Leveraging Biomaterial Platforms to Study Aging-Related Neural and Muscular Degeneration
Abstract
1. Introduction
2. Biomaterials as Models of the Aging Extracellular Environment
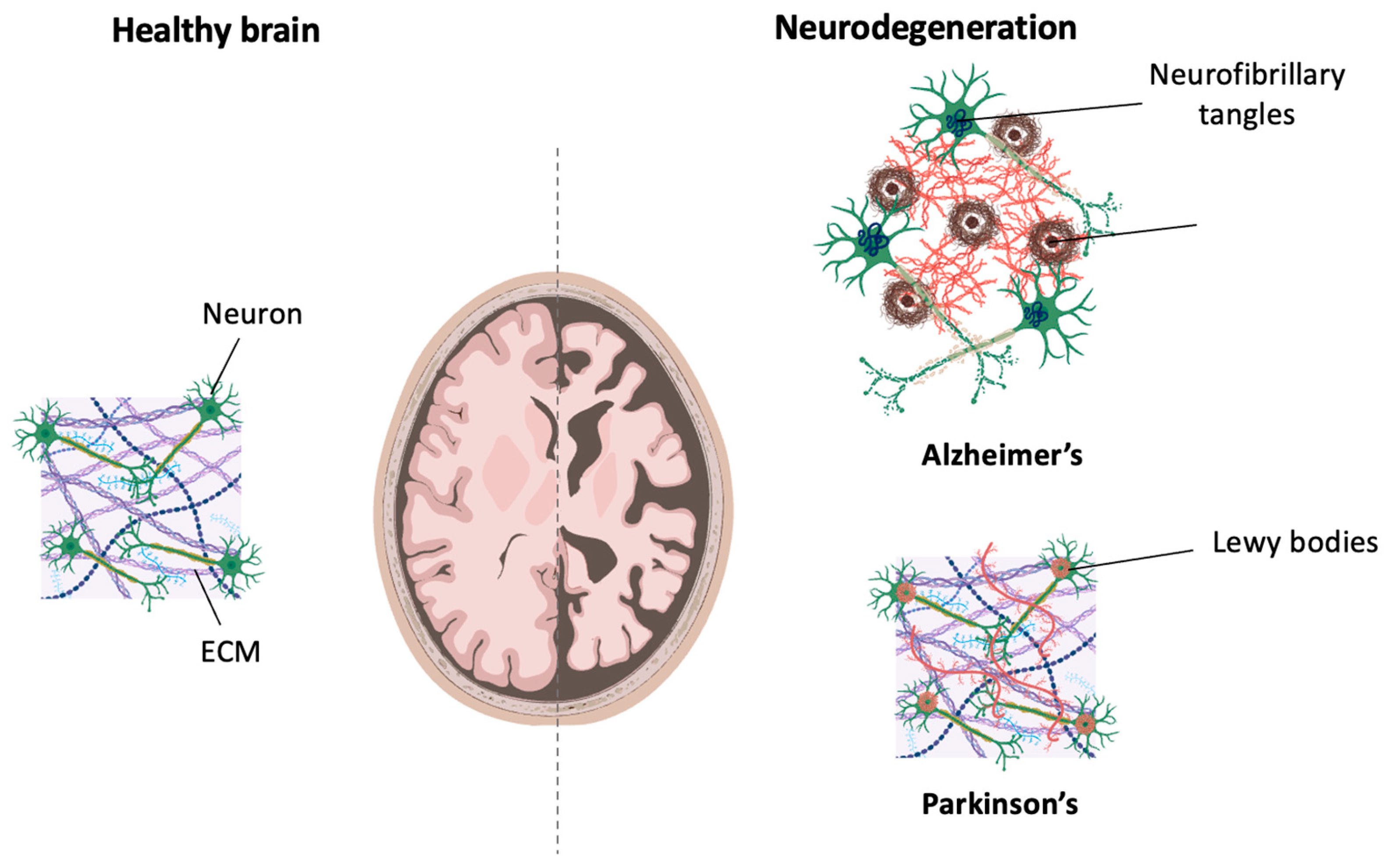
3. Modeling Neurodegeneration
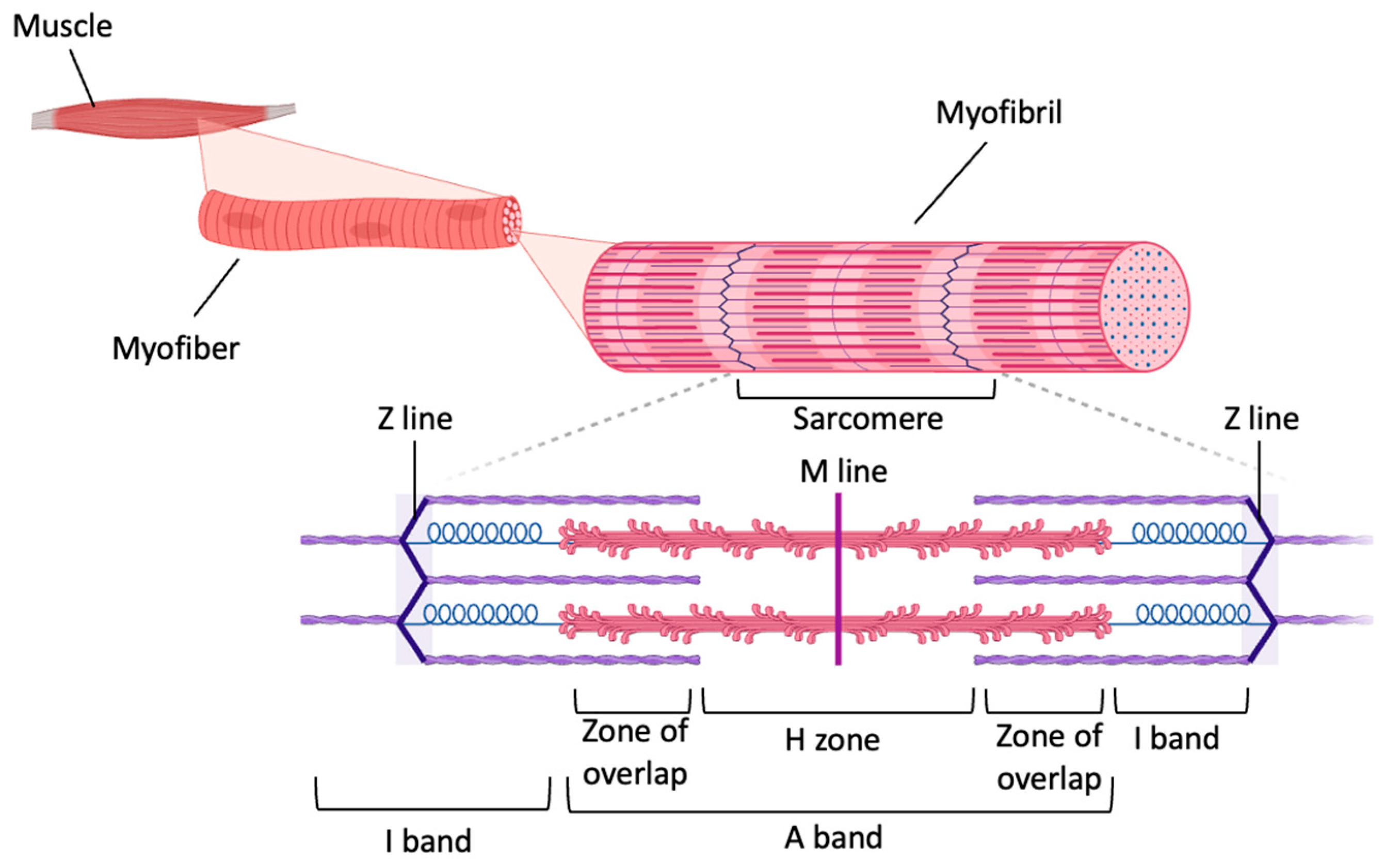
4. Modeling Aging-Related Muscle Degeneration
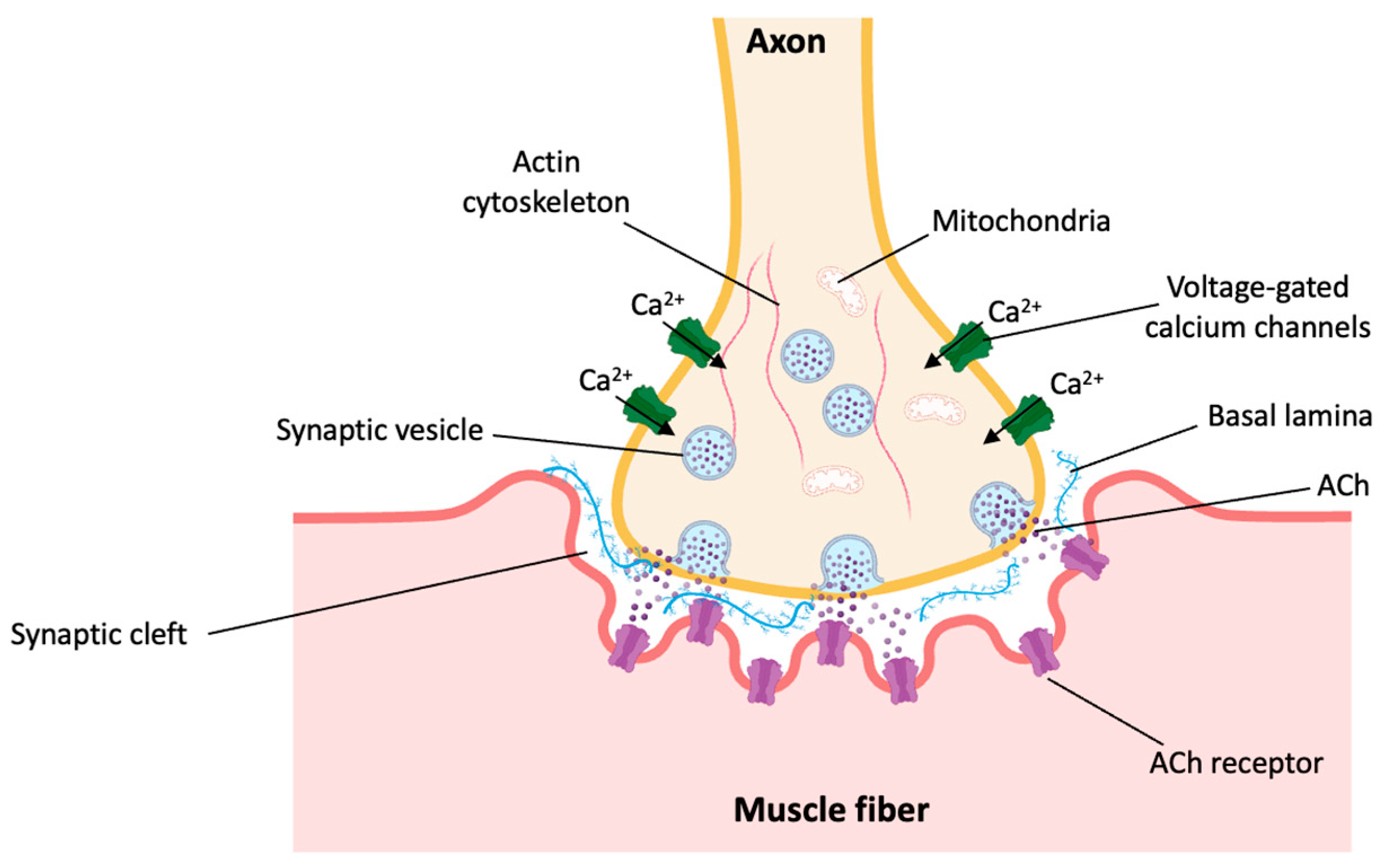
5. Modeling Aging-Related Neuromuscular Junction Degeneration
6. Conclusions and Future Directions
Author Contributions
Funding
Conflicts of Interest
References
- Maldonado, E.; Morales-Pison, S.; Urbina, F.; Solari, A. Aging Hallmarks and the Role of Oxidative Stress. Antioxidants 2023, 12, 651. [Google Scholar] [CrossRef] [PubMed]
- Goh, J.; Wong, E.; Soh, J.; Maier, A.B.; Kennedy, B.K. Targeting the Molecular & Cellular Pillars of Human Aging with Exercise. FEBS J. 2023, 290, 649–668. [Google Scholar] [CrossRef] [PubMed]
- Volpi, E.; Nazemi, R.; Fujita, S. Muscle Tissue Changes with Aging. Curr. Opin. Clin. Nutr. Metab. Care 2004, 7, 405–410. [Google Scholar] [CrossRef] [PubMed]
- Naruse, M.; Trappe, S.; Trappe, T.A. Human Skeletal Muscle-Specific Atrophy with Aging: A Comprehensive Review. J. Appl. Physiol. 2023, 134, 900–914. [Google Scholar] [CrossRef] [PubMed]
- Wilson, R.S.; Leurgans, S.E.; Boyle, P.A.; Schneider, J.A.; Bennett, D.A. Neurodegenerative Basis of Age-Related Cognitive Decline. Neurology 2010, 75, 1070–1078. [Google Scholar] [CrossRef] [PubMed]
- Trojsi, F.; Christidi, F.; Migliaccio, R.; Santamaría-García, H.; Santangelo, G. Behavioural and Cognitive Changes in Neurodegenerative Diseases and Brain Injury. Behav. Neurol. 2018, 2018, 4935915. [Google Scholar] [CrossRef] [PubMed]
- Smith, M.; Barker, R.; Williams, G.; Carr, J.; Gunnarsson, R. The Effect of Exercise on High-Level Mobility in Individuals with Neurodegenerative Disease: A Systematic Literature Review. Physiotherapy 2020, 106, 174–193. [Google Scholar] [CrossRef]
- Lamptey, R.N.L.; Chaulagain, B.; Trivedi, R.; Gothwal, A.; Layek, B.; Singh, J. A Review of the Common Neurodegenerative Disorders: Current Therapeutic Approaches and the Potential Role of Nanotherapeutics. Int. J. Mol. Sci. 2022, 23, 1851. [Google Scholar] [CrossRef]
- Hebisch, M.; Klostermeier, S.; Wolf, K.; Boccaccini, A.R.; Wolf, S.E.; Tanzi, R.E.; Kim, D.Y. The Impact of the Cellular Environment and Aging on Modeling Alzheimer’s Disease in 3D Cell Culture Models. Adv. Sci. 2023, 10, 2205037. [Google Scholar] [CrossRef]
- Irvine, G.B.; El-Agnaf, O.M.; Shankar, G.M.; Walsh, D.M. Protein Aggregation in the Brain: The Molecular Basis for Alzheimer’s and Parkinson’s Diseases. Mol. Med. 2008, 14, 451–464. [Google Scholar] [CrossRef]
- Preston, A.R.; Eichenbaum, H. Interplay of Hippocampus and Prefrontal Cortex in Memory. Curr. Biol. 2013, 23, R764–R773. [Google Scholar] [CrossRef] [PubMed]
- Wakabayashi, K.; Tanji, K.; Odagiri, S.; Miki, Y.; Mori, F.; Takahashi, H. The Lewy Body in Parkinson’s Disease and Related Neurodegenerative Disorders. Mol. Neurobiol. 2013, 47, 495–508. [Google Scholar] [CrossRef]
- Rike, W.A.; Stern, S. Proteins and Transcriptional Dysregulation of the Brain Extracellular Matrix in Parkinson’s Disease: A Systematic Review. Int. J. Mol. Sci. 2023, 24, 7435. [Google Scholar] [CrossRef] [PubMed]
- El-Agnaf, O.M.A.; Salem, S.A.; Paleologou, K.E.; Curran, M.D.; Gibson, M.J.; Court, J.A.; Schlossmacher, M.G.; Allsop, D. Detection of Oligomeric Forms of α-synuclein Protein in Human Plasma as a Potential Biomarker for Parkinson’s Disease. FASEB J. 2006, 20, 419–425. [Google Scholar] [CrossRef] [PubMed]
- Jang, Y.C.; Van Remmen, H. Age-Associated Alterations of the Neuromuscular Junction. Exp. Gerontol. 2011, 46, 193–198. [Google Scholar] [CrossRef] [PubMed]
- Colomar, A.; Robitaille, R. Glial Modulation of Synaptic Transmission at the Neuromuscular Junction. Glia 2004, 47, 284–289. [Google Scholar] [CrossRef]
- Rodríguez Cruz, P.M.; Cossins, J.; Beeson, D.; Vincent, A. The Neuromuscular Junction in Health and Disease: Molecular Mechanisms Governing Synaptic Formation and Homeostasis. Front. Mol. Neurosci. 2020, 13, 610964. [Google Scholar] [CrossRef]
- Slater, C. The Structure of Human Neuromuscular Junctions: Some Unanswered Molecular Questions. Int. J. Mol. Sci. 2017, 18, 2183. [Google Scholar] [CrossRef]
- Gonzalez-Freire, M.; De Cabo, R.; Studenski, S.A.; Ferrucci, L. The Neuromuscular Junction: Aging at the Crossroad between Nerves and Muscle. Front. Aging Neurosci. 2014, 6, 208. [Google Scholar] [CrossRef]
- Heikkinen, A.; Härönen, H.; Norman, O.; Pihlajaniemi, T. Collagen XIII and Other ECM Components in the Assembly and Disease of the Neuromuscular Junction. Anat. Rec. 2020, 303, 1653–1663. [Google Scholar] [CrossRef]
- Chen, W.-J.; Lin, I.-H.; Lee, C.-W.; Chen, Y.-F. Aged Skeletal Muscle Retains the Ability to Remodel Extracellular Matrix for Degradation of Collagen Deposition after Muscle Injury. Int. J. Mol. Sci. 2021, 22, 2123. [Google Scholar] [CrossRef] [PubMed]
- Gerou, M.; Hall, B.; Woof, R.; Allsop, J.; Kolb, S.J.; Meyer, K.; Shaw, P.J.; Allen, S.P. Amyotrophic Lateral Sclerosis Alters the Metabolic Aging Profile in Patient Derived Fibroblasts. Neurobiol. Aging 2021, 105, 64–77. [Google Scholar] [CrossRef] [PubMed]
- Cappello, V.; Francolini, M. Neuromuscular Junction Dismantling in Amyotrophic Lateral Sclerosis. Int. J. Mol. Sci. 2017, 18, 2092. [Google Scholar] [CrossRef] [PubMed]
- Birch, H.L. Extracellular Matrix and Ageing. In Biochemistry and Cell Biology of Ageing: Part I Biomedical Science; Harris, J.R., Korolchuk, V.I., Eds.; Subcellular Biochemistry; Springer: Singapore, 2018; Volume 90, pp. 169–190. ISBN 9789811328343. [Google Scholar]
- Natale, B.V.; Kotadia, R.; Gustin, K.; Harihara, A.; Min, S.; Kreisman, M.J.; Breen, K.M.; Natale, D.R.C. Extracellular Matrix Influences Gene Expression and Differentiation of Mouse Trophoblast Stem Cells. Stem Cells Dev. 2023, 32, 622–637. [Google Scholar] [CrossRef] [PubMed]
- Morawski, M.; Filippov, M.; Tzinia, A.; Tsilibary, E.; Vargova, L. ECM in Brain Aging and Dementia. In Progress in Brain Research; Elsevier: Amsterdam, The Netherlands, 2014; Volume 214, pp. 207–227. ISBN 978-0-444-63486-3. [Google Scholar]
- Yang, L.; Wei, M.; Xing, B.; Zhang, C. Extracellular Matrix and Synapse Formation. Biosci. Rep. 2023, 43, BSR20212411. [Google Scholar] [CrossRef] [PubMed]
- Downs, M.; Sethi, M.K.; Raghunathan, R.; Layne, M.D.; Zaia, J. Matrisome Changes in Parkinson’s Disease. Anal. Bioanal. Chem. 2022, 414, 3005–3015. [Google Scholar] [CrossRef] [PubMed]
- Samuel, M.A.; Valdez, G.; Tapia, J.C.; Lichtman, J.W.; Sanes, J.R. Agrin and Synaptic Laminin Are Required to Maintain Adult Neuromuscular Junctions. PLoS ONE 2012, 7, e46663. [Google Scholar] [CrossRef]
- Stearns-Reider, K.M.; D’Amore, A.; Beezhold, K.; Rothrauff, B.; Cavalli, L.; Wagner, W.R.; Vorp, D.A.; Tsamis, A.; Shinde, S.; Zhang, C.; et al. Aging of the Skeletal Muscle Extracellular Matrix Drives a Stem Cell Fibrogenic Conversion. Aging Cell 2017, 16, 518–528. [Google Scholar] [CrossRef]
- Mann, C.J.; Perdiguero, E.; Kharraz, Y.; Aguilar, S.; Pessina, P.; Serrano, A.L.; Muñoz-Cánoves, P. Aberrant Repair and Fibrosis Development in Skeletal Muscle. Skelet. Muscle 2011, 1, 21. [Google Scholar] [CrossRef]
- Pavan, P.; Monti, E.; Bondí, M.; Fan, C.; Stecco, C.; Narici, M.; Reggiani, C.; Marcucci, L. Alterations of Extracellular Matrix Mechanical Properties Contribute to Age-Related Functional Impairment of Human Skeletal Muscles. Int. J. Mol. Sci. 2020, 21, 3992. [Google Scholar] [CrossRef]
- Edmondson, R.; Broglie, J.J.; Adcock, A.F.; Yang, L. Three-Dimensional Cell Culture Systems and Their Applications in Drug Discovery and Cell-Based Biosensors. ASSAY Drug Dev. Technol. 2014, 12, 207–218. [Google Scholar] [CrossRef] [PubMed]
- Antoni, D.; Burckel, H.; Josset, E.; Noel, G. Three-Dimensional Cell Culture: A Breakthrough in Vivo. Int. J. Mol. Sci. 2015, 16, 5517–5527. [Google Scholar] [CrossRef] [PubMed]
- Alemany-Ribes, M.; Semino, C.E. Bioengineering 3D Environments for Cancer Models. Adv. Drug Deliv. Rev. 2014, 79–80, 40–49. [Google Scholar] [CrossRef] [PubMed]
- Moysidou, C.-M.; Barberio, C.; Owens, R.M. Advances in Engineering Human Tissue Models. Front. Bioeng. Biotechnol. 2021, 8, 620962. [Google Scholar] [CrossRef]
- Kajtez, J.; Nilsson, F.; Fiorenzano, A.; Parmar, M.; Emnéus, J. 3D Biomaterial Models of Human Brain Disease. Neurochem. Int. 2021, 147, 105043. [Google Scholar] [CrossRef] [PubMed]
- Tibbitt, M.W.; Anseth, K.S. Hydrogels as Extracellular Matrix Mimics for 3D Cell Culture. Biotechnol. Bioeng. 2009, 103, 655–663. [Google Scholar] [CrossRef] [PubMed]
- Hidalgo-Alvarez, V.; Dhowre, H.S.; Kingston, O.A.; Sheridan, C.M.; Levis, H.J. Biofabrication of Artificial Stem Cell Niches in the Anterior Ocular Segment. Bioengineering 2021, 8, 135. [Google Scholar] [CrossRef] [PubMed]
- Park, Y.; Huh, K.M.; Kang, S.-W. Applications of Biomaterials in 3D Cell Culture and Contributions of 3D Cell Culture to Drug Development and Basic Biomedical Research. Int. J. Mol. Sci. 2021, 22, 2491. [Google Scholar] [CrossRef]
- Madl, C.M. Accelerating Aging with Dynamic Biomaterials: Recapitulating Aged Tissue Phenotypes in Engineered Platforms. iScience 2023, 26, 106825. [Google Scholar] [CrossRef]
- Caprio, N.D.; Burdick, J.A. Engineered Biomaterials to Guide Spheroid Formation, Function, and Fabrication into 3D Tissue Constructs. Acta Biomater. 2023, 165, 4–18. [Google Scholar] [CrossRef]
- Teli, P.; Kale, V.; Vaidya, A. Beyond Animal Models: Revolutionizing Neurodegenerative Disease Modeling Using 3D in Vitro Organoids, Microfluidic Chips, and Bioprinting. Cell Tissue Res. 2023, 394, 75–91. [Google Scholar] [CrossRef] [PubMed]
- Han, S.; Cruz, S.H.; Park, S.; Shin, S.R. Nano-Biomaterials and Advanced Fabrication Techniques for Engineering Skeletal Muscle Tissue Constructs in Regenerative Medicine. Nano Converg. 2023, 10, 48. [Google Scholar] [CrossRef] [PubMed]
