Route to Measure Exact Parameters of Bio-Nanostructures Self-Assembly
Abstract
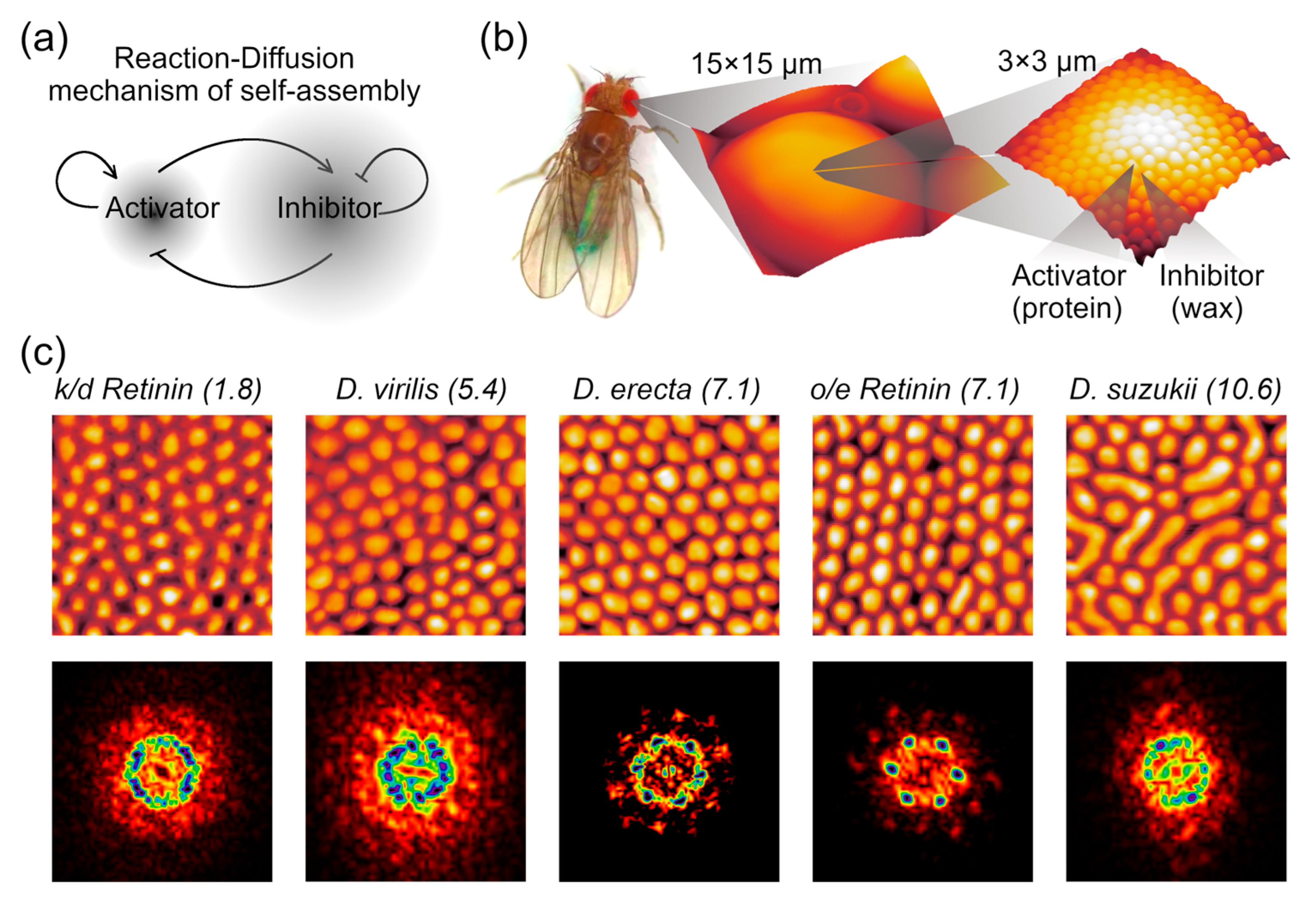
1. Introduction
2. Materials and Methods
3. Results
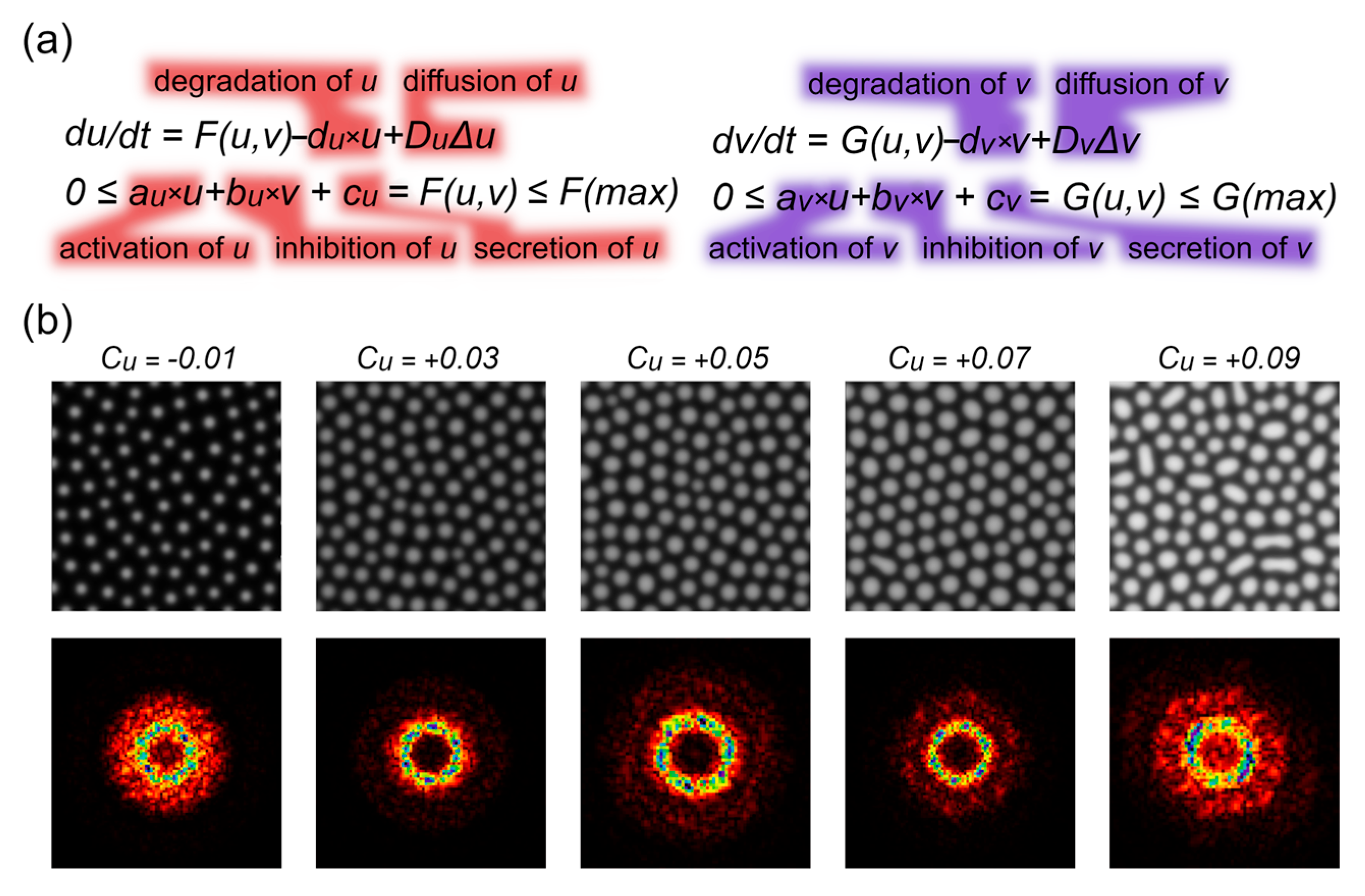
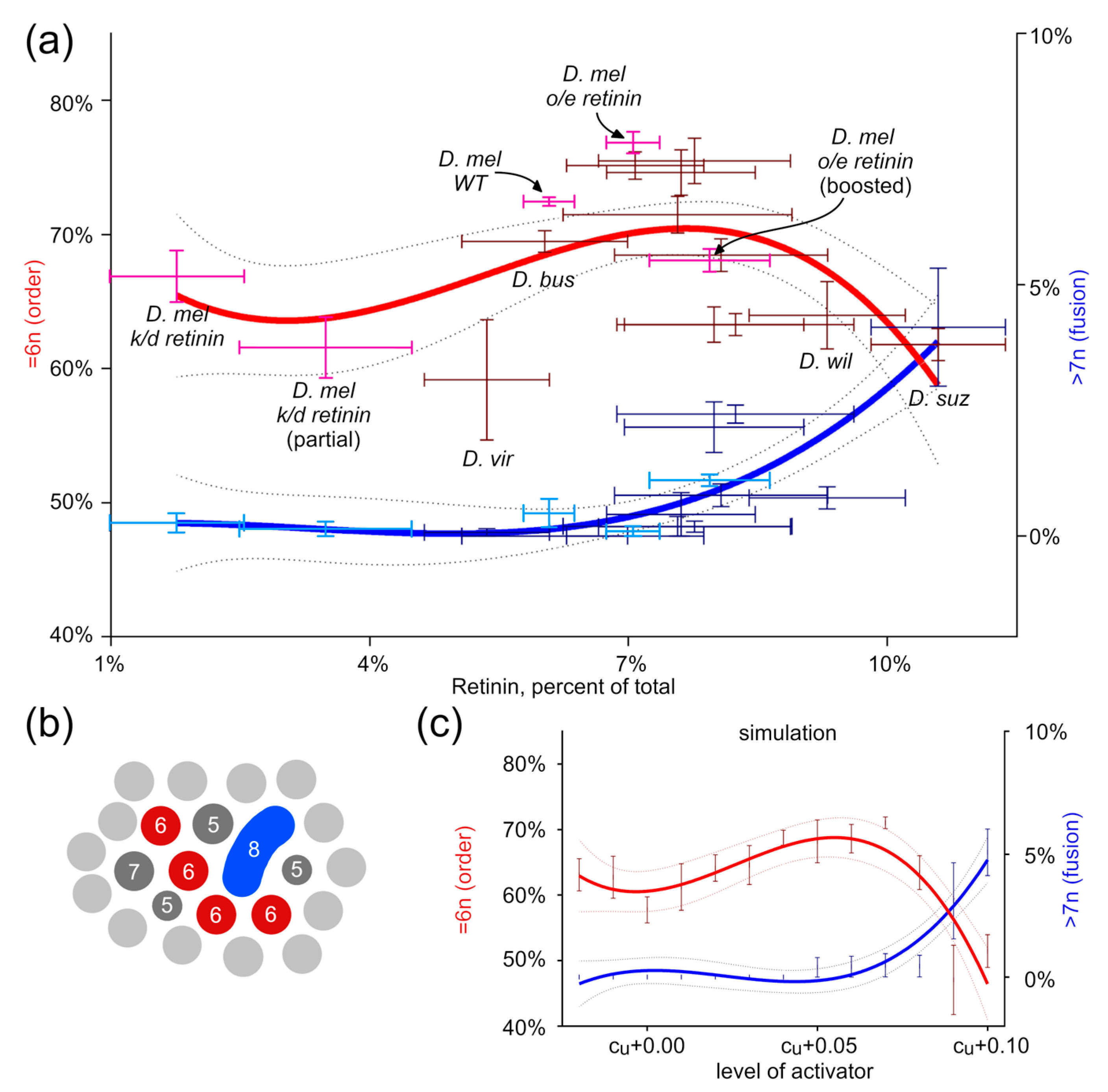
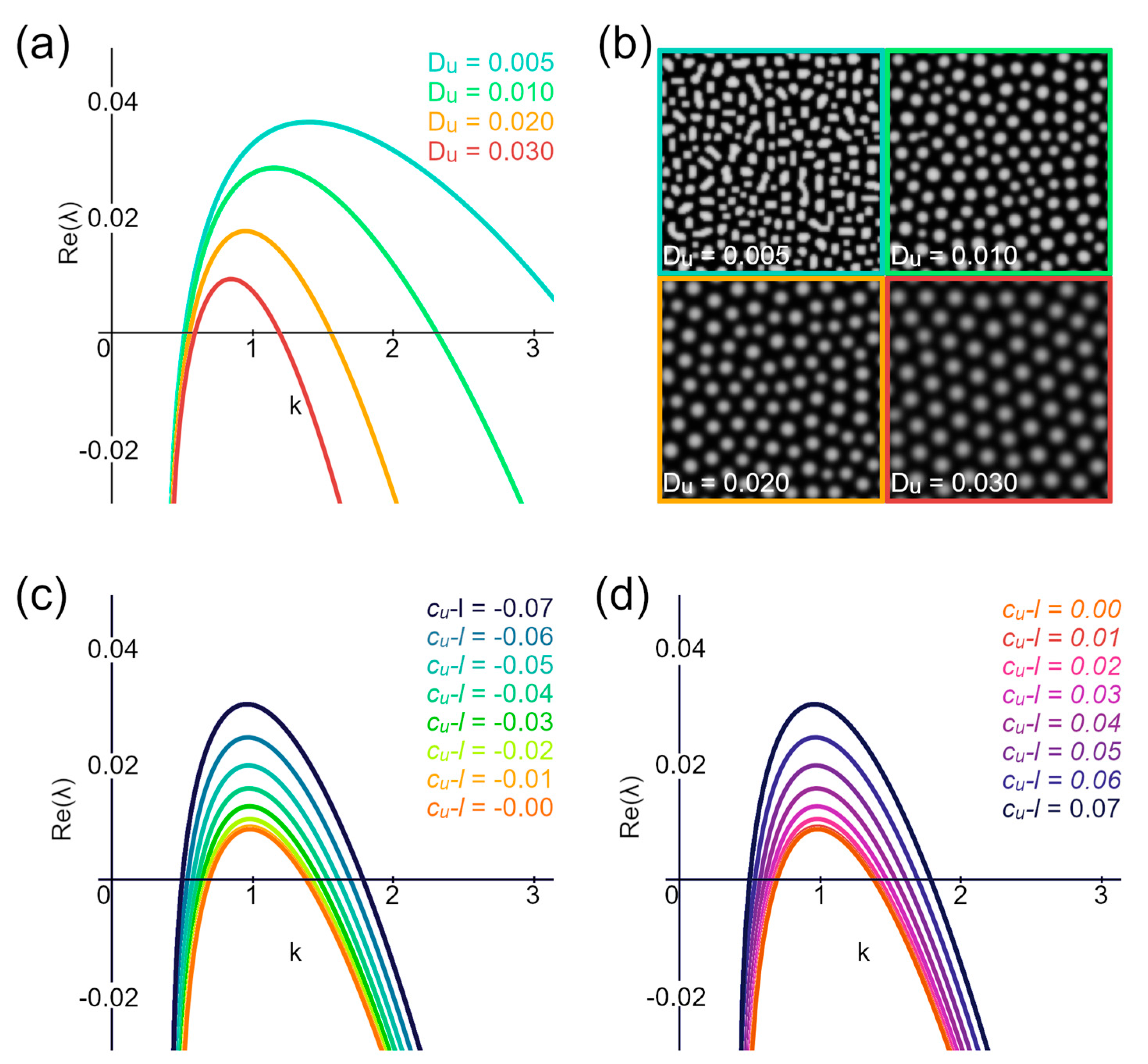
3.1. Order as a Function of the Activator’s Level
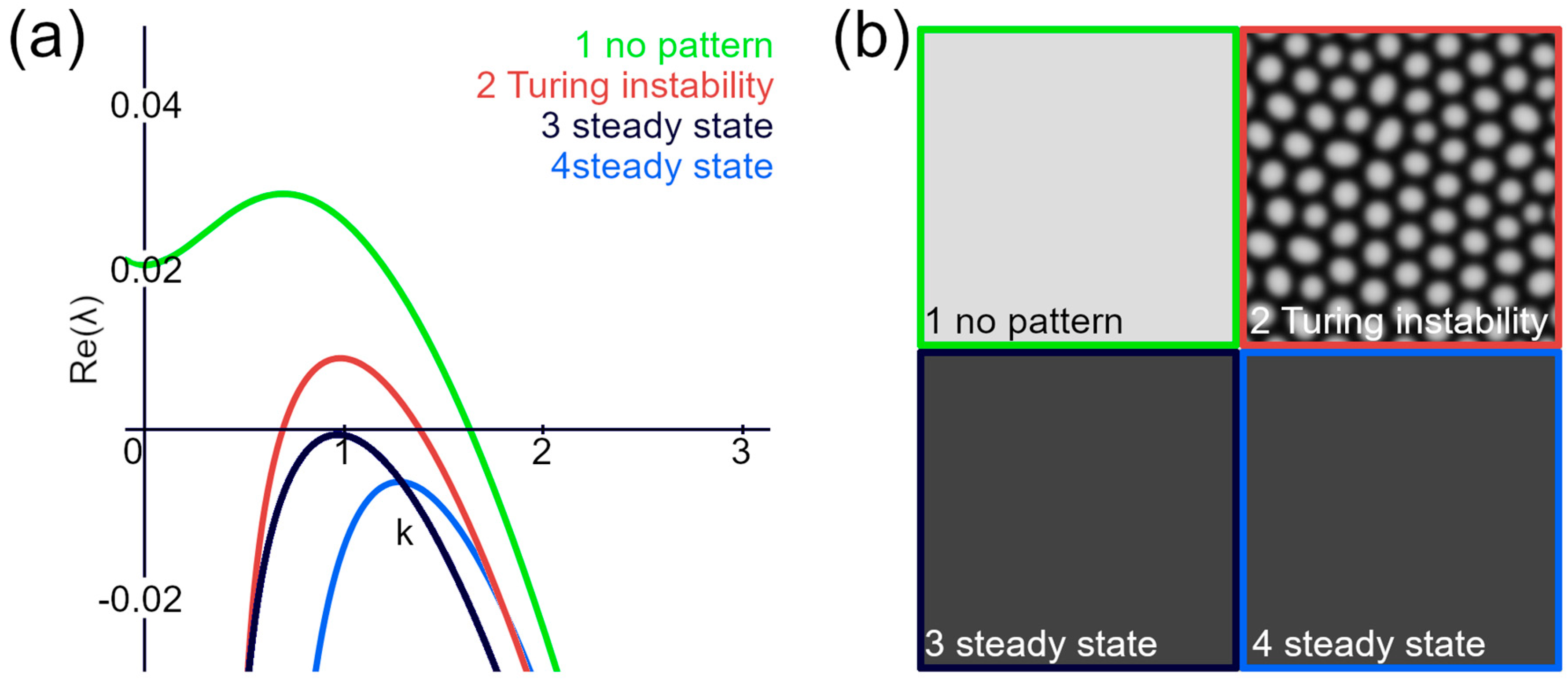
3.2. Spread of the Lyapunov Exponent’s Positive Part Is Minimized for Ordered Structures
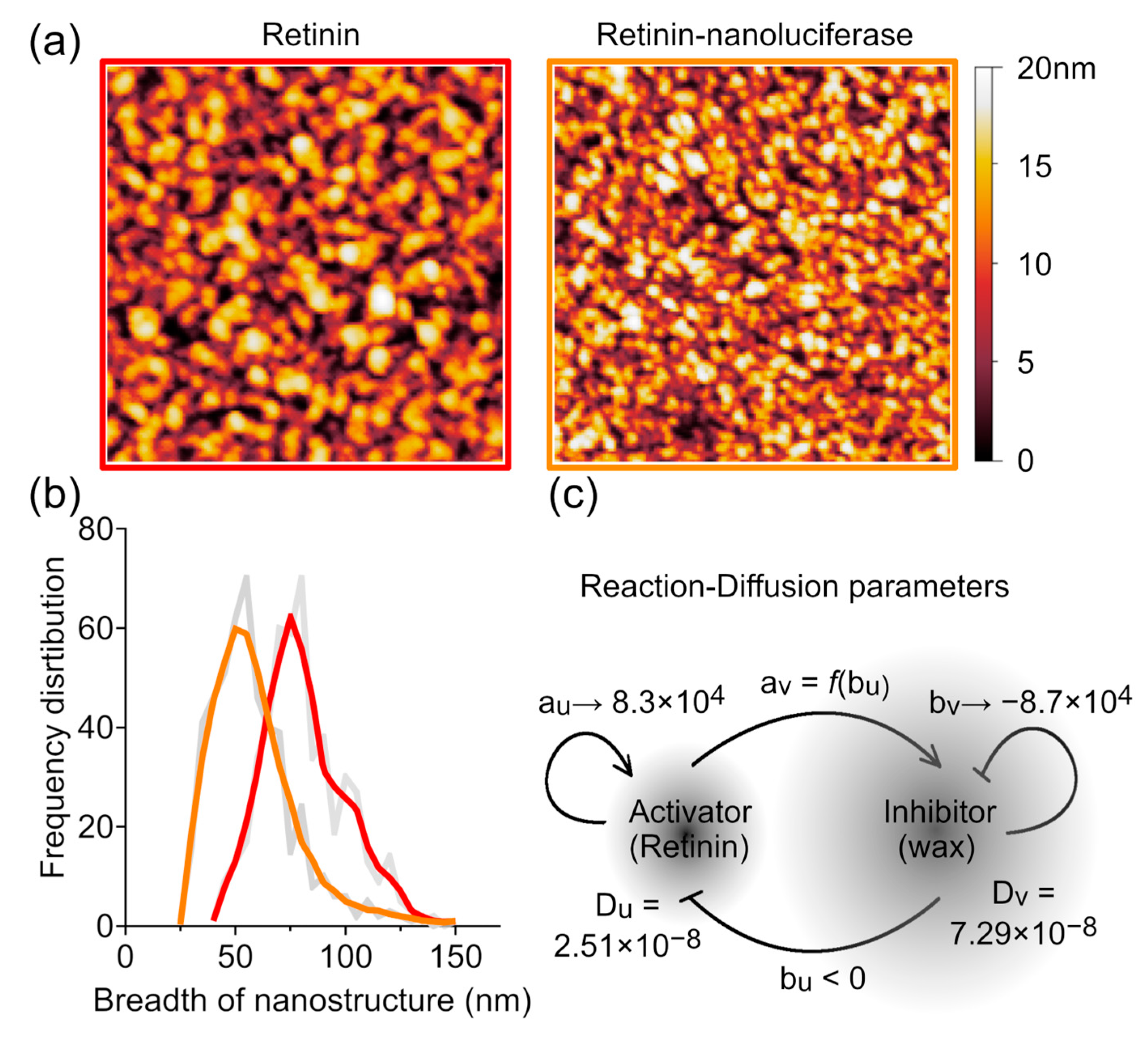
3.3. Reaction Rate Determination Method
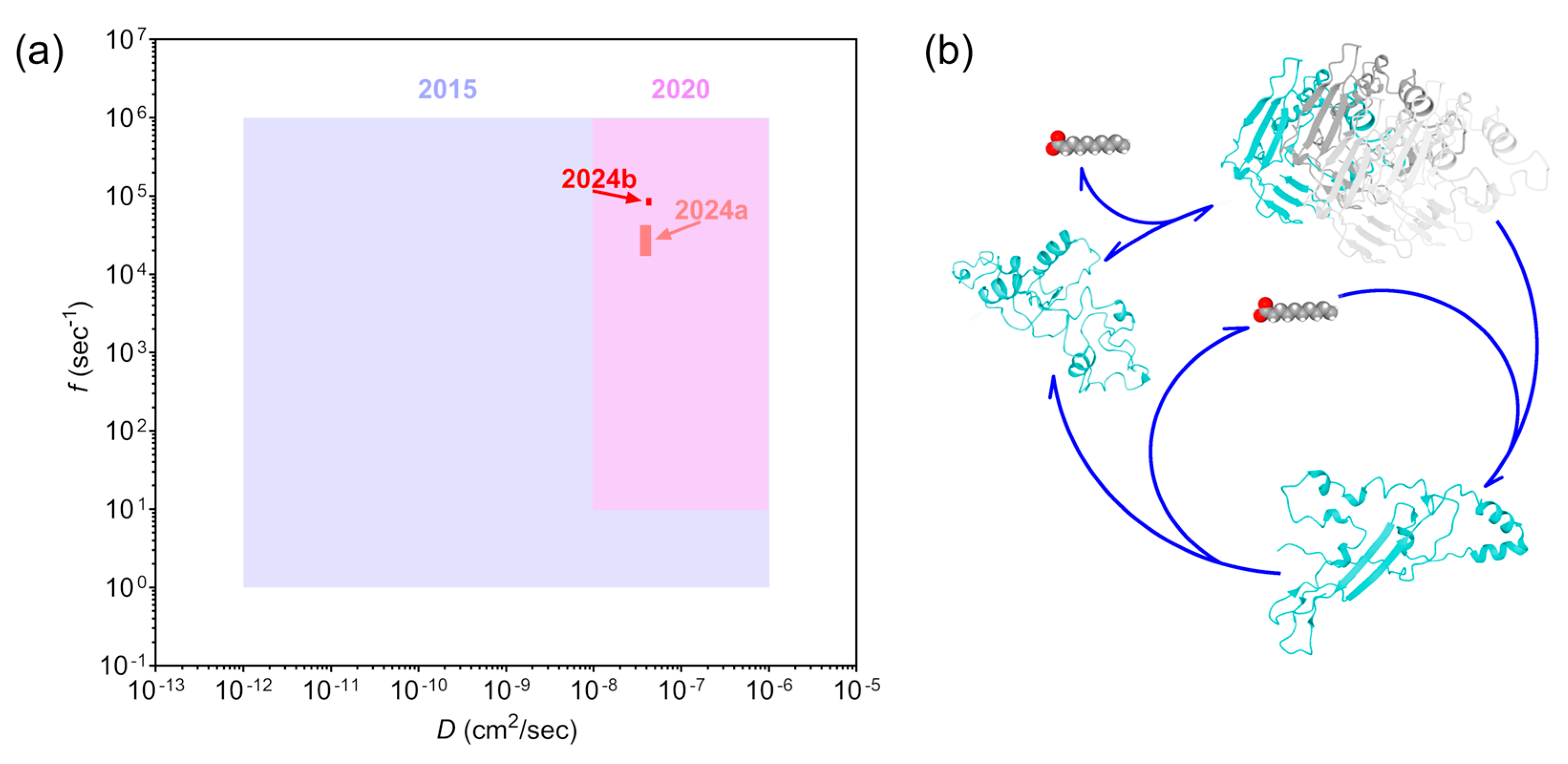
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Turing, A.M. The Chemical Basis of Morphogenesis. Philos. T Roy. Soc. B 1952, 237, 37–72. [Google Scholar]
- Fofonjka, A.; Milinkovitch, M.C. Reaction-diffusion in a growing 3D domain of skin scales generates a discrete cellular automaton. Nat. Commun. 2021, 12, 2433. [Google Scholar] [CrossRef] [PubMed]
- Nakamasu, A.; Takahashi, G.; Kanbe, A.; Kondo, S. Interactions between zebrafish pigment cells responsible for the generation of Turing patterns. Proc. Natl. Acad. Sci. USA 2009, 106, 8429–8434. [Google Scholar] [CrossRef] [PubMed]
- Sheth, R.; Marcon, L.; Bastida, M.F.; Junco, M.; Quintana, L.; Dahn, R.; Kmita, M.; Sharpe, J.; Ros, M.A. Hox Genes Regulate Digit Patterning by Controlling the Wavelength of a Turing-Type Mechanism. Science 2012, 338, 1476–1480. [Google Scholar] [CrossRef]
- Green, J.B.A.; Sharpe, J. Positional information and reaction-diffusion: Two big ideas in developmental biology combine. Development 2015, 142, 1203–1211. [Google Scholar] [CrossRef]
