Answering the Cell Stress Call: Satellite Non-Coding Transcription as a Response Mechanism
Abstract
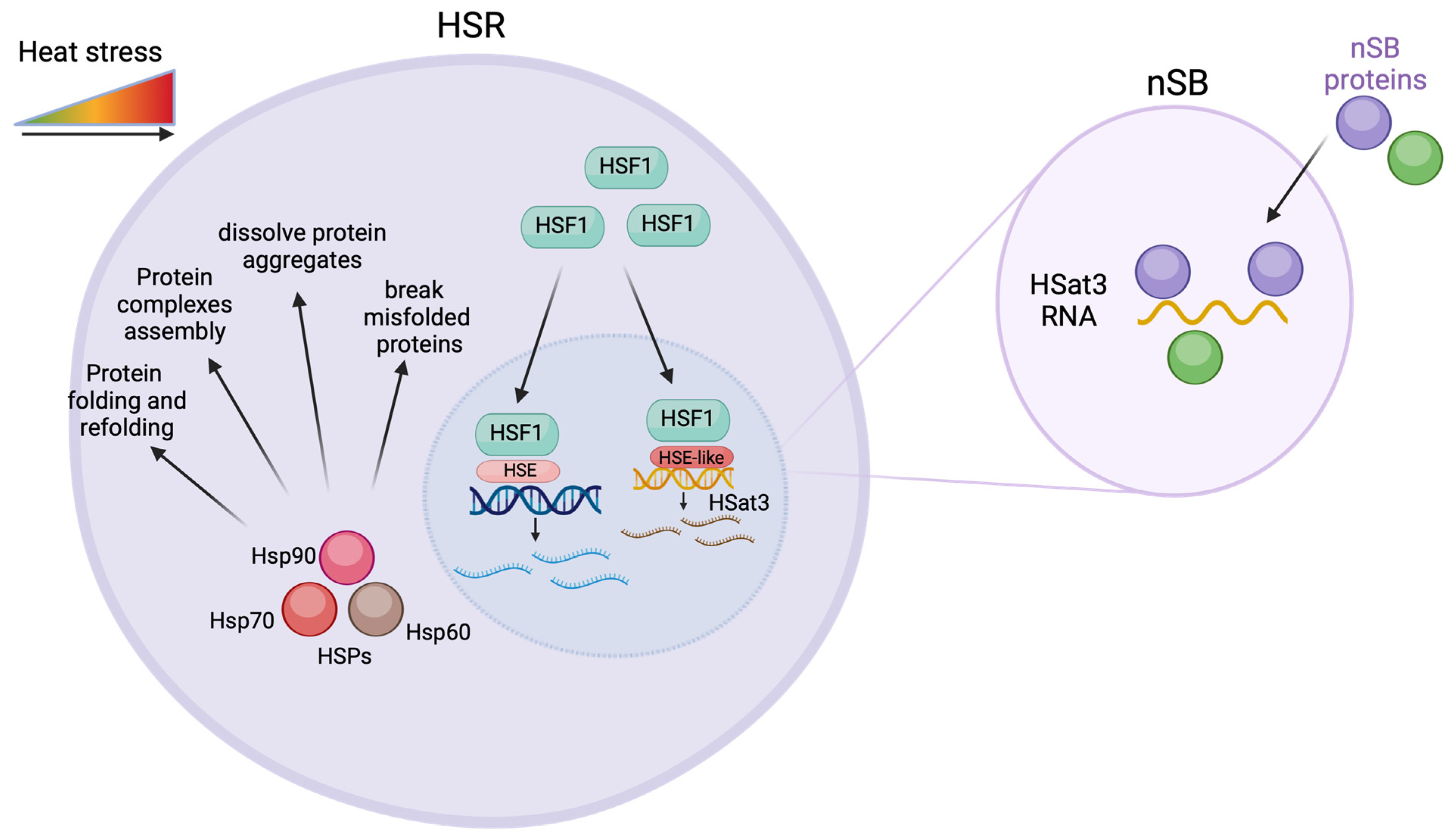
1. Introduction
2. Heat Shock Response Mechanisms as a Way to Counterpart Thermal and Oxidative Stress
2.1. Satellite DNA Transcription as a Shared Response to Thermal Stress among Different Species
| Stress | Condition | Satellite | Species | References |
|---|---|---|---|---|
| Heat stress | High temperature | HSat3 | Homo sapiens | [12,41] |
| HSat2 | Homo sapiens | [53,54] | ||
| αSat | Homo sapiens | [32] | ||
| γ-satellite | Mus musculus | [55] | ||
| TCAST1 | Tribolium castaneum | [56] | ||
| hsrω | Drosophila melanogaster | [38,39,40] | ||
| Oxidative Stress | H2O2 | HSat3 | Homo sapiens | [12] |
| PDX5 knockdown | HSat3 | Homo sapiens | [57] | |
| αSat | Homo sapiens | [57] | ||
| Osmotic Stress | Sorbitol | HSat3 | Homo sapiens | [12] |
| DNA damage response | UV-C | HSat3 | Homo sapiens | [12] |
| 5-FU, Cisplatin | HSat3 | Homo sapiens | [33] | |
| Zeocin, Etoposide | HSat2 | Homo sapiens | [58] | |
| Cytostatic drugs | γ-satellite | Mus musculus | [55] | |
| Zeocin, Etoposide | MiSat MaSat | Mus musculus | [34] |
2.2. Satellite DNA Transcription in the Response to Oxidative Stress
3. The Relation between Osmotic Stress and Satellite DNA Transcription
4. The Pathways of DNA Damage Response (DDR) Encompass Satellite DNA Transcription
5. Beyond Satellite Transcription: Satellite DNA Copy Number Variation in Response to Emotional Stress during Aging and Disease
6. Concluding Remarks
Author Contributions
Funding
Conflicts of Interest
List of Abbreviations
| 5-FU | Fluorouracil |
| ALL | Acute lymphoblastic leukemia |
| AP-1 | Activator protein-1 |
| ARPE-19 | Arising retinal pigment epithelia |
| ATM | Ataxia-telangiectasia mutated |
| CIN | Chromosomal instability |
| CLK1 | Protein kinase 1 |
| CPT | Camptothecin |
| CREBBP | CREB-binding protein |
| Daxx | Death domain associated protein |
| DDR | DNA damage repair |
| DSBs | Double-stranded DNA breaks |
| H3K9me2/3 | Histone 3 lysine 9 trimethylation |
| hnRNPs | Heterogeneous nuclear ribonucleoproteins |
| HP1 | Heterochromatin protein 1 |
| HS | Heat shock |
| HSat2 | Human Satellite 2 |
| HSat3 | Human Satellite 3 |
| HSE | Heat shock elements |
| HSF1 | Heat shock transcription factor 1 |
| HSFs | Heat shock factors |
| HSPs | Heat shock proteins |
| HSR | Heat shock response |
| Hsrω | Heat-shock RNA omega |
| MAPK | Mitogen-activated protein kinases |
| MaSat | Major satellite |
| MiSat | Minor satellite |
| ncRNAs | Non-coding RNAs |
| NF-κB | Nuclear factor-κB |
| NFAT5 | Nuclear factor of activated T-cells 5 |
| Nrf2 | Nuclear factor erythroid 2 related factor 2 |
| nSBs | Nuclear stress bodies |
| P53 | Protein 53 |
| PRDX5 | Peroxiredoxin-5 |
| ROS | Reactive oxygen species |
| satDNA | Satellite DNA |
| satncRNAs | Satellite non-coding RNAs |
| SRSF1 | Serine/arginine-rich splicing factors 1 |
| SRSF9 | Serine/arginine-rich splicing factors 9 |
| SRSFs | Serine/arginine-rich splicing factors |
| TCAST1 | Tribolium castaneum the major satellite |
| TonEBP | Transcription factor tone enhancer |
| TOP2A | Topoisomerase II alpha |
| αSat | Alpha satellite DNA |
| γH2AX | γ phosphorylated form of the histone H2AX |
References
- Porokhovnik, L.N.; Veiko, N.N.; Ershova, E.S.; Kostyuk, S.V. The Role of Human Satellite III (1q12) Copy Number Variation in the Adaptive Response during Aging, Stress, and Pathology: A Pendulum Model. Genes 2021, 12, 1524. [Google Scholar] [CrossRef]
