tRNA and tsRNA: From Heterogeneity to Multifaceted Regulators
Abstract
1. Introduction
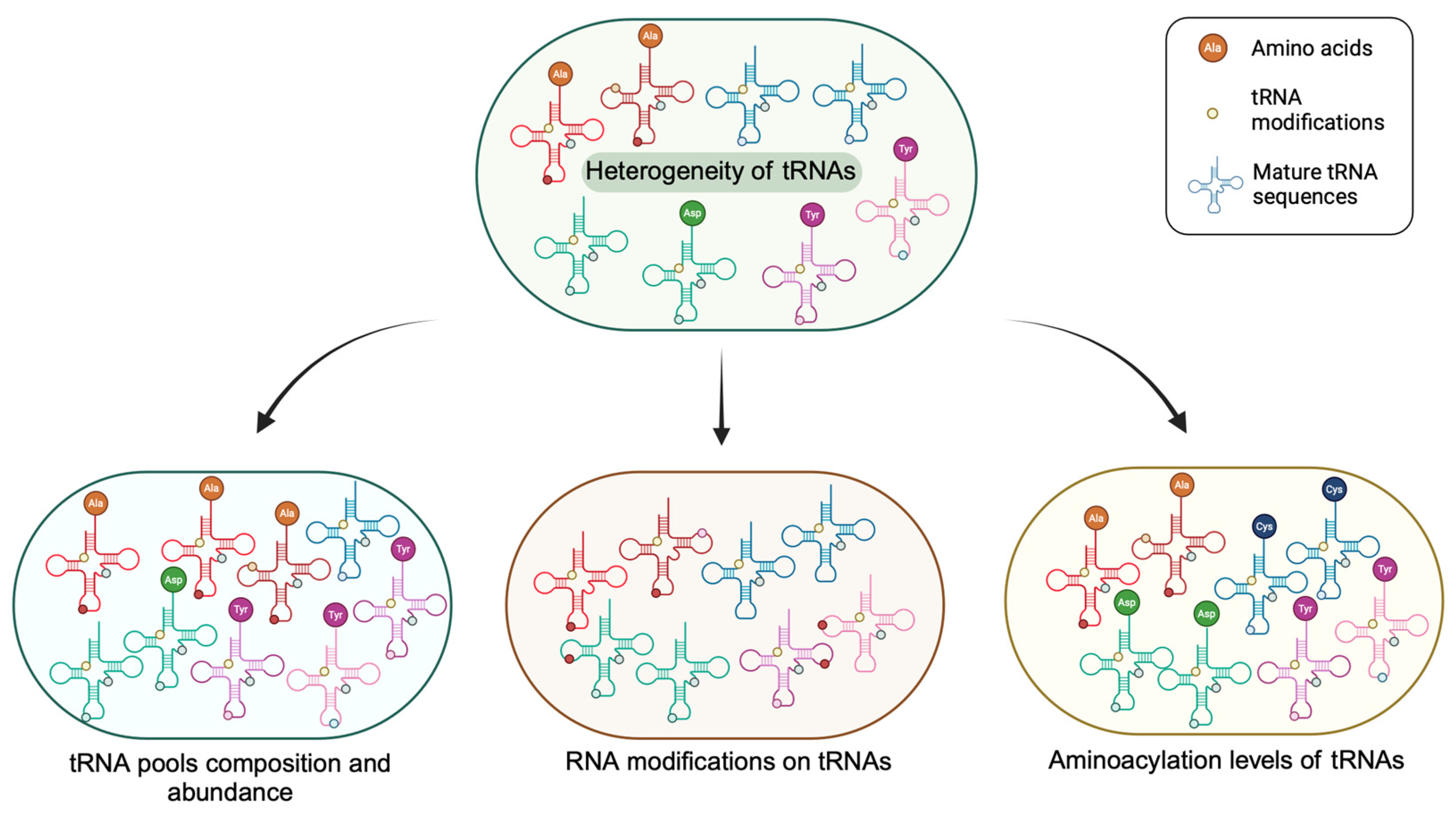
2. The Heterogeneity of the Mature tRNA Pool
2.1. Total Abundance Variation
2.2. Composition Ratio Difference
2.3. Charging Level Sufficiency
3. Chemical Modifications: Guardians of tRNA Integrity and Function
3.1. RNA Modification on tRNA
3.1.1. 5-Methylcytidine (m5C)
3.1.2. N1-Methyladenosine (m1A)
3.1.3. N1-Methylguanosine (m1G)
3.1.4. N7-Methylguanosine (m7G)
3.1.5. N3-Methylcytidine (m3C)
3.1.6. Pseudouridine (Ψ)
3.1.7. Queuosine (Q)
| tRNA Modification | Associated Pathology | References |
|---|---|---|
| m5C | Male germ cell differentiation Tumor metastasis Paternally acquired metabolic disorders | [57] [122] [52] |
| m1A | T cell proliferation post-antigen stimulation Mitochondrial disorders: MERRF (myoclonus epilepsy, ragged-red fibers) | [123] [124] [125] |
| m1G | Ehrlich ascites tumor; neuroblastoma Hereditary spastic paraparesis Neuropathy syndromes Combined oxidative phosphorylation deficiency 26 (COXPD26) Idiopathic non-cirrhotic portal hypertension; hepatopulmonary syndrome | [126] [78] [79] [80] [81] |
| m7G | Acute myeloid leukemia Bladder cancer Hepatocellular carcinoma Prostate cancer Stem cell fate determination Aging | [86,92] [93] [94] [95] [16] [97] |
| m3C | Human developmental delay and epileptic encephalopathy | [103] |
| Ψ | Mitochondrial disease | [127] |
| Q | Sex-dependent learning and memory formation in the hippocampus | [121] |
3.2. Rebirth of tsRNA from tRNA
4. Revisit the Regulatory Mechanisms of tRNA and tsRNA
4.1. tRNA: Focus on Both Main and Side Businesses
4.1.1. Being Primers for Reverse Transcription Transposons
4.1.2. Binding to DNA/RNA
4.1.3. Interacting with Proteins
4.2. tsRNA Regulation: Structure Dictates Function
4.2.1. Sequence Complementarity
4.2.2. tsRNA Secondary Structure
5. Regulators in Cell Fate Determination and Embryonic Development
6. tsRNA: From Sensor of Environment to Epigenetic Inheritance Messenger
7. Emerging Stars in Disease and Therapeutic Modulation
8. Conclusions and Perspectives
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Parisien, M.; Wang, X.; Pan, T. Diversity of human tRNA genes from the 1000-genomes project. RNA Biol. 2013, 10, 1853–1867. [Google Scholar] [CrossRef] [PubMed]
- Goodenbour, J.M.; Pan, T. Diversity of tRNA genes in eukaryotes. Nucleic Acids Res. 2006, 34, 6137–6146. [Google Scholar] [CrossRef] [PubMed]
- Kirchner, S.; Ignatova, Z. Emerging roles of tRNA in adaptive translation, signalling dynamics and disease. Nat. Rev. Genet. 2015, 16, 98–112. [Google Scholar] [CrossRef] [PubMed]
- Behrens, A.; Rodschinka, G.; Nedialkova, D.D. High-resolution quantitative profiling of tRNA abundance and modification status in eukaryotes by mim-tRNAseq. Mol. Cell 2021, 81, 1802–1815.e7. [Google Scholar] [CrossRef]
- Boccaletto, P.; Stefaniak, F.; Ray, A.; Cappannini, A.; Mukherjee, S.; Purta, E.; Kurkowska, M.; Shirvanizadeh, N.; Destefanis, E.; Groza, P.; et al. MODOMICS: A database of RNA modification pathways. 2021 update. Nucleic Acids Res. 2022, 50, D231–D235. [Google Scholar] [CrossRef]
- Zhang, W.; Foo, M.; Eren, A.M.; Pan, T. tRNA modification dynamics from individual organisms to metaepitranscriptomics of microbiomes. Mol. Cell 2022, 82, 891–906. [Google Scholar] [CrossRef]
- Nguyen, H.A.; Hoffer, E.D.; Dunham, C.M. Importance of a tRNA anticodon loop modification and a conserved, noncanonical anticodon stem pairing in tRNACGGProfor decoding. J. Biol. Chem. 2019, 294, 5281–5291. [Google Scholar] [CrossRef]
- Rapino, F.; Zhou, Z.; Roncero Sanchez, A.M.; Joiret, M.; Seca, C.; El Hachem, N.; Valenti, G.; Latini, S.; Shostak, K.; Geris, L.; et al. Wobble tRNA modification and hydrophilic amino acid patterns dictate protein fate. Nat. Commun. 2021, 12, 2170. [Google Scholar] [CrossRef]
- Motorin, Y.; Helm, M. tRNA stabilization by modified nucleotides. Biochemistry 2010, 49, 4934–4944. [Google Scholar] [CrossRef]
- Kwon, N.H.; Fox, P.L.; Kim, S. Aminoacyl-tRNA synthetases as therapeutic targets. Nat. Rev. Drug Discov. 2019, 18, 629–650. [Google Scholar] [CrossRef]
- Vo, M.N.; Terrey, M.; Lee, J.W.; Roy, B.; Moresco, J.J.; Sun, L.; Fu, H.; Liu, Q.; Weber, T.G.; Yates, J.R., 3rd; et al. ANKRD16 prevents neuron loss caused by an editing-defective tRNA synthetase. Nature 2018, 557, 510–515. [Google Scholar] [CrossRef] [PubMed]
- Pinzaru, A.M.; Tavazoie, S.F. Transfer RNAs as dynamic and critical regulators of cancer progression. Nat. Rev. Cancer 2023, 23, 746–761. [Google Scholar] [CrossRef] [PubMed]
- Thandapani, P.; Kloetgen, A.; Witkowski, M.T.; Glytsou, C.; Lee, A.K.; Wang, E.; Wang, J.; LeBoeuf, S.E.; Avrampou, K.; Papagiannakopoulos, T.; et al. Valine tRNA levels and availability regulate complex I assembly in leukaemia. Nature 2022, 601, 428–433. [Google Scholar] [CrossRef] [PubMed]
- Huh, D.; Passarelli, M.C.; Gao, J.; Dusmatova, S.N.; Goin, C.; Fish, L.; Pinzaru, A.M.; Molina, H.; Ren, Z.; McMillan, E.A.; et al. A stress-induced tyrosine-tRNA depletion response mediates codon-based translational repression and growth suppression. EMBO J. 2021, 40, e106696. [Google Scholar] [CrossRef]
- Saba, J.A.; Liakath-Ali, K.; Green, R.; Watt, F.M. Translational control of stem cell function. Nat. Rev. Mol. Cell Biol. 2021, 22, 671–690. [Google Scholar] [CrossRef]
- Lin, S.; Liu, Q.; Lelyveld, V.S.; Choe, J.; Szostak, J.W.; Gregory, R.I. Mettl1/Wdr4-Mediated m7G tRNA Methylome Is Required for Normal mRNA Translation and Embryonic Stem Cell Self-Renewal and Differentiation. Mol. Cell 2018, 71, 244–255.e5. [Google Scholar] [CrossRef]
- Hughes, L.A.; Rudler, D.L.; Siira, S.J.; McCubbin, T.; Raven, S.A.; Browne, J.M.; Ermer, J.A.; Rientjes, J.; Rodger, J.; Marcellin, E.; et al. Copy number variation in tRNA isodecoder genes impairs mammalian development and balanced translation. Nat. Commun. 2023, 14, 2210. [Google Scholar] [CrossRef]
- Dedon, P.C.; Begley, T.J. Dysfunctional tRNA reprogramming and codon-biased translation in cancer. Trends Mol. Med. 2022, 28, 964–978. [Google Scholar] [CrossRef]
- Passarelli, M.C.; Pinzaru, A.M.; Asgharian, H.; Liberti, M.V.; Heissel, S.; Molina, H.; Goodarzi, H.; Tavazoie, S.F. Leucyl-tRNA synthetase is a tumour suppressor in breast cancer and regulates codon-dependent translation dynamics. Nat. Cell Biol. 2022, 24, 307–315. [Google Scholar] [CrossRef]