- Nakamura, T.; Mine, N.; Nakaguchi, E.; Mochizuki, A.; Yamamoto, M.; Yashiro, K.; Meno, C.; Hamada, H. Generation of Robust Left-Right Asymmetry in the Mouse Embryo Requires a Self-Enhancement and Lateral-Inhibition System. Dev. Cell 2006, 11, 495–504. [Google Scholar] [CrossRef]
- Kryuchkov, M.; Bilousov, O.; Lehmann, J.; Fiebig, M.; Katanaev, V.L. Reverse and forward engineering of Drosophila corneal nanocoatings. Nature 2020, 585, 383–389. [Google Scholar] [CrossRef]
- Abd-Elsalam, K.A. Chapter 1—Multifunctional hybrid nanomaterials for sustainable agri-food and ecosystems: A note from the editor. In Multifunctional Hybrid Nanomaterials for Sustainable Agri-Food and Ecosystems; Abd-Elsalam, K.A., Ed.; Elsevier: Amsterdam, The Netherlands, 2020; pp. 1–19. [Google Scholar]
- Johnson, B.J.; Russ Algar, W.; Malanoski, A.P.; Ancona, M.G.; Medintz, I.L. Understanding enzymatic acceleration at nanoparticle interfaces: Approaches and challenges. Nano Today 2014, 9, 102–131. [Google Scholar] [CrossRef]
- Ghanta, S.R.; Muralidharan, K. Chemical synthesis of aluminum nanoparticles. J. Nanoparticle Res. 2013, 15, 1715. [Google Scholar] [CrossRef]
- Din, M.I.; Rehan, R. Synthesis, Characterization, and Applications of Copper Nanoparticles. Anal. Lett. 2017, 50, 50–62. [Google Scholar] [CrossRef]
- Zhang, W. Protein Corona of Nanoparticles and Its Application in Drug Delivery. In Nanotechnology in Regenerative Medicine and Drug Delivery Therapy; Xu, H., Gu, N., Eds.; Springer: Singapore, 2020; pp. 389–419. [Google Scholar]