- Åkerfelt, M.; Morimoto, R.I.; Sistonen, L. Heat shock factors: Integrators of cell stress, development and lifespan. Nat. Rev. Mol. Cell Biol. 2010, 11, 545–555. [Google Scholar] [CrossRef] [PubMed]
- Valadkhan, S.; Hipólito, A.V. lncRNAs in Stress Response. In Long Non-Coding RNAs in Human Disease; Current Topics in Microbiology and Immunology; Springer: Cham, Switzerland, 2016; Volume 394. [Google Scholar] [CrossRef]
- Ferreira, D.; Meles, S.; Escudeiro, A.; Mendes-da-Silva, A.; Adega, F.; Chaves, R. Satellite non-coding RNAs: The emerging players in cells, cellular pathways and cancer. Chromosome Res. Int. J. Mol. Supramol. Evol. Asp. Chromosome Biol. 2015, 23, 479–493. [Google Scholar] [CrossRef] [PubMed]
- Kit, S. Equilibrium sedimentation in density gradients of DNA preparations from animal tissues. J. Mol. Biol. 1961, 3, 711–716. [Google Scholar] [CrossRef] [PubMed]
- Sueoka, N.; Cheng, T.a.-Y. Natural occurrence of a deoxyribonucleic acid resembling the deoxyadenylate-deoxythymidylate polymer. Proc. Natl. Acad. Sci. USA 1962, 48, 1851–1856. [Google Scholar] [CrossRef] [PubMed]
- Gregory, T.R.; Nicol, J.A.; Tamm, H.; Kullman, B.; Kullman, K.; Leitch, I.J.; Murray, B.G.; Kapraun, D.F.; Greilhuber, J.; Bennett, M.D. Eukaryotic genome size databases. Nucleic Acids Res. 2007, 35, D332–D338. [Google Scholar] [CrossRef] [PubMed]
- Arney, K.L.; Fisher, A.G. Epigenetic aspects of differentiation. J. Cell Sci. 2004, 117, 4355–4363. [Google Scholar] [CrossRef][Green Version]
- Plohl, M.; Luchetti, A.; Mestrovic, N.; Mantovani, B. Satellite DNAs between selfishness and functionality: Structure, genomics and evolution of tandem repeats in centromeric (hetero)chromatin. Gene 2008, 409, 72–82. [Google Scholar] [CrossRef]
- Chaves, R.; Ferreira, D.; Mendes-da-Silva, A.; Meles, S.; Adega, F. FA-SAT Is an Old Satellite DNA Frozen in Several Bilateria Genomes. Genome Biol. Evol. 2017, 9, 3073–3087. [Google Scholar] [CrossRef]
- Feliciello, I.; Pezer, Ž.; Sermek, A.; Mađarić, B.B.; Ljubić, S.; Ugarković, Đ. Satellite DNA-Mediated Gene Expression Regulation: Physiological and Evolutionary Implication. In Satellite DNAs in Physiology and Evolution; Progress in Molecular and Subcellular Biology; Springer: Cham, Switzerland, 2021; Volume 60. [Google Scholar] [CrossRef]
- Valgardsdottir, R.; Chiodi, I.; Giordano, M.; Rossi, A.; Bazzini, S.; Ghigna, C.; Riva, S.; Biamonti, G. Transcription of Satellite III non-coding RNAs is a general stress response in human cells. Nucleic Acids Res. 2008, 36, 423–434. [Google Scholar] [CrossRef]
- Biscotti, M.A.; Canapa, A.; Forconi, M.; Olmo, E.; Barucca, M. Transcription of tandemly repetitive DNA: Functional roles. Chromosome Res. 2015, 23, 463–477. [Google Scholar] [CrossRef] [PubMed]
- Lopes, M.; Louzada, S.; Ferreira, D.; Veríssimo, G.; Eleutério, D.; Gama-Carvalho, M.; Chaves, R. Human Satellite 1A analysis provides evidence of pericentromeric transcription. BMC Biol. 2023, 21, 28. [Google Scholar] [CrossRef] [PubMed]
- Palomeque, T.; Lorite, P. Satellite DNA in insects: A review. Heredity 2008, 100, 564–573. [Google Scholar] [CrossRef] [PubMed]
- Pezer, Z.; Ugarkovic, D. Satellite DNA-associated siRNAs as mediators of heat shock response in insects. RNA Biol. 2012, 9, 587–595. [Google Scholar] [CrossRef]
- Jolly, C.; Metz, A.; Govin, J.; Vigneron, M.; Turner, B.M.; Khochbin, S.; Vourc’h, C. Stress-induced transcription of satellite III repeats. J. Cell Biol. 2004, 164, 25–33. [Google Scholar] [CrossRef]