- Kolotyeva, N.A.; Gilmiyarova, F.N.; Averchuk, A.S.; Baranich, T.I.; Rozanova, N.A.; Kukla, M.V.; Tregub, P.P.; Salmina, A.B. Novel Approaches to the Establishment of Local Microenvironment from Resorbable Biomaterials in the Brain In Vitro Models. Int. J. Mol. Sci. 2023, 24, 14709. [Google Scholar] [CrossRef] [PubMed]
- Basit, R.; Wiseman, J.; Chowdhury, F.; Chari, D. Simulating Traumatic Brain Injury in Vitro: Developing High Throughput Models to Test Biomaterial Based Therapies. Neural Regen. Res. 2023, 18, 289. [Google Scholar] [CrossRef]
- Sun, L.; Bian, F.; Xu, D.; Luo, Y.; Wang, Y.; Zhao, Y. Tailoring Biomaterials for Biomimetic Organs-on-Chips. Mater. Horiz. 2023, 10, 4724–4745. [Google Scholar] [CrossRef] [PubMed]
- Pires, P.C.; Mascarenhas-Melo, F.; Pedrosa, K.; Lopes, D.; Lopes, J.; Macário-Soares, A.; Peixoto, D.; Giram, P.S.; Veiga, F.; Paiva-Santos, A.C. Polymer-Based Biomaterials for Pharmaceutical and Biomedical Applications: A Focus on Topical Drug Administration. Eur. Polym. J. 2023, 187, 111868. [Google Scholar] [CrossRef]
- Catoira, M.C.; Fusaro, L.; Di Francesco, D.; Ramella, M.; Boccafoschi, F. Overview of Natural Hydrogels for Regenerative Medicine Applications. J. Mater. Sci. Mater. Med. 2019, 30, 115. [Google Scholar] [CrossRef]
- Geckil, H.; Xu, F.; Zhang, X.; Moon, S.; Demirci, U. Engineering Hydrogels as Extracellular Matrix Mimics. Nanomedicine 2010, 5, 469–484. [Google Scholar] [CrossRef]
- Ornaghi, H.L.; Monticeli, F.M.; Agnol, L.D. A Review on Polymers for Biomedical Applications on Hard and Soft Tissues and Prosthetic Limbs. Polymers 2023, 15, 4034. [Google Scholar] [CrossRef]
- Castellote-Borrell, M.; Merlina, F.; Rodríguez, A.R.; Guasch, J. Biohybrid Hydrogels for Tumoroid Culture. Adv. Biol. 2023, 7, 2300118. [Google Scholar] [CrossRef]
- Boso, D.; Maghin, E.; Carraro, E.; Giagante, M.; Pavan, P.; Piccoli, M. Extracellular Matrix-Derived Hydrogels as Biomaterial for Different Skeletal Muscle Tissue Replacements. Materials 2020, 13, 2483. [Google Scholar] [CrossRef] [PubMed]
- Kaliaraj, G.; Shanmugam, D.; Dasan, A.; Mosas, K. Hydrogels—A Promising Materials for 3D Printing Technology. Gels 2023, 9, 260. [Google Scholar] [CrossRef] [PubMed]
- Madduma-Bandarage, U.S.K.; Madihally, S.V. Synthetic Hydrogels: Synthesis, Novel Trends, and Applications. J. Appl. Polym. Sci. 2021, 138, 50376. [Google Scholar] [CrossRef]
- Burdick, J.A.; Murphy, W.L. Moving from Static to Dynamic Complexity in Hydrogel Design. Nat. Commun. 2012, 3, 1269. [Google Scholar] [CrossRef] [PubMed]
- Morris, E.; Chavez, M.; Tan, C. Dynamic Biomaterials: Toward Engineering Autonomous Feedback. Curr. Opin. Biotechnol. 2016, 39, 97–104. [Google Scholar] [CrossRef] [PubMed]
- Tang, G.; Zhou, B.; Li, F.; Wang, W.; Liu, Y.; Wang, X.; Liu, C.; Ye, X. Advances of Naturally Derived and Synthetic Hydrogels for Intervertebral Disk Regeneration. Front. Bioeng. Biotechnol. 2020, 8, 745. [Google Scholar] [CrossRef] [PubMed]
- Prete, S.; Dattilo, M.; Patitucci, F.; Pezzi, G.; Parisi, O.I.; Puoci, F. Natural and Synthetic Polymeric Biomaterials for Application in Wound Management. J. Funct. Biomater. 2023, 14, 455. [Google Scholar] [CrossRef]
- Revete, A.; Aparicio, A.; Cisterna, B.A.; Revete, J.; Luis, L.; Ibarra, E.; Segura González, E.A.; Molino, J.; Reginensi, D. Advancements in the Use of Hydrogels for Regenerative Medicine: Properties and Biomedical Applications. Int. J. Biomater. 2022, 2022, 3606765. [Google Scholar] [CrossRef]
- Reyes-Martínez, J.E.; Ruiz-Pacheco, J.A.; Flores-Valdéz, M.A.; Elsawy, M.A.; Vallejo-Cardona, A.A.; Castillo-Díaz, L.A. Advanced Hydrogels for Treatment of Diabetes. J. Tissue Eng. Regen. Med. 2019, 13, 1375–1393. [Google Scholar] [CrossRef]
- Ramires, P.A.; Miccoli, M.A.; Panzarini, E.; Dini, L.; Protopapa, C. In Vitro and in Vivo Biocompatibility Evaluation of a Polyalkylimide Hydrogel for Soft Tissue Augmentation. J. Biomed. Mater. Res. B Appl. Biomater. 2005, 72B, 230–238. [Google Scholar] [CrossRef]
- Reddy, M.S.B.; Ponnamma, D.; Choudhary, R.; Sadasivuni, K.K. A Comparative Review of Natural and Synthetic Biopolymer Composite Scaffolds. Polymers 2021, 13, 1105. [Google Scholar] [CrossRef] [PubMed]
- Tang, S.; Richardson, B.M.; Anseth, K.S. Dynamic Covalent Hydrogels as Biomaterials to Mimic the Viscoelasticity of Soft Tissues. Prog. Mater. Sci. 2021, 120, 100738. [Google Scholar] [CrossRef]
- Ooi, H.W.; Hafeez, S.; Van Blitterswijk, C.A.; Moroni, L.; Baker, M.B. Hydrogels That Listen to Cells: A Review of Cell-Responsive Strategies in Biomaterial Design for Tissue Regeneration. Mater. Horiz. 2017, 4, 1020–1040. [Google Scholar] [CrossRef]
- Jia, X.; Chen, J.; Lv, W.; Li, H.; Ariga, K. Engineering Dynamic and Interactive Biomaterials Using Material Nanoarchitectonics for Modulation of Cellular Behaviors. Cell Rep. Phys. Sci. 2023, 4, 101251. [Google Scholar] [CrossRef]
- Özkale, B.; Sakar, M.S.; Mooney, D.J. Active Biomaterials for Mechanobiology. Biomaterials 2021, 267, 120497. [Google Scholar] [CrossRef]
- Bonnans, C.; Chou, J.; Werb, Z. Remodelling the Extracellular Matrix in Development and Disease. Nat. Rev. Mol. Cell Biol. 2014, 15, 786–801. [Google Scholar] [CrossRef]
- Xie, W.; Wei, X.; Kang, H.; Jiang, H.; Chu, Z.; Lin, Y.; Hou, Y.; Wei, Q. Static and Dynamic: Evolving Biomaterial Mechanical Properties to Control Cellular Mechanotransduction. Adv. Sci. 2023, 10, 2204594. [Google Scholar] [CrossRef]
- Cox, T.R.; Erler, J.T. Remodeling and Homeostasis of the Extracellular Matrix: Implications for Fibrotic Diseases and Cancer. Dis. Models Mech. 2011, 4, 165–178. [Google Scholar] [CrossRef]
- Xu, X.; Jia, Z.; Zheng, Y.; Wang, Y. Bioadaptability of Biomaterials: Aiming at Precision Medicine. Matter 2021, 4, 2648–2650. [Google Scholar] [CrossRef]
- Neumann, M.; Di Marco, G.; Iudin, D.; Viola, M.; Van Nostrum, C.F.; Van Ravensteijn, B.G.P.; Vermonden, T. Stimuli-Responsive Hydrogels: The Dynamic Smart Biomaterials of Tomorrow. Macromolecules 2023, 56, 8377–8392. [Google Scholar] [CrossRef]
- Braegelman, A.S.; Webber, M.J. Integrating Stimuli-Responsive Properties in Host-Guest Supramolecular Drug Delivery Systems. Theranostics 2019, 9, 3017–3040. [Google Scholar] [CrossRef] [PubMed]
- Rizwan, M.; Baker, A.E.G.; Shoichet, M.S. Designing Hydrogels for 3D Cell Culture Using Dynamic Covalent Crosslinking. Adv. Healthc. Mater. 2021, 10, 2100234. [Google Scholar] [CrossRef]
- Yang, B.; Wei, K.; Loebel, C.; Zhang, K.; Feng, Q.; Li, R.; Wong, S.H.D.; Xu, X.; Lau, C.; Chen, X.; et al. Enhanced Mechanosensing of Cells in Synthetic 3D Matrix with Controlled Biophysical Dynamics. Nat. Commun. 2021, 12, 3514. [Google Scholar] [CrossRef]
- Ma, X.; Zhao, Y. Biomedical Applications of Supramolecular Systems Based on Host–Guest Interactions. Chem. Rev. 2015, 115, 7794–7839. [Google Scholar] [CrossRef] [PubMed]
- Takashima, Y.; Sawa, Y.; Iwaso, K.; Nakahata, M.; Yamaguchi, H.; Harada, A. Supramolecular Materials Cross-Linked by Host–Guest Inclusion Complexes: The Effect of Side Chain Molecules on Mechanical Properties. Macromolecules 2017, 50, 3254–3261. [Google Scholar] [CrossRef]
- Sayed, M.; Pal, H. An Overview from Simple Host–Guest Systems to Progressively Complex Supramolecular Assemblies. Phys. Chem. Chem. Phys. 2021, 23, 26085–26107. [Google Scholar] [CrossRef]
- Wang, H.; Heilshorn, S.C. Adaptable Hydrogel Networks with Reversible Linkages for Tissue Engineering. Adv. Mater. 2015, 27, 3717–3736. [Google Scholar] [CrossRef]
- Liu, Y.; Li, Z.; Xu, J.; Wang, B.; Liu, F.; Na, R.; Guan, S.; Liu, F. Effects of Amphiphilic Monomers and Their Hydrophilic Spacers on Polyacrylamide Hydrogels. RSC Adv. 2019, 9, 3462–3468. [Google Scholar] [CrossRef]
- Jiang, H.; Duan, L.; Ren, X.; Gao, G. Hydrophobic Association Hydrogels with Excellent Mechanical and Self-Healing Properties. Eur. Polym. J. 2019, 112, 660–669. [Google Scholar] [CrossRef]
- Lin, M.; Dai, Y.; Xia, F.; Zhang, X. Advances in Non-Covalent Crosslinked Polymer Micelles for Biomedical Applications. Mater. Sci. Eng. C 2021, 119, 111626. [Google Scholar] [CrossRef]
- Yu, H.; Xiao, Q.; Qi, G.; Chen, F.; Tu, B.; Zhang, S.; Li, Y.; Chen, Y.; Yu, H.; Duan, P. A Hydrogen Bonds-Crosslinked Hydrogels with Self-Healing and Adhesive Properties for Hemostatic. Front. Bioeng. Biotechnol. 2022, 10, 855013. [Google Scholar] [CrossRef] [PubMed]
- Tong, Z.; Jin, L.; Oliveira, J.M.; Reis, R.L.; Zhong, Q.; Mao, Z.; Gao, C. Adaptable Hydrogel with Reversible Linkages for Regenerative Medicine: Dynamic Mechanical Microenvironment for Cells. Bioact. Mater. 2021, 6, 1375–1387. [Google Scholar] [CrossRef] [PubMed]
- Rowley, J.A.; Madlambayan, G.; Mooney, D.J. Alginate Hydrogels as Synthetic Extracellular Matrix Materials. Biomaterials 1999, 20, 45–53. [Google Scholar] [CrossRef] [PubMed]
- Novikova, L.N.; Mosahebi, A.; Wiberg, M.; Terenghi, G.; Kellerth, J.; Novikov, L.N. Alginate Hydrogel and Matrigel as Potential Cell Carriers for Neurotransplantation. J. Biomed. Mater. Res. A 2006, 77A, 242–252. [Google Scholar] [CrossRef] [PubMed]
- Beaupre, D.M.; Weiss, R.G. Thiol- and Disulfide-Based Stimulus-Responsive Soft Materials and Self-Assembling Systems. Molecules 2021, 26, 3332. [Google Scholar] [CrossRef] [PubMed]
- Deng, C.C.; Brooks, W.L.A.; Abboud, K.A.; Sumerlin, B.S. Boronic Acid-Based Hydrogels Undergo Self-Healing at Neutral and Acidic pH. ACS Macro Lett. 2015, 4, 220–224. [Google Scholar] [CrossRef] [PubMed]
- McKinnon, D.D.; Domaille, D.W.; Cha, J.N.; Anseth, K.S. Biophysically Defined and Cytocompatible Covalently Adaptable Networks as Viscoelastic 3D Cell Culture Systems. Adv. Mater. 2014, 26, 865–872. [Google Scholar] [CrossRef]
- Liu, A.; Wu, K.; Chen, S.; Wu, C.; Gao, D.; Chen, L.; Wei, D.; Luo, H.; Sun, J.; Fan, H. Tunable Fast Relaxation in Imine-Based Nanofibrillar Hydrogels Stimulates Cell Response through TRPV4 Activation. Biomacromolecules 2020, 21, 3745–3755. [Google Scholar] [CrossRef]
- Sánchez-Morán, H.; Ahmadi, A.; Vogler, B.; Roh, K.-H. Oxime Cross-Linked Alginate Hydrogels with Tunable Stress Relaxation. Biomacromolecules 2019, 20, 4419–4429. [Google Scholar] [CrossRef]