- Kryuchkov, M.; Savitsky, V.; Wilts, B.D.; Gray, E.; Katanaev, V.L. Light Polarization by Biological Nanocoatings. ACS Appl. Mater. Interfaces 2021, 13, 23481–23488. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Wang, Z.; Sun, D.; Li, S.; Deng, Q.; Xin, X. Supramolecular Self-Assembly of Atomically Precise Silver Nanoclusters with Chiral Peptide for Temperature Sensing and Detection of Arginine. Nanomaterials 2022, 12, 424. [Google Scholar] [CrossRef] [PubMed]
- Kryuchkov, M.; Adamcik, J.; Katanaev, V.L. Bactericidal and Antiviral Bionic Metalized Nanocoatings. Nanomaterials 2022, 12, 1868. [Google Scholar] [CrossRef] [PubMed]
- Urca, T.; Lehmann, F.-O.; Gorb, E.V.; Gorb, S.N. Nanoscale mesh acts as anti-adhesive surface against particulate contamination in eyes of whiteflies. Sci. Rep. 2024, 14, 18267. [Google Scholar] [CrossRef]
- Hensel, R.; Helbig, R.; Aland, S.; Voigt, A.; Neinhuis, C.; Werner, C. Tunable nano-replication to explore the omniphobic characteristics of springtail skin. NPG Asia Mater. 2013, 5, e37. [Google Scholar] [CrossRef]
- Wolff, J.O.; Schwaha, T.; Seiter, M.; Gorb, S.N. Whip spiders (Amblypygi) become water-repellent by a colloidal secretion that self-assembles into hierarchical microstructures. Zool. Lett. 2016, 2, 23. [Google Scholar] [CrossRef]
- Peng, Q.; Huang, Z.; Sun, K.; Liu, Y.; Yoon, C.W.; Harrison, R.E.S.; Schmitt, D.L.; Zhu, L.; Wu, Y.; Tasan, I.; et al. Engineering inducible biomolecular assemblies for genome imaging and manipulation in living cells. Nat. Commun. 2022, 13, 7933. [Google Scholar] [CrossRef]