- Connerty, P.; Lock, R.B.; de Bock, C.E. Long Non-coding RNAs: Major Regulators of Cell Stress in Cancer. Front. Oncol. 2020, 10, 285. [Google Scholar] [CrossRef]
- Amaral, P.P.; Dinger, M.E.; Mattick, J.S. Non-coding RNAs in homeostasis, disease and stress responses: An evolutionary perspective. Brief. Funct. Genom. 2013, 12, 254–278. [Google Scholar] [CrossRef]
- Macario, A.J.L.; Conway de Macario, E. Heat Shock Response, Overview. In Encyclopedia of Stress, 2nd ed.; Fink, G., Ed.; Academic Press: New York, NY, USA, 2007; pp. 292–299. [Google Scholar] [CrossRef]
- Vourc’h, C.; Dufour, S.; Timcheva, K.; Seigneurin-Berny, D.; Verdel, A. HSF1-Activated Non-Coding Stress Response: Satellite lncRNAs and beyond, an Emerging Story with a Complex Scenario. Genes 2022, 13, 597. [Google Scholar] [CrossRef]
- Ugarković, Đ.; Sermek, A.; Ljubić, S.; Feliciello, I. Satellite DNAs in Health and Disease. Genes 2022, 13, 1154. [Google Scholar] [CrossRef]
- Sengupta, S.; Parihar, R.; Ganesh, S. Satellite III non-coding RNAs show distinct and stress-specific patterns of induction. Biochem. Biophys. Res. Commun. 2009, 382, 102–107. [Google Scholar] [CrossRef]
- Zheng, X.; Krakowiak, J.; Patel, N.; Beyzavi, A.; Ezike, J.; Khalil, A.S.; Pincus, D. Dynamic control of Hsf1 during heat shock by a chaperone switch and phosphorylation. eLife 2016, 5, e18638. [Google Scholar] [CrossRef] [PubMed]
- Hu, C.; Yang, J.; Qi, Z.; Wu, H.; Wang, B.; Zou, F.; Mei, H.; Liu, J.; Wang, W.; Liu, Q. Heat shock proteins: Biological functions, pathological roles, and therapeutic opportunities. MedComm 2022, 3, e161. [Google Scholar] [CrossRef] [PubMed]
- Richter, K.; Haslbeck, M.; Buchner, J. The Heat Shock Response: Life on the Verge of Death. Mol. Cell 2010, 40, 253–266. [Google Scholar] [CrossRef] [PubMed]
- Onoguchi-Mizutani, R.; Kishi, Y.; Ogura, Y.; Nishimura, Y.; Imamachi, N.; Suzuki, Y.; Miyazaki, S.; Akimitsu, N. Identification of novel heat shock-induced long non-coding RNA in human cells. J. Biochem. 2021, 169, 497–505. [Google Scholar] [CrossRef]
- Somero, G.N. The cellular stress response and temperature: Function, regulation, and evolution. J. Exp. Zool. Part A Ecol. Integr. Physiol. 2020, 333, 379–397. [Google Scholar] [CrossRef] [PubMed]
- Feliciello, I.; Akrap, I.; Brajkovic, J.; Zlatar, I.; Ugarkovic, D. Satellite DNA as a driver of population divergence in the red flour beetle Tribolium castaneum. Genome Biol. Evol. 2015, 7, 228–239. [Google Scholar] [CrossRef]
- Brajković, J.; Pezer, Ž.; Bruvo-Mađarić, B.; Sermek, A.; Feliciello, I.; Ugarković, Đ. Dispersion Profiles and Gene Associations of Repetitive DNAs in the Euchromatin of the Beetle Tribolium castaneum. G3 Genes Genomes Genet. 2018, 8, 875–886. [Google Scholar] [CrossRef] [PubMed]
- Rizzi, N.; Denegri, M.; Chiodi, I.; Corioni, M.; Valgardsdottir, R.; Cobianchi, F.; Riva, S.; Biamonti, G. Transcriptional Activation of a Constitutive Heterochromatic Domain of the Human Genome in Response to Heat Shock. Mol. Biol. Cell 2003, 15, 543–551. [Google Scholar] [CrossRef]
- Feliciello, I.; Sermek, A.; Pezer, Ž.; Matulić, M.; Ugarković, Đ. Heat Stress Affects H3K9me3 Level at Human Alpha Satellite DNA Repeats. Genes 2020, 11, 663. [Google Scholar] [CrossRef]
- Chatterjee, M.; Sengupta, S. Human satellite III long noncoding RNA imparts survival benefits to cancer cells. Cell Biol. Int. 2022, 46, 611–627. [Google Scholar] [CrossRef]
