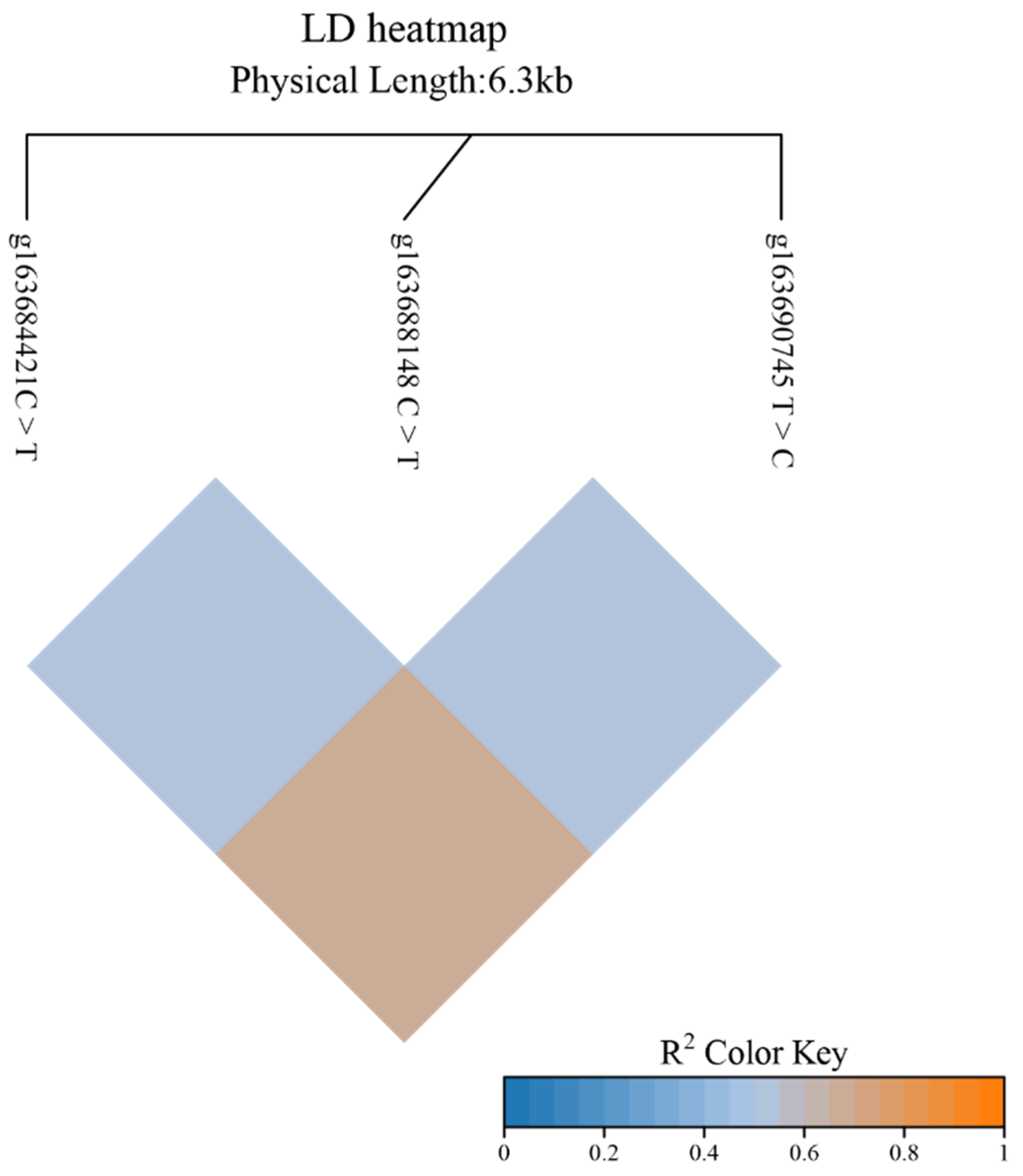
3.2. Linkage Disequilibrium Analysis of KCNJ6 Gene SNP in Yaks
3.3. Association Analysis Between Immune Indicators and SNP Genotypes in Yaks
4. Discussion
4.1. Genetic Polymorphism Analysis of KCNJ6 Gene in Yaks
4.2. Correlation Analysis of KCNJ6 Gene Polymorphisms and Immune Indicators in Yaks
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hibino, H.; Inanobe, A.; Furutani, K.; Murakami, S.; Findlay, I.; Kurachi, Y. Inwardly rectifying potassium channels: Their structure, function, and physiological roles. Physiol. Rev. 2010, 90, 291–366. [Google Scholar] [CrossRef] [PubMed]
- Luescher, C.; Slesinger, P.A. Emerging roles for G protein-gated inwardly rectifying potassium (GIRK) channels in health and disease. Nat. Rev. Neurosci. 2010, 11, 301–315. [Google Scholar] [CrossRef] [PubMed]
- Cooper, A.; Grigoryan, G.; Guy-David, L.; Tsoory, M.M.; Chen, A.; Reuveny, E. Trisomy of the G protein-coupled K+ channel gene, Kcnj6, affects reward mechanisms, cognitive functions, and synaptic plasticity in mice. Proc. Natl. Acad. Sci. USA 2012, 109, 2642–2647. [Google Scholar] [CrossRef] [PubMed]
- Horvath, G.A.; Zhao, Y.; Tarailo-Graovac, M.; Boelman, C.; Gill, H.; Shyr, C.; Lee, J.; Blydt-Hansen, I.; Drogemoller, B.I.; Moreland, J.; et al. Gain-of-function KCNJ6 Mutation in a Severe Hyperkinetic Movement Disorder Phenotype. Neuroscience 2018, 384, 152–164. [Google Scholar] [CrossRef]
- Doupnik, C.A.; Davidson, N.; Lester, H.A. The inward rectifier potassium channel family. Curr. Opin. Neurobiol. 1995, 5, 268–277. [Google Scholar] [CrossRef]
- Patil, N.; Cox, D.R.; Bhat, D.; Faham, M.; Myers, R.M.; Peterson, A.S. A potassium channel mutation in weaver mice implicates membrane excitability in granule cell differentiation. Nat. Genet. 1995, 11, 126–129. [Google Scholar] [CrossRef]
- Askew, E.W. Environmental and physical stress and nutrient requirements. Am. J. Clin. Nutr. 1995, 61, 631S–637S. [Google Scholar] [CrossRef]