- Chen, M.; Ren, X.; Dong, L.; Li, X.; Cheng, H. Preparation of Dynamic Covalently Crosslinking Keratin Hydrogels Based on Thiol/Disulfide Bonds Exchange Strategy. Int. J. Biol. Macromol. 2021, 182, 1259–1267. [Google Scholar] [CrossRef]
- Wei, Z.; Yang, J.H.; Du, X.J.; Xu, F.; Zrinyi, M.; Osada, Y.; Li, F.; Chen, Y.M. Dextran-Based Self-Healing Hydrogels Formed by Reversible Diels–Alder Reaction under Physiological Conditions. Macromol. Rapid Commun. 2013, 34, 1464–1470. [Google Scholar] [CrossRef] [PubMed]
- Bhagawati, M. Biofunctionalization of Hydrogels for Engineering the Cellular Microenvironment. In Micro-and Nanoengineering of the Cell Surface; William Andrew Publishing: Norwich, NY, USA, 2014; Chapter 14. [Google Scholar]
- Young, J.L.; Engler, A.J. Hydrogels with Time-Dependent Material Properties Enhance Cardiomyocyte Differentiation in Vitro. Biomaterials 2011, 32, 1002–1009. [Google Scholar] [CrossRef] [PubMed]
- Deshayes, S.; Kasko, A.M. Polymeric Biomaterials with Engineered Degradation. J. Polym. Sci. Part Polym. Chem. 2013, 51, 3531–3566. [Google Scholar] [CrossRef]
- Zustiak, S.P.; Leach, J.B. Hydrolytically Degradable Poly(Ethylene Glycol) Hydrogel Scaffolds with Tunable Degradation and Mechanical Properties. Biomacromolecules 2010, 11, 1348–1357. [Google Scholar] [CrossRef] [PubMed]
- Azevedo, H.S.; Reis, R.L. Enzymatic Degradation of Biodegradable Polymers and Strategies to Control Their Degradation Rate. In Biodegradable Systems in Tissue Engineering and Regenerative Medicine; CRC Press: Boca Raton, FL, USA, 2004. [Google Scholar]
- Lutolf, M.P.; Lauer-Fields, J.L.; Schmoekel, H.G.; Metters, A.T.; Weber, F.E.; Fields, G.B.; Hubbell, J.A. Synthetic Matrix Metalloproteinase-Sensitive Hydrogels for the Conduction of Tissue Regeneration: Engineering Cell-Invasion Characteristics. Proc. Natl. Acad. Sci. USA 2003, 100, 5413–5418. [Google Scholar] [CrossRef]
- Li, Z.; Zhou, Y.; Li, T.; Zhang, J.; Tian, H. Stimuli-responsive Hydrogels: Fabrication and Biomedical Applications. VIEW 2022, 3, 20200112. [Google Scholar] [CrossRef]
- Bril, M.; Fredrich, S.; Kurniawan, N.A. Stimuli-Responsive Materials: A Smart Way to Study Dynamic Cell Responses. Smart Mater. Med. 2022, 3, 257–273. [Google Scholar] [CrossRef]
- Doberenz, F.; Zeng, K.; Willems, C.; Zhang, K.; Groth, T. Thermoresponsive Polymers and Their Biomedical Application in Tissue Engineering—A Review. J. Mater. Chem. B 2020, 8, 607–628. [Google Scholar] [CrossRef]
- Roy, D.; Cambre, J.N.; Sumerlin, B.S. Future Perspectives and Recent Advances in Stimuli-Responsive Materials. Prog. Polym. Sci. 2010, 35, 278–301. [Google Scholar] [CrossRef]
- Chandan, R.; Mehta, S.; Banerjee, R. Ultrasound-Responsive Carriers for Therapeutic Applications. ACS Biomater. Sci. Eng. 2020, 6, 4731–4747. [Google Scholar] [CrossRef]
- Gao, Y.; Xu, S.; Wu, R.; Wang, J.; Wei, J. Preparation and Characteristic of Electric Stimuli Responsive Hydrogel Composed of Polyvinyl Alcohol/Poly (Sodium Maleate-Co-sodium Acrylate). J. Appl. Polym. Sci. 2008, 107, 391–395. [Google Scholar] [CrossRef]
- Rotjanasuworapong, K.; Thummarungsan, N.; Lerdwijitjarud, W.; Sirivat, A. Facile Formation of Agarose Hydrogel and Electromechanical Responses as Electro-Responsive Hydrogel Materials in Actuator Applications. Carbohydr. Polym. 2020, 247, 116709. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.; Titushkin, I.; Cho, M. Regulation of Mesenchymal Stem Cell Adhesion and Orientation in 3D Collagen Scaffold by Electrical Stimulus. Bioelectrochemistry 2006, 69, 133–141. [Google Scholar] [CrossRef] [PubMed]
- Kiang, J.D.; Wen, J.H.; Del Álamo, J.C.; Engler, A.J. Dynamic and Reversible Surface Topography Influences Cell Morphology. J. Biomed. Mater. Res. A 2013, 101A, 2313–2321. [Google Scholar] [CrossRef]
- Testa, P.; Style, R.W.; Cui, J.; Donnelly, C.; Borisova, E.; Derlet, P.M.; Dufresne, E.R.; Heyderman, L.J. Magnetically Addressable Shape-Memory and Stiffening in a Composite Elastomer. Adv. Mater. 2019, 31, 1900561. [Google Scholar] [CrossRef] [PubMed]
- Tran, K.A.; Kraus, E.; Clark, A.T.; Bennett, A.; Pogoda, K.; Cheng, X.; Cēbers, A.; Janmey, P.A.; Galie, P.A. Dynamic Tuning of Viscoelastic Hydrogels with Carbonyl Iron Microparticles Reveals the Rapid Response of Cells to Three-Dimensional Substrate Mechanics. ACS Appl. Mater. Interfaces 2021, 13, 20947–20959. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.; Wiendels, M.; Yuan, H.; Ruan, C.; Kouwer, P.H.J. Cell-Matrix Reciprocity in 3D Culture Models with Nonlinear Elasticity. Bioact. Mater. 2022, 9, 316–331. [Google Scholar] [CrossRef]
- Piechocka, I.K.; Bacabac, R.G.; Potters, M.; MacKintosh, F.C.; Koenderink, G.H. Structural Hierarchy Governs Fibrin Gel Mechanics. Biophys. J. 2010, 98, 2281–2289. [Google Scholar] [CrossRef]
- Piechocka, I.K.; Jansen, K.A.; Broedersz, C.P.; Kurniawan, N.A.; MacKintosh, F.C.; Koenderink, G.H. Multi-Scale Strain-Stiffening of Semiflexible Bundle Networks. Soft Matter 2016, 12, 2145–2156. [Google Scholar] [CrossRef]
- Van Helvert, S.; Friedl, P. Strain Stiffening of Fibrillar Collagen during Individual and Collective Cell Migration Identified by AFM Nanoindentation. ACS Appl. Mater. Interfaces 2016, 8, 21946–21955. [Google Scholar] [CrossRef]
- Van Helvert, S.; Storm, C.; Friedl, P. Mechanoreciprocity in Cell Migration. Nat. Cell Biol. 2018, 20, 8–20. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Castaño Romera, M.; Lou, X.; Schill, J.; Ter Huurne, G.; Fransen, P.-P.K.H.; Voets, I.K.; Storm, C.; Sijbesma, R.P. Strain-Stiffening in Dynamic Supramolecular Fiber Networks. J. Am. Chem. Soc. 2018, 140, 17547–17555. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Xu, Z.; Lovrak, M.; Le Sage, V.A.A.; Zhang, K.; Guo, X.; Eelkema, R.; Mendes, E.; Van Esch, J.H. Biomimetic Strain-Stiffening Self-Assembled Hydrogels. Angew. Chem. Int. Ed. 2020, 59, 4830–4834. [Google Scholar] [CrossRef] [PubMed]
- Pereira, I.; Lopez-Martinez, M.J.; Samitier, J. Advances in Current in Vitro Models on Neurodegenerative Diseases. Front. Bioeng. Biotechnol. 2023, 11, 1260397. [Google Scholar] [CrossRef] [PubMed]
- Mullis, A.S.; Kaplan, D.L. Functional Bioengineered Tissue Models of Neurodegenerative Diseases. Biomaterials 2023, 298, 122143. [Google Scholar] [CrossRef] [PubMed]
- Metaxas, A.; Kempf, S. Neurofibrillary Tangles in Alzheimer’s Disease: Elucidation of the Molecular Mechanism by Immunohistochemistry and Tau Protein Phospho-Proteomics. Neural Regen. Res. 2016, 11, 1579. [Google Scholar] [CrossRef]
- Gouras, G.K.; Olsson, T.T.; Hansson, O. β-Amyloid Peptides and Amyloid Plaques in Alzheimer’s Disease. Neurotherapeutics 2015, 12, 3–11. [Google Scholar] [CrossRef]
- DeMaagd, G.; Philip, A. Parkinson’s Disease and Its Management: Part 1: Disease Entity, Risk Factors, Pathophysiology, Clinical Presentation, and Diagnosis. Pharm. Ther. 2015, 40, 504. [Google Scholar]
- Del Bigio, M.R. Ependymal Cells: Biology and Pathology. Acta Neuropathol. 2010, 119, 55–73. [Google Scholar] [CrossRef]
- Maldonado-Soto, A.R.; Oakley, D.H.; Wichterle, H.; Stein, J.; Doetsch, F.K.; Henderson, C.E. Stem Cells in the Nervous System. Am. J. Phys. Med. Rehabil. 2014, 93, S132–S144. [Google Scholar] [CrossRef]
- Fields, R.D.; Stevens-Graham, B. New Insights into Neuron-Glia Communication. Science 2002, 298, 556–562. [Google Scholar] [CrossRef] [PubMed]
- Barros, C.S.; Franco, S.J.; Muller, U. Extracellular Matrix: Functions in the Nervous System. Cold Spring Harb. Perspect. Biol. 2011, 3, a005108. [Google Scholar] [CrossRef] [PubMed]
- Ferrer-Ferrer, M.; Dityatev, A. Shaping Synapses by the Neural Extracellular Matrix. Front. Neuroanat. 2018, 12, 40. [Google Scholar] [CrossRef] [PubMed]
- Long, K.R.; Huttner, W.B. How the Extracellular Matrix Shapes Neural Development. Open Biol. 2019, 9, 180216. [Google Scholar] [CrossRef] [PubMed]
- Suttkus, A.; Morawski, M.; Arendt, T. Protective Properties of Neural Extracellular Matrix. Mol. Neurobiol. 2016, 53, 73–82. [Google Scholar] [CrossRef]
- Lau, L.W.; Cua, R.; Keough, M.B.; Haylock-Jacobs, S.; Yong, V.W. Pathophysiology of the Brain Extracellular Matrix: A New Target for Remyelination. Nat. Rev. Neurosci. 2013, 14, 722–729. [Google Scholar] [CrossRef]
- Roth, J.G.; Huang, M.S.; Li, T.L.; Feig, V.R.; Jiang, Y.; Cui, B.; Greely, H.T.; Bao, Z.; Paşca, S.P.; Heilshorn, S.C. Advancing Models of Neural Development with Biomaterials. Nat. Rev. Neurosci. 2021, 22, 593–615. [Google Scholar] [CrossRef]
- Mouw, J.K.; Ou, G.; Weaver, V.M. Extracellular Matrix Assembly: A Multiscale Deconstruction. Nat. Rev. Mol. Cell Biol. 2014, 15, 771–785. [Google Scholar] [CrossRef]
- Zimmermann, D.R.; Dours-Zimmermann, M.T. Extracellular Matrix of the Central Nervous System: From Neglect to Challenge. Histochem. Cell Biol. 2008, 130, 635–653. [Google Scholar] [CrossRef]
- Dityatev, A. Remodeling of Extracellular Matrix and Epileptogenesis. Epilepsia 2010, 51, 61–65. [Google Scholar] [CrossRef]
- Nirwane, A.; Yao, Y. Laminins and Their Receptors in the CNS. Biol. Rev. 2019, 94, 283–306. [Google Scholar] [CrossRef] [PubMed]
- Dauth, S.; Grevesse, T.; Pantazopoulos, H.; Campbell, P.H.; Maoz, B.M.; Berretta, S.; Parker, K.K. Extracellular Matrix Protein Expression Is Brain Region Dependent. J. Comp. Neurol. 2016, 524, 1309–1336. [Google Scholar] [CrossRef] [PubMed]
- Streitberger, K.-J.; Sack, I.; Krefting, D.; Pfüller, C.; Braun, J.; Paul, F.; Wuerfel, J. Brain Viscoelasticity Alteration in Chronic-Progressive Multiple Sclerosis. PLoS ONE 2012, 7, e29888. [Google Scholar] [CrossRef] [PubMed]
- Soria, F.N.; Paviolo, C.; Doudnikoff, E.; Arotcarena, M.-L.; Lee, A.; Danné, N.; Mandal, A.K.; Gosset, P.; Dehay, B.; Groc, L.; et al. Synucleinopathy Alters Nanoscale Organization and Diffusion in the Brain Extracellular Space through Hyaluronan Remodeling. Nat. Commun. 2020, 11, 3440. [Google Scholar] [CrossRef]