- Hédouin, S.; Grillo, G.; Ivkovic, I.; Velasco, G.; Francastel, C. CENP-A chromatin disassembly in stressed and senescent murine cells. Sci. Rep. 2017, 7, 42520. [Google Scholar] [CrossRef] [PubMed]
- Eymery, A.; Callanan, M.; Vourc’h, C. The secret message of heterochromatin: New insights into the mechanisms and function of centromeric and pericentric repeat sequence transcription. Int. J. Dev. Biol. 2009, 53, 259–268. [Google Scholar] [CrossRef] [PubMed]
- Feliciello, I.; Akrap, I.; Ugarković, Đ. Satellite DNA Modulates Gene Expression in the Beetle Tribolium castaneum after Heat Stress. PLoS Genet. 2015, 11, e1005547. [Google Scholar] [CrossRef]
- Sermek, A.; Feliciello, I.; Ugarković, Đ. Distinct Regulation of the Expression of Satellite DNAs in the Beetle Tribolium castaneum. Int. J. Mol. Sci. 2021, 22, 296. [Google Scholar] [CrossRef] [PubMed]
- Jolly, C.; Lakhotia, S.C. Human sat III and Drosophila hsrω transcripts: A common paradigm for regulation of nuclear RNA processing in stressed cells. Nucleic Acids Res. 2006, 34, 5508–5514. [Google Scholar] [CrossRef] [PubMed]
- Lakhotia, S.C. Long non-coding RNAs coordinate cellular responses to stress. Wiley Interdiscip. Rev. 2012, 3, 779–796. [Google Scholar] [CrossRef]
- Lakhotia, S.C.; Mallik, M.; Singh, A.K.; Ray, M. The large noncoding hsrω-n transcripts are essential for thermotolerance and remobilization of hnRNPs, HP1 and RNA polymerase II during recovery from heat shock in Drosophila. Chromosoma 2011, 121, 49–70. [Google Scholar] [CrossRef] [PubMed]
- Giordano, M.; Infantino, L.; Biggiogera, M.; Montecucco, A.; Biamonti, G. Heat Shock Affects Mitotic Segregation of Human Chromosomes Bound to Stress-Induced Satellite III RNAs. Int. J. Mol. Sci. 2020, 21, 2812. [Google Scholar] [CrossRef]
- Penin, J.; Dufour, S.; Faure, V.; Fritah, S.; Seigneurin-Berny, D.; Col, E.; Verdel, A.; Vourc’h, C. Chromosome Y pericentric heterochromatin is a primary target of HSF1 in male cells. Chromosoma 2021, 130, 53–60. [Google Scholar] [CrossRef]
- Nakahori, Y.; Mitani, K.; Yamada, M.; Nakagome, Y. A human Y-chromosome specific repeated DNA family (DYZ1) consists of a tandem array of pentanucleotides. Nucleic Acids Res. 1986, 14, 7569–7580. [Google Scholar] [CrossRef]
- Schwarzacher-Robinson, T.; Cram, L.S.; Meyne, J.; Moyzis, R.K. Characterization of human heterochromatin by in situ hybridization with satellite DNA clones. Cytogenet. Cell Genet. 1988, 47, 192–196. [Google Scholar] [CrossRef]
- Moyzis, R.K.; Torney, D.C.; Meyne, J.; Buckingham, J.M.; Wu, J.R.; Burks, C.; Sirotkin, K.M.; Goad, W.B. The distribution of interspersed repetitive DNA sequences in the human genome. Genomics 1989, 4, 273–289. [Google Scholar] [CrossRef]
- Cotto, J.; Fox, S.; Morimoto, R. HSF1 granules: A novel stress-induced nuclear compartment of human cells. J. Cell Sci. 1997, 110, 2925–2934. [Google Scholar] [CrossRef]
- Metz, A.; Soret, J.; Vourc’h, C.; Tazi, J.; Jolly, C. A key role for stress-induced satellite III transcripts in the relocalization of splicing factors into nuclear stress granules. J. Cell Sci. 2004, 117, 4551–4558. [Google Scholar] [CrossRef] [PubMed]
- Goenka, A.; Sengupta, S.; Pandey, R.; Parihar, R.; Mohanta, G.C.; Mukerji, M.; Ganesh, S. Human satellite-III non-coding RNAs modulate heat-shock-induced transcriptional repression. J. Cell Sci. 2016, 129, 3541–3552. [Google Scholar] [CrossRef]