- Cui, Q.; Yin, K.; Zhang, X.; Ye, P.; Chen, X.; Chao, J.; Meng, H.; Wei, J.; Roeth, D.; Li, L.; et al. Targeting PUS7 suppresses tRNA pseudouridylation and glioblastoma tumorigenesis. Nat. Cancer 2021, 2, 932–949. [Google Scholar] [CrossRef]
- Marshall, L.; Kenneth, N.S.; White, R.J. Elevated tRNA(iMet) synthesis can drive cell proliferation and oncogenic transformation. Cell 2008, 133, 78–89. [Google Scholar] [CrossRef] [PubMed]
- Schorn, A.J.; Martienssen, R. Tie-Break: Host and Retrotransposons Play tRNA. Trends Cell Biol. 2018, 28, 793–806. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Xu, W.; Liu, K.; Jiang, Z.; Han, Y.; Jin, H.; Zhang, L.; Shen, W.; Jia, S.; Sun, Q.; et al. 5′ Half of specific tRNAs feeds back to promote corresponding tRNA gene transcription in vertebrate embryos. Sci. Adv. 2021, 7, eabh0494. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Zhang, Y.; Zhou, T.; Chen, Q. tsRNAs: The Swiss Army Knife for Translational Regulation. Trends Biochem. Sci. 2019, 44, 185–189. [Google Scholar] [CrossRef]
- Muthukumar, S.; Li, C.T.; Liu, R.J.; Bellodi, C. Roles and regulation of tRNA-derived small RNAs in animals. Nat. Rev. Mol. Cell Biol. 2024, 25, 359–378. [Google Scholar] [CrossRef]
- Kuhle, B.; Chen, Q.; Schimmel, P. tRNA renovatio: Rebirth through fragmentation. Mol. Cell 2023, 83, 3953–3971. [Google Scholar] [CrossRef]
- Goodarzi, H.; Nguyen, H.C.B.; Zhang, S.; Dill, B.D.; Molina, H.; Tavazoie, S.F. Modulated Expression of Specific tRNAs Drives Gene Expression and Cancer Progression. Cell 2016, 165, 1416–1427. [Google Scholar] [CrossRef]
- Xia, L.; Guo, H.; Wu, X.; Xu, Y.; Zhao, P.; Yan, B.; Zeng, Y.; He, Y.; Chen, D.; Gale, R.P.; et al. Human circulating small non-coding RNA signature as a non-invasive biomarker in clinical diagnosis of acute myeloid leukaemia. Theranostics 2023, 13, 1289–1301. [Google Scholar] [CrossRef]
- Albers, S.; Allen, E.C.; Bharti, N.; Davyt, M.; Joshi, D.; Perez-Garcia, C.G.; Santos, L.; Mukthavaram, R.; Delgado-Toscano, M.A.; Molina, B.; et al. Engineered tRNAs suppress nonsense mutations in cells and in vivo. Nature 2023, 618, 842–848. [Google Scholar] [CrossRef]
- Wang, J.; Zhang, Y.; Mendonca, C.A.; Yukselen, O.; Muneeruddin, K.; Ren, L.; Liang, J.; Zhou, C.; Xie, J.; Li, J.; et al. AAV-delivered suppressor tRNA overcomes a nonsense mutation in mice. Nature 2022, 604, 343–348. [Google Scholar] [CrossRef]
- Dittmar, K.A.; Goodenbour, J.M.; Pan, T. Tissue-specific differences in human transfer RNA expression. PLoS Genet. 2006, 2, e221. [Google Scholar] [CrossRef] [PubMed]
- Gingold, H.; Tehler, D.; Christoffersen, N.R.; Nielsen, M.M.; Asmar, F.; Kooistra, S.M.; Christophersen, N.S.; Christensen, L.L.; Borre, M.; Sørensen, K.D.; et al. A dual program for translation regulation in cellular proliferation and differentiation. Cell 2014, 158, 1281–1292. [Google Scholar] [CrossRef] [PubMed]
- Willis, I.M.; Moir, R.D.; Hernandez, N. Metabolic programming a lean phenotype by deregulation of RNA polymerase III. Proc. Natl. Acad. Sci. USA 2018, 115, 12182–12187. [Google Scholar] [CrossRef] [PubMed]
- Gao, L.; Behrens, A.; Rodschinka, G.; Forcelloni, S.; Wani, S.; Strasser, K.; Nedialkova, D.D. Selective gene expression maintains human tRNA anticodon pools during differentiation. Nat. Cell Biol. 2024, 26, 100–112. [Google Scholar] [CrossRef]
- Pinkard, O.; McFarland, S.; Sweet, T.; Coller, J. Quantitative tRNA-sequencing uncovers metazoan tissue-specific tRNA regulation. Nat. Commun. 2020, 11, 4104. [Google Scholar] [CrossRef]
- Yu, P.; Zhou, S.; Gao, Y.; Liang, Y.; Guo, W.; Wang, D.O.; Ding, S.; Lin, S.; Wang, J.; Cun, Y. Dynamic Landscapes of tRNA Transcriptomes and Translatomes in Diverse Mouse Tissues. Genom. Proteom. Bioinform. 2023, 21, 834–849. [Google Scholar] [CrossRef]
- Ferro, I.; Liebeton, K.; Ignatova, Z. Growth-Rate Dependent Regulation of tRNA Level and Charging in Bacillus licheniformis. J. Mol. Biol. 2017, 429, 3102–3112. [Google Scholar] [CrossRef]
- He, Q.; He, X.; Xiao, Y.; Zhao, Q.; Ye, Z.; Cui, L.; Chen, Y.; Guan, M.X. Tissue-specific expression atlas of murine mitochondrial tRNAs. J. Biol. Chem. 2021, 297, 100960. [Google Scholar] [CrossRef]
- Yamagami, R.; Hori, H. Functional analysis of tRNA modification enzymes using mutational profiling. Methods Enzymol. 2023, 692, 69–101. [Google Scholar] [CrossRef]
- Wang, Y.; Tao, E.W.; Tan, J.; Gao, Q.Y.; Chen, Y.X.; Fang, J.Y. tRNA modifications: Insights into their role in human cancers. Trends Cell Biol. 2023, 33, 1035–1048. [Google Scholar] [CrossRef]
- Suzuki, T. The expanding world of tRNA modifications and their disease relevance. Nat. Rev. Mol. Cell Biol. 2021, 22, 375–392. [Google Scholar] [CrossRef] [PubMed]
- Yasukawa, T.; Suzuki, T.; Ueda, T.; Ohta, S.; Watanabe, K. Modification defect at anticodon wobble nucleotide of mitochondrial tRNAs(Leu)(UUR) with pathogenic mutations of mitochondrial myopathy, encephalopathy, lactic acidosis, and stroke-like episodes. J. Biol. Chem. 2000, 275, 4251–4257. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, T.; Nagao, A.; Suzuki, T. Human mitochondrial tRNAs: Biogenesis, function, structural aspects, and diseases. Annu. Rev. Genet. 2011, 45, 299–329. [Google Scholar] [CrossRef] [PubMed]
- Frye, M.; Watt, F.M. The RNA methyltransferase Misu (NSun2) mediates Myc-induced proliferation and is upregulated in tumors. Curr. Biol. 2006, 16, 971–981. [Google Scholar] [CrossRef]
- Navarro, I.C.; Tuorto, F.; Jordan, D.; Legrand, C.; Price, J.; Braukmann, F.; Hendrick, A.G.; Akay, A.; Kotter, A.; Helm, M.; et al. Translational adaptation to heat stress is mediated by RNA 5-methylcytosine in Caenorhabditis elegans. EMBO J. 2021, 40, e105496. [Google Scholar] [CrossRef]
- Li, J.; Li, H.; Long, T.; Dong, H.; Wang, E.D.; Liu, R.J. Archaeal NSUN6 catalyzes m5C72 modification on a wide-range of specific tRNAs. Nucleic Acids Res. 2019, 47, 2041–2055. [Google Scholar] [CrossRef]
- Goll, M.G.; Kirpekar, F.; Maggert, K.A.; Yoder, J.A.; Hsieh, C.L.; Zhang, X.; Golic, K.G.; Jacobsen, S.E.; Bestor, T.H. Methylation of tRNAAsp by the DNA methyltransferase homolog Dnmt2. Science 2006, 311, 395–398. [Google Scholar] [CrossRef]
- Shen, H.; Ontiveros, R.J.; Owens, M.C.; Liu, M.Y.; Ghanty, U.; Kohli, R.M.; Liu, K.F. TET-mediated 5-methylcytosine oxidation in tRNA promotes translation. J. Biol. Chem. 2021, 296, 100087. [Google Scholar] [CrossRef]
- Arguello, A.E.; Li, A.; Sun, X.; Eggert, T.W.; Mairhofer, E.; Kleiner, R.E. Reactivity-dependent profiling of RNA 5-methylcytidine dioxygenases. Nat. Commun. 2022, 13, 4176. [Google Scholar] [CrossRef]