- Choi, S.H.; Kim, Y.H.; Hebisch, M.; Sliwinski, C.; Lee, S.; D’Avanzo, C.; Chen, H.; Hooli, B.; Asselin, C.; Muffat, J.; et al. A Three-Dimensional Human Neural Cell Culture Model of Alzheimer’s Disease. Nature 2014, 515, 274–278. [Google Scholar] [CrossRef] [PubMed]
- Nicholson, C.; Hrabětová, S. Brain Extracellular Space: The Final Frontier of Neuroscience. Biophys. J. 2017, 113, 2133–2142. [Google Scholar] [CrossRef]
- Shetty, A.K.; Zanirati, G. The Interstitial System of the Brain in Health and Disease. Aging Dis. 2020, 11, 200. [Google Scholar] [CrossRef] [PubMed]
- Esparza, T.J.; Wildburger, N.C.; Jiang, H.; Gangolli, M.; Cairns, N.J.; Bateman, R.J.; Brody, D.L. Soluble Amyloid-Beta Aggregates from Human Alzheimer’s Disease Brains. Sci. Rep. 2016, 6, 38187. [Google Scholar] [CrossRef]
- Dubnovitsky, A.; Sandberg, A.; Rahman, M.M.; Benilova, I.; Lendel, C.; Härd, T. Amyloid-β Protofibrils: Size, Morphology and Synaptotoxicity of an Engineered Mimic. PLoS ONE 2013, 8, e66101. [Google Scholar] [CrossRef]
- De Calignon, A.; Polydoro, M.; Suárez-Calvet, M.; William, C.; Adamowicz, D.H.; Kopeikina, K.J.; Pitstick, R.; Sahara, N.; Ashe, K.H.; Carlson, G.A.; et al. Propagation of Tau Pathology in a Model of Early Alzheimer’s Disease. Neuron 2012, 73, 685–697. [Google Scholar] [CrossRef]
- Liu, L.; Drouet, V.; Wu, J.W.; Witter, M.P.; Small, S.A.; Clelland, C.; Duff, K. Trans-Synaptic Spread of Tau Pathology In Vivo. PLoS ONE 2012, 7, e31302. [Google Scholar] [CrossRef] [PubMed]
- Pooler, A.M.; Polydoro, M.; Maury, E.A.; Nicholls, S.B.; Reddy, S.M.; Wegmann, S.; William, C.; Saqran, L.; Cagsal-Getkin, O.; Pitstick, R.; et al. Amyloid Accelerates Tau Propagation and Toxicity in a Model of Early Alzheimer’s Disease. Acta Neuropathol. Commun. 2015, 3, 14. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.W.; Hussaini, S.A.; Bastille, I.M.; Rodriguez, G.A.; Mrejeru, A.; Rilett, K.; Sanders, D.W.; Cook, C.; Fu, H.; Boonen, R.A.C.M.; et al. Neuronal Activity Enhances Tau Propagation and Tau Pathology in Vivo. Nat. Neurosci. 2016, 19, 1085–1092. [Google Scholar] [CrossRef] [PubMed]
- Li, K.; Li, J.; Zheng, J.; Qin, S. Reactive Astrocytes in Neurodegenerative Diseases. Aging Dis. 2019, 10, 664. [Google Scholar] [CrossRef] [PubMed]
- East, E.; Golding, J.P.; Phillips, J.B. A Versatile 3D Culture Model Facilitates Monitoring of Astrocytes Undergoing Reactive Gliosis. J. Tissue Eng. Regen. Med. 2009, 3, 634–646. [Google Scholar] [CrossRef] [PubMed]
- Watson, P.M.D.; Kavanagh, E.; Allenby, G.; Vassey, M. Bioengineered 3D Glial Cell Culture Systems and Applications for Neurodegeneration and Neuroinflammation. SLAS Discov. 2017, 22, 583–601. [Google Scholar] [CrossRef]
- Calà, G.; Sina, B.; De Coppi, P.; Giobbe, G.G.; Gerli, M.F.M. Primary Human Organoids Models: Current Progress and Key Milestones. Front. Bioeng. Biotechnol. 2023, 11, 1058970. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Koo, B.-K.; Knoblich, J.A. Human Organoids: Model Systems for Human Biology and Medicine. Nat. Rev. Mol. Cell Biol. 2020, 21, 571–584. [Google Scholar] [CrossRef]
- Cembran, A.; Bruggeman, K.F.; Williams, R.J.; Parish, C.L.; Nisbet, D.R. Biomimetic Materials and Their Utility in Modeling the 3-Dimensional Neural Environment. iScience 2020, 23, 100788. [Google Scholar] [CrossRef]
- Xu, Z.; Fang, P.; Xu, B.; Lu, Y.; Xiong, J.; Gao, F.; Wang, X.; Fan, J.; Shi, P. High-Throughput Three-Dimensional Chemotactic Assays Reveal Steepness-Dependent Complexity in Neuronal Sensation to Molecular Gradients. Nat. Commun. 2018, 9, 4745. [Google Scholar] [CrossRef]
- Sordini, L.; Garrudo, F.F.F.; Rodrigues, C.A.V.; Linhardt, R.J.; Cabral, J.M.S.; Ferreira, F.C.; Morgado, J. Effect of Electrical Stimulation Conditions on Neural Stem Cells Differentiation on Cross-Linked PEDOT:PSS Films. Front. Bioeng. Biotechnol. 2021, 9, 591838. [Google Scholar] [CrossRef] [PubMed]
- Madhusudanan, P.; Raju, G.; Shankarappa, S. Hydrogel Systems and Their Role in Neural Tissue Engineering. J. R. Soc. Interface 2020, 17, 20190505. [Google Scholar] [CrossRef] [PubMed]
- Vallejo-Giraldo, C.; Genta, M.; Cauvi, O.; Goding, J.; Green, R. Hydrogels for 3D Neural Tissue Models: Understanding Cell-Material Interactions at a Molecular Level. Front. Bioeng. Biotechnol. 2020, 8, 601704. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.-W.; Lee, H.J.; Hwang, H.S.; Ko, K.; Han, D.W.; Ko, K. Optimization of Matrigel-Based Culture for Expansion of Neural Stem Cells. Anim. Cells Syst. 2015, 19, 175–180. [Google Scholar] [CrossRef]
- Terek, J.C.; Hebb, M.O.; Flynn, L.E. Development of Brain-Derived Bioscaffolds for Neural Progenitor Cell Culture. ACS Pharmacol. Transl. Sci. 2023, 6, 320–333. [Google Scholar] [CrossRef] [PubMed]
- Hughes, C.S.; Postovit, L.M.; Lajoie, G.A. Matrigel: A Complex Protein Mixture Required for Optimal Growth of Cell Culture. Proteomics 2010, 10, 1886–1890. [Google Scholar] [CrossRef]
- Aisenbrey, E.A.; Murphy, W.L. Synthetic Alternatives to Matrigel. Nat. Rev. Mater. 2020, 5, 539–551. [Google Scholar] [CrossRef]
- Murphy, A.R.; Laslett, A.; O’Brien, C.M.; Cameron, N.R. Scaffolds for 3D in Vitro Culture of Neural Lineage Cells. Acta Biomater. 2017, 54, 1–20. [Google Scholar] [CrossRef]
- Ngo, M.T.; Harley, B.A.C. Progress in Mimicking Brain Microenvironments to Understand and Treat Neurological Disorders. APL Bioeng. 2021, 5, 020902. [Google Scholar] [CrossRef]
- Liu, J.; Liu, S.; Zeng, L.; Tsilioni, I. Amyloid Beta Peptides Lead to Mast Cell Activation in a Novel 3D Hydrogel Model. Int. J. Mol. Sci. 2023, 24, 12002. [Google Scholar] [CrossRef]
- Baron-Van Evercooren, A.; Kleinman, H.K.; Ohno, S.; Marangos, P.; Schwartz, J.P.; Dubois-Dalcq, M.E. Nerve Growth Factor, Laminin, and Fibronectin Promote Neurite Growth in Human Fetal Sensory Ganglia Cultures. J. Neurosci. Res. 1982, 8, 179–193. [Google Scholar] [CrossRef] [PubMed]
- Rogers, S.L.; Letourneau, P.C.; Palm, S.L.; McCarthy, J.; Furcht, L.T. Neurite Extension by Peripheral and Central Nervous System Neurons in Response to Substratum-Bound Fibronectin and Laminin. Dev. Biol. 1983, 98, 212–220. [Google Scholar] [CrossRef] [PubMed]
- Manthorpe, M.; Engvall, E.; Ruoslahti, E.; Longo, F.M.; Davis, G.E.; Varon, S. Laminin Promotes Neuritic Regeneration from Cultured Peripheral and Central Neurons. J. Cell Biol. 1983, 97, 1882–1890. [Google Scholar] [CrossRef]
- Cantley, W.L.; Du, C.; Lomoio, S.; DePalma, T.; Peirent, E.; Kleinknecht, D.; Hunter, M.; Tang-Schomer, M.D.; Tesco, G.; Kaplan, D.L. Functional and Sustainable 3D Human Neural Network Models from Pluripotent Stem Cells. ACS Biomater. Sci. Eng. 2018, 4, 4278–4288. [Google Scholar] [CrossRef] [PubMed]
- Frampton, J.P.; Hynd, M.R.; Shuler, M.L.; Shain, W. Fabrication and Optimization of Alginate Hydrogel Constructs for Use in 3D Neural Cell Culture. Biomed. Mater. 2011, 6, 015002. [Google Scholar] [CrossRef] [PubMed]
- Martău, G.A.; Mihai, M.; Vodnar, D.C. The Use of Chitosan, Alginate, and Pectin in the Biomedical and Food Sector—Biocompatibility, Bioadhesiveness, and Biodegradability. Polymers 2019, 11, 1837. [Google Scholar] [CrossRef] [PubMed]
- Tedesco, M.T.; Di Lisa, D.; Massobrio, P.; Colistra, N.; Pesce, M.; Catelani, T.; Dellacasa, E.; Raiteri, R.; Martinoia, S.; Pastorino, L. Soft Chitosan Microbeads Scaffold for 3D Functional Neuronal Networks. Biomaterials 2018, 156, 159–171. [Google Scholar] [CrossRef] [PubMed]
- Almari, B.; Brough, D.; Harte, M.; Tirella, A. Fabrication of Amyloid-β-Secreting Alginate Microbeads for Use in Modelling Alzheimer’s Disease. J. Vis. Exp. 2019, 59597. [Google Scholar] [CrossRef]
- Ranjan, V.D.; Qiu, L.; Lee, J.W.-L.; Chen, X.; Jang, S.E.; Chai, C.; Lim, K.-L.; Tan, E.-K.; Zhang, Y.; Huang, W.M.; et al. A Microfiber Scaffold-Based 3D in Vitro Human Neuronal Culture Model of Alzheimer’s Disease. Biomater. Sci. 2020, 8, 4861–4874. [Google Scholar] [CrossRef]
- Cormier, A.R.; Pang, X.; Zimmerman, M.I.; Zhou, H.-X.; Paravastu, A.K. Molecular Structure of RADA16-I Designer Self-Assembling Peptide Nanofibers. ACS Nano 2013, 7, 7562–7572. [Google Scholar] [CrossRef]
- Cigognini, D.; Satta, A.; Colleoni, B.; Silva, D.; Donegà, M.; Antonini, S.; Gelain, F. Evaluation of Early and Late Effects into the Acute Spinal Cord Injury of an Injectable Functionalized Self-Assembling Scaffold. PLoS ONE 2011, 6, e19782. [Google Scholar] [CrossRef] [PubMed]
- Gelain, F.; Bottai, D.; Vescovi, A.; Zhang, S. Designer Self-Assembling Peptide Nanofiber Scaffolds for Adult Mouse Neural Stem Cell 3-Dimensional Cultures. PLoS ONE 2006, 1, e119. [Google Scholar] [CrossRef] [PubMed]
- Ni, N.; Hu, Y.; Ren, H.; Luo, C.; Li, P.; Wan, J.-B.; Su, H. Self-Assembling Peptide Nanofiber Scaffolds Enhance Dopaminergic Differentiation of Mouse Pluripotent Stem Cells in 3-Dimensional Culture. PLoS ONE 2013, 8, e84504. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Rouleau, N.; Cantley, W.L.; Liaudanskaya, V.; Berk, A.; Du, C.; Rusk, W.; Peirent, E.; Koester, C.; Nieland, T.J.F.; Kaplan, D.L. A Long-Living Bioengineered Neural Tissue Platform to Study Neurodegeneration. Macromol. Biosci. 2020, 20, 2000004. [Google Scholar] [CrossRef]
- Sood, D.; Chwalek, K.; Stuntz, E.; Pouli, D.; Du, C.; Tang-Schomer, M.; Georgakoudi, I.; Black, L.D.; Kaplan, D.L. Fetal Brain Extracellular Matrix Boosts Neuronal Network Formation in 3D Bioengineered Model of Cortical Brain Tissue. ACS Biomater. Sci. Eng. 2016, 2, 131–140. [Google Scholar] [CrossRef] [PubMed]
- Baiguera, S.; Del Gaudio, C.; Lucatelli, E.; Kuevda, E.; Boieri, M.; Mazzanti, B.; Bianco, A.; Macchiarini, P. Electrospun Gelatin Scaffolds Incorporating Rat Decellularized Brain Extracellular Matrix for Neural Tissue Engineering. Biomaterials 2014, 35, 1205–1214. [Google Scholar] [CrossRef] [PubMed]
- Béduer, A.; Braschler, T.; Peric, O.; Fantner, G.E.; Mosser, S.; Fraering, P.C.; Benchérif, S.; Mooney, D.J.; Renaud, P. A Compressible Scaffold for Minimally Invasive Delivery of Large Intact Neuronal Networks. Adv. Healthc. Mater. 2015, 4, 301–312. [Google Scholar] [CrossRef] [PubMed]