- Wang, Y.J.; Wang, Z.S.; Hu, R.; Peng, Q.H.; Xia, K.; Zhao, S.N.; Zhou, Y.Q. Effects of Different Nutritional Regulations on Growth and Development, Blood Routine, and Plasma Antioxidant and Immune Indexes of Yaks with Growth Retardation. Chin. J. Anim. Nutr. 2018, 30, 1344–1352. [Google Scholar]
- Wang, Y.X.; Hu, R.; Wang, H.Z.; Wei, X.Q.; Jing, X.P.; Xue, B.; Cao, B.H.; Wang, Z.S.; Bao, S.K.; Zhao, S.N.; et al. Effects of Different Nutritional Regulations on Body Measurements and Gastrointestinal Tract Development of Yaks with Growth Retardation. Chin. J. Anim. Nutr. 2015, 27, 1690–1697. [Google Scholar]
- Xu, J.G.; Wang, C.L.; Che, C.Y.; Cai, Z.H.; Li, S.H.; Zhang, Q. Expression Profile and Polymorphism of CD8B Gene and Its Association with Blood Immune Traits in Pigs (Sus scrofa). J. Agric. Biotechnol. 2016, 24, 357–365. [Google Scholar]
- Gui, L.; Raza, S.; Sun, Y.; Sabek, A.; Abbas, S.Q.; Shah, M.A.; Khan, R.; Abdelnour, S.A. Molecular characterization and analysis of the association of growth hormone 1 gene with growth traits in Chinese indigenous yak (Bos grunniens). Trop. Anim. Health Prod. 2021, 53, 221. [Google Scholar] [CrossRef] [PubMed]
- Lefevre, F.; Gallais, A. Partitioning heterozygosity in subdivided populations: Some misuses of Nei’s decomposition and an alternative probabilistic approach. Mol. Ecol. 2020, 29, 2957–2962. [Google Scholar] [CrossRef] [PubMed]
- Serrote, C.; Reiniger, L.; Silva, K.B.; Rabaiolli, S.; Stefanel, C.M. Determining the Polymorphism Information Content of a molecular marker. Gene 2020, 726, 144175. [Google Scholar] [CrossRef] [PubMed]
- Zintzaras, E. Impact of Hardy-Weinberg equilibrium deviation on allele-based risk effect of genetic association studies and meta-analysis. Eur. J. Epidemiol. 2010, 25, 553–560. [Google Scholar] [CrossRef] [PubMed]
- Bionaz, M.; Loor, J.J. ACSL1, AGPAT6, FABP3, LPIN1, and SLC27A6 are the most abundant isoforms in bovine mammary tissue and their expression is affected by stage of lactation. J. Nutr. 2008, 138, 1019–1024. [Google Scholar] [CrossRef]
