Resveratrol Inhibits Nucleosome Binding and Catalytic Activity of PARP1
Abstract
1. Introduction
2. Materials and Methods
2.1. Reagents
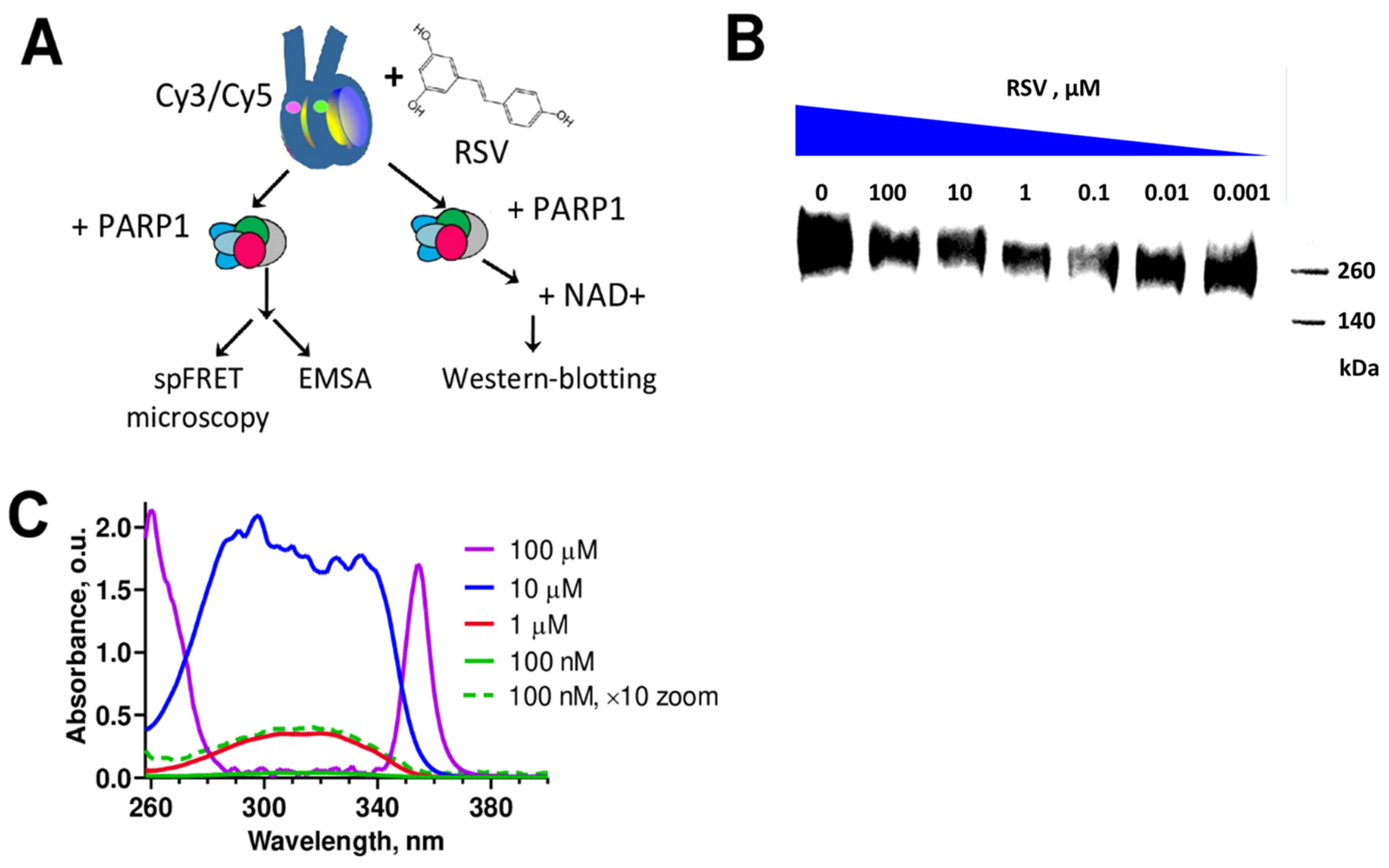
2.2. spFRET Microscopy
2.3. Electrophoretic Mobility Shift Assay (EMSA)
2.4. Western Blot Analysis
2.5. UV–VIS Spectrophotometry
3. Results
3.1. RSV Inhibits Catalytic Activity of PARP1
3.2. RSV Aggregates in an Aqueous Solution
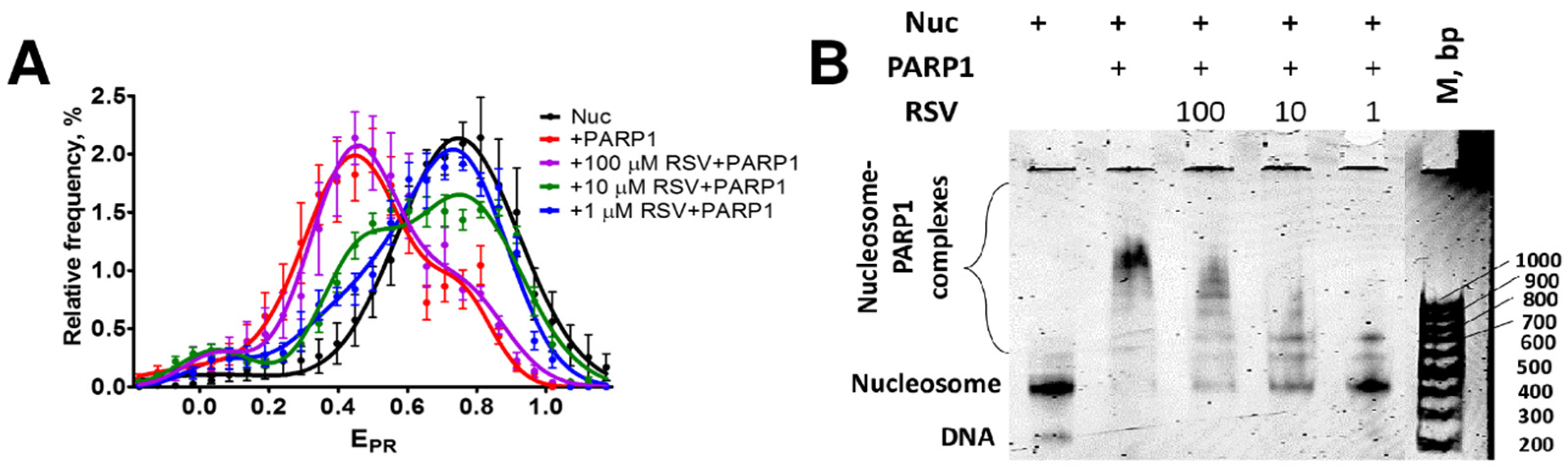
3.3. RSV Reduces the Binding of PARP1 to Nucleosomes
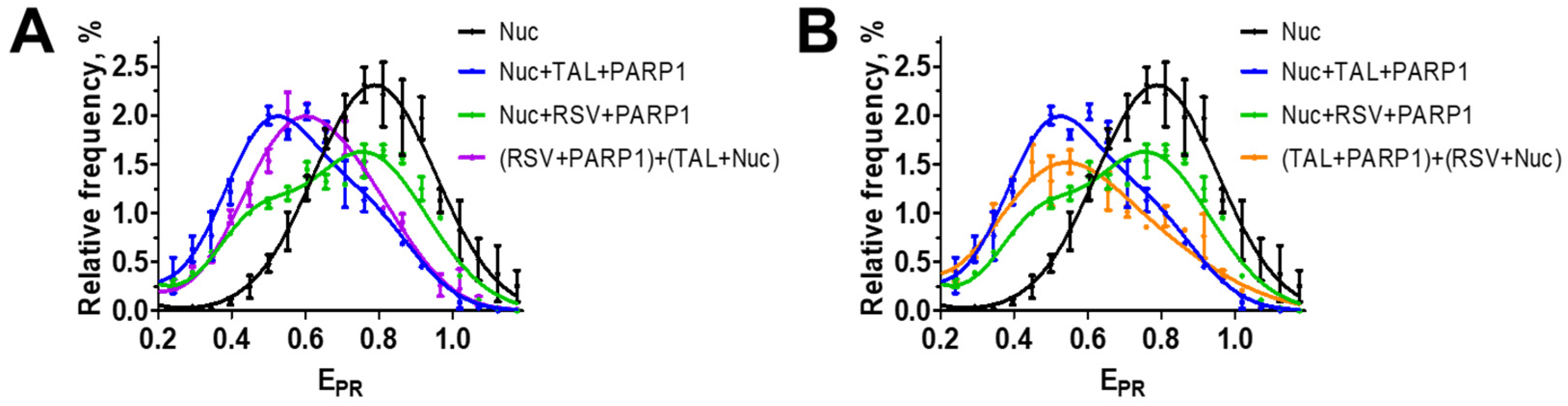
3.4. RSV Binds to the Catalytic Center of PARP1
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Marti, J.M.; Fernandez-Cortes, M.; Serrano-Saenz, S.; Zamudio-Martinez, E.; Delgado-Bellido, D.; Garcia-Diaz, A.; Oliver, F.J. The Multifactorial Role of PARP-1 in Tumor Microenvironment. Cancers 2020, 12, 739. [Google Scholar] [CrossRef] [PubMed]
- Fröhlich, L.M.; Villar-Miyar, A.; Heintze, T.; Sauer, B.; Schittek, B. PARP1 Expression Predicts PARP Inhibitor Sensitivity and Correlates with Metastatic Potential and Overall Survival in Melanoma. Int. J. Cancer 2024, 155, 203–210. [Google Scholar] [CrossRef] [PubMed]
- Murnyák, B.; Kouhsari, M.C.; Hershkovitch, R.; Kálmán, B.; Marko-Varga, G.; Klekner, Á.; Hortobágyi, T. PARP1 Expression and Its Correlation with Survival Is Tumour Molecular Subtype Dependent in Glioblastoma. Oncotarget 2017, 8, 46348–46362. [Google Scholar] [CrossRef]
- Rojo, F.; García-Parra, J.; Zazo, S.; Tusquets, I.; Ferrer-Lozano, J.; Menendez, S.; Eroles, P.; Chamizo, C.; Servitja, S.; Ramírez-Merino, N.; et al. Nuclear PARP-1 Protein Overexpression Is Associated with Poor Overall Survival in Early Breast Cancer. Ann. Oncol. 2012, 23, 1156–1164. [Google Scholar] [CrossRef]
- Qiao, W.; Pan, L.; Kou, C.; Li, K.; Yang, M. Prognostic and Clinicopathological Value of Poly (Adenosine Diphosphate-Ribose) Polymerase Expression in Breast Cancer: A Meta-Analysis. PLoS ONE 2017, 12, e0172413. [Google Scholar] [CrossRef]
- Jarrar, A.; Lotti, F.; DeVecchio, J.; Ferrandon, S.; Gantt, G.; Mace, A.; Karagkounis, G.; Orloff, M.; Venere, M.; Hitomi, M.; et al. Poly(ADP-Ribose) Polymerase Inhibition Sensitizes Colorectal Cancer-Initiating Cells to Chemotherapy. Stem Cells 2019, 37, 42–53. [Google Scholar] [CrossRef]