- Rouleau, N.; Murugan, N.J.; Kaplan, D.L. Functional Bioengineered Models of the Central Nervous System. Nat. Rev. Bioeng. 2023, 1, 252–270. [Google Scholar] [CrossRef]
- Vanaei, S.; Parizi, M.S.; Vanaei, S.; Salemizadehparizi, F.; Vanaei, H.R. An Overview on Materials and Techniques in 3D Bioprinting Toward Biomedical Application. Eng. Regen. 2021, 2, 1–18. [Google Scholar] [CrossRef]
- Kačarević, Ž.; Rider, P.; Alkildani, S.; Retnasingh, S.; Smeets, R.; Jung, O.; Ivanišević, Z.; Barbeck, M. An Introduction to 3D Bioprinting: Possibilities, Challenges and Future Aspects. Materials 2018, 11, 2199. [Google Scholar] [CrossRef]
- Joung, D.; Truong, V.; Neitzke, C.C.; Guo, S.; Walsh, P.J.; Monat, J.R.; Meng, F.; Park, S.H.; Dutton, J.R.; Parr, A.M.; et al. 3D Printed Stem-Cell Derived Neural Progenitors Generate Spinal Cord Scaffolds. Adv. Funct. Mater. 2018, 28, 1801850. [Google Scholar] [CrossRef] [PubMed]
- Benwood, C.; Walters-Shumka, J.; Scheck, K.; Willerth, S.M. 3D Bioprinting Patient-Derived Induced Pluripotent Stem Cell Models of Alzheimer’s Disease Using a Smart Bioink. Bioelectron. Med. 2023, 9, 10. [Google Scholar] [CrossRef] [PubMed]
- Abdelrahman, S.; Alsanie, W.F.; Khan, Z.N.; Albalawi, H.I.; Felimban, R.I.; Moretti, M.; Steiner, N.; Chaudhary, A.G.; Hauser, C.A.E. A Parkinson’s Disease Model Composed of 3D Bioprinted Dopaminergic Neurons within a Biomimetic Peptide Scaffold. Biofabrication 2022, 14, 044103. [Google Scholar] [CrossRef] [PubMed]
- Cadena, M.; Ning, L.; King, A.; Hwang, B.; Jin, L.; Serpooshan, V.; Sloan, S.A. 3D Bioprinting of Neural Tissues. Adv. Healthc. Mater. 2021, 10, 2001600. [Google Scholar] [CrossRef] [PubMed]
- Bhatia, S.N.; Ingber, D.E. Microfluidic Organs-on-Chips. Nat. Biotechnol. 2014, 32, 760–772. [Google Scholar] [CrossRef]
- Koo, Y.; Hawkins, B.T.; Yun, Y. Three-Dimensional (3D) Tetra-Culture Brain on Chip Platform for Organophosphate Toxicity Screening. Sci. Rep. 2018, 8, 2841. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Boddeda, A.; Parker, B.; Samanipour, R.; Ghosh, S.; Menard, F.; Kim, K. A High-Resolution Minimicroscope System for Wireless Real-Time Monitoring. IEEE Trans. Biomed. Eng. 2018, 65, 1524–1531. [Google Scholar] [CrossRef] [PubMed]
- Moutaux, E.; Charlot, B.; Genoux, A.; Saudou, F.; Cazorla, M. An Integrated Microfluidic/Microelectrode Array for the Study of Activity-Dependent Intracellular Dynamics in Neuronal Networks. Lab Chip 2018, 18, 3425–3435. [Google Scholar] [CrossRef]
- Virlogeux, A.; Moutaux, E.; Christaller, W.; Genoux, A.; Bruyère, J.; Fino, E.; Charlot, B.; Cazorla, M.; Saudou, F. Reconstituting Corticostriatal Network On-a-Chip Reveals the Contribution of the Presynaptic Compartment to Huntington’s Disease. Cell Rep. 2018, 22, 110–122. [Google Scholar] [CrossRef]
- Li, W.; Xu, Z.; Xu, B.; Chan, C.Y.; Lin, X.; Wang, Y.; Chen, G.; Wang, Z.; Yuan, Q.; Zhu, G.; et al. Investigation of the Subcellular Neurotoxicity of Amyloid-β Using a Device Integrating Microfluidic Perfusion and Chemotactic Guidance. Adv. Healthc. Mater. 2017, 6, 1600895. [Google Scholar] [CrossRef]
- Park, J.; Wetzel, I.; Marriott, I.; Dréau, D.; D’Avanzo, C.; Kim, D.Y.; Tanzi, R.E.; Cho, H. A 3D Human Triculture System Modeling Neurodegeneration and Neuroinflammation in Alzheimer’s Disease. Nat. Neurosci. 2018, 21, 941–951. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Wang, Y.; Liu, J. Bioprinting of 3D Tissues/Organs Combined with Microfluidics. RSC Adv. 2018, 8, 21712–21727. [Google Scholar] [CrossRef] [PubMed]
- Chliara, M.A.; Elezoglou, S.; Zergioti, I. Bioprinting on Organ-on-Chip: Development and Applications. Biosensors 2022, 12, 1135. [Google Scholar] [CrossRef] [PubMed]
- Mukund, K.; Subramaniam, S. Skeletal Muscle: A Review of Molecular Structure and Function, in Health and Disease. WIREs Syst. Biol. Med. 2020, 12, e1462. [Google Scholar] [CrossRef] [PubMed]
- Plotkin, D.L.; Roberts, M.D.; Haun, C.T.; Schoenfeld, B.J. Muscle Fiber Type Transitions with Exercise Training: Shifting Perspectives. Sports 2021, 9, 127. [Google Scholar] [CrossRef]
- Cai, L.; Shi, L.; Peng, Z.; Sun, Y.; Chen, J. Ageing of Skeletal Muscle Extracellular Matrix and Mitochondria: Finding a Potential Link. Ann. Med. 2023, 55, 2240707. [Google Scholar] [CrossRef] [PubMed]
- Herzog, W. What Can We Learn from Single Sarcomere and Myofibril Preparations? Front. Physiol. 2022, 13, 837611. [Google Scholar] [CrossRef]
- Parmentier, E.; Thiry, M. A New Organisational Design in Skeletal Muscle Fibres. Cell Tissue Res. 2023, 393, 111–117. [Google Scholar] [CrossRef]
- Irving, M. Regulation of Contraction by the Thick Filaments in Skeletal Muscle. Biophys. J. 2017, 113, 2579–2594. [Google Scholar] [CrossRef]
- Rassier, D.E. Sarcomere Mechanics in Striated Muscles: From Molecules to Sarcomeres to Cells. Am. J. Physiol.-Cell Physiol. 2017, 313, C134–C145. [Google Scholar] [CrossRef]
- Craig, R.W.; Padrón, R. Molecular Structure of the Sarcomere. Myology 2004, 3, 129–166. [Google Scholar]
- Chaturvedi, V.; Dye, D.E.; Kinnear, B.F.; Van Kuppevelt, T.H.; Grounds, M.D.; Coombe, D.R. Interactions between Skeletal Muscle Myoblasts and Their Extracellular Matrix Revealed by a Serum Free Culture System. PLoS ONE 2015, 10, e0127675. [Google Scholar] [CrossRef] [PubMed]
- Fry, C.S.; Kirby, T.J.; Kosmac, K.; McCarthy, J.J.; Peterson, C.A. Myogenic Progenitor Cells Control Extracellular Matrix Production by Fibroblasts during Skeletal Muscle Hypertrophy. Cell Stem Cell 2017, 20, 56–69. [Google Scholar] [CrossRef] [PubMed]
- Chazaud, B. Inflammation during Skeletal Muscle Regeneration and Tissue Remodeling: Application to Exercise-induced Muscle Damage Management. Immunol. Cell Biol. 2016, 94, 140–145. [Google Scholar] [CrossRef] [PubMed]
- Almada, A.E.; Wagers, A.J. Molecular Circuitry of Stem Cell Fate in Skeletal Muscle Regeneration, Ageing and Disease. Nat. Rev. Mol. Cell Biol. 2016, 17, 267–279. [Google Scholar] [CrossRef] [PubMed]
- Yin, H.; Price, F.; Rudnicki, M.A. Satellite Cells and the Muscle Stem Cell Niche. Physiol. Rev. 2013, 93, 23–67. [Google Scholar] [CrossRef]
- Gillies, A.R.; Lieber, R.L. Structure and Function of the Skeletal Muscle Extracellular Matrix. Muscle Nerve 2011, 44, 318–331. [Google Scholar] [CrossRef]
- Zhang, W.; Liu, Y.; Zhang, H. Extracellular Matrix: An Important Regulator of Cell Functions and Skeletal Muscle Development. Cell Biosci. 2021, 11, 65. [Google Scholar] [CrossRef]
- Purslow, P.P. The Structure and Role of Intramuscular Connective Tissue in Muscle Function. Front. Physiol. 2020, 11, 495. [Google Scholar] [CrossRef]
- Carraro, E.; Rossi, L.; Maghin, E.; Canton, M.; Piccoli, M. 3D in Vitro Models of Pathological Skeletal Muscle: Which Cells and Scaffolds to Elect? Front. Bioeng. Biotechnol. 2022, 10, 941623. [Google Scholar] [CrossRef]
- Grounds, M.D. Complexity of Extracellular Matrix and Skeletal Muscle Regeneration. In Skeletal Muscle Repair and Regeneration; Advances in Muscle Research; Springer: Dordrecht, The Netherlands, 2008; Volume 3, pp. 269–302. ISBN 978-1-4020-6767-9. [Google Scholar]
- Gumpenberger, M.; Wessner, B.; Graf, A.; Narici, M.V.; Fink, C.; Braun, S.; Hoser, C.; Blazevich, A.J.; Csapo, R. Remodeling the Skeletal Muscle Extracellular Matrix in Older Age—Effects of Acute Exercise Stimuli on Gene Expression. Int. J. Mol. Sci. 2020, 21, 7089. [Google Scholar] [CrossRef] [PubMed]
- Csapo, R.; Gumpenberger, M.; Wessner, B. Skeletal Muscle Extracellular Matrix–What Do We Know About Its Composition, Regulation, and Physiological Roles? A Narrative Review. Front. Physiol. 2020, 11, 253. [Google Scholar] [CrossRef] [PubMed]
- Light, N.; Champion, A.E. Characterization of Muscle Epimysium, Perimysium and Endomysium Collagens. Biochem. J. 1984, 219, 1017–1026. [Google Scholar] [CrossRef] [PubMed]
- Sleboda, D.A.; Stover, K.K.; Roberts, T.J. Diversity of Extracellular Matrix Morphology in Vertebrate Skeletal Muscle. J. Morphol. 2020, 281, 160–169. [Google Scholar] [CrossRef] [PubMed]
- Hantaï, D.; Labat-robert, J.; Grimaud, J.-A.; Fardeau, M. Fibronectin, Laminin, Type I, III and IV Collagens in Duchenne’s Muscular Dystrophy, Congenital Muscular Dystrophies and Congenital Myopathies: An Immunocytochemical Study. Connect. Tissue Res. 1985, 13, 273–281. [Google Scholar] [CrossRef]
- DiMario, J.; Buffinger, N.; Yamada, S.; Strohman, R.C. Fibroblast Growth Factor in the Extracellular Matrix of Dystrophic (Mdx) Mouse Muscle. Science 1989, 244, 688–690. [Google Scholar] [CrossRef] [PubMed]
- Kjær, M. Role of Extracellular Matrix in Adaptation of Tendon and Skeletal Muscle to Mechanical Loading. Physiol. Rev. 2004, 84, 649–698. [Google Scholar] [CrossRef]
- Kragstrup, T.W.; Kjaer, M.; Mackey, A.L. Structural, Biochemical, Cellular, and Functional Changes in Skeletal Muscle Extracellular Matrix with Aging. Scand. J. Med. Sci. Sports 2011, 21, 749–757. [Google Scholar] [CrossRef]
- Csapo, R.; Malis, V.; Sinha, U.; Du, J.; Sinha, S. Age-Associated Differences in Triceps Surae Muscle Composition and Strength–An MRI-Based Cross-Sectional Comparison of Contractile, Adipose and Connective Tissue. BMC Musculoskelet. Disord. 2014, 15, 209. [Google Scholar] [CrossRef]
- Haus, J.M.; Carrithers, J.A.; Trappe, S.W.; Trappe, T.A. Collagen, Cross-Linking, and Advanced Glycation End Products in Aging Human Skeletal Muscle. J. Appl. Physiol. 2007, 103, 2068–2076. [Google Scholar] [CrossRef]
- Olson, L.C.; Redden, J.T.; Schwartz, Z.; Cohen, D.J.; McClure, M.J. Advanced Glycation End-Products in Skeletal Muscle Aging. Bioengineering 2021, 8, 168. [Google Scholar] [CrossRef] [PubMed]
- Hindle, A.G.; Horning, M.; Mellish, J.-A.E.; Lawler, J.M. Diving into Old Age: Muscular Senescence in a Large-Bodied, Long-Lived Mammal, the Weddell Seal (Leptonychotes weddellii). J. Exp. Biol. 2009, 212, 790–796. [Google Scholar] [CrossRef] [PubMed]
- Kovanen, V.; Suominen, H.; Risteli, J.; Risteli, L. Type IV Collagen and Laminin in Slow and Fast Skeletal Muscle in Rats—Effects of Age and Life-Time Endurance Training. Coll. Relat. Res. 1988, 8, 145–153. [Google Scholar] [CrossRef] [PubMed]