- Gupta, S.K.; Carmi, S.; Waldman, B.H.; Tkacz, I.D.; Naboishchikov, I.; Michaeli, S. Basal splicing factors regulate the stability of mature mRNAs in trypanosomes. J. Biol. Chem. 2013, 288, 4991–5006. [Google Scholar] [CrossRef] [PubMed]
- Sharma, Y.; Miladi, M.; Dukare, S.; Boulay, K.; Caudron-Herger, M.; Gross, M.; Backofen, R.; Diederichs, S. A pan-cancer analysis of synonymous mutations. Nat. Commun. 2019, 10, 2569. [Google Scholar] [CrossRef]
- Zhang, Z.; Liu, C.; Hao, W.; Yin, W.; Ai, S.; Zhao, Y.; Duan, Z. Novel Single Nucleotide Polymorphisms and Haplotype of MYF5 Gene Are Associated with Body Measurements and Ultrasound Traits in Grassland Short-Tailed Sheep. Genes 2022, 13, 483. [Google Scholar] [CrossRef]
- Ma, X.; Guan, L.; Xuan, J.; Wang, H.; Yuan, Z.; Wu, M.; Liu, R.; Zhu, C.; Wei, C.; Zhao, F.; et al. Effect of polymorphisms in the CAMKMT gene on growth traits in Ujumqin sheep. Anim. Genet. 2016, 47, 618–622. [Google Scholar] [CrossRef]
- Cui, P.; Wang, W.; Zhang, D.; Li, C.; Huang, Y.; Ma, Z.; Wang, X.; Zhao, L.; Zhang, Y.; Yang, X.; et al. Identification of TRAPPC9 and BAIAP2 Gene Polymorphisms and Their Association With Fat Deposition-Related Traits in Hu Sheep. Front. Vet. Sci. 2022, 9, 928375. [Google Scholar] [CrossRef]
- Ma, Z.; Wang, W.; Zhang, D.; Zhang, Y.; Zhao, Y.; Li, X.; Zhao, L.; Cheng, J.; Xu, D.; Yang, X.; et al. Polymorphisms of PLIN1 and MOGAT1 genes and their association with feed efficiency in Hu sheep. Gene 2024, 897, 148072. [Google Scholar] [CrossRef] [PubMed]
- Xia, R.W.; Sun, L.; Yin, X.M.; Cheng, H.Q.; Wu, S.L.; Bao, W.B. Assessment Model of General Disease Resistance in Yorkshire Based on Principal Component Analysis. China Anim. Husb. Vet. Med. 2015, 42, 986–990. [Google Scholar] [CrossRef]
- Lei, Y.C.; Wang, Z.T.; Jiang, J.Y.; Wang, S.H.; Sun, X.Z. Advances on Expression Profile and Diversity Mechanisms of Immunoglobulins in Herbivore. China Anim. Husb. Vet. Med. 2023, 50, 3313–3324. [Google Scholar] [CrossRef]