- Ramnarine-Sanchez, R.S.; Kanczler, J.M.; Evans, N.D.; Oreffo, R.O.C.; Dawson, J.I. Self-Assembly of Structured Colloidal Gels for High-Resolution 3D Micropatterning of Proteins at Scale. Adv. Mater. 2023, 35, 2304461. [Google Scholar] [CrossRef]
- Kryuchkov, M.; Jobin, M.; Savitsky, V.; Smirnov, S.; Katanaev, V.L. Evolutionary Bet-Hedging at the Nanoscale, 1st ed.; Preprint; Research Square: Durham, NC, USA, 2024. [Google Scholar]
- Miura, T.; Maini, P.K. Speed of pattern appearance in reaction-diffusion models: Implications in the pattern formation of limb bud mesenchyme cells. Bull. Math. Biol. 2004, 66, 627–649. [Google Scholar] [CrossRef]
- Villar-Sepúlveda, E.; Champneys, A.R. General conditions for Turing and wave instabilities in reaction -diffusion systems. J. Math. Biol. 2023, 86, 39. [Google Scholar] [CrossRef]
- Schnörr, D.; Schnörr, C. Learning system parameters from turing patterns. Mach. Learn. 2023, 112, 3151–3190. [Google Scholar] [CrossRef] [PubMed]
- Ho, C.; Morsut, L. Novel synthetic biology approaches for developmental systems. Stem Cell Rep. 2021, 16, 1051–1064. [Google Scholar] [CrossRef] [PubMed]
- Kondo, S.; Miura, T. Reaction-Diffusion Model as a Framework for Understanding Biological Pattern Formation. Science 2010, 329, 1616–1620. [Google Scholar] [CrossRef] [PubMed]
- Stavroulakis, P.; Boden, S.; Johnson, T.; Bagnall, D.M. Suppression of backscattered diffraction from sub-wavelength ‘moth-eye’ arrays. Opt. Express 2013, 21, 1–11. [Google Scholar] [CrossRef]