- Jolly, C.; Konecny, L.; Grady, D.L.; Kutskova, Y.A.; Cotto, J.J.; Morimoto, R.I.; Vourc’h, C. In vivo binding of active heat shock transcription factor 1 to human chromosome 9 heterochromatin during stress. J. Cell Biol. 2002, 156, 775–781. [Google Scholar] [CrossRef]
- Chiodi, I.; Corioni, M.; Giordano, M.; Valgardsdottir, R.; Ghigna, C.; Cobianchi, F.; Xu, R.M.; Riva, S.; Biamonti, G. RNA recognition motif 2 directs the recruitment of SF2/ASF to nuclear stress bodies. Nucleic Acids Res. 2004, 32, 4127–4136. [Google Scholar] [CrossRef] [PubMed]
- Ninomiya, K.; Adachi, S.; Natsume, T.; Iwakiri, J.; Terai, G.; Asai, K.; Hirose, T. LncRNA-dependent nuclear stress bodies promote intron retention through SR protein phosphorylation. EMBO J. 2020, 39, e102729. [Google Scholar] [CrossRef]
- Ninomiya, K.; Yamazaki, T.; Hirose, T. Satellite RNAs: Emerging players in subnuclear architecture and gene regulation. EMBO J. 2023, 42, e114331. [Google Scholar] [CrossRef]
- Ting, D.T.; Lipson, D.; Paul, S.; Brannigan, B.W.; Akhavanfard, S.; Coffman, E.J.; Contino, G.; Deshpande, V.; Iafrate, A.J.; Letovsky, S.; et al. Aberrant overexpression of satellite repeats in pancreatic and other epithelial cancers. Science 2011, 331, 593–596. [Google Scholar] [CrossRef]
- Tilman, G.; Arnoult, N.; Lenglez, S.; Beneden, A.V.; Loriot, A.; Smet, C.D.; Decottignies, A. Cancer-linked satellite 2 DNA hypomethylation does not regulate Sat2 non-coding RNA expression and is initiated by heat shock pathway activation. Epigenetics 2012, 7, 903–913. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Arutyunyan, A.; Stoddart, S.; Yi, S.-j.; Fei, F.; Lim, M.; Groffen, P.; Feldhahn, N.; Groffen, J.; Heisterkamp, N. Expression of cassini, a murine gamma-satellite sequence conserved in evolution, is regulated in normal and malignant hematopoietic cells. BMC Genom. 2012, 13, 418. [Google Scholar] [CrossRef]
- Feliciello, I.; Parazajder, J.; Akrap, I.; Ugarković, D. First evidence of DNA methylation in insect Tribolium castaneum: Environmental regulation of DNA methylation within heterochromatin. Epigenetics 2013, 8, 534–541. [Google Scholar] [CrossRef]
- Kropotov, A.; Serikov, V.; Suh, J.; Smirnova, A.; Bashkirov, V.; Zhivotovsky, B.; Tomilin, N. Constitutive expression of the human peroxiredoxin V gene contributes to protection of the genome from oxidative DNA lesions and to suppression of transcription of noncoding DNA. FEBS J. 2006, 273, 2607–2617. [Google Scholar] [CrossRef]
- Nogalski, M.T.; Shenk, T. HSATII RNA is induced via a noncanonical ATM-regulated DNA damage response pathway and promotes tumor cell proliferation and movement. Proc. Natl. Acad. Sci. USA 2020, 117, 31891–31901. [Google Scholar] [CrossRef] [PubMed]
- Morozov, V.M.; Gavrilova, E.V.; Ogryzko, V.V.; Ishov, A.M. Dualistic function of Daxx at centromeric and pericentromeric heterochromatin in normal and stress conditions. Nucleus 2012, 3, 276–285. [Google Scholar] [CrossRef] [PubMed]
- Prasanth, K.V.; Rajendra, T.K.; Lal, A.K.; Lakhotia, S.C. Omega speckles—A novel class of nuclear speckles containing hnRNPs associated with noncoding hsr-omega RNA in Drosophila. J. Cell Sci. 2000, 113, 3375–3386. [Google Scholar] [CrossRef]
- Weighardt, F.; Cobianchi, F.; Cartegni, L.; Chiodi, I.; Villa, A.; Riva, S.; Biamonti, G. A novel hnRNP protein (HAP/SAF-B) enters a subset of hnRNP complexes and relocates in nuclear granules in response to heat shock. J. Cell Sci. 1999, 112, 1465–1476. [Google Scholar] [CrossRef]