- Woof, J.M. Immunoglobulins and their receptors, and subversion of their protective roles by bacterial pathogens. Biochem. Soc. Trans. 2016, 44, 1651–1658. [Google Scholar] [CrossRef] [PubMed]
- Zhu, W.H. Poylmorphisms of Porcine TLR4 Gene and Their Associations with Immune Indexes in Weaned Piglets. Master’s Thesis, Anhui Agricultural University, Hefei, China, 2014. [Google Scholar]
- Baeuerle, P.A.; Henkel, T. Function and activation of NF-kappa B in the immune system. Annu. Rev. Immunol. 1994, 12, 141–179. [Google Scholar] [CrossRef] [PubMed]
- Campbell, K.J.; Perkins, N.D. Regulation of NF-kappaB function. Biochem. Soc. Symp. 2006, 73, 165–180. [Google Scholar] [CrossRef]
- Lv, Q.X.; Zhang, S.X.; Zhao, R.Q. Effects of transportation stress on transcription of IL-2, IL-6 and IL-10 and the relative receptors mRNA of lymph nodes in pig. J. Nanjing Agric. Univ. 2011, 34, 95–100. [Google Scholar]
- Stein, M.P.; Edberg, J.C.; Kimberly, R.P.; Mangan, E.K.; Bharadwaj, D.; Mold, C.; Du Clos, T.W. C-reactive protein binding to FcgammaRIIa on human monocytes and neutrophils is allele-specific. J. Clin. Investig. 2000, 105, 369–376. [Google Scholar] [CrossRef]
- Pepys, M.B. C-reactive protein: The role of an ancient protein in modern rheumatology. Clin. Exp. Rheumatol. 1983, 1, 3–7. [Google Scholar]
- Heegaard, P.M.; Godson, D.L.; Toussaint, M.J.; Tjornehoj, K.; Larsen, L.E.; Viuff, B.; Ronsholt, L. The acute phase response of haptoglobin and serum amyloid A (SAA) in cattle undergoing experimental infection with bovine respiratory syncytial virus. Vet. Immunol. Immunopathol. 2000, 77, 151–159. [Google Scholar] [CrossRef]
- Tan, L.N.; Huang, J.H. Recent advances in the investigation of haptoglobin. J. Clin. Pathol. Res. 2006, 26, 43–47. [Google Scholar]
- Hao, J.F.; Wang, J.G.; Zhu, X.Y.; Yang, W.T.; Ding, H.Y.; Shi, X.X.; Song, Y.X.; Deng, Q.H.; Liu, Z.X.; Wang, L.; et al. Comparative study of haptoglobin and serum amyloid A protein in the serum of healthy dairy cows and cows with ketosis. Chin. J. Vet. Med. 2013, 49, 9–11. [Google Scholar]