- Li, H.; Zhu, D.; Wu, J.; Ma, Y.; Cai, C.; Chen, Y.; Qin, M.; Dai, H. New substrates and determinants for tRNA recognition of RNA methyltransferase DNMT2/TRDMT1. RNA Biol. 2021, 18, 2531–2545. [Google Scholar] [CrossRef]
- Schaefer, M.; Pollex, T.; Hanna, K.; Tuorto, F.; Meusburger, M.; Helm, M.; Lyko, F. RNA methylation by Dnmt2 protects transfer RNAs against stress-induced cleavage. Genes Dev. 2010, 24, 1590–1595. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhang, X.; Shi, J.; Tuorto, F.; Li, X.; Liu, Y.; Liebers, R.; Zhang, L.; Qu, Y.; Qian, J.; et al. Dnmt2 mediates intergenerational transmission of paternally acquired metabolic disorders through sperm small non-coding RNAs. Nat. Cell Biol. 2018, 20, 535–540. [Google Scholar] [CrossRef] [PubMed]
- Van Haute, L.; Lee, S.Y.; McCann, B.J.; Powell, C.A.; Bansal, D.; Vasiliauskaite, L.; Garone, C.; Shin, S.; Kim, J.S.; Frye, M.; et al. NSUN2 introduces 5-methylcytosines in mammalian mitochondrial tRNAs. Nucleic Acids Res. 2019, 47, 8720–8733. [Google Scholar] [CrossRef]
- Long, T.; Li, J.; Li, H.; Zhou, M.; Zhou, X.-L.; Liu, R.-J.; Wang, E.-D. Sequence-specific and Shape-selective RNA Recognition by the Human RNA 5-Methylcytosine Methyltransferase NSun6. J. Biol. Chem. 2016, 291, 24293–24303. [Google Scholar] [CrossRef] [PubMed]
- Nakano, S.; Suzuki, T.; Kawarada, L.; Iwata, H.; Asano, K.; Suzuki, T. NSUN3 methylase initiates 5-formylcytidine biogenesis in human mitochondrial tRNA(Met). Nat. Chem. Biol. 2016, 12, 546–551. [Google Scholar] [CrossRef] [PubMed]
- Haag, S.; Sloan, K.E.; Ranjan, N.; Warda, A.S.; Kretschmer, J.; Blessing, C.; Hubner, B.; Seikowski, J.; Dennerlein, S.; Rehling, P.; et al. NSUN3 and ABH1 modify the wobble position of mt-tRNAMet to expand codon recognition in mitochondrial translation. EMBO J. 2016, 35, 2104–2119. [Google Scholar] [CrossRef]
- Hussain, S.; Tuorto, F.; Menon, S.; Blanco, S.; Cox, C.; Flores, J.V.; Watt, S.; Kudo, N.R.; Lyko, F.; Frye, M. The mouse cytosine-5 RNA methyltransferase NSun2 is a component of the chromatoid body and required for testis differentiation. Mol. Cell Biol. 2013, 33, 1561–1570. [Google Scholar] [CrossRef]
- Oerum, S.; Degut, C.; Barraud, P.; Tisne, C. m1A Post-Transcriptional Modification in tRNAs. Biomolecules 2017, 7, 20. [Google Scholar] [CrossRef]
- Cui, L.; Ma, R.; Cai, J.; Guo, C.; Chen, Z.; Yao, L.; Wang, Y.; Fan, R.; Wang, X.; Shi, Y. RNA modifications: Importance in immune cell biology and related diseases. Signal Transduct. Target. Ther. 2022, 7, 334. [Google Scholar] [CrossRef]
- Ozanick, S.G.; Bujnicki, J.M.; Sem, D.S.; Anderson, J.T. Conserved amino acids in each subunit of the heteroligomeric tRNA m1A58 Mtase from Saccharomyces cerevisiae contribute to tRNA binding. Nucleic Acids Res. 2007, 35, 6808–6819. [Google Scholar] [CrossRef]
- Liu, Y.; Zhou, J.; Li, X.; Zhang, X.; Shi, J.; Wang, X.; Li, H.; Miao, S.; Chen, H.; He, X.; et al. tRNA-m(1)A modification promotes T cell expansion via efficient MYC protein synthesis. Nat. Immunol. 2022, 23, 1433–1444. [Google Scholar] [CrossRef] [PubMed]
- Pei, W.; Kuchroo, V.K. tRNA-m(1)A modification: A translational checkpoint for T cell expansion. Cell Res. 2023, 33, 271–272. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Clark, W.; Luo, G.; Wang, X.; Fu, Y.; Wei, J.; Wang, X.; Hao, Z.; Dai, Q.; Zheng, G.; et al. ALKBH1-Mediated tRNA Demethylation Regulates Translation. Cell 2016, 167, 816–828.e816. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Duan, H.C.; Paduch, M.; Hu, J.; Zhang, C.; Mu, Y.; Lin, H.; He, C.; Kossiakoff, A.A.; Jia, G.; et al. The Molecular Basis of Human ALKBH3 Mediated RNA N1-methyladenosine (m1A) Demethylation. Angew. Chem. Int. Ed. Engl. 2024, 63, e202313900. [Google Scholar] [CrossRef] [PubMed]
- Gu, X.; Zhuang, A.; Yu, J.; Yang, L.; Ge, S.; Ruan, J.; Jia, R.; Fan, X.; Chai, P. Histone lactylation-boosted ALKBH3 potentiates tumor progression and diminished promyelocytic leukemia protein nuclear condensates by m1A demethylation of SP100A. Nucleic Acids Res. 2024, 52, 2273–2289. [Google Scholar] [CrossRef] [PubMed]
- Kuang, W.; Jin, H.; Yang, F.; Chen, X.; Liu, J.; Li, T.; Chang, Y.; Liu, M.; Xu, Z.; Huo, C.; et al. ALKBH3-dependent m1A demethylation of Aurora A mRNA inhibits ciliogenesis. Cell Discov. 2022, 8, 25. [Google Scholar] [CrossRef]
- Sun, Y.; Dai, H.; Dai, X.; Yin, J.; Cui, Y.; Liu, X.; Gonzalez, G.; Yuan, J.; Tang, F.; Wang, N.; et al. m1A in CAG repeat RNA binds to TDP-43 and induces neurodegeneration. Nature 2023, 623, 580–587. [Google Scholar] [CrossRef]
- Chen, Z.; Qi, M.; Shen, B.; Luo, G.; Wu, Y.; Li, J.; Lu, Z.; Zheng, Z.; Dai, Q.; Wang, H. Transfer RNA demethylase ALKBH3 promotes cancer progression via induction of tRNA-derived small RNAs. Nucleic Acids Res. 2019, 47, 2533–2545. [Google Scholar] [CrossRef]
- Zhang, L.S.; Xiong, Q.P.; Pena Perez, S.; Liu, C.; Wei, J.; Le, C.; Zhang, L.; Harada, B.T.; Dai, Q.; Feng, X.; et al. ALKBH7-mediated demethylation regulates mitochondrial polycistronic RNA processing. Nat. Cell Biol. 2021, 23, 684–691. [Google Scholar] [CrossRef]
- Fu, Y.; Jia, G.; Pang, X.; Wang, R.N.; Wang, X.; Li, C.J.; Smemo, S.; Dai, Q.; Bailey, K.A.; Nobrega, M.A.; et al. FTO-mediated formation of N6-hydroxymethyladenosine and N6-formyladenosine in mammalian RNA. Nat. Commun. 2013, 4, 1798. [Google Scholar] [CrossRef]
- Wei, J.; Liu, F.; Lu, Z.; Fei, Q.; Ai, Y.; He, P.C.; Shi, H.; Cui, X.; Su, R.; Klungland, A.; et al. Differential m6A, m6Am, and m1A Demethylation Mediated by FTO in the Cell Nucleus and Cytoplasm. Mol. Cell 2018, 71, 973–985.e5. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Wei, L.H.; Wang, Y.; Xiao, Y.; Liu, J.; Zhang, W.; Yan, N.; Amu, G.; Tang, X.; Zhang, L.; et al. Structural insights into FTO’s catalytic mechanism for the demethylation of multiple RNA substrates. Proc. Natl. Acad. Sci. USA 2019, 116, 2919–2924. [Google Scholar] [CrossRef]
- Lei, H.T.; Wang, Z.H.; Li, B.; Sun, Y.; Mei, S.Q.; Yang, J.H.; Qu, L.H.; Zheng, L.L. tModBase: Deciphering the landscape of tRNA modifications and their dynamic changes from epitranscriptome data. Nucleic Acids Res. 2023, 51, D315–D327. [Google Scholar] [CrossRef] [PubMed]
- Droogmans, L.; Grosjean, H. Enzymatic conversion of guanosine 3′ adjacent to the anticodon of yeast tRNAPhe to N1-methylguanosine and the wye nucleoside: Dependence on the anticodon sequence. EMBO J. 1987, 6, 477–483. [Google Scholar] [CrossRef] [PubMed]