- Pizzino, G.; Irrera, N.; Cucinotta, M.; Pallio, G.; Mannino, F.; Arcoraci, V.; Squadrito, F.; Altavilla, D.; Bitto, A. Oxidative Stress: Harms and Benefits for Human Health. Oxidative Med. Cell. Longev. 2017, 2017, 8416763. [Google Scholar] [CrossRef]
- Aranda-Rivera, A.K.; Cruz-Gregorio, A.; Arancibia-Hernández, Y.L.; Hernández-Cruz, E.Y.; Pedraza-Chaverri, J. RONS and Oxidative Stress: An Overview of Basic Concepts. Oxygen 2022, 2, 437–478. [Google Scholar] [CrossRef]
- Daenen, K.; Andries, A.; Mekahli, D.; Schepdael, A.V.; Jouret, F.; Bammen, B. Oxidative stress in chronic kidney disease. Pediatr. Nephrol. 2019, 34, 975–991. [Google Scholar] [CrossRef]
- Wu, Y.-Z.; Su, Y.H.; Kuo, C.Y. Stressing the Regulatory Role of Long Non-Coding RNA in the Cellular Stress Response during Cancer Progression and Therapy. Biomedicines 2022, 10, 1212. [Google Scholar] [CrossRef] [PubMed]
- Pascual-Ahuir, A.; Manzanares-Estreder, S.; Proft, M. Pro- and Antioxidant Functions of the Peroxisome-Mitochondria Connection and Its Impact on Aging and Disease. Oxidative Med. Cell. Longev. 2017, 2017, 9860841. [Google Scholar] [CrossRef] [PubMed]
- Juan, C.A.; Pérez de la Lastra, J.M.; Plou, F.J.; Pérez-Lebeña, E. The Chemistry of Reactive Oxygen Species (ROS) Revisited: Outlining Their Role in Biological Macromolecules (DNA, Lipids and Proteins) and Induced Pathologies. Int. J. Mol. Sci. 2021, 22, 4642. [Google Scholar] [CrossRef] [PubMed]
- Zorov, D.B.; Juhaszova, M.; Sollott, S.J. Mitochondrial reactive oxygen species (ROS) and ROS-induced ROS release. Physiol. Rev. 2014, 94, 909–950. [Google Scholar] [CrossRef]
- Lü, J.-M.; Lin, P.H.; Yao, Q.; Chen, C. Chemical and molecular mechanisms of antioxidants: Experimental approaches and model systems. J. Cell. Mol. Med. 2010, 14, 840–860. [Google Scholar] [CrossRef]
- Nousis, L.; Kanavaros, P.; Barbouti, A. Oxidative Stress-Induced Cellular Senescence: Is Labile Iron the Connecting Link? Antioxidants 2023, 12, 1250. [Google Scholar] [CrossRef]
- Szyller, J.; Bil-Lula, I. Heat Shock Proteins in Oxidative Stress and Ischemia/Reperfusion Injury and Benefits from Physical Exercises: A Review to the Current Knowledge. Oxidative Med. Cell. Longev. 2021, 2021, 6678457. [Google Scholar] [CrossRef]
- Karunatilleke, N.C.; Fast, C.S.; Ngo, V.; Brickenden, A.; Duennwald, M.L.; Konermann, L.; Choy, W.-Y. Nrf2, the Major Regulator of the Cellular Oxidative Stress Response, is Partially Disordered. Int. J. Mol. Sci. 2021, 22, 7434. [Google Scholar] [CrossRef]
- Fulda, S.; Gorman, A.M.; Hori, O.; Samali, A. Cellular stress responses: Cell survival and cell death. Int. J. Cell Biol. 2010, 2010, 214074. [Google Scholar] [CrossRef]
- Poole, L.B.; Hall, A.; Nelson, K.J. Overview of peroxiredoxins in oxidant defense and redox regulation. Curr. Protoc. Toxicol. 2011, 49, 7–9. [Google Scholar] [CrossRef] [PubMed]
- Enukashvily, N.I.; Malashicheva, A.B.; Waisertreiger, I.S.R. Satellite DNA Spatial Localization and Transcriptional Activity in Mouse Embryonic E-14 and IOUD2 Stem Cells. Cytogenet. Genome Res. 2009, 124, 277–287. [Google Scholar] [CrossRef]
- Tan, K.P.; Kosuge, K.; Yang, M.; Ito, S. NRF2 as a determinant of cellular resistance in retinoic acid cytotoxicity. Free. Radic. Biol. Med. 2008, 45, 1663–1673. [Google Scholar] [CrossRef]
- Colin, L.; Ruhnow, F.; Zhu, J.-K.; Zhao, C.; Zhao, Y.; Persson, S. The cell biology of primary cell walls during salt stress. Plant Cell 2023, 35, 201–217. [Google Scholar] [CrossRef]