- Du, X.M.; Sheng, S.S.; Qiao, S.W.; Chen, X.L.; Dong, L.Y.; Wu, Z.C. Detection of new mutation sites of TLR5 gene in Large White pigs and its association with some immune indexes. Swine Ind. Sci. 2022, 39, 103–105. [Google Scholar]
- Malek, T.R. The main function of IL-2 is to promote the development of T regulatory cells. J. Leukoc. Biol. 2003, 74, 961–965. [Google Scholar] [CrossRef]
- Nishizawa, D.; Fukuda, K.; Kasai, S.; Ogai, Y.; Hasegawa, J.; Sato, N.; Yamada, H.; Tanioka, F.; Sugimura, H.; Hayashida, M.; et al. Association between KCNJ6 (GIRK2) gene polymorphism rs2835859 and post-operative analgesia, pain sensitivity, and nicotine dependence. J. Pharmacol. Sci. 2014, 126, 253–263. [Google Scholar] [CrossRef]
- Kober, K.M.; Dodd, M.J.; Miaskowski, C.; Aouizerat, B.E. Variations in potassium channel genes are associated with distinct trajectories of persistent breast pain after breast cancer surgery. Pain 2015, 156, 371–380. [Google Scholar] [CrossRef]
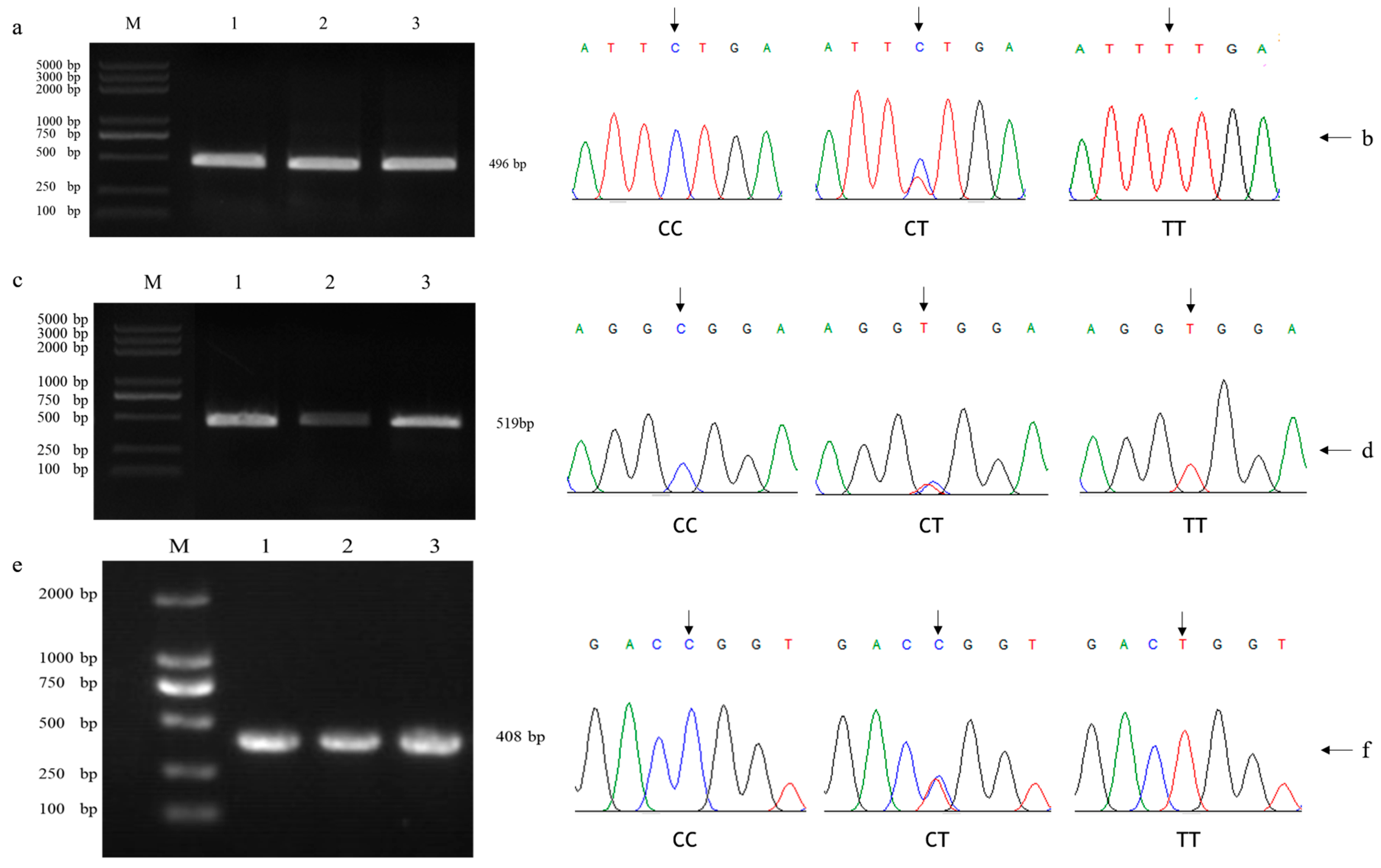
| SNPs | Primer Pair Sequences 1 (5′-3′) | Amplicon Length (bp) | Tm (°C) |
|---|---|---|---|
| g163684421 C > T | F: GCAGCAGGTCCGTCCACAT | 496 | 61.2 |
| R: GTAGCCCATCCAGGCCACA | |||
| g163688148 C > T | F: ACTAAGATCCCATGAGCCA | 519 | 52.4 |
| R: ATTGAATGAGACACCACCAG | |||
| g163690745 T > C | F: GCGACCTCCTCATTCTCCT | 408 | 54.3 |
| R: TGTACCTTCTTCCACCCAC |
| Trait | Mean | Standard Error | Maximum | Minimum |
|---|---|---|---|---|
| IgA (μg/mL) | 1155.22 | 13.98 | 2163.35 | 538.09 |
| IgG (mg/mL) | 2.38 | 0.05 | 5.13 | 0.19 |
| IgM (μg/mL) | 2112.42 | 20.91 | 3247.17 | 929.93 |
| CRP (mg/L) | 13.50 | 0.16 | 21.04 | 3.67 |
| HP (ng/mL) | 399.70 | 4.37 | 641.62 | 167.36 |
| IL-2 (pg/mL) | 310.37 | 4.06 | 542.28 | 132.27 |
| IL-4 (pg/mL) | 67.32 | 1.11 | 100.10 | 13.27 |
| IL-6 (pg/mL) | 433.27 | 5.23 | 706.11 | 162.48 |
| IFN-γ (pg/mL) | 1742.22 | 18.37 | 2664.85 | 567.67 |
| TNF-α (pg/mL) | 80.73 | 0.90 | 117.10 | 29.43 |
| SNPs | Position | Genotypic Frequencies | Allelic Frequencies | χ2 | p | PIC | He | Ne | |||
|---|---|---|---|---|---|---|---|---|---|---|---|