- Quinonero, F.; Cepero, A.; Urbano, D.; Munoz-Gamez, J.A.; Martin-Guerrero, S.M.; Martin-Oliva, D.; Prados, J.; Melguizo, C.; Ortiz, R. Identification of PARP-1 in Cancer Stem Cells of Gastrointestinal Cancers: A Preliminary Study. J. Biosci. 2021, 46, 6. [Google Scholar] [CrossRef]
- Puentes-Pardo, J.D.; Moreno-SanJuan, S.; Casado, J.; Escudero-Feliu, J.; López-Pérez, D.; Sánchez-Uceta, P.; González-Novoa, P.; Gálvez, J.; Carazo, Á.; León, J. PARP-1 Expression Influences Cancer Stem Cell Phenotype in Colorectal Cancer Depending on P53. Int. J. Mol. Sci. 2023, 24, 4787. [Google Scholar] [CrossRef] [PubMed]
- Weaver, A.N.; Yang, E.S. Beyond DNA Repair: Additional Functions of PARP-1 in Cancer. Front. Oncol. 2013, 3, 290. [Google Scholar] [CrossRef]
- Lawrence, L.M.; Russell, R.; Denning, C.E.; Zgheib, N.B.; Salisbury, T.; Lirette, S.T.; Valluri, J.; Claudio, P.P.; Denning, K.L. Expression of Poly-ADP-Ribose Polymerase (PARP) in Endometrial Adenocarcinoma: Prognostic Potential. Pathol. Res. Pract. 2020, 216, 152965. [Google Scholar] [CrossRef]
- Bai, P.; Cantó, C.; Oudart, H.; Brunyánszki, A.; Cen, Y.; Thomas, C.; Yamamoto, H.; Huber, A.; Kiss, B.; Houtkooper, R.H.; et al. PARP-1 Inhibition Increases Mitochondrial Metabolism through SIRT1 Activation. Cell Metab. 2011, 13, 461–468. [Google Scholar] [CrossRef] [PubMed]
- Luna, A.; Aladjem, M.I.; Kohn, K.W. SIRT1/PARP1 Crosstalk: Connecting DNA Damage and Metabolism. Genome Integr. 2013, 4, 6. [Google Scholar] [CrossRef] [PubMed]
- Mohamed, J.S.; Wilson, J.C.; Myers, M.J.; Sisson, K.J.; Alway, S.E. Dysregulation of SIRT-1 in Aging Mice Increases Skeletal Muscle Fatigue by a PARP-1-Dependent Mechanism. Aging 2014, 6, 820–834. [Google Scholar] [CrossRef] [PubMed]
- Edatt, L.; Poyyakkara, A.; Raji, G.R.; Ramachandran, V.; Shankar, S.S.; Kumar, V.B.S. Role of Sirtuins in Tumor Angiogenesis. Front. Oncol. 2019, 9, 1516. [Google Scholar] [CrossRef] [PubMed]
- Maluchenko, N.V.; Feofanov, A.V.; Studitsky, V.M. PARP-1-Associated Pathological Processes: Inhibition by Natural Polyphenols. Int. J. Mol. Sci. 2021, 22, 11441. [Google Scholar] [CrossRef]
- Koehler, R.C.; Dawson, V.L.; Dawson, T.M. Targeting Parthanatos in Ischemic Stroke. Front. Neurol. 2021, 12, 662034. [Google Scholar] [CrossRef]