- Christian, T.; Gamper, H.; Hou, Y.M. Conservation of structure and mechanism by Trm5 enzymes. RNA 2013, 19, 1192–1199. [Google Scholar] [CrossRef] [PubMed]
- Bjork, G.R.; Wikstrom, P.M.; Bystrom, A.S. Prevention of translational frameshifting by the modified nucleoside 1-methylguanosine. Science 1989, 244, 986–989. [Google Scholar] [CrossRef]
- Ito, T.; Masuda, I.; Yoshida, K.; Goto-Ito, S.; Sekine, S.; Suh, S.W.; Hou, Y.M.; Yokoyama, S. Structural basis for methyl-donor-dependent and sequence-specific binding to tRNA substrates by knotted methyltransferase TrmD. Proc. Natl. Acad. Sci. USA 2015, 112, E4197–E4205. [Google Scholar] [CrossRef]
- Tarnopolsky, M.A.; Brady, L.; Tetreault, M.; Care4Rare Canada, C. TRMT5 mutations are associated with features of complex hereditary spastic paraparesis. Neurology 2017, 89, 2210–2211. [Google Scholar] [CrossRef]
- Argente-Escrig, H.; Vilchez, J.J.; Frasquet, M.; Muelas, N.; Azorin, I.; Vilchez, R.; Millet-Sancho, E.; Pitarch, I.; Tomas-Vila, M.; Vazquez-Costa, J.F.; et al. A novel TRMT5 mutation causes a complex inherited neuropathy syndrome: The role of nerve pathology in defining a demyelinating neuropathy. Neuropathol. Appl. Neurobiol. 2022, 48, e12817. [Google Scholar] [CrossRef]
- Wu, S.; Li, W.; Bai, Z.; Huang, S.; Yang, D.; Chen, H.; Li, Y.; Liu, Y.; Lv, H. Novel heterozygous compound TRMT5 mutations associated with combined oxidative phosphorylation deficiency 26 in a Chinese family: A case report. BMC Pediatr. 2022, 22, 74. [Google Scholar] [CrossRef]
- Warasnhe, K.; Ozcay, F.; Aydin, H.I.; Ozgun, G.; Ceylaner, S. A novel mutation in TRMT5 associated with idiopathic non-cirrhotic portal hypertension and hepatopulmonary syndrome: Case report of two siblings. Clin. Res. Hepatol. Gastroenterol. 2022, 46, 101928. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Q.; Zhang, L.; He, Q.; Chang, H.; Wang, Z.; Cao, H.; Zhou, Y.; Pan, R.; Chen, Y. Targeting TRMT5 suppresses hepatocellular carcinoma progression via inhibiting the HIF-1alpha pathways. J. Zhejiang Univ. Sci. B 2023, 24, 50–63. [Google Scholar] [CrossRef] [PubMed]
- Meng, F.; Zhou, M.; Xiao, Y.; Mao, X.; Zheng, J.; Lin, J.; Lin, T.; Ye, Z.; Cang, X.; Fu, Y.; et al. A deafness-associated tRNA mutation caused pleiotropic effects on the m1G37 modification, processing, stability and aminoacylation of tRNAIle and mitochondrial translation. Nucleic Acids Res. 2021, 49, 1075–1093. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.; Xue, L.; Chen, Y.; Li, H.; He, Q.; Wang, B.; Meng, F.; Wang, M.; Guan, M.X. A hypertension-associated mitochondrial DNA mutation introduces an m1G37 modification into tRNAMet, altering its structure and function. J. Biol. Chem. 2018, 293, 1425–1438. [Google Scholar] [CrossRef]
- Dai, Z.; Liu, H.; Liao, J.; Huang, C.; Ren, X.; Zhu, W.; Zhu, S.; Peng, B.; Li, S.; Lai, J.; et al. N7-Methylguanosine tRNA modification enhances oncogenic mRNA translation and promotes intrahepatic cholangiocarcinoma progression. Mol. Cell 2021, 81, 3339–3355.e8. [Google Scholar] [CrossRef]
- Orellana, E.A.; Liu, Q.; Yankova, E.; Pirouz, M.; De Braekeleer, E.; Zhang, W.; Lim, J.; Aspris, D.; Sendinc, E.; Garyfallos, D.A.; et al. METTL1-mediated m7G modification of Arg-TCT tRNA drives oncogenic transformation. Mol. Cell 2021, 81, 3323–3338.e14. [Google Scholar] [CrossRef]
- Ma, J.; Han, H.; Huang, Y.; Yang, C.; Zheng, S.; Cai, T.; Bi, J.; Huang, X.; Liu, R.; Huang, L.; et al. METTL1/WDR4-mediated m7G tRNA modifications and m7G codon usage promote mRNA translation and lung cancer progression. Mol. Ther. 2021, 29, 3422–3435. [Google Scholar] [CrossRef]
- Chen, Z.; Zhu, W.; Zhu, S.; Sun, K.; Liao, J.; Liu, H.; Dai, Z.; Han, H.; Ren, X.; Yang, Q.; et al. METTL1 promotes hepatocarcinogenesis via m7G tRNA modification-dependent translation control. Clin. Transl. Med. 2021, 11, e661. [Google Scholar] [CrossRef]
- Ruiz-Arroyo, V.M.; Raj, R.; Babu, K.; Onolbaatar, O.; Roberts, P.H.; Nam, Y. Structures and mechanisms of tRNA methylation by METTL1-WDR4. Nature 2023, 613, 383–390. [Google Scholar] [CrossRef]
- Li, J.; Wang, L.; Hahn, Q.; Nowak, R.P.; Viennet, T.; Orellana, E.A.; Roy Burman, S.S.; Yue, H.; Hunkeler, M.; Fontana, P.; et al. Structural basis of regulated m7G tRNA modification by METTL1-WDR4. Nature 2023, 613, 391–397. [Google Scholar] [CrossRef]
- Li, Q.; Liu, H.; Li, L.; Guo, H.; Xie, Z.; Kong, X.; Xu, J.; Zhang, J.; Chen, Y.; Zhang, Z.; et al. Mettl1-mediated internal m7G methylation of Sptbn2 mRNA elicits neurogenesis and anti-alzheimer’s disease. Cell Biosci. 2023, 13, 183. [Google Scholar] [CrossRef] [PubMed]
- Zhao, P.; Xia, L.; Chen, D.; Xu, W.; Guo, H.; Xu, Y.; Yan, B.; Wu, X.; Li, Y.; Zhang, Y.; et al. METTL1 mediated tRNA m7G modification promotes leukaemogenesis of AML via tRNA regulated translational control. Exp. Hematol. Oncol. 2024, 13, 8. [Google Scholar] [CrossRef] [PubMed]
- Ying, X.; Liu, B.; Yuan, Z.; Huang, Y.; Chen, C.; Jiang, X.; Zhang, H.; Qi, D.; Yang, S.; Lin, S.; et al. METTL1-m7G-EGFR/EFEMP1 axis promotes the bladder cancer development. Clin. Transl. Med. 2021, 11, e675. [Google Scholar] [CrossRef] [PubMed]
- Huang, M.; Long, J.; Yao, Z.; Zhao, Y.; Zhao, Y.; Liao, J.; Lei, K.; Xiao, H.; Dai, Z.; Peng, S.; et al. METTL1-Mediated m7G tRNA Modification Promotes Lenvatinib Resistance in Hepatocellular Carcinoma. Cancer Res. 2023, 83, 89–102. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Vilchez, R.; Anazco-Guenkova, A.M.; Dietmann, S.; Lopez, J.; Moron-Calvente, V.; D’Ambrosi, S.; Nombela, P.; Zamacola, K.; Mendizabal, I.; Garcia-Longarte, S.; et al. METTL1 promotes tumorigenesis through tRNA-derived fragment biogenesis in prostate cancer. Mol. Cancer 2023, 22, 119. [Google Scholar] [CrossRef]
- Luo, Y.; Yao, Y.; Wu, P.; Zi, X.; Sun, N.; He, J. The potential role of N7-methylguanosine (m7G) in cancer. J. Hematol. Oncol. 2022, 15, 63. [Google Scholar] [CrossRef]
- Fu, Y.; Jiang, F.; Zhang, X.; Pan, Y.; Xu, R.; Liang, X.; Wu, X.; Li, X.; Lin, K.; Shi, R.; et al. Perturbation of METTL1-mediated tRNA N7-methylguanosine modification induces senescence and aging. Nat. Commun. 2024, 15, 5713. [Google Scholar] [CrossRef]
- Bohnsack, K.E.; Kleiber, N.; Lemus-Diaz, N.; Bohnsack, M.T. Roles and dynamics of 3-methylcytidine in cellular RNAs. Trends Biochem. Sci. 2022, 47, 596–608. [Google Scholar] [CrossRef]