- Nederveen, J.P.; Joanisse, S.; Thomas, A.C.Q.; Snijders, T.; Manta, K.; Bell, K.E.; Phillips, S.M.; Kumbhare, D.; Parise, G. Age-related Changes to the Satellite Cell Niche Are Associated with Reduced Activation Following Exercise. FASEB J. 2020, 34, 8975–8989. [Google Scholar] [CrossRef] [PubMed]
- Garg, K.; Boppart, M.D. Influence of Exercise and Aging on Extracellular Matrix Composition in the Skeletal Muscle Stem Cell Niche. J. Appl. Physiol. 2016, 121, 1053–1058. [Google Scholar] [CrossRef] [PubMed]
- Shi, X.; Garry, D.J. Muscle Stem Cells in Development, Regeneration, and Disease. Genes Dev. 2006, 20, 1692–1708. [Google Scholar] [CrossRef]
- Labat-Robert, J. Age-Dependent Remodeling of Connective Tissue: Role of Fibronectin and Laminin. Pathol. Biol. 2003, 51, 563–568. [Google Scholar] [CrossRef]
- Aslam, M.A.; Ma, E.B.; Huh, J.Y. Pathophysiology of Sarcopenia: Genetic Factors and Their Interplay with Environmental Factors. Metabolism 2023, 149, 155711. [Google Scholar] [CrossRef]
- Wilkinson, D.J.; Piasecki, M.; Atherton, P.J. The Age-Related Loss of Skeletal Muscle Mass and Function: Measurement and Physiology of Muscle Fibre Atrophy and Muscle Fibre Loss in Humans. Ageing Res. Rev. 2018, 47, 123–132. [Google Scholar] [CrossRef]
- Cho, M.-R.; Lee, S.; Song, S.-K. A Review of Sarcopenia Pathophysiology, Diagnosis, Treatment and Future Direction. J. Korean Med. Sci. 2022, 37, e146. [Google Scholar] [CrossRef]
- Arango-Lopera, V.E.; Arroyo, P.; Gutiérrez-Robledo, L.M.; Perez-Zepeda, M.U.; Cesari, M. Mortality as an Adverse Outcome of Sarcopenia. J. Nutr. Health Aging 2013, 17, 259–262. [Google Scholar] [CrossRef] [PubMed]
- Ali, S.; Garcia, J.M. Sarcopenia, Cachexia and Aging: Diagnosis, Mechanisms and Therapeutic Options—A Mini-Review. Gerontology 2014, 60, 294–305. [Google Scholar] [CrossRef] [PubMed]
- Deschenes, M.R. Effects of Aging on Muscle Fibre Type and Size. Sports Med. 2004, 34, 809–824. [Google Scholar] [CrossRef] [PubMed]
- McCormick, R.; Vasilaki, A. Age-Related Changes in Skeletal Muscle: Changes to Life-Style as a Therapy. Biogerontology 2018, 19, 519–536. [Google Scholar] [CrossRef] [PubMed]
- Chondrogianni, N.; Petropoulos, I.; Franceschi, C.; Friguet, B.; Gonos, E.S. Fibroblast Cultures from Healthy Centenarians Have an Active Proteasome. Exp. Gerontol. 2000, 35, 721–728. [Google Scholar] [CrossRef] [PubMed]
- Shefer, G.; Van De Mark, D.P.; Richardson, J.B.; Yablonka-Reuveni, Z. Satellite-Cell Pool Size Does Matter: Defining the Myogenic Potency of Aging Skeletal Muscle. Dev. Biol. 2006, 294, 50–66. [Google Scholar] [CrossRef] [PubMed]
- Verdijk, L.B.; Snijders, T.; Drost, M.; Delhaas, T.; Kadi, F.; Van Loon, L.J.C. Satellite Cells in Human Skeletal Muscle; from Birth to Old Age. AGE 2014, 36, 545–557. [Google Scholar] [CrossRef]
- Frontera, W.R.; Rodriguez Zayas, A.; Rodriguez, N. Aging of Human Muscle: Understanding Sarcopenia at the Single Muscle Cell Level. Phys. Med. Rehabil. Clin. N. Am. 2012, 23, 201–207. [Google Scholar] [CrossRef]
- Ciciliot, S.; Rossi, A.C.; Dyar, K.A.; Blaauw, B.; Schiaffino, S. Muscle Type and Fiber Type Specificity in Muscle Wasting. Int. J. Biochem. Cell Biol. 2013, 45, 2191–2199. [Google Scholar] [CrossRef]
- Edström, E.; Altun, M.; Bergman, E.; Johnson, H.; Kullberg, S.; Ramírez-León, V.; Ulfhake, B. Factors Contributing to Neuromuscular Impairment and Sarcopenia during Aging. Physiol. Behav. 2007, 92, 129–135. [Google Scholar] [CrossRef]
- Fulop, T.; Larbi, A.; Dupuis, G.; Le Page, A.; Frost, E.H.; Cohen, A.A.; Witkowski, J.M.; Franceschi, C. Immunosenescence and Inflamm-Aging As Two Sides of the Same Coin: Friends or Foes? Front. Immunol. 2018, 8, 1960. [Google Scholar] [CrossRef] [PubMed]
- Walrand, S.; Zangarelli, A.; Guillet, C.; Salles, J.; Soulier, K.; Giraudet, C.; Patrac, V.; Boirie, Y. Effect of Fast Dietary Proteins on Muscle Protein Synthesis Rate and Muscle Strength in Ad Libitum-Fed and Energy-Restricted Old Rats. Br. J. Nutr. 2011, 106, 1683–1690. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.H.; Hood, D.A. Age-associated Mitochondrial Dysfunction in Skeletal Muscle: Contributing Factors and Suggestions for Long-term Interventions. IUBMB Life 2009, 61, 201–214. [Google Scholar] [CrossRef] [PubMed]
- Ferri, E.; Marzetti, E.; Calvani, R.; Picca, A.; Cesari, M.; Arosio, B. Role of Age-Related Mitochondrial Dysfunction in Sarcopenia. Int. J. Mol. Sci. 2020, 21, 5236. [Google Scholar] [CrossRef] [PubMed]
- Ji, L.L. Exercise at Old Age: Does It Increase or Alleviate Oxidative Stress? Ann. N. Y. Acad. Sci. 2001, 928, 236–247. [Google Scholar] [CrossRef] [PubMed]
- Dhillon, R.J.S.; Hasni, S. Pathogenesis and Management of Sarcopenia. Clin. Geriatr. Med. 2017, 33, 17–26. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.; Sances, S.; Workman, M.J.; Svendsen, C.N. Multi-Lineage Human iPSC-Derived Platforms for Disease Modeling and Drug Discovery. Cell Stem Cell 2020, 26, 309–329. [Google Scholar] [CrossRef]
- Maffioletti, S.M.; Sarcar, S.; Henderson, A.B.H.; Mannhardt, I.; Pinton, L.; Moyle, L.A.; Steele-Stallard, H.; Cappellari, O.; Wells, K.E.; Ferrari, G.; et al. Three-Dimensional Human iPSC-Derived Artificial Skeletal Muscles Model Muscular Dystrophies and Enable Multilineage Tissue Engineering. Cell Rep. 2018, 23, 899–908. [Google Scholar] [CrossRef]
- Dessauge, F.; Schleder, C.; Perruchot, M.-H.; Rouger, K. 3D in Vitro Models of Skeletal Muscle: Myopshere, Myobundle and Bioprinted Muscle Construct. Vet. Res. 2021, 52, 72. [Google Scholar] [CrossRef]
- Dalmao-Fernandez, A.; Aizenshtadt, A.; Bakke, H.G.; Krauss, S.; Rustan, A.C.; Thoresen, G.H.; Kase, E.T. Development of Three-Dimensional Primary Human Myospheres as Culture Model of Skeletal Muscle Cells for Metabolic Studies. Front. Bioeng. Biotechnol. 2023, 11, 1130693. [Google Scholar] [CrossRef]
- Thomas, K.; Engler, A.J.; Meyer, G.A. Extracellular Matrix Regulation in the Muscle Satellite Cell Niche. Connect. Tissue Res. 2015, 56, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Khodabukus, A.; Baar, K. Factors That Affect Tissue-Engineered Skeletal Muscle Function and Physiology. Cells Tissues Organs 2016, 202, 159–168. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Smith, S.H.; Iijima, H.; Hettinger, Z.R.; Mallepally, A.; Shroff, S.G.; Ambrosio, F. Bioengineered 3D Skeletal Muscle Model Reveals Complement 4b as a Cell-Autonomous Mechanism of Impaired Regeneration with Aging. Adv. Mater. 2023, 35, 2207443. [Google Scholar] [CrossRef] [PubMed]
- Alave Reyes-Furrer, A.; De Andrade, S.; Bachmann, D.; Jeker, H.; Steinmann, M.; Accart, N.; Dunbar, A.; Rausch, M.; Bono, E.; Rimann, M.; et al. Matrigel 3D Bioprinting of Contractile Human Skeletal Muscle Models Recapitulating Exercise and Pharmacological Responses. Commun. Biol. 2021, 4, 1183. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Ju, Y.M.; Kim, I.; Elsangeedy, E.; Lee, J.H.; Yoo, J.J.; Atala, A.; Lee, S.J. A Novel Decellularized Skeletal Muscle-Derived ECM Scaffolding System for in Situ Muscle Regeneration. Methods 2020, 171, 77–85. [Google Scholar] [CrossRef] [PubMed]
- Choi, Y.; Kim, T.G.; Jeong, J.; Yi, H.; Park, J.W.; Hwang, W.; Cho, D. 3D Cell Printing of Functional Skeletal Muscle Constructs Using Skeletal Muscle-Derived Bioink. Adv. Healthc. Mater. 2016, 5, 2636–2645. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Garibay, X.; Ortega, M.A.; Cerro-Herreros, E.; Comelles, J.; Martínez, E.; Artero, R.; Fernández-Costa, J.M.; Ramón-Azcón, J. Bioengineered in Vitro 3D Model of Myotonic Dystrophy Type 1 Human Skeletal Muscle. Biofabrication 2021, 13, 035035. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Costa, J.M.; Fernández-Garibay, X.; Velasco-Mallorquí, F.; Ramón-Azcón, J. Bioengineered in Vitro Skeletal Muscles as New Tools for Muscular Dystrophies Preclinical Studies. J. Tissue Eng. 2021, 12, 204173142098133. [Google Scholar] [CrossRef]
- Zhuang, P.; An, J.; Chua, C.K.; Tan, L.P. Bioprinting of 3D in Vitro Skeletal Muscle Models: A Review. Mater. Des. 2020, 193, 108794. [Google Scholar] [CrossRef]
- Beldjilali-Labro, M.; Jellali, R.; Brown, A.D.; Garcia Garcia, A.; Lerebours, A.; Guenin, E.; Bedoui, F.; Dufresne, M.; Stewart, C.; Grosset, J.-F.; et al. Multiscale-Engineered Muscle Constructs: PEG Hydrogel Micro-Patterning on an Electrospun PCL Mat Functionalized with Gold Nanoparticles. Int. J. Mol. Sci. 2021, 23, 260. [Google Scholar] [CrossRef]
- Charest, J.L.; García, A.J.; King, W.P. Myoblast Alignment and Differentiation on Cell Culture Substrates with Microscale Topography and Model Chemistries. Biomaterials 2007, 28, 2202–2210. [Google Scholar] [CrossRef] [PubMed]
- Tran, K.T.M.; Nguyen, T.D. Lithography-Based Methods to Manufacture Biomaterials at Small Scales. J. Sci. Adv. Mater. Devices 2017, 2, 1–14. [Google Scholar] [CrossRef]
- LaFratta, C.N.; Li, L.; Fourkas, J.T. Soft-Lithographic Replication of 3D Microstructures with Closed Loops. Proc. Natl. Acad. Sci. USA 2006, 103, 8589–8594. [Google Scholar] [CrossRef] [PubMed]
- Lim, Y.C.; Johnson, J.; Fei, Z.; Wu, Y.; Farson, D.F.; Lannutti, J.J.; Choi, H.W.; Lee, L.J. Micropatterning and Characterization of Electrospun Poly(Ε-caprolactone)/Gelatin Nanofiber Tissue Scaffolds by Femtosecond Laser Ablation for Tissue Engineering Applications. Biotechnol. Bioeng. 2011, 108, 116–126. [Google Scholar] [CrossRef] [PubMed]
- Okano, K.; Matsui, A.; Maezawa, Y.; Hee, P.-Y.; Matsubara, M.; Yamamoto, H.; Hosokawa, Y.; Tsubokawa, H.; Li, Y.-K.; Kao, F.-J.; et al. In Situ Laser Micropatterning of Proteins for Dynamically Arranging Living Cells. Lab Chip 2013, 13, 4078. [Google Scholar] [CrossRef] [PubMed]
- Koo, S.; Santoni, S.M.; Gao, B.Z.; Grigoropoulos, C.P.; Ma, Z. Laser-Assisted Biofabrication in Tissue Engineering and Regenerative Medicine. J. Mater. Res. 2017, 32, 128–142. [Google Scholar] [CrossRef]
- Yi, H.-G.; Kim, H.; Kwon, J.; Choi, Y.-J.; Jang, J.; Cho, D.-W. Application of 3D Bioprinting in the Prevention and the Therapy for Human Diseases. Signal Transduct. Target. Ther. 2021, 6, 177. [Google Scholar] [CrossRef]