- Gariani, K.; Ryu, D.; Menzies, K.J.; Yi, H.-S.; Stein, S.; Zhang, H.; Perino, A.; Lemos, V.; Katsyuba, E.; Jha, P.; et al. Inhibiting Poly ADP-Ribosylation Increases Fatty Acid Oxidation and Protects against Fatty Liver Disease. J. Hepatol. 2017, 66, 132–141. [Google Scholar] [CrossRef]
- Sriram, C.S.; Jangra, A.; Kasala, E.R.; Bodduluru, L.N.; Bezbaruah, B.K. Targeting Poly(ADP-Ribose)Polymerase1 in Neurological Diseases: A Promising Trove for New Pharmacological Interventions to Enter Clinical Translation. Neurochem. Int. 2014, 76, 70–81. [Google Scholar] [CrossRef]
- Castedo, M.; Lafarge, A.; Kroemer, G. Poly(ADP-Ribose) Polymerase-1 and Its Ambiguous Role in Cellular Life and Death. Cell Stress. 2023, 7, 1–6. [Google Scholar] [CrossRef]
- Henning, R.J.; Bourgeois, M.; Harbison, R.D. Poly(ADP-Ribose) Polymerase (PARP) and PARP Inhibitors: Mechanisms of Action and Role in Cardiovascular Disorders. Cardiovasc. Toxicol. 2018, 18, 493–506. [Google Scholar] [CrossRef]
- Pazzaglia, S.; Pioli, C. Multifaceted Role of PARP-1 in DNA Repair and Inflammation: Pathological and Therapeutic Implications in Cancer and Non-Cancer Diseases. Cells 2019, 9, 41. [Google Scholar] [CrossRef]
- Mekhaeil, M.; Dev, K.K.; Conroy, M.J. Existing Evidence for the Repurposing of PARP-1 Inhibitors in Rare Demyelinating Diseases. Cancers 2022, 14, 687. [Google Scholar] [CrossRef] [PubMed]
- LaFargue, C.J.; Dal Molin, G.Z.; Sood, A.K.; Coleman, R.L. Exploring and Comparing Adverse Events between PARP Inhibitors. Lancet Oncol. 2019, 20, e15–e28. [Google Scholar] [CrossRef] [PubMed]
- Geraets, L.; Moonen, H.J.; Brauers, K.; Wouters, E.F.; Bast, A.; Hageman, G.J. Dietary Flavones and Flavonoles Are Inhibitors of Poly(ADP-Ribose)Polymerase-1 in Pulmonary Epithelial Cells. J. Nutr. 2007, 137, 2190–2195. [Google Scholar] [CrossRef]
- Dal Piaz, F.; Ferro, P.; Vassallo, A.; Vasaturo, M.; Forte, G.; Chini, M.G.; Bifulco, G.; Tosco, A.; De Tommasi, N. Identification and Mechanism of Action Analysis of the New PARP-1 Inhibitor 2’’-Hydroxygenkwanol A. Biochim. Biophys. Acta 2015, 1850, 1806–1814. [Google Scholar] [CrossRef]
- Alqahtani, S.; Welton, K.; Gius, J.P.; Elmegerhi, S.; Kato, T.A. The Effect of Green and Black Tea Polyphenols on BRCA2 Deficient Chinese Hamster Cells by Synthetic Lethality through PARP Inhibition. Int. J. Mol. Sci. 2019, 20, 1274. [Google Scholar] [CrossRef] [PubMed]