- Mao, X.L.; Li, Z.H.; Huang, M.H.; Wang, J.T.; Zhou, J.B.; Li, Q.R.; Xu, H.; Wang, X.J.; Zhou, X.L. Mutually exclusive substrate selection strategy by human m3C RNA transferases METTL2A and METTL6. Nucleic Acids Res. 2021, 49, 8309–8323. [Google Scholar] [CrossRef]
- Mao, S.; Haruehanroengra, P.; Ranganathan, S.V.; Shen, F.; Begley, T.J.; Sheng, J. Base Pairing and Functional Insights into N3-Methylcytidine (m3C) in RNA. ACS Chem. Biol. 2021, 16, 76–85. [Google Scholar] [CrossRef]
- Watkins, C.P.; Zhang, W.; Wylder, A.C.; Katanski, C.D.; Pan, T. A multiplex platform for small RNA sequencing elucidates multifaceted tRNA stress response and translational regulation. Nat. Commun. 2022, 13, 2491. [Google Scholar] [CrossRef] [PubMed]
- Cui, J.; Sendinc, E.; Liu, Q.; Kim, S.; Fang, J.Y.; Gregory, R.I. m3C32 tRNA modification controls serine codon-biased mRNA translation, cell cycle, and DNA-damage response. Nat. Commun. 2024, 15, 5775. [Google Scholar] [CrossRef] [PubMed]
- Lentini, J.M.; Alsaif, H.S.; Faqeih, E.; Alkuraya, F.S.; Fu, D. DALRD3 encodes a protein mutated in epileptic encephalopathy that targets arginine tRNAs for 3-methylcytosine modification. Nat. Commun. 2020, 11, 2510. [Google Scholar] [CrossRef] [PubMed]
- Huang, M.H.; Peng, G.X.; Mao, X.L.; Wang, J.T.; Zhou, J.B.; Zhang, J.H.; Chen, M.; Wang, E.D.; Zhou, X.L. Molecular basis for human mitochondrial tRNA m3C modification by alternatively spliced METTL8. Nucleic Acids Res. 2022, 50, 4012–4028. [Google Scholar] [CrossRef]
- Huang, M.H.; Wang, J.T.; Zhang, J.H.; Mao, X.L.; Peng, G.X.; Lin, X.; Lv, D.; Yuan, C.; Lin, H.; Wang, E.D.; et al. Mitochondrial RNA m3C methyltransferase METTL8 relies on an isoform-specific N-terminal extension and modifies multiple heterogenous tRNAs. Sci. Bull. 2023, 68, 2094–2105. [Google Scholar] [CrossRef]
- Lentini, J.M.; Bargabos, R.; Chen, C.; Fu, D. Methyltransferase METTL8 is required for 3-methylcytosine modification in human mitochondrial tRNAs. J. Biol. Chem. 2022, 298, 101788. [Google Scholar] [CrossRef]
- Zhang, F.; Yoon, K.; Zhang, D.Y.; Kim, N.S.; Ming, G.L.; Song, H. Epitranscriptomic regulation of cortical neurogenesis via Mettl8-dependent mitochondrial tRNA m3C modification. Cell Stem Cell 2023, 30, 300–311.e311. [Google Scholar] [CrossRef]
- Lee, B.W.L.; Chuah, Y.H.; Yoon, J.; Grinchuk, O.V.; Liang, Y.; Hirpara, J.L.; Shen, Y.; Wang, L.C.; Lim, Y.T.; Zhao, T.; et al. METTL8 links mt-tRNA m3C modification to the HIF1alpha/RTK/Akt axis to sustain GBM stemness and tumorigenicity. Cell Death Dis. 2024, 15, 338. [Google Scholar] [CrossRef]
- Song, J.; Zhuang, Y.; Zhu, C.; Meng, H.; Lu, B.; Xie, B.; Peng, J.; Li, M.; Yi, C. Differential roles of human PUS10 in miRNA processing and tRNA pseudouridylation. Nat. Chem. Biol. 2020, 16, 160–169. [Google Scholar] [CrossRef]
- Jia, Z.; Meng, F.; Chen, H.; Zhu, G.; Li, X.; He, Y.; Zhang, L.; He, X.; Zhan, H.; Chen, M.; et al. Human TRUB1 is a highly conserved pseudouridine synthase responsible for the formation of Psi55 in mitochondrial tRNAAsn, tRNAGln, tRNAGlu and tRNAPro. Nucleic Acids Res. 2022, 50, 9368–9381. [Google Scholar] [CrossRef]
- Mukhopadhyay, S.; Deogharia, M.; Gupta, R. Mammalian nuclear TRUB1, mitochondrial TRUB2, and cytoplasmic PUS10 produce conserved pseudouridine 55 in different sets of tRNA. RNA 2021, 27, 66–79. [Google Scholar] [CrossRef] [PubMed]
- Borchardt, E.K.; Martinez, N.M.; Gilbert, W.V. Regulation and Function of RNA Pseudouridylation in Human Cells. Annu. Rev. Genet. 2020, 54, 309–336. [Google Scholar] [CrossRef] [PubMed]
- Guzzi, N.; Ciesla, M.; Ngoc, P.C.T.; Lang, S.; Arora, S.; Dimitriou, M.; Pimkova, K.; Sommarin, M.N.E.; Munita, R.; Lubas, M.; et al. Pseudouridylation of tRNA-Derived Fragments Steers Translational Control in Stem Cells. Cell 2018, 173, 1204–1216.e26. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Matuszek, Z.; Huang, Y.; Parisien, M.; Dai, Q.; Clark, W.; Schwartz, M.H.; Pan, T. Queuosine modification protects cognate tRNAs against ribonuclease cleavage. RNA 2018, 24, 1305–1313. [Google Scholar] [CrossRef]
- Muller, M.; Legrand, C.; Tuorto, F.; Kelly, V.P.; Atlasi, Y.; Lyko, F.; Ehrenhofer-Murray, A.E. Queuine links translational control in eukaryotes to a micronutrient from bacteria. Nucleic Acids Res. 2019, 47, 3711–3727. [Google Scholar] [CrossRef]
- Hung, S.H.; Elliott, G.I.; Ramkumar, T.R.; Burtnyak, L.; McGrenaghan, C.J.; Alkuzweny, S.; Quaiyum, S.; Iwata-Reuyl, D.; Pan, X.; Green, B.D.; et al. Structural basis of Qng1-mediated salvage of the micronutrient queuine from queuosine-5′-monophosphate as the biological substrate. Nucleic Acids Res. 2023, 51, 935–951. [Google Scholar] [CrossRef]
- Chen, Y.C.; Brooks, A.F.; Goodenough-Lashua, D.M.; Kittendorf, J.D.; Showalter, H.D.; Garcia, G.A. Evolution of eukaryal tRNA-guanine transglycosylase: Insight gained from the heterocyclic substrate recognition by the wild-type and mutant human and Escherichia coli tRNA-guanine transglycosylases. Nucleic Acids Res. 2011, 39, 2834–2844. [Google Scholar] [CrossRef]
- Kang, M.; Peterson, R.; Feigon, J. Structural Insights into riboswitch control of the biosynthesis of queuosine, a modified nucleotide found in the anticodon of tRNA. Mol. Cell 2009, 33, 784–790. [Google Scholar] [CrossRef]
- Tuorto, F.; Legrand, C.; Cirzi, C.; Federico, G.; Liebers, R.; Muller, M.; Ehrenhofer-Murray, A.E.; Dittmar, G.; Grone, H.J.; Lyko, F. Queuosine-modified tRNAs confer nutritional control of protein translation. EMBO J. 2018, 37, e99777. [Google Scholar] [CrossRef]
- Ehrenhofer-Murray, A.E. Cross-Talk between Dnmt2-Dependent tRNA Methylation and Queuosine Modification. Biomolecules 2017, 7, 14. [Google Scholar] [CrossRef]
- Cirzi, C.; Dyckow, J.; Legrand, C.; Schott, J.; Guo, W.; Perez Hernandez, D.; Hisaoka, M.; Parlato, R.; Pitzer, C.; van der Hoeven, F.; et al. Queuosine-tRNA promotes sex-dependent learning and memory formation by maintaining codon-biased translation elongation speed. EMBO J. 2023, 42, e112507. [Google Scholar] [CrossRef] [PubMed]
- Delaunay, S.; Pascual, G.; Feng, B.; Klann, K.; Behm, M.; Hotz-Wagenblatt, A.; Richter, K.; Zaoui, K.; Herpel, E.; Munch, C.; et al. Mitochondrial RNA modifications shape metabolic plasticity in metastasis. Nature 2022, 607, 593–603. [Google Scholar] [CrossRef] [PubMed]
- m1A tRNA modification facilitates rapid T cell proliferation. Nat. Immunol. 2022, 23, 1408–1409. [CrossRef] [PubMed]