- Mestre, R.; García, N.; Patiño, T.; Guix, M.; Fuentes, J.; Valerio-Santiago, M.; Almiñana, N.; Sánchez, S. 3D-Bioengineered Model of Human Skeletal Muscle Tissue with Phenotypic Features of Aging for Drug Testing Purposes. Biofabrication. 2021, 13. [Google Scholar] [CrossRef]
- Kim, W.; Lee, H.; Lee, J.; Atala, A.; Yoo, J.J.; Lee, S.J.; Kim, G.H. Efficient Myotube Formation in 3D Bioprinted Tissue Construct by Biochemical and Topographical Cues. Biomaterials 2020, 230, 119632. [Google Scholar] [CrossRef]
- García-Lizarribar, A.; Villasante, A.; Lopez-Martin, J.A.; Flandez, M.; Soler-Vázquez, M.C.; Serra, D.; Herrero, L.; Sagrera, A.; Efeyan, A.; Samitier, J. 3D Bioprinted Functional Skeletal Muscle Models Have Potential Applications for Studies of Muscle Wasting in Cancer Cachexia. Biomater. Adv. 2023, 150, 213426. [Google Scholar] [CrossRef]
- Huang, N.F.; Patel, S.; Thakar, R.G.; Wu, J.; Hsiao, B.S.; Chu, B.; Lee, R.J.; Li, S. Myotube Assembly on Nanofibrous and Micropatterned Polymers. Nano Lett. 2006, 6, 537–542. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.-X.; Yap, C.C.; He, J.; Chen, C.; Wong, S.Y.; Li, X. Electrospinning: A Facile Technique for Fabricating Functional Nanofibers for Environmental Applications. Nanotechnol. Rev. 2016, 5, 51–73. [Google Scholar] [CrossRef]
- Zulkifli, M.Z.A.; Nordin, D.; Shaari, N.; Kamarudin, S.K. Overview of Electrospinning for Tissue Engineering Applications. Polymers 2023, 15, 2418. [Google Scholar] [CrossRef] [PubMed]
- Politi, S.; Carotenuto, F.; Rinaldi, A.; Di Nardo, P.; Manzari, V.; Albertini, M.C.; Araneo, R.; Ramakrishna, S.; Teodori, L. Smart ECM-Based Electrospun Biomaterials for Skeletal Muscle Regeneration. Nanomaterials 2020, 10, 1781. [Google Scholar] [CrossRef] [PubMed]
- Soliman, E.; Bianchi, F.; Sleigh, J.N.; George, J.H.; Cader, M.Z.; Cui, Z.; Ye, H. Engineered Method for Directional Growth of Muscle Sheets on Electrospun Fibers. J. Biomed. Mater. Res. A 2018, 106, 1165–1176. [Google Scholar] [CrossRef]
- Fattahi, P.; Dover, J.T.; Brown, J.L. 3D Near-Field Electrospinning of Biomaterial Microfibers with Potential for Blended Microfiber-Cell-Loaded Gel Composite Structures. Adv. Healthc. Mater. 2017, 6, 1700456. [Google Scholar] [CrossRef]
- Reizabal, A.; Kangur, T.; Saiz, P.G.; Menke, S.; Moser, C.; Brugger, J.; Dalton, P.D.; Luposchainsky, S. MEWron: An Open-Source Melt Electrowriting Platform. Addit. Manuf. 2023, 71, 103604. [Google Scholar] [CrossRef]
- Auluck, A.; Mudera, V.; Hunt, N.P.; Lewis, M.P. A Three-dimensional in Vitro Model System to Study the Adaptation of Craniofacial Skeletal Muscle Following Mechanostimulation. Eur. J. Oral Sci. 2005, 113, 218–224. [Google Scholar] [CrossRef]
- Maghin, E.; Carraro, E.; Boso, D.; Dedja, A.; Giagante, M.; Caccin, P.; Barna, R.A.-M.; Bresolin, S.; Cani, A.; Borile, G.; et al. Customized Bioreactor Enables the Production of 3D Diaphragmatic Constructs Influencing Matrix Remodeling and Fibroblast Overgrowth. Npj Regen. Med. 2022, 7, 25. [Google Scholar] [CrossRef]
- Powell, C.A.; Smiley, B.L.; Mills, J.; Vandenburgh, H.H. Mechanical Stimulation Improves Tissue-Engineered Human Skeletal Muscle. Am. J. Physiol.-Cell Physiol. 2002, 283, C1557–C1565. [Google Scholar] [CrossRef]
- Juhas, M.; Ye, J.; Bursac, N. Design, Evaluation, and Application of Engineered Skeletal Muscle. Methods 2016, 99, 81–90. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, T.; Sasaki, J.-I.; Alsberg, E.; Egusa, H.; Yatani, H.; Sohmura, T. Three-Dimensional Cell and Tissue Patterning in a Strained Fibrin Gel System. PLoS ONE 2007, 2, e1211. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Liu, J.; Wang, X.; Feng, L.; Wu, J.; Zhu, X.; Wen, W.; Gong, X. Organ-on-a-Chip: Recent Breakthroughs and Future Prospects. BioMed. Eng. OnLine 2020, 19, 9. [Google Scholar] [CrossRef] [PubMed]
- Kaarj, K.; Yoon, J.-Y. Methods of Delivering Mechanical Stimuli to Organ-on-a-Chip. Micromachines 2019, 10, 700. [Google Scholar] [CrossRef] [PubMed]
- Smoak, M.M.; Pearce, H.A.; Mikos, A.G. Microfluidic Devices for Disease Modeling in Muscle Tissue. Biomaterials 2019, 198, 250–258. [Google Scholar] [CrossRef] [PubMed]
- Khodabukus, A.; Madden, L.; Prabhu, N.K.; Koves, T.R.; Jackman, C.P.; Muoio, D.M.; Bursac, N. Electrical Stimulation Increases Hypertrophy and Metabolic Flux in Tissue-Engineered Human Skeletal Muscle. Biomaterials 2019, 198, 259–269. [Google Scholar] [CrossRef] [PubMed]
- Pedrotty, D.M.; Koh, J.; Davis, B.H.; Taylor, D.A.; Wolf, P.; Niklason, L.E. Engineering Skeletal Myoblasts: Roles of Three-Dimensional Culture and Electrical Stimulation. Am. J. Physiol.-Heart Circ. Physiol. 2005, 288, H1620–H1626. [Google Scholar] [CrossRef]
- McKeon-Fischer, K.D.; Browe, D.P.; Olabisi, R.M.; Freeman, J.W. Poly(3,4-ethylenedioxythiophene) Nanoparticle and Poly(Ɛ-caprolactone) Electrospun Scaffold Characterization for Skeletal Muscle Regeneration. J. Biomed. Mater. Res. A 2015, 103, 3633–3641. [Google Scholar] [CrossRef]
- Jing, Q.; Law, J.Y.; Tan, L.P.; Silberschmidt, V.V.; Li, L.; Dong, Z. Preparation, Characterization and Properties of Polycaprolactone Diol-Functionalized Multi-Walled Carbon Nanotube/Thermoplastic Polyurethane Composite. Compos. Part Appl. Sci. Manuf. 2015, 70, 8–15. [Google Scholar] [CrossRef]
- Abedi, A.; Hasanzadeh, M.; Tayebi, L. Conductive Nanofibrous Chitosan/PEDOT:PSS Tissue Engineering Scaffolds. Mater. Chem. Phys. 2019, 237, 121882. [Google Scholar] [CrossRef]
- Chaudhuri, B.; Mondal, B.; Kumar, S.; Sarkar, S.C. Myoblast Differentiation and Protein Expression in Electrospun Graphene Oxide (GO)-Poly (ε-Caprolactone, PCL) Composite Meshes. Mater. Lett. 2016, 182, 194–197. [Google Scholar] [CrossRef]
- Robin, G.; Allard, B. Dihydropyridine Receptors Actively Control Gating of Ryanodine Receptors in Resting Mouse Skeletal Muscle Fibres. J. Physiol. 2012, 590, 6027–6036. [Google Scholar] [CrossRef] [PubMed]
- Kuo, I.Y.; Ehrlich, B.E. Signaling in Muscle Contraction. Cold Spring Harb. Perspect. Biol. 2015, 7, a006023. [Google Scholar] [CrossRef] [PubMed]
- Patton, B.L. Basal Lamina and the Organization of Neuromuscular Synapses. J. Neurocytol. 2003, 32, 883–903. [Google Scholar] [CrossRef]
- Sanes, J.R. The Basement Membrane/Basal Lamina of Skeletal Muscle. J. Biol. Chem. 2003, 278, 12601–12604. [Google Scholar] [CrossRef] [PubMed]
- Dobrowolny, G.; Barbiera, A.; Sica, G.; Scicchitano, B.M. Age-Related Alterations at Neuromuscular Junction: Role of Oxidative Stress and Epigenetic Modifications. Cells 2021, 10, 1307. [Google Scholar] [CrossRef] [PubMed]
- Arnold, W.D.; Clark, B.C. Neuromuscular Junction Transmission Failure in Aging and Sarcopenia: The Nexus of the Neurological and Muscular Systems. Ageing Res. Rev. 2023, 89, 101966. [Google Scholar] [CrossRef] [PubMed]
- Vila, O.F.; Qu, Y.; Vunjak-Novakovic, G. In Vitro Models of Neuromuscular Junctions and Their Potential for Novel Drug Discovery and Development. Expert Opin. Drug Discov. 2020, 15, 307–317. [Google Scholar] [CrossRef]
- Zhou, L.; Chen, W.; Jiang, S.; Xu, R. In Vitro Models of Amyotrophic Lateral Sclerosis. Cell. Mol. Neurobiol. 2023, 43, 3783–3799. [Google Scholar] [CrossRef]
- Osaki, T.; Uzel, S.G.M.; Kamm, R.D. Microphysiological 3D Model of Amyotrophic Lateral Sclerosis (ALS) from Human iPS-Derived Muscle Cells and Optogenetic Motor Neurons. Sci. Adv. 2018, 4, eaat5847. [Google Scholar] [CrossRef]
- Massih, B.; Veh, A.; Schenke, M.; Mungwa, S.; Seeger, B.; Selvaraj, B.T.; Chandran, S.; Reinhardt, P.; Sterneckert, J.; Hermann, A.; et al. A 3D Cell Culture System for Bioengineering Human Neuromuscular Junctions to Model ALS. Front. Cell Dev. Biol. 2023, 11, 996952. [Google Scholar] [CrossRef] [PubMed]
- Harley, P.; Paredes-Redondo, A.; Grenci, G.; Viasnoff, V.; Lin, Y.-; Lieberam, I. 3D Compartmentalised Human Pluripotent Stem Cell–Derived Neuromuscular Co-Cultures. Bio-Protocol 2023, 13, e4624. [Google Scholar] [CrossRef]
- Uzel, S.G.M.; Platt, R.J.; Subramanian, V.; Pearl, T.M.; Rowlands, C.J.; Chan, V.; Boyer, L.A.; So, P.T.C.; Kamm, R.D. Microfluidic Device for the Formation of Optically Excitable, Three-Dimensional, Compartmentalized Motor Units. Sci. Adv. 2016, 2, e1501429. [Google Scholar] [CrossRef] [PubMed]
- Leng, Y.; Li, X.; Zheng, F.; Liu, H.; Wang, C.; Wang, X.; Liao, Y.; Liu, J.; Meng, K.; Yu, J.; et al. Advances in In Vitro Models of Neuromuscular Junction: Focusing on Organ-on-a-Chip, Organoids, and Biohybrid Robotics. Adv. Mater. 2023, 35, 2211059. [Google Scholar] [CrossRef]
- Arjmand, B.; Kokabi Hamidpour, S.; Rabbani, Z.; Tayanloo-Beik, A.; Rahim, F.; Aghayan, H.R.; Larijani, B. Organ on a Chip: A Novel in Vitro Biomimetic Strategy in Amyotrophic Lateral Sclerosis (ALS) Modeling. Front. Neurol. 2022, 12, 788462. [Google Scholar] [CrossRef] [PubMed]
- Turksen, K. (Ed.) Stem Cells and Lineage Commitment: Methods and Protocols; Methods in Molecular Biology; Springer: New York, NY, USA, 2023; Volume 2736, ISBN 978-1-07-163536-0. [Google Scholar]
- Park, H.S.; Liu, S.; McDonald, J.; Thakor, N.; Yang, I.H. Neuromuscular Junction in a Microfluidic Device. In Proceedings of the 2013 35th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Osaka, Japan, 3–7 July 2013; IEEE: Piscataway, NJ, USA, 2013; pp. 2833–2835. [Google Scholar]
- Gazzola, M.; Martinat, C. Unlocking the Complexity of Neuromuscular Diseases: Insights from Human Pluripotent Stem Cell-Derived Neuromuscular Junctions. Int. J. Mol. Sci. 2023, 24, 15291. [Google Scholar] [CrossRef] [PubMed]
- Southam, K.A.; King, A.E.; Blizzard, C.A.; McCormack, G.H.; Dickson, T.C. Microfluidic Primary Culture Model of the Lower Motor Neuron–Neuromuscular Junction Circuit. J. Neurosci. Methods 2013, 218, 164–169. [Google Scholar] [CrossRef]
- Stoklund Dittlau, K.; Terrie, L.; Baatsen, P.; Kerstens, A.; De Swert, L.; Janky, R.; Corthout, N.; Masrori, P.; Van Damme, P.; Hyttel, P.; et al. FUS-ALS hiPSC-Derived Astrocytes Impair Human Motor Units through Both Gain-of-Toxicity and Loss-of-Support Mechanisms. Mol. Neurodegener. 2023, 18, 5. [Google Scholar] [CrossRef]