| CC | CT | TT | C | T | |||||||
| g163684421 C > T | Intron | 0.239 | 0.426 | 0.335 | 0.452 | 0.548 | 3.741 | 0.053 | 0.373 | 0.495 | 1.982 |
| g163688148 C > T | Intron | 0.448 | 0.422 | 0.130 | 0.659 | 0.341 | 0.727 | 0.394 | 0.348 | 0.449 | 1.816 |
| g163690745 T > C | Intron | 0.311 | 0.442 | 0.247 | 0.532 | 0.468 | 2.394 | 0.122 | 0.374 | 0.498 | 1.992 |
| Trait | Genotype | p-Value | ||
|---|---|---|---|---|
| CC | CT | TT | ||
| g163684421 C > T | ||||
| IgA (μg/mL) | 1232.96 ± 34.57 a | 1155.69 ± 18.71 b | 1096.92 ± 22.46 b | 0.001 ** |
| IgG (mg/mL) | 2.67 ± 0.10 a | 2.29 ± 0.07 b | 2.26 ± 0.08 b | 0.002 ** |
| IgM (μg/mL) | 2207.93 ± 42.35 a | 2112.79 ± 28.10 ab | 2041.07 ± 40.25 b | 0.013 ** |
| CRP (mg/L) | 14.32 ± 0.32 a | 13.60 ± 0.20 a | 12.76 ± 0.29 b | 0.001 ** |
| HP (ng/mL) | 426.39 ± 9.26 a | 399.08 ± 5.37 b | 379.95 ± 8.62 b | <0.001 ** |
| IL-2 (pg/mL) | 320.35 ± 10.18 a | 315.57 ± 5.52 a | 294.49 ± 6.24 b | 0.025 * |
| IL-4 (pg/mL) | 70.51 ± 2.32 a | 70.47 ± 1.46 a | 61.25 ± 2.06 b | <0.001 ** |
| IL-6 (pg/mL) | 446.14 ± 12.58 | 434.21 ± 7.31 | 421.68 ± 8.87 | 0.218 |
| IFN-γ (pg/mL) | 1894.67 ± 38.02 a | 1732.04 ± 21.35 b | 1648.00 ± 35.62 b | <0.001 ** |
| TNF-α (pg/mL) | 83.28 ± 1.83 a | 82.94 ± 1.22 a | 75.98 ± 1.66 b | 0.001 ** |
| g163688148 C > T | ||||
| IgA (μg/mL) | 1119.21 ± 19.01 b | 1169.21 ± 22.05 ab | 1233.76 ± 42.62 a | 0.023 * |
| IgG (mg/mL) | 2.28 ± 0.07 b | 2.35 ± 0.06 b | 2.78 ± 0.16 a | 0.003 ** |
| IgM (μg/mL) | 2069.59 ± 32.82 | 2129.82 ± 30.65 | 2203.38 ± 52.87 | 0.098 |
| CRP(mg/L) | 13.05 ± 0.22 b | 13.70 ± 0.24 ab | 14.38 ± 0.43 a | 0.013 * |
| HP (ng/mL) | 389.72 ± 6.51 b | 401.84 ± 6.86 b | 426.72 ± 9.89 a | 0.024 * |
| IL-2 (pg/mL) | 300.68 ± 4.98 | 320.81 ± 6.86 | 309.85 ± 13.04 | 0.069 |
| IL-4 (pg/mL) | 63.57 ± 1.61 b | 70.47 ± 1.65 a | 69.99 ± 3.25 a | 0.009 ** |
| IL-6 (pg/mL) | 429.15 ± 7.32 | 432.87 ± 8.39 | 448.56 ± 15.63 | 0.500 |
| IFN-γ (pg/mL) | 1671.79 ± 27.44 b | 1763.28 ± 26.55 b | 1916.24 ± 42.43 a | <0.001 ** |