- Tirapegui, E.; Zeller, W. Instabilities and Nonequilibrium Structures V; Springer: Dordrecht, The Netherlands, 2012. [Google Scholar]
- Arnold, L.; Crauel, H.; Eckmann, J.P. Lyapunov Exponents; Springer: Berlin/Heidelberg, Germany, 2014. [Google Scholar]
- Cooper, S.B.; Hodges, A. The Once and Future Turing: Computing the World; Cambridge University Press: Cambridge, UK, 2016. [Google Scholar]
- Muolo, R.; Gallo, L.; Latora, V.; Frasca, M.; Carletti, T. Turing patterns in systems with high-order interactions. Chaos Solitons Fractals 2023, 166, 112912. [Google Scholar] [CrossRef]
- Blagodatski, A.; Sergeev, A.; Kryuchkov, M.; Lopatina, Y.; Katanaev, V.L. Diverse set of Turing nanopatterns coat corneae across insect lineages. Proc. Natl. Acad. Sci. USA 2015, 112, 10750–10755. [Google Scholar] [CrossRef]
- Sun, Z.; Inagaki, S.; Miyoshi, K.; Saito, K.; Hayashi, S. Osiris gene family defines the cuticle nanopatterns of Drosophila. Genetics 2024, 227, iyae065. [Google Scholar] [CrossRef]
- Ghosh, N.; Treisman, J.E. Apical cell expansion maintained by Dusky-like establishes a scaffold for corneal lens morphogenesis. Sci. Adv. 2024, 10, eado4167. [Google Scholar] [CrossRef]
- Deryusheva, E.; Nemashkalova, E.; Galloux, M.; Richard, C.A.; Eléouët, J.F.; Kovacs, D.; Van Belle, K.; Tompa, P.; Uversky, V.; Permyakov, S. Does Intrinsic Disorder in Proteins Favor Their Interaction with Lipids? Proteomics 2019, 19, e1800098. [Google Scholar] [CrossRef]
- Hunter, A.W.; Caplow, M.; Coy, D.L.; Hancock, W.O.; Diez, S.; Wordeman, L.; Howard, J. The kinesin-related protein MCAK is a microtubule depolymerase that forms an ATP-hydrolyzing complex at microtubule ends. Mol. Cell 2003, 11, 445–457. [Google Scholar] [CrossRef]