- Chen, C.; Ye, L. The m1A modification of tRNAs: A translational accelerator of T-cell activation. Cell Mol. Immunol. 2022, 19, 1328–1329. [Google Scholar] [CrossRef] [PubMed]
- Richter, U.; Evans, M.E.; Clark, W.C.; Marttinen, P.; Shoubridge, E.A.; Suomalainen, A.; Wredenberg, A.; Wedell, A.; Pan, T.; Battersby, B.J. RNA modification landscape of the human mitochondrial tRNA(Lys) regulates protein synthesis. Nat. Commun. 2018, 9, 3966. [Google Scholar] [CrossRef]
- Kuchino, Y.; Borek, E.; Grunberger, D.; Mushinski, J.F.; Nishimura, S. Changes of post-transcriptional modification of wye base in tumor-specific tRNAPhe. Nucleic Acids Res. 1982, 10, 6421–6432. [Google Scholar] [CrossRef][Green Version]
- Wang, B.; Shi, D.; Yang, S.; Lian, Y.; Li, H.; Cao, M.; He, Y.; Zhang, L.; Qiu, C.; Liu, T.; et al. Mitochondrial tRNA pseudouridylation governs erythropoiesis. Blood 2024, 144, 657–671. [Google Scholar] [CrossRef]
- Liu, B.; Cao, J.; Wang, X.; Guo, C.; Liu, Y.; Wang, T. Deciphering the tRNA-derived small RNAs: Origin, development, and future. Cell Death Dis. 2021, 13, 24. [Google Scholar] [CrossRef]
- Su, Z.; Wilson, B.; Kumar, P.; Dutta, A. Noncanonical Roles of tRNAs: tRNA Fragments and Beyond. Annu. Rev. Genet. 2020, 54, 47–69. [Google Scholar] [CrossRef]
- Kikuchi, Y.; Sasaki, N.; Ando-Yamagami, Y. Cleavage of tRNA within the mature tRNA sequence by the catalytic RNA of RNase P: Implication for the formation of the primer tRNA fragment for reverse transcription in copia retrovirus-like particles. Proc. Natl. Acad. Sci. USA 1990, 87, 8105–8109. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, Y.; Shi, J.; Zhang, H.; Cao, Z.; Gao, X.; Ren, W.; Ning, Y.; Ning, L.; Cao, Y.; et al. Identification and characterization of an ancient class of small RNAs enriched in serum associating with active infection. J. Mol. Cell Biol. 2014, 6, 172–174. [Google Scholar] [CrossRef] [PubMed]
- Peng, H.; Shi, J.; Zhang, Y.; Zhang, H.; Liao, S.; Li, W.; Lei, L.; Han, C.; Ning, L.; Cao, Y.; et al. A novel class of tRNA-derived small RNAs extremely enriched in mature mouse sperm. Cell Res. 2012, 22, 1609–1612. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Zhang, Y.; Tan, D.; Zhang, X.; Yan, M.; Zhang, Y.; Franklin, R.; Shahbazi, M.; Mackinlay, K.; Liu, S.; et al. PANDORA-seq expands the repertoire of regulatory small RNAs by overcoming RNA modifications. Nat. Cell Biol. 2021, 23, 424–436. [Google Scholar] [CrossRef] [PubMed]
- Shen, Z.; Naveed, M.; Bao, J. Untacking small RNA profiling and RNA fragment footprinting: Approaches and challenges in library construction. Wiley Interdiscip. Rev. RNA 2024, 15, e1852. [Google Scholar] [CrossRef] [PubMed]
- Billmeier, M.; Xu, P. Small RNA Profiling by Next-Generation Sequencing Using High-Definition Adapters. Methods Mol. Biol. 2017, 1580, 45–57. [Google Scholar] [CrossRef]
- Slotkin, R.K.; Martienssen, R. Transposable elements and the epigenetic regulation of the genome. Nat. Rev. Genet. 2007, 8, 272–285. [Google Scholar] [CrossRef]
- Jin, D.; Musier-Forsyth, K. Role of host tRNAs and aminoacyl-tRNA synthetases in retroviral replication. J. Biol. Chem. 2019, 294, 5352–5364. [Google Scholar] [CrossRef]
- Zhu, B.; Lee, S.J.; Tan, M.; Wang, E.D.; Richardson, C.C. Gene 5.5 protein of bacteriophage T7 in complex with Escherichia coli nucleoid protein H-NS and transfer RNA masks transfer RNA priming in T7 DNA replication. Proc. Natl. Acad. Sci. USA 2012, 109, 8050–8055. [Google Scholar] [CrossRef]
- Kamhi, E.; Raitskin, O.; Sperling, R.; Sperling, J. A potential role for initiator-tRNA in pre-mRNA splicing regulation. Proc. Natl. Acad. Sci. USA 2010, 107, 11319–11324. [Google Scholar] [CrossRef]
- Rudinger-Thirion, J.; Lescure, A.; Paulus, C.; Frugier, M. Misfolded human tRNA isodecoder binds and neutralizes a 3′ UTR-embedded Alu element. Proc. Natl. Acad. Sci. USA 2011, 108, E794–E802. [Google Scholar] [CrossRef]
- Ronneau, S.; Hallez, R. Make and break the alarmone: Regulation of (p)ppGpp synthetase/hydrolase enzymes in bacteria. FEMS Microbiol. Rev. 2019, 43, 389–400. [Google Scholar] [CrossRef] [PubMed]
- Dong, J.; Qiu, H.; Garcia-Barrio, M.; Anderson, J.; Hinnebusch, A.G. Uncharged tRNA activates GCN2 by displacing the protein kinase moiety from a bipartite tRNA-binding domain. Mol. Cell 2000, 6, 269–279. [Google Scholar] [CrossRef] [PubMed]
- Mei, Y.; Yong, J.; Liu, H.; Shi, Y.; Meinkoth, J.; Dreyfuss, G.; Yang, X. tRNA binds to cytochrome c and inhibits caspase activation. Mol. Cell 2010, 37, 668–678. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.T.; Strugatsky, D.; Liu, W.; Zhou, Z.H. Structure of human cytomegalovirus virion reveals host tRNA binding to capsid-associated tegument protein pp150. Nat. Commun. 2021, 12, 5513. [Google Scholar] [CrossRef]
- Li, M.; Kao, E.; Malone, D.; Gao, X.; Wang, J.Y.J.; David, M. DNA damage-induced cell death relies on SLFN11-dependent cleavage of distinct type II tRNAs. Nat. Struct. Mol. Biol. 2018, 25, 1047–1058. [Google Scholar] [CrossRef]
- Yang, J.Y.; Deng, X.Y.; Li, Y.S.; Ma, X.C.; Feng, J.X.; Yu, B.; Chen, Y.; Luo, Y.L.; Wang, X.; Chen, M.L.; et al. Structure of Schlafen13 reveals a new class of tRNA/rRNA- targeting RNase engaged in translational control. Nat. Commun. 2018, 9, 1165. [Google Scholar] [CrossRef]
- Katibah, G.E.; Lee, H.J.; Huizar, J.P.; Vogan, J.M.; Alber, T.; Collins, K. tRNA binding, structure, and localization of the human interferon-induced protein IFIT5. Mol. Cell 2013, 49, 743–750. [Google Scholar] [CrossRef]
- Keller, P.; Freund, I.; Marchand, V.; Bec, G.; Huang, R.; Motorin, Y.; Eigenbrod, T.; Dalpke, A.; Helm, M. Double methylation of tRNA-U54 to 2′-O-methylthymidine (Tm) synergistically decreases immune response by Toll-like receptor 7. Nucleic Acids Res. 2018, 46, 9764–9775. [Google Scholar] [CrossRef]
- Schorn, A.J.; Gutbrod, M.J.; LeBlanc, C.; Martienssen, R. LTR-Retrotransposon Control by tRNA-Derived Small RNAs. Cell 2017, 170, 61–71.e11. [Google Scholar] [CrossRef]
- Kim, H.K.; Fuchs, G.; Wang, S.; Wei, W.; Zhang, Y.; Park, H.; Roy-Chaudhuri, B.; Li, P.; Xu, J.; Chu, K.; et al. A transfer-RNA-derived small RNA regulates ribosome biogenesis. Nature 2017, 552, 57–62. [Google Scholar] [CrossRef]
- Di Fazio, A.; Schlackow, M.; Pong, S.K.; Alagia, A.; Gullerova, M. Dicer dependent tRNA derived small RNAs promote nascent RNA silencing. Nucleic Acids Res. 2022, 50, 1734–1752. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Hannon, G.J. MicroRNAs: Small RNAs with a big role in gene regulation. Nat. Rev. Genet. 2004, 5, 522–531. [Google Scholar] [CrossRef] [PubMed]