- Arora, I.; Sharma, M.; Tollefsbol, T.O. Combinatorial Epigenetics Impact of Polyphenols and Phytochemicals in Cancer Prevention and Therapy. Int. J. Mol. Sci. 2019, 20, 4567. [Google Scholar] [CrossRef]
- Almatroodi, S.A.; Alsahli, M.A.; Aljohani, A.S.M.; Alhumaydhi, F.A.; Babiker, A.Y.; Khan, A.A.; Rahmani, A.H. Potential Therapeutic Targets of Resveratrol, a Plant Polyphenol, and Its Role in the Therapy of Various Types of Cancer. Molecules 2022, 27, 2665. [Google Scholar] [CrossRef]
- Kumar, A.; Kurmi, B.D.; Singh, A.; Singh, D. Potential Role of Resveratrol and Its Nano-Formulation as Anti-Cancer Agent. Explor. Target. Antitumor Ther. 2022, 3, 643–658. [Google Scholar] [CrossRef]
- Pan, P.; Li, J.; Lin, W.; Long, G. Effects of Resveratrol on Hepatitis B Virus Replication: In Vitro and in Vivo Experiments. Intervirology 2022, 65, 206–214. [Google Scholar] [CrossRef]
- Maluchenko, N.V.; Andreeva, T.V.; Geraskina, O.V.; Gerasimova, N.S.; Lubitelev, A.V.; Feofanov, A.V.; Studitsky, V.M. On the Interaction of Resveratrol with Nucleosomes. Biophysics 2023, 68, 369–375. [Google Scholar] [CrossRef]
- Islam, F.; Nafady, M.H.; Islam, M.R.; Saha, S.; Rashid, S.; Akter, A.; Or-Rashid, M.H.; Akhtar, M.F.; Perveen, A.; Md Ashraf, G.; et al. Resveratrol and Neuroprotection: An Insight into Prospective Therapeutic Approaches against Alzheimer’s Disease from Bench to Bedside. Mol. Neurobiol. 2022, 59, 4384–4404. [Google Scholar] [CrossRef] [PubMed]
- Fan, D.; Liu, C.; Zhang, Z.; Huang, K.; Wang, T.; Chen, S.; Li, Z. Progress in the Preclinical and Clinical Study of Resveratrol for Vascular Metabolic Disease. Molecules 2022, 27, 7524. [Google Scholar] [CrossRef] [PubMed]
- Pangeni, R.; Sahni, J.K.; Ali, J.; Sharma, S.; Baboota, S. Resveratrol: Review on Therapeutic Potential and Recent Advances in Drug Delivery. Expert. Opin. Drug Deliv. 2014, 11, 1285–1298. [Google Scholar] [CrossRef]
- Zhang, L.-X.; Li, C.-X.; Kakar, M.U.; Khan, M.S.; Wu, P.-F.; Amir, R.M.; Dai, D.-F.; Naveed, M.; Li, Q.-Y.; Saeed, M.; et al. Resveratrol (RV): A Pharmacological Review and Call for Further Research. Biomed. Pharmacother. 2021, 143, 112164. [Google Scholar] [CrossRef]