- Maney, T.; Wagenbach, M.; Wordeman, L. Molecular Dissection of the Microtubule Depolymerizing Activity of Mitotic Centromere-associated Kinesin. J. Biol. Chem. 2001, 276, 34753–34758. [Google Scholar] [CrossRef] [PubMed]
- Weisenberg, R.C.; Deery, W.J.; Dickinson, P.J. Tubulin-nucleotide interactions during the polymerization and depolymerization of microtubules. Biochemistry 1976, 15, 4248–4254. [Google Scholar] [CrossRef] [PubMed]
- Sirimungkala, A.; Försterling, H.-D.; Dlask, V.; Field, R.J. Bromination Reactions Important in the Mechanism of the Belousov−Zhabotinsky System. J. Phys. Chem. A 1999, 103, 1038–1043. [Google Scholar] [CrossRef]
- Lengyel, I.; Epstein, I.R. A chemical approach to designing Turing patterns in reaction-diffusion systems. Proc. Natl. Acad. Sci. USA 1992, 89, 3977–3979. [Google Scholar] [CrossRef]
- Morris, A.M.; Watzky, M.A.; Finke, R.G. Protein aggregation kinetics, mechanism, and curve-fitting: A review of the literature. Biochim. Et. Biophys. Acta (BBA)—Proteins Proteom. 2009, 1794, 375–397. [Google Scholar] [CrossRef]
- Cukalevski, R.; Yang, X.; Meisl, G.; Weininger, U.; Bernfur, K.; Frohm, B.; Knowles, T.P.J.; Linse, S. The Aβ40 and Aβ42 peptides self-assemble into separate homomolecular fibrils in binary mixtures but cross-react during primary nucleation. Chem. Sci. 2015, 6, 4215–4233. [Google Scholar] [CrossRef]