- Maute, R.L.; Schneider, C.; Sumazin, P.; Holmes, A.; Califano, A.; Basso, K.; Dalla-Favera, R. tRNA-derived microRNA modulates proliferation and the DNA damage response and is down-regulated in B cell lymphoma. Proc. Natl. Acad. Sci. USA 2013, 110, 1404–1409. [Google Scholar] [CrossRef] [PubMed]
- Luo, S.; He, F.; Luo, J.; Dou, S.; Wang, Y.; Guo, A.; Lu, J. Drosophila tsRNAs preferentially suppress general translation machinery via antisense pairing and participate in cellular starvation response. Nucleic Acids Res. 2018, 46, 5250–5268. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Zhou, T. Emerging functional principles of tRNA-derived small RNAs and other regulatory small RNAs. J. Biol. Chem. 2023, 299, 105225. [Google Scholar] [CrossRef]
- Couvillion, M.T.; Bounova, G.; Purdom, E.; Speed, T.P.; Collins, K. A Tetrahymena Piwi bound to mature tRNA 3′ fragments activates the exonuclease Xrn2 for RNA processing in the nucleus. Mol. Cell 2012, 48, 509–520. [Google Scholar] [CrossRef]
- Couvillion, M.T.; Sachidanandam, R.; Collins, K. A growth-essential Tetrahymena Piwi protein carries tRNA fragment cargo. Genes Dev. 2010, 24, 2742–2747. [Google Scholar] [CrossRef]
- Lyons, S.M.; Gudanis, D.; Coyne, S.M.; Gdaniec, Z.; Ivanov, P. Identification of functional tetramolecular RNA G-quadruplexes derived from transfer RNAs. Nat. Commun. 2017, 8, 1127. [Google Scholar] [CrossRef]
- Goodarzi, H.; Liu, X.; Nguyen, H.C.; Zhang, S.; Fish, L.; Tavazoie, S.F. Endogenous tRNA-Derived Fragments Suppress Breast Cancer Progression via YBX1 Displacement. Cell 2015, 161, 790–802. [Google Scholar] [CrossRef]
- Bornelov, S.; Selmi, T.; Flad, S.; Dietmann, S.; Frye, M. Codon usage optimization in pluripotent embryonic stem cells. Genome Biol. 2019, 20, 119. [Google Scholar] [CrossRef]
- Aharon-Hefetz, N.; Frumkin, I.; Mayshar, Y.; Dahan, O.; Pilpel, Y.; Rak, R. Manipulation of the human tRNA pool reveals distinct tRNA sets that act in cellular proliferation or cell cycle arrest. eLife 2020, 9, e58461. [Google Scholar] [CrossRef] [PubMed]
- Rappol, T.; Waldl, M.; Chugunova, A.; Hofacker, I.L.; Pauli, A.; Vilardo, E. tRNA expression and modification landscapes, and their dynamics during zebrafish embryo development. Nucleic Acids Res. 2024, 52, 10575–10594. [Google Scholar] [CrossRef] [PubMed]
- Murakami, Y.; Wei, F.Y.; Kawamura, Y.; Horiguchi, H.; Kadomatsu, T.; Miyata, K.; Miura, K.; Oike, Y.; Ando, Y.; Ueda, M.; et al. NSUN3-mediated mitochondrial tRNA 5-formylcytidine modification is essential for embryonic development and respiratory complexes in mice. Commun. Biol. 2023, 6, 307. [Google Scholar] [CrossRef] [PubMed]
- Yoo, H.; Son, D.; Jang, Y.J.; Hong, K. Indispensable role for mouse ELP3 in embryonic stem cell maintenance and early development. Biochem. Biophys. Res. Commun. 2016, 478, 631–636. [Google Scholar] [CrossRef]
- Yang, Q.; Lin, J.; Liu, M.; Li, R.; Tian, B.; Zhang, X.; Xu, B.; Liu, M.; Zhang, X.; Li, Y.; et al. Highly sensitive sequencing reveals dynamic modifications and activities of small RNAs in mouse oocytes and early embryos. Sci. Adv. 2016, 2, e1501482. [Google Scholar] [CrossRef]
- Fan, Y.; Pavani, K.C.; Smits, K.; Van Soom, A.; Peelman, L. tRNA(Glu)-derived fragments from embryonic extracellular vesicles modulate bovine embryo hatching. J. Anim. Sci. Biotechnol. 2024, 15, 23. [Google Scholar] [CrossRef]
- Krishna, S.; Yim, D.G.; Lakshmanan, V.; Tirumalai, V.; Koh, J.L.; Park, J.E.; Cheong, J.K.; Low, J.L.; Lim, M.J.; Sze, S.K.; et al. Dynamic expression of tRNA-derived small RNAs define cellular states. EMBO Rep. 2019, 20, e47789. [Google Scholar] [CrossRef]
- Chen, Q. Sperm RNA-mediated epigenetic inheritance in mammals: Challenges and opportunities. Reprod. Fertil. Dev. 2022, 35, 118–124. [Google Scholar] [CrossRef]
- Chen, Q.; Yan, M.; Cao, Z.; Li, X.; Zhang, Y.; Shi, J.; Feng, G.H.; Peng, H.; Zhang, X.; Zhang, Y.; et al. Sperm tsRNAs contribute to intergenerational inheritance of an acquired metabolic disorder. Science 2016, 351, 397–400. [Google Scholar] [CrossRef]
- Cai, C.; Chen, Q. Father’s diet influences son’s metabolic health through sperm RNA. Nature 2024, 630, 571–573. [Google Scholar] [CrossRef]
- Zhang, Y.; Chen, Q. Human sperm RNA code senses dietary sugar. Nat. Rev. Endocrinol. 2020, 16, 200–201. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Bai, D.; Liu, W.; Liu, Y.; Zhang, Y.; Kou, X.; Chen, J.; Wang, H.; Teng, X.; Zuo, J.; et al. Altered sperm tsRNAs in aged male contribute to anxiety-like behavior in offspring. Aging Cell 2021, 20, e13466. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.; Xia, L.; Wang, W.; Xu, W.; Shen, X.; Wu, X.; He, T.; Jiang, X.; Xu, Y.; Zhao, P.; et al. Hypoxia induces alterations in tRNA modifications involved in translational control. BMC Biol. 2023, 21, 39. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Li, J.; Liu, J.C.; Zeng, Q.; Yeung, W.S.; Chiu, P.C.; Duan, Y.G. Alterations of small non-coding RNA in the spermatozoa of mice with paternal experimental autoimmune epididymo-orchitis are associated with metabolic dysfunction in offspring. Andrology 2023, 1–11. [Google Scholar] [CrossRef]
- Argaw-Denboba, A.; Schmidt, T.S.B.; Di Giacomo, M.; Ranjan, B.; Devendran, S.; Mastrorilli, E.; Lloyd, C.T.; Pugliese, D.; Paribeni, V.; Dabin, J.; et al. Paternal microbiome perturbations impact offspring fitness. Nature 2024, 629, 652–659. [Google Scholar] [CrossRef]
- Natt, D.; Kugelberg, U.; Casas, E.; Nedstrand, E.; Zalavary, S.; Henriksson, P.; Nijm, C.; Jaderquist, J.; Sandborg, J.; Flinke, E.; et al. Human sperm displays rapid responses to diet. PLoS Biol. 2019, 17, e3000559. [Google Scholar] [CrossRef]
- He, T.; Guo, H.; Shen, X.; Wu, X.; Xia, L.; Jiang, X.; Xu, Y.; Chen, D.; Zhang, Y.; Tan, D.; et al. Hypoxia-induced alteration of RNA modifications in the mouse testis and spermdagger. Biol. Reprod. 2021, 105, 1171–1178. [Google Scholar] [CrossRef]
- Burgess, R.W.; Storkebaum, E. tRNA Dysregulation in Neurodevelopmental and Neurodegenerative Diseases. Annu. Rev. Cell Dev. Biol. 2023, 39, 223–252. [Google Scholar] [CrossRef]
- Patton, J.R.; Bykhovskaya, Y.; Mengesha, E.; Bertolotto, C.; Fischel-Ghodsian, N. Mitochondrial myopathy and sideroblastic anemia (MLASA): Missense mutation in the pseudouridine synthase 1 (PUS1) gene is associated with the loss of tRNA pseudouridylation. J. Biol. Chem. 2005, 280, 19823–19828. [Google Scholar] [CrossRef]