- Shaito, A.; Posadino, A.M.; Younes, N.; Hasan, H.; Halabi, S.; Alhababi, D.; Al-Mohannadi, A.; Abdel-Rahman, W.M.; Eid, A.H.; Nasrallah, G.K.; et al. Potential Adverse Effects of Resveratrol: A Literature Review. Int. J. Mol. Sci. 2020, 21, 2084. [Google Scholar] [CrossRef]
- Howitz, K.T.; Bitterman, K.J.; Cohen, H.Y.; Lamming, D.W.; Lavu, S.; Wood, J.G.; Zipkin, R.E.; Chung, P.; Kisielewski, A.; Zhang, L.L.; et al. Small Molecule Activators of Sirtuins Extend Saccharomyces Cerevisiae Lifespan. Nature 2003, 425, 191–196. [Google Scholar] [CrossRef]
- Gatz, S.A.; Wiesmuller, L. Take a Break--Resveratrol in Action on DNA. Carcinogenesis 2008, 29, 321–332. [Google Scholar] [CrossRef]
- Zhang, S.; Sun, X.; Jing, Z.; Qu, F. Spectroscopic Analysis on the Resveratrol-DNA Binding Interactions at Physiological pH. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2011, 82, 213–216. [Google Scholar] [CrossRef]
- N’Soukpoe-Kossi, C.N.; Bourassa, P.; Mandeville, J.S.; Bekale, L.; Tajmir-Riahi, H.A. Structural Modeling for DNA Binding to Antioxidants Resveratrol, Genistein and Curcumin. J. Photochem. Photobiol. B 2015, 151, 69–75. [Google Scholar] [CrossRef]
- Kumar, S.; Kumar, P.; Nair, M.S. Exploring the Binding of Resveratrol to a Promoter DNA Sequence d(CCAATTGG)(2) through Multispectroscopic, Nuclear Magnetic Resonance and Molecular Dynamics Studies. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2021, 252, 119488. [Google Scholar] [CrossRef] [PubMed]
- Andreeva, T.V.; Maluchenko, N.V.; Efremenko, A.V.; Lyubitelev, A.V.; Korovina, A.N.; Afonin, D.A.; Kirpichnikov, M.P.; Studitsky, V.M.; Feofanov, A.V. Epigallocatechin Gallate Affects the Structure of Chromatosomes, Nucleosomes and Their Complexes with PARP1. Int. J. Mol. Sci. 2023, 24, 14187. [Google Scholar] [CrossRef] [PubMed]
- Thåström, A.; Lowary, P.T.; Widlund, H.R.; Cao, H.; Kubista, M.; Widom, J. Sequence Motifs and Free Energies of Selected Natural and Non-Natural Nucleosome Positioning DNA Sequences. J. Mol. Biol. 1999, 288, 213–229. [Google Scholar] [CrossRef] [PubMed]
- Malyuchenko, N.V. The Effect of Gossypol on the Structure of Nucleosomes. Moscow Univ. Biol.Sci. Bull. 2020, 75, 142–146. [Google Scholar] [CrossRef]