- Peyrin, J.-M.; Deleglise, B.; Saias, L.; Vignes, M.; Gougis, P.; Magnifico, S.; Betuing, S.; Pietri, M.; Caboche, J.; Vanhoutte, P.; et al. Axon Diodes for the Reconstruction of Oriented Neuronal Networks in Microfluidic Chambers. Lab Chip 2011, 11, 3663. [Google Scholar] [CrossRef]
- Yamamoto, K.; Yamaoka, N.; Imaizumi, Y.; Nagashima, T.; Furutani, T.; Ito, T.; Okada, Y.; Honda, H.; Shimizu, K. Development of a Human Neuromuscular Tissue-on-a-Chip Model on a 24-Well-Plate-Format Compartmentalized Microfluidic Device. Lab Chip 2021, 21, 1897–1907. [Google Scholar] [CrossRef]
- Natarajan, A.; Sethumadhavan, A.; Krishnan, U.M. Toward Building the Neuromuscular Junction: In Vitro Models To Study Synaptogenesis and Neurodegeneration. ACS Omega 2019, 4, 12969–12977. [Google Scholar] [CrossRef] [PubMed]
- Cvetkovic, C.; Rich, M.H.; Raman, R.; Kong, H.; Bashir, R. A 3D-Printed Platform for Modular Neuromuscular Motor Units. Microsyst. Nanoeng. 2017, 3, 17015. [Google Scholar] [CrossRef] [PubMed]
| Material Re-Modeling Mechanism | Type of Interaction | Crosslinking Mechanism | Molecular Mechanism | Advantages | Examples | References |
|---|---|---|---|---|---|---|
Reversible crosslinks Reversible crosslinks | Non-covalent interactions | Host–guest complexes | Macrocyclic hosts with hydrophobic cavities and hydrophilic external surfaces (cyclodextrins, curcubit[n]urils, and calix[n]arenes) act as host molecules that encapsulate hydrophobic guest molecules, thus forming stable host–guest complexes | Specificity of the host–guest complex Ease of reaction Applied to a diverse range of materials | Functionalization of hyaluronic acid (HA) with mono-acryloyl cyclodextrin and subsequent complexation with either adamantane or cholic acid via host–guest chemistry. The studies performed with these materials showed that crosslinks with a large dissociation rate constant facilitated cell spreading and mechanosensing. | [75,76,77,78] |
| Hydrophobic interactions | Driven by the repulsion between hydrophobic groups and the aqueous environment | Ease of preparation Excellent mechanical and self-healing properties | α-helical coiled-coil peptide hydrogels Amphiphilic block copolymers | [79,80,81,82] | ||
| Hydrogen bonds | Secondary interactions that are weak in isolation but that lead to the formation of hydrogels with dynamic reversible crosslinks when present at significant numbers | Reliable and adaptable Self-healing Toughening effect through dissipation of external energy | Injectable four-arm PEG functionalized with either adenine or thymine. After mixing, a hydrogel was formed through hydrogen bonding between the nucleobases. Hydrogel formation via β-sheet assembly | [79,83,84] | ||
| Ionic interactions | Attractive or repulsive forces between charged molecules | Good solubility Rapid gelation | Alginate hydrogels crosslinked with divalent ions such as calcium | [79,84,85,86] | ||
| Covalent interactions | Dynamic covalent networks |
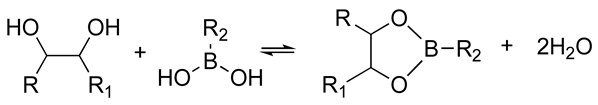
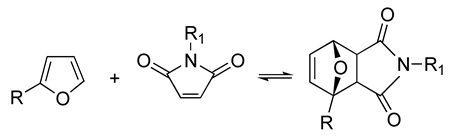
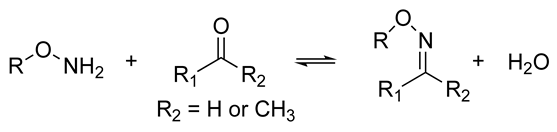
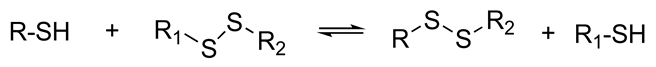
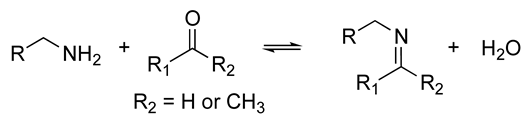
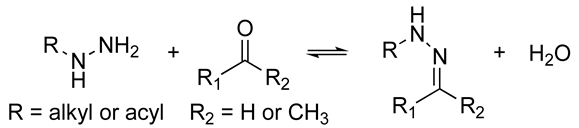 | The resulting hydrogels can be tuned to have a similar viscoelasticity to that of the native human ECM Dynamic covalent bonds can break and form on timescales that are comparable to those of cell-based matrix remodeling These reactions can be carried out at physiological pH and temperature These reactions proceed at relatively fast speeds, and their kinetics can be tailored to produce hydrogels with pre-determined viscoelastic properties | Self-healing hydrogel formed of a copolymer of 2-acrylamidophenylboronic acid (2-APBA) and N,N-dimethylacrylamide (DMA) mixed with poly (vinyl alcohol) (PVA) Self-healing dextran hydrogels formed via reaction between fulvene-modified hydrophilic dextran (diene) and dichloromaleic-acid-modified poly(ethylene glycol) (PEG) Sodium alginate hydrogel with tunable stress relaxation via reaction between alkoxyamine-functionalized alginate and aldehyde-containing oxidized alginate. The resulting hydrogel had calcium-mediated and oxime crosslinking, which led to a greater degree of tunability. Dynamic covalently crosslinked keratin hydrogels formed via thiol–disulfide exchange. The hydrogels showed injectability, self-healing, and redox-responsive capacity. Collagen hydrogels crosslinked with imine bonds had greater stress relaxation rates than collagen crosslinked with methacrylate bonds. The faster stress relaxation promoted cell spreading within one day. Formation of hydrazone bond between an aliphatic aldehyde-terminated multi-arm PEG and an aliphatic hydrazine-terminated multi-arm PEG, resulting in highly viscoelastic gels that promoted 3D cell spreading and the formation of multinucleate structures with a myotube-like morphology | [74,79,87,88,89,90,91,92,93] | |
| Chemically responsive moieties | Hydrolyzable ester linkages | Ester bonds are spontaneously hydrolyzed in water | Not dependent on the levels of enzyme present in the sample | Hyaluronic acid crosslinked with PEGDA to make the hydrogel susceptible to hydrolysis Polyesters, polyethers, polycarbonates | [94,95,96,97] | |
| Enzymatically-degradable peptide crosslinks | Degradation of hydrogels by cell-secreted enzymes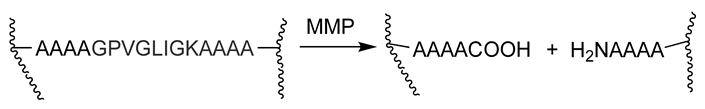 | Restricts degradation to regions of cell invasion Cell-mediated mechanism can couple the degradation rate with the rate of tissue formation The rate of degradation can be tuned by altering the peptide sequence | MMP-degradable hydrogels formed via reaction between 4arm-PEG tetravinyl sulfone and bis-cysteine peptide with an MMP-sensitive sequence | [98,99] |
| Stimuli | Molecular Mechanism | Principle | Advantages | Disadvantages | References |
|---|---|---|---|---|---|
| Temperature | Thermoresponsive polymers with upper or lower critical solution temperature (UCST or LCST), below which they are either insoluble or soluble, respectively | At the critical temperature, a change in the polymer solubility occurs and causes a change in molecular conformation | Easy to control the culture temperature | Temperature changes can affect cell viability and metabolic processes | [101,102] |
| Light | Incorporation of a photosensitive molecule such as azobenzene Photocleavable crosslinkers | Illumination changes the molecular conformation or induces a chemical reaction in the photoactivated moiety | Contact-free, easy, and precise on-demand control of stimulation | Possible chromophore toxicity if not covalently bound to the polymer Potential phototoxicity | [101] |
| Ultrasound | Crosslinker cleavage that causes changes in stiffness Disassembly of vesicles that leads to cargo release Gel transitions | High-frequency waves cause a rise in temperature and cavitation effects (growth and shrinkage or implosion of micro bubbles). The resulting pressure causes an alteration in the mechanical properties of the material. | Ease of application using an ultrasound transducer May be able to use existing material design without incorporating additives | Limited range of parameters that can be tuned in response to ultrasound | [101,103,104] |
| Electric field | Conductive polymers or incorporation of conductive materials into the polymer The deformation of a material in an electric field is influenced by variations in osmotic pressure, pH, electrode position, and the applied voltage | The application of an external electric field causes changes in the structural and mechanical properties of the material | Allows the generation of soft robotic materials that closely mimic human motor function Wide variety of electroresponsive materials | Difficulties optimizing the magnitude of the electric current Some of these materials have a low biocompatibility Many of the materials have poor mechanical strength and are brittle Cells respond to the electric field | [101,103,105,106,107] |
| Magnetic field | Materials that contain ferromagnetic structures | The application of a magnetic field triggers a reorganization in the magnetic structures that leads to changes in polymer structure, viscosity, or stiffness | Fast and reversible material modulation Magnets are easily included in culture systems | Possible leakage and toxicity of magnetic particles Materials containing magnetic particles are often opaque, making observing real-time changes in cell behavior challenging | [101,103,108,109,110] |
| Strain | Fiber reorganization and alignment, non-covalent interactions between fibers | The application of a mechanical strain leads to changes in the mechanical properties of the material | The strain can be externally applied or generated by cells Wide variety of materials with these properties | The application of strain can cause alterations in the topography of the material, such as the generation of wrinkles, which may serve as confounding variables It is difficult to separate the effects of cell-generated vs. user-generated strain effects | [101,103,111,112,113,114,115,116,117] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hidalgo-Alvarez, V.; Madl, C.M. Leveraging Biomaterial Platforms to Study Aging-Related Neural and Muscular Degeneration. Biomolecules 2024, 14, 69. https://doi.org/10.3390/biom14010069
Hidalgo-Alvarez V, Madl CM. Leveraging Biomaterial Platforms to Study Aging-Related Neural and Muscular Degeneration. Biomolecules. 2024; 14(1):69. https://doi.org/10.3390/biom14010069
Chicago/Turabian StyleHidalgo-Alvarez, Veronica, and Christopher M. Madl. 2024. "Leveraging Biomaterial Platforms to Study Aging-Related Neural and Muscular Degeneration" Biomolecules 14, no. 1: 69. https://doi.org/10.3390/biom14010069
APA StyleHidalgo-Alvarez, V., & Madl, C. M. (2024). Leveraging Biomaterial Platforms to Study Aging-Related Neural and Muscular Degeneration. Biomolecules, 14(1), 69. https://doi.org/10.3390/biom14010069