- Ho, S.N. Intracellular water homeostasis and the mammalian cellular osmotic stress response. J. Cell. Physiol. 2006, 206, 9–15. [Google Scholar] [CrossRef] [PubMed]
- Sies, H.; Jones, D.P. Reactive oxygen species (ROS) as pleiotropic physiological signalling agents. Nat. Rev. Mol. Cell Biol. 2020, 21, 363–383. [Google Scholar] [CrossRef] [PubMed]
- Kumar, R.R.; Arora, K.; Goswami, S.; Sakhare, A.; Singh, B.; Chinnusamy, V.; Praveen, S. MAPK Enzymes: A ROS Activated Signaling Sensors Involved in Modulating Heat Stress Response, Tolerance and Grain Stability of Wheat under Heat Stress. 3 Biotech 2020, 10, 380. [Google Scholar] [CrossRef]
- Villalpando-Rodriguez, G.E.; Gibson, S.B. Reactive Oxygen Species (ROS) Regulates Different Types of Cell Death by Acting as a Rheostat. Oxidative Med. Cell. Longev. 2021, 2021, 9912436. [Google Scholar] [CrossRef]
- Cen, L.; Xing, F.; Xu, L.; Cao, Y. Potential Role of Gene Regulator NFAT5 in the Pathogenesis of Diabetes Mellitus. J. Diabetes Res. 2020, 2020, 6927429. [Google Scholar] [CrossRef]
- Kumar, R.; DuMond, J.F.; Khan, S.H.; Thompson, E.B.; He, Y.; Burg, M.B.; Ferraris, J.D. NFAT5, which protects against hypertonicity, is activated by that stress via structuring of its intrinsically disordered domain. Proc. Natl. Acad. Sci. USA 2020, 117, 20292–20297. [Google Scholar] [CrossRef]
- Chernyakov, D.; Fischer, A.; Brandau, M.; Petrillo, F.; Fenton, R.A.; Edemir, B. The nuclear factor of activated T cells 5 (NFAT5) contributes to the renal corticomedullary differences in gene expression. Sci. Rep. 2022, 12, 20304. [Google Scholar] [CrossRef]
- Iyama, T.; Wilson, D.M. DNA repair mechanisms in dividing and non-dividing cells. DNA Repair 2013, 12, 620–636. [Google Scholar] [CrossRef]
- Goldstine, J.V.; Nahas, S.; Gamo, K.; Gartler, S.M.; Hansen, R.S.; Roelfsema, J.H.; Gatti, R.A.; Marahrens, Y. Constitutive phosphorylation of ATM in lymphoblastoid cell lines from patients with ICF syndrome without downstream kinase activity. DNA Repair 2006, 5, 432–443. [Google Scholar] [CrossRef] [PubMed]
- May, J.M.; Bylicky, M.; Chopra, S.; Coleman, C.N.; Aryankalayil, M.J. Long and short non-coding RNA and radiation response: A review. Transl. Res. 2021, 233, 162–179. [Google Scholar] [CrossRef]
- Kumar, A.; Raj, A.; Gupta, A.; Gautam, S.; Kumar, M.; Bherwani, H.; Anshul, A. Pollution free UV-C radiation to mitigate COVID-19 transmission. Gondwana Res. 2023, 114, 78–86. [Google Scholar] [CrossRef]
- Batista, L.F.Z.; Kaina, B.; Meneghini, R.; Menck, C.F.M. How DNA lesions are turned into powerful killing structures: Insights from UV-induced apoptosis. Mutat. Res. 2009, 681, 197–208. [Google Scholar] [CrossRef]
- Latonen, L.; Laiho, M. Cellular UV damage responses—Functions of tumor suppressor p53. Biochim. Biophys. Acta BBA Rev. Cancer 2005, 1755, 71–89. [Google Scholar] [CrossRef] [PubMed]
- Shiloh, Y.; Ziv, Y. The ATM protein kinase: Regulating the cellular response to genotoxic stress, and more. Nat. Rev. Mol. Cell Biol. 2013, 14, 197–210. [Google Scholar] [CrossRef]
- Zhu, Q.; Pao, G.M.; Huynh, A.M.; Suh, H.; Tonnu, N.; Nederlof, P.M.; Gage, F.H.; Verma, I.M. BRCA1 tumour suppression occurs via heterochromatin-mediated silencing. Nature 2011, 477, 179–184. [Google Scholar] [CrossRef]
- Zhu, Q.; Hoong, N.; Aslanian, A.; Hara, T.; Benner, C.; Heinz, S.; Miga, K.H.; Ke, E.; Verma, S.; Soroczynski, J.; et al. Heterochromatin-Encoded Satellite RNAs Induce Breast Cancer. Mol. Cell 2018, 70, 842–853.e847. [Google Scholar] [CrossRef]