- Gaykalova, D.A.; Kulaeva, O.I.; Bondarenko, V.A.; Studitsky, V.M. Preparation and Analysis of Uniquely Positioned Mononucleosomes. In Chromatin Protocols; Chellappan, S.P., Ed.; Humana Press: Totowa, NJ, USA, 2009; Volume 523, pp. 109–123. ISBN 978-1-58829-873-7. [Google Scholar]
- Kudryashova, K.S.; Nikitin, D.V. Preparation of Mononucleosomal Templates for Analysis of Transcription with RNA Polymerase Using spFRET. Methods Mol. Biol. 2015, 1288, 395–412. [Google Scholar] [CrossRef]
- Thomas, C.; Ji, Y.; Wu, C.; Datz, H.; Boyle, C.; MacLeod, B.; Patel, S.; Ampofo, M.; Currie, M.; Harbin, J.; et al. Hit and Run versus Long-Term Activation of PARP-1 by Its Different Domains Fine-Tunes Nuclear Processes. Proc. Natl. Acad. Sci. USA 2019, 116, 9941–9946. [Google Scholar] [CrossRef]
- Patel, A.G.; Sarkaria, J.N.; Kaufmann, S.H. Nonhomologous End Joining Drives Poly(ADP-Ribose) Polymerase (PARP) Inhibitor Lethality in Homologous Recombination-Deficient Cells. Proc. Natl. Acad. Sci. USA 2011, 108, 3406–3411. [Google Scholar] [CrossRef]
- Chiruvella, K.K.; Liang, Z.; Wilson, T.E. Repair of Double-Strand Breaks by End Joining. Cold Spring Harb. Perspect. Biol. 2013, 5, a012757. [Google Scholar] [CrossRef]
- Clark, N.J.; Kramer, M.; Muthurajan, U.M.; Luger, K. Alternative Modes of Binding of Poly(ADP-Ribose) Polymerase 1 to Free DNA and Nucleosomes. J. Biol. Chem. 2012, 287, 32430–32439. [Google Scholar] [CrossRef]
- Maluchenko, N.V.; Nilov, D.K.; Pushkarev, S.V.; Kotova, E.Y.; Gerasimova, N.S.; Kirpichnikov, M.P.; Langelier, M.F.; Pascal, J.M.; Akhtar, M.S.; Feofanov, A.V.; et al. Mechanisms of Nucleosome Reorganization by PARP1. Int. J. Mol. Sci. 2021, 22, 12127. [Google Scholar] [CrossRef]
- Zandarashvili, L.; Langelier, M.F.; Velagapudi, U.K.; Hancock, M.A.; Steffen, J.D.; Billur, R.; Hannan, Z.M.; Wicks, A.J.; Krastev, D.B.; Pettitt, S.J.; et al. Structural Basis for Allosteric PARP-1 Retention on DNA Breaks. Science 2020, 368, eaax6367. [Google Scholar] [CrossRef] [PubMed]
- Chan, C.Y.; Tan, K.V.; Cornelissen, B. PARP Inhibitors in Cancer Diagnosis and Therapy. Clin. Cancer Res. 2021, 27, 1585–1594. [Google Scholar] [CrossRef] [PubMed]
- Ferraris, D.V. Evolution of Poly(ADP-Ribose) Polymerase-1 (PARP-1) Inhibitors. From Concept to Clinic. J. Med. Chem. 2010, 53, 4561–4584. [Google Scholar] [CrossRef] [PubMed]