| TNF-α (pg/mL) | 77.71 ± 1.33 b | 83.42 ± 1.34 ab | 82.42 ± 2.44 a | 0.009 ** |
| g163690745 T > C | ||||
| IgA (μg/mL) | 1225.55 ± 32.19 a | 1153.40 ± 20.09 b | 1100.64 ± 22.19 b | 0.004 ** |
| IgG (mg/mL) | 2.61 ± 0.10 a | 2.30 ± 0.07 b | 2.30 ± 0.08 b | 0.018 * |
| IgM (μg/mL) | 2197.82 ± 39.71 a | 2103.37 ± 30.74 ab | 2051.51 ± 39.65 b | 0.034 * |
| CRP (mg/L) | 14.36 ± 0.30 a | 13.49 ± 0.23 b | 12.82 ± 0.28 b | 0.001 ** |
| HP (ng/mL) | 423.74 ± 7.70 a | 396.82 ± 6.47 b | 385.48 ± 8.36 b | 0.004 ** |
| IL-2 (pg/mL) | 324.88 ± 10.04 a | 312.49 ± 5.88 ab | 295.09 ± 5.96 b | 0.023 * |
| IL-4 (pg/mL) | 69.93 ± 2.15 a | 69.40 ± 1.67 a | 61.74 ± 1.94 b | 0.004 ** |
| IL-6 (pg/mL) | 449.09 ± 11.40 | 430.54 ± 7.76 | 425.15 ± 9.05 | 0.215 |
| IFN-γ(pg/mL) | 1890.97 ± 33.34 a | 1721.55 ± 24.24 b | 1658.28 ± 35.59 b | <0.001 ** |
| TNF-α(pg/mL) | 84.49 ± 1.68 a | 81.65 ± 1.34 a | 76.20 ± 1.58 b | 0.002 ** |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ren, W.; Ma, X.; Yu, D.; Wu, X.; La, Y.; Guo, X.; Chu, M.; Yan, P.; Lan, X.; Liang, C. Polymorphisms of KCNJ6 Gene and Their Correlation with Immune Indicators in Yaks (Bos grunniens). Biomolecules 2024, 14, 1576. https://doi.org/10.3390/biom14121576
Ren W, Ma X, Yu D, Wu X, La Y, Guo X, Chu M, Yan P, Lan X, Liang C. Polymorphisms of KCNJ6 Gene and Their Correlation with Immune Indicators in Yaks (Bos grunniens). Biomolecules. 2024; 14(12):1576. https://doi.org/10.3390/biom14121576
Chicago/Turabian StyleRen, Wenwen, Xiaoming Ma, Daoning Yu, Xiaoyun Wu, Yongfu La, Xian Guo, Min Chu, Ping Yan, Xianyong Lan, and Chunnian Liang. 2024. "Polymorphisms of KCNJ6 Gene and Their Correlation with Immune Indicators in Yaks (Bos grunniens)" Biomolecules 14, no. 12: 1576. https://doi.org/10.3390/biom14121576
APA StyleRen, W., Ma, X., Yu, D., Wu, X., La, Y., Guo, X., Chu, M., Yan, P., Lan, X., & Liang, C. (2024). Polymorphisms of KCNJ6 Gene and Their Correlation with Immune Indicators in Yaks (Bos grunniens). Biomolecules, 14(12), 1576. https://doi.org/10.3390/biom14121576