- Diaferia, C.; Gallo, E.; Cimmino, L.; Laurenzi, V.; De Marco, A.; Morelli, G.; Stornaiuolo, M.; Accardo, A. Fluorescence of Aggregated Aromatic Peptides for Studying the Kinetics of Aggregation and Hardening of Amyloid-like Structures. Chem.—A Eur. J. 2024, 30, e202401998. [Google Scholar] [CrossRef]
- Ruey-Tarng, R.-T.; Liaw, S.-S.; Maini, P. Oscillatory Turing Patterns in a Simple Reaction-Diffusion System. J. Korean Phys. Soc. 2007, 50, 234–238. [Google Scholar] [CrossRef]
- Mukherjee, N.; Volpert, V. Bifurcation scenario of Turing patterns in prey-predator model with nonlocal consumption in the prey dynamics. Commun. Nonlinear Sci. Numer. Simul. 2021, 96, 105677. [Google Scholar] [CrossRef]
- Ahmed, N.; Yasin, M.W.; Baleanu, D.; Tintareanu-Mircea, O.; Iqbal, M.S.; Akgül, A. Pattern Formation and analysis of reaction–diffusion ratio-dependent prey–predator model with harvesting in predator. Chaos Solitons Fractals 2024, 186, 115164. [Google Scholar] [CrossRef]
- Mondal, A.; Upadhyay, R.K.; Mondal, A.; Sharma, S.K. Emergence of Turing patterns and dynamic visualization in excitable neuron model. Appl. Math. Comput. 2022, 423, 127010. [Google Scholar] [CrossRef]
- Steyn-Ross, M.; Steyn-Ross, D.; Sleigh, J.; Wilson, M. A Mechanism for Ultra-Slow Oscillations in the Cortical Default Network. Bull. Math. Biol. 2011, 73, 398–416. [Google Scholar] [CrossRef] [PubMed]
- Fuseya, Y.; Katsuno, H.; Behnia, K.; Kapitulnik, A. Nanoscale Turing patterns in a bismuth monolayer. Nat. Phys. 2021, 17, 1031–1036. [Google Scholar] [CrossRef]
- Getzin, S.; Erickson, T.E.; Yizhaq, H.; Muñoz-Rojas, M.; Huth, A.; Wiegand, K. Bridging ecology and physics: Australian fairy circles regenerate following model assumptions on ecohydrological feedbacks. J. Ecol. 2021, 109, 399–416. [Google Scholar] [CrossRef]