- Maluchenko, N.; Koshkina, D.; Korovina, A.; Studitsky, V.; Feofanov, A. Interactions of PARP1 Inhibitors with PARP1-Nucleosome Complexes. Cells 2022, 11, 3343. [Google Scholar] [CrossRef]
- Weseler, A.R.; Geraets, L.; Moonen, H.J.; Manders, R.J.; van Loon, L.J.; Pennings, H.J.; Wouters, E.F.; Bast, A.; Hageman, G.J. Poly (ADP-Ribose) Polymerase-1-Inhibiting Flavonoids Attenuate Cytokine Release in Blood from Male Patients with Chronic Obstructive Pulmonary Disease or Type 2 Diabetes. J. Nutr. 2009, 139, 952–957. [Google Scholar] [CrossRef] [PubMed]
- Engen, A.; Maeda, J.; Wozniak, D.E.; Brents, C.A.; Bell, J.J.; Uesaka, M.; Aizawa, Y.; Kato, T.A. Induction of Cytotoxic and Genotoxic Responses by Natural and Novel Quercetin Glycosides. Mutat. Res. Genet. Toxicol. Environ. Mutagen. 2015, 784–785, 15–22. [Google Scholar] [CrossRef]
- Brady, P.N.; Goel, A.; Johnson, M.A. Correction for Brady et al., “Poly(ADP-Ribose) Polymerases in Host-Pathogen Interactions, Inflammation, and Immunity”. Microbiol. Mol. Biol. Rev. 2019, 83, e00010-19. [Google Scholar] [CrossRef]
- Pai Bellare, G.; Sankar Patro, B. Resveratrol Sensitizes Breast Cancer to PARP Inhibitor, Talazoparib through Dual Inhibition of AKT and Autophagy Flux. Biochem. Pharmacol. 2022, 199, 115024. [Google Scholar] [CrossRef]
- Jhanji, M.; Rao, C.N.; Sajish, M. Towards Resolving the Enigma of the Dichotomy of Resveratrol: Cis- and Trans-Resveratrol Have Opposite Effects on TyrRS-Regulated PARP1 Activation. Geroscience 2021, 43, 1171–1200. [Google Scholar] [CrossRef]
- Jhanji, M.; Rao, C.N.; Massey, J.C.; Hope, M.C.; Zhou, X.; Keene, C.D.; Ma, T.; Wyatt, M.D.; Stewart, J.A.; Sajish, M. Cis- and Trans-Resveratrol Have Opposite Effects on Histone Serine-ADP-Ribosylation and Tyrosine Induced Neurodegeneration. Nat. Commun. 2022, 13, 3244. [Google Scholar] [CrossRef]
- Sajish, M.; Schimmel, P. A Human tRNA Synthetase Is a Potent PARP1-Activating Effector Target for Resveratrol. Nature 2015, 519, 370–373. [Google Scholar] [CrossRef] [PubMed]
- Turner, R.S.; Thomas, R.G.; Craft, S.; Van Dyck, C.H.; Mintzer, J.; Reynolds, B.A.; Brewer, J.B.; Rissman, R.A.; Raman, R.; Aisen, P.S.; et al. A Randomized, Double-Blind, Placebo-Controlled Trial of Resveratrol for Alzheimer Disease. Neurology 2015, 85, 1383–1391. [Google Scholar] [CrossRef] [PubMed]