- Tamaki, S.; Suzuki, K.; Abe, I.; Endo, Y.; Kakizawa, N.; Watanabe, F.; Saito, M.; Tsujinaka, S.; Miyakura, Y.; Ohta, S.; et al. Overexpression of satellite RNAs in heterochromatin induces chromosomal instability and reflects drug sensitivity in mouse cancer cells. Sci. Rep. 2022, 12, 10999. [Google Scholar] [CrossRef] [PubMed]
- Ichida, K.; Suzuki, K.; Fukui, T.; Takayama, Y.; Kakizawa, N.; Watanabe, F.; Ishikawa, H.; Muto, Y.; Kato, T.; Saito, M.; et al. Overexpression of satellite alpha transcripts leads to chromosomal instability via segregation errors at specific chromosomes. Int. J. Oncol. 2018, 52, 1685–1693. [Google Scholar] [CrossRef]
- Kanne, J.; Hussong, M.; Isensee, J.; Muñoz-López, Á.; Wolffgramm, J.; Heß, F.; Grimm, C.; Bessonov, S.; Meder, L.; Wang, J.; et al. Pericentromeric Satellite III transcripts induce etoposide resistance. Cell Death Dis. 2021, 12, 530. [Google Scholar] [CrossRef]
- Ershova, E.S.; Malinovskaya, E.M.; Konkova, M.S.; Veiko, R.V.; Umriukhin, P.E.; Martynov, A.V.; Kutsev, S.I.; Veiko, N.N.; Kostyuk, S.V. Copy Number Variation of Human Satellite III (1q12) with Aging. Front. Genet. 2019, 10, 704. [Google Scholar] [CrossRef] [PubMed]
- Soria, V.; Uribe, J.; Salvat-Pujol, N.; Palao, D.; Menchón, J.M.; Labad, J. Psiconeuroinmunología de los trastornos mentales. Rev. Psiquiatr. Salud Ment. 2018, 11, 115–124. [Google Scholar] [CrossRef]
- Ershova, E.S.; Malinovskaya, E.M.; Golimbet, V.E.; Lezheiko, T.V.; Zakharova, N.V.; Shmarina, G.V.; Veiko, R.V.; Umriukhin, P.E.; Kostyuk, G.P.; Kutsev, S.I.; et al. Copy number variations of satellite III (1q12) and ribosomal repeats in health and schizophrenia. Schizophr. Res. 2020, 223, 199–212. [Google Scholar] [CrossRef]
- Ershova, E.S.; Agafonova, O.N.; Zakharova, N.V.; Bravve, L.V.; Jestkova, E.M.; Golimbet, V.E.; Lezheiko, T.V.; Morozova, A.Y.; Martynov, A.V.; Veiko, R.V.; et al. Copy Number Variation of Satellite III(1q12) in Patients with Schizophrenia. Front. Genet. 2019, 10, 1132. [Google Scholar] [CrossRef]
- Konkova, M.S.; Ershova, E.S.; Savinova, E.A.; Malinovskaya, E.M.; Shmarina, G.V.; Martynov, A.V.; Veiko, R.V.; Zakharova, N.V.; Umriukhin, P.; Kostyuk, G.P.; et al. 1Q12 Loci Movement in the Interphase Nucleus Under the Action of ROS Is an Important Component of the Mechanism That Determines Copy Number Variation of Satellite III (1q12) in Health and Schizophrenia. Front. Cell Dev. Biol. 2020, 8, 386. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fonseca-Carvalho, M.; Veríssimo, G.; Lopes, M.; Ferreira, D.; Louzada, S.; Chaves, R. Answering the Cell Stress Call: Satellite Non-Coding Transcription as a Response Mechanism. Biomolecules 2024, 14, 124. https://doi.org/10.3390/biom14010124
Fonseca-Carvalho M, Veríssimo G, Lopes M, Ferreira D, Louzada S, Chaves R. Answering the Cell Stress Call: Satellite Non-Coding Transcription as a Response Mechanism. Biomolecules. 2024; 14(1):124. https://doi.org/10.3390/biom14010124
Chicago/Turabian StyleFonseca-Carvalho, Marisa, Gabriela Veríssimo, Mariana Lopes, Daniela Ferreira, Sandra Louzada, and Raquel Chaves. 2024. "Answering the Cell Stress Call: Satellite Non-Coding Transcription as a Response Mechanism" Biomolecules 14, no. 1: 124. https://doi.org/10.3390/biom14010124
APA StyleFonseca-Carvalho, M., Veríssimo, G., Lopes, M., Ferreira, D., Louzada, S., & Chaves, R. (2024). Answering the Cell Stress Call: Satellite Non-Coding Transcription as a Response Mechanism. Biomolecules, 14(1), 124. https://doi.org/10.3390/biom14010124