- Zeng, R.; Lv, C.; Wang, C.; Zhao, G. Bionanomaterials based on protein self-assembly: Design and applications in biotechnology. Biotechnol. Adv. 2021, 52, 107835. [Google Scholar] [CrossRef]
- Kulikowski, J.; Wang, S.; Aitken, Z.; Grimm, J.; Gao, B.; Wang, M.M.; Doan, D.; Lee, A.C.; Shen, L.; Huang, W.; et al. DNA-silica nanolattices as mechanical metamaterials. Matter 2024, 7, 2144–2160. [Google Scholar] [CrossRef]
- Liljeström, V.; Mikkilä, J.; Kostiainen, M.A. Self-assembly and modular functionalization of three-dimensional crystals from oppositely charged proteins. Nat. Commun. 2014, 5, 4445. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kryuchkov, M.; Valnohova, J.; Katanaev, V.L. Route to Measure Exact Parameters of Bio-Nanostructures Self-Assembly. Biomolecules 2024, 14, 1388. https://doi.org/10.3390/biom14111388
Kryuchkov M, Valnohova J, Katanaev VL. Route to Measure Exact Parameters of Bio-Nanostructures Self-Assembly. Biomolecules. 2024; 14(11):1388. https://doi.org/10.3390/biom14111388
Chicago/Turabian StyleKryuchkov, Mikhail, Jana Valnohova, and Vladimir L. Katanaev. 2024. "Route to Measure Exact Parameters of Bio-Nanostructures Self-Assembly" Biomolecules 14, no. 11: 1388. https://doi.org/10.3390/biom14111388
APA StyleKryuchkov, M., Valnohova, J., & Katanaev, V. L. (2024). Route to Measure Exact Parameters of Bio-Nanostructures Self-Assembly. Biomolecules, 14(11), 1388. https://doi.org/10.3390/biom14111388