- Zhu, C.W.; Grossman, H.; Neugroschl, J.; Parker, S.; Burden, A.; Luo, X.; Sano, M. A Randomized, Double-blind, Placebo-controlled Trial of Resveratrol with Glucose and Malate (RGM) to Slow the Progression of Alzheimer’s Disease: A Pilot Study. A&D Transl. Res. Clin. Interv. 2018, 4, 609–616. [Google Scholar] [CrossRef]
- Mankowski, R.T.; You, L.; Buford, T.W.; Leeuwenburgh, C.; Manini, T.M.; Schneider, S.; Qiu, P.; Anton, S.D. Higher Dose of Resveratrol Elevated Cardiovascular Disease Risk Biomarker Levels in Overweight Older Adults—A Pilot Study. Exp. Gerontol. 2020, 131, 110821. [Google Scholar] [CrossRef] [PubMed]
- Gal, R.; Deres, L.; Horvath, O.; Eros, K.; Sandor, B.; Urban, P.; Soos, S.; Marton, Z.; Sumegi, B.; Toth, K.; et al. Resveratrol Improves Heart Function by Moderating Inflammatory Processes in Patients with Systolic Heart Failure. Antioxidants 2020, 9, 1108. [Google Scholar] [CrossRef]
- Zortea, K.; Franco, V.C.; Guimarães, P.; Belmonte-de-Abreu, P.S. Resveratrol Supplementation Did Not Improve Cognition in Patients with Schizophrenia: Results from a Randomized Clinical Trial. Front. Psychiatry 2016, 7, 159. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Koshkina, D.O.; Maluchenko, N.V.; Korovina, A.N.; Lobanova, A.A.; Feofanov, A.V.; Studitsky, V.M. Resveratrol Inhibits Nucleosome Binding and Catalytic Activity of PARP1. Biomolecules 2024, 14, 1398. https://doi.org/10.3390/biom14111398
Koshkina DO, Maluchenko NV, Korovina AN, Lobanova AA, Feofanov AV, Studitsky VM. Resveratrol Inhibits Nucleosome Binding and Catalytic Activity of PARP1. Biomolecules. 2024; 14(11):1398. https://doi.org/10.3390/biom14111398
Chicago/Turabian StyleKoshkina, Daria O., Natalya V. Maluchenko, Anna N. Korovina, Angelina A. Lobanova, Alexey V. Feofanov, and Vasily M. Studitsky. 2024. "Resveratrol Inhibits Nucleosome Binding and Catalytic Activity of PARP1" Biomolecules 14, no. 11: 1398. https://doi.org/10.3390/biom14111398
APA StyleKoshkina, D. O., Maluchenko, N. V., Korovina, A. N., Lobanova, A. A., Feofanov, A. V., & Studitsky, V. M. (2024). Resveratrol Inhibits Nucleosome Binding and Catalytic Activity of PARP1. Biomolecules, 14(11), 1398. https://doi.org/10.3390/biom14111398