- Ren, D.; Mo, Y.; Yang, M.; Wang, D.; Wang, Y.; Yan, Q.; Guo, C.; Xiong, W.; Wang, F.; Zeng, Z. Emerging roles of tRNA in cancer. Cancer Lett. 2023, 563, 216170. [Google Scholar] [CrossRef]
- Ren, B.; Guan, M.X.; Zhou, T.; Cai, X.; Shan, G. Emerging functions of mitochondria-encoded noncoding RNAs. Trends Genet. 2023, 39, 125–139. [Google Scholar] [CrossRef] [PubMed]
- Orellana, E.A.; Siegal, E.; Gregory, R.I. tRNA dysregulation and disease. Nat. Rev. Genet. 2022, 23, 651–664. [Google Scholar] [CrossRef] [PubMed]
- Turvey, A.K.; Horvath, G.A.; Cavalcanti, A.R.O. Aminoacyl-tRNA synthetases in human health and disease. Front. Physiol. 2022, 13, 1029218. [Google Scholar] [CrossRef]
- Chomyn, A.; Enriquez, J.A.; Micol, V.; Fernandez-Silva, P.; Attardi, G. The mitochondrial myopathy, encephalopathy, lactic acidosis, and stroke-like episode syndrome-associated human mitochondrial tRNALeu(UUR) mutation causes aminoacylation deficiency and concomitant reduced association of mRNA with ribosomes. J. Biol. Chem. 2000, 275, 19198–19209. [Google Scholar] [CrossRef] [PubMed]
- Hogg, M.C.; Raoof, R.; El Naggar, H.; Monsefi, N.; Delanty, N.; O’Brien, D.F.; Bauer, S.; Rosenow, F.; Henshall, D.C.; Prehn, J.H. Elevation in plasma tRNA fragments precede seizures in human epilepsy. J. Clin. Investig. 2019, 129, 2946–2951. [Google Scholar] [CrossRef]
- Karousi, P.; Adamopoulos, P.G.; Papageorgiou, S.G.; Pappa, V.; Scorilas, A.; Kontos, C.K. A novel, mitochondrial, internal tRNA-derived RNA fragment possesses clinical utility as a molecular prognostic biomarker in chronic lymphocytic leukemia. Clin. Biochem. 2020, 85, 20–26. [Google Scholar] [CrossRef]
- Gu, W.; Shi, J.; Liu, H.; Zhang, X.; Zhou, J.J.; Li, M.; Zhou, D.; Li, R.; Lv, J.; Wen, G.; et al. Peripheral blood non-canonical small non-coding RNAs as novel biomarkers in lung cancer. Mol. Cancer 2020, 19, 159. [Google Scholar] [CrossRef]
- Wang, J.; Ma, G.; Ge, H.; Han, X.; Mao, X.; Wang, X.; Veeramootoo, J.S.; Xia, T.; Liu, X.; Wang, S. Circulating tRNA-derived small RNAs (tsRNAs) signature for the diagnosis and prognosis of breast cancer. NPJ Breast Cancer 2021, 7, 4. [Google Scholar] [CrossRef]
- Li, D.; Gao, X.; Ma, X.; Wang, M.; Cheng, C.; Xue, T.; Gao, F.; Shen, Y.; Zhang, J.; Liu, Q. Aging-induced tRNA(Glu)-derived fragment impairs glutamate biosynthesis by targeting mitochondrial translation-dependent cristae organization. Cell Metab. 2024, 36, 1059–1075.e9. [Google Scholar] [CrossRef]
- Buvoli, M.; Buvoli, A.; Leinwand, L.A. Suppression of nonsense mutations in cell culture and mice by multimerized suppressor tRNA genes. Mol. Cell Biol. 2000, 20, 3116–3124. [Google Scholar] [CrossRef]
- Bordeira-Carrico, R.; Ferreira, D.; Mateus, D.D.; Pinheiro, H.; Pego, A.P.; Santos, M.A.; Oliveira, C. Rescue of wild-type E-cadherin expression from nonsense-mutated cancer cells by a suppressor-tRNA. Eur. J. Hum. Genet. 2014, 22, 1085–1092. [Google Scholar] [CrossRef] [PubMed]
- Lueck, J.D.; Yoon, J.S.; Perales-Puchalt, A.; Mackey, A.L.; Infield, D.T.; Behlke, M.A.; Pope, M.R.; Weiner, D.B.; Skach, W.R.; McCray, P.B., Jr.; et al. Engineered transfer RNAs for suppression of premature termination codons. Nat. Commun. 2019, 10, 822. [Google Scholar] [CrossRef] [PubMed]
- Zuko, A.; Mallik, M.; Thompson, R.; Spaulding, E.L.; Wienand, A.R.; Been, M.; Tadenev, A.L.D.; van Bakel, N.; Sijlmans, C.; Santos, L.A.; et al. tRNA overexpression rescues peripheral neuropathy caused by mutations in tRNA synthetase. Science 2021, 373, 1161–1166. [Google Scholar] [CrossRef] [PubMed]
- Chan, P.P.; Lowe, T.M. GtRNAdb 2.0: An expanded database of transfer RNA genes identified in complete and draft genomes. Nucleic Acids Res. 2016, 44, D184–D189. [Google Scholar] [CrossRef]
- Pang, Y.L.; Abo, R.; Levine, S.S.; Dedon, P.C. Diverse cell stresses induce unique patterns of tRNA up- and down-regulation: tRNA-seq for quantifying changes in tRNA copy number. Nucleic Acids Res. 2014, 42, e170. [Google Scholar] [CrossRef]
- Phizicky, E.M.; Hopper, A.K. The life and times of a tRNA. RNA 2023, 29, 898–957. [Google Scholar] [CrossRef]
- Krutyholowa, R.; Zakrzewski, K.; Glatt, S. Charging the code—tRNA modification complexes. Curr. Opin. Struct. Biol. 2019, 55, 138–146. [Google Scholar] [CrossRef]
- Han, L.; Phizicky, E.M. A rationale for tRNA modification circuits in the anticodon loop. RNA 2018, 24, 1277–1284. [Google Scholar] [CrossRef]
- Hernandez-Alias, X.; Katanski, C.D.; Zhang, W.; Assari, M.; Watkins, C.P.; Schaefer, M.H.; Serrano, L.; Pan, T. Single-read tRNA-seq analysis reveals coordination of tRNA modification and aminoacylation and fragmentation. Nucleic Acids Res. 2023, 51, e17. [Google Scholar] [CrossRef]
- Chen, Q.; Zhang, X.; Shi, J.; Yan, M.; Zhou, T. Origins and evolving functionalities of tRNA-derived small RNAs. Trends Biochem. Sci. 2021, 46, 790–804. [Google Scholar] [CrossRef]
- Padhiar, N.H.; Katneni, U.; Komar, A.A.; Motorin, Y.; Kimchi-Sarfaty, C. Advances in methods for tRNA sequencing and quantification. Trends Genet. 2024, 40, 276–290. [Google Scholar] [CrossRef] [PubMed]
- Lucas, M.C.; Pryszcz, L.P.; Medina, R.; Milenkovic, I.; Camacho, N.; Marchand, V.; Motorin, Y.; Ribas de Pouplana, L.; Novoa, E.M. Quantitative analysis of tRNA abundance and modifications by nanopore RNA sequencing. Nat. Biotechnol. 2024, 42, 72–86. [Google Scholar] [CrossRef] [PubMed]
- Coller, J.; Ignatova, Z. tRNA therapeutics for genetic diseases. Nat. Rev. Drug Discov. 2024, 23, 108–125. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Y.; Yu, Z.; Jiang, W.; Lyu, X.; Guo, A.; Sun, X.; Yang, Y.; Zhang, Y. tRNA and tsRNA: From Heterogeneity to Multifaceted Regulators. Biomolecules 2024, 14, 1340. https://doi.org/10.3390/biom14101340
Li Y, Yu Z, Jiang W, Lyu X, Guo A, Sun X, Yang Y, Zhang Y. tRNA and tsRNA: From Heterogeneity to Multifaceted Regulators. Biomolecules. 2024; 14(10):1340. https://doi.org/10.3390/biom14101340
Chicago/Turabian StyleLi, Yun, Zongyu Yu, Wenlin Jiang, Xinyi Lyu, Ailian Guo, Xiaorui Sun, Yiting Yang, and Yunfang Zhang. 2024. "tRNA and tsRNA: From Heterogeneity to Multifaceted Regulators" Biomolecules 14, no. 10: 1340. https://doi.org/10.3390/biom14101340
APA StyleLi, Y., Yu, Z., Jiang, W., Lyu, X., Guo, A., Sun, X., Yang, Y., & Zhang, Y. (2024). tRNA and tsRNA: From Heterogeneity to Multifaceted Regulators. Biomolecules, 14(10), 1340. https://doi.org/10.3390/biom14101340







