Current Technologies Unraveling the Significance of Post-Translational Modifications (PTMs) as Crucial Players in Neurodegeneration
Abstract
1. Introduction
2. Biogenesis of PTMS
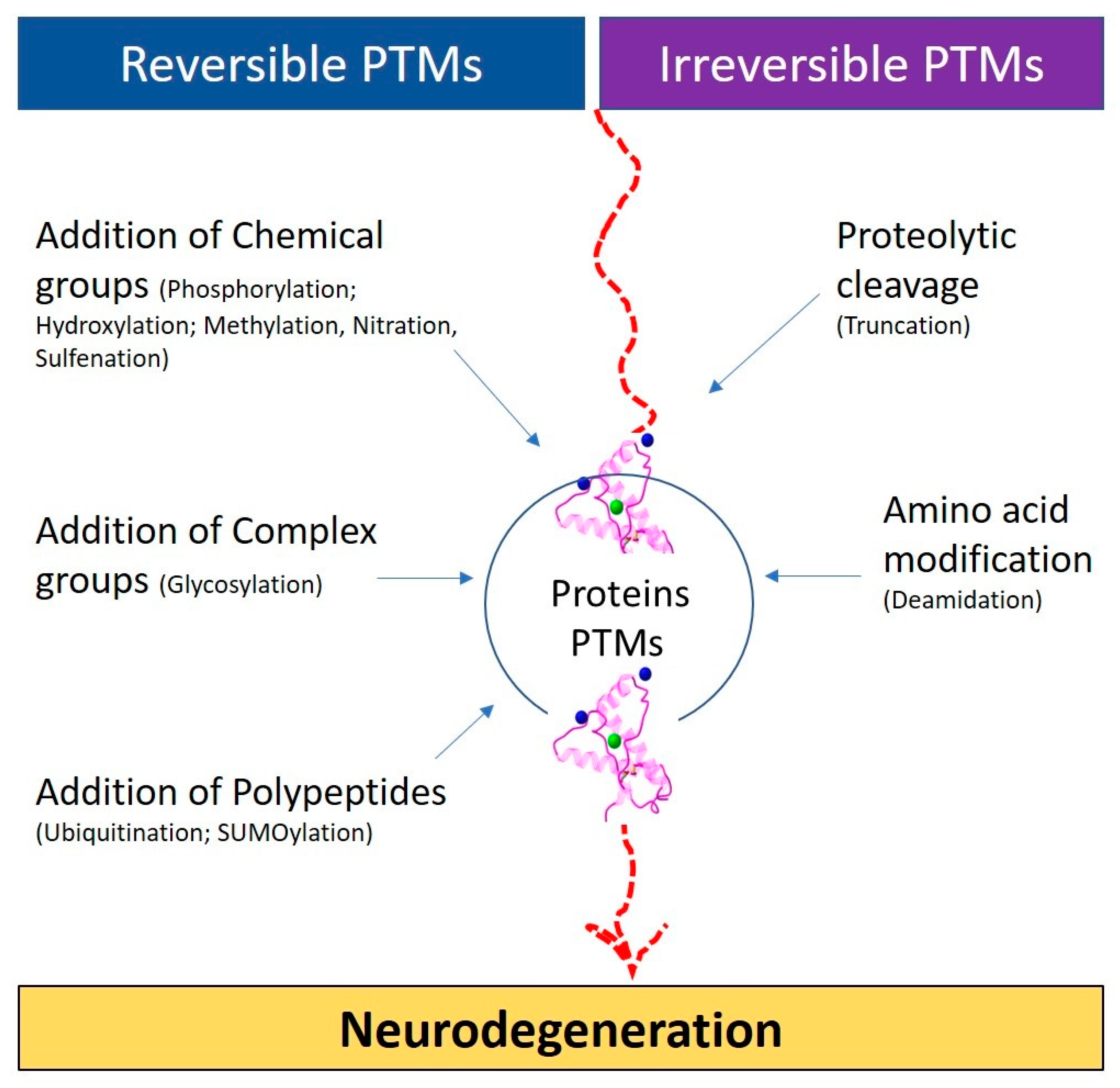
3. Types of PTMS
- 3.1
- Addition of chemical groups (phosphorylation; hydroxylation; methylation, nitration, sulfenation);
- 3.2
- Addition of complex groups (glycosylation);
- 3.3
- Addition of polypeptides (ubiquitination; SUMOylation);
- 3.4
- Proteolytic cleavage (truncation);
- 3.5
- Deamidation.
3.1. Addition of Chemical Groups (Phosphorylation; Hydroxylation; Methylation, Nitration, Sulfenation)
3.1.1. Phosphorylation
3.1.2. Hydroxylation
3.1.3. Methylation
3.1.4. Nitration
3.1.5. Sulfenation
3.2. Addition of Complex Groups (O-GlcNAcylation)
O-GlcNAcylation
3.3. Addition of Polypeptides (Ubiquitination; SUMOylation)
3.3.1. Ubiquitination
3.3.2. SUMOylation
3.4. Proteolytic Cleavage (Truncation)
3.5. Deamidation
4. Current Technologies to Decipher PTMs
4.1. Methylation
Mass Spectrometry (MS)
4.2. Phosphorylation
4.2.1. Proximity Ligation Assay (PLA)
4.2.2. Phosphoproteomics
4.3. SUMOylation
4.3.1. SUMOylation Site Identification by Mass Spectrometry (SIMS)
4.3.2. SUMO Protease Protection Assay (SuPrPA)
4.4. Ubiquitination
4.4.1. Ubiquitin Enrichment
4.4.2. Tandem Ubiquitin Binding Entities (TUBEs)
4.5. Nitration
4.5.1. Immunoprecipitation Combined with Mass Spectrometry (IP-MS)
4.5.2. Liquid Chromatography-Tandem Mass Spectrometry (LC-MS/MS)
4.6. Truncation
4.6.1. N-Terminal COFRADIC (Combined Fractional Diagonal Chromatography)
4.6.2. C-Terminal Sequencing
4.7. O-GlcNAcylation
4.7.1. Chemoenzymatic Labeling
4.7.2. Metabolic Labeling with Azide-Modified GlcNAc (GlcNAz)
4.8. Hydroxylation
4.8.1. Proximity Ligation Assay (PLA)
4.8.2. Stable Isotope Labeling by Amino Acids in Cell Culture (SILAC) Combined with Mass Spectrometry
4.9. Sulfenation
4.9.1. Biotin Switch Technique
4.9.2. Resin-Assisted Capture (RAC)
5. Emerging Role of PTMS towards Therapeutics
| PTM Technique | Analytical Method | Applications | Example | Limitations | Recent Developments | References |
|---|---|---|---|---|---|---|
| Methylation | Mass Spectrometry (MS) | Identifying and quantifying methylation sites; epigenetics and gene regulation studies. Comprehensive coverage of DNA methylation across the genome. | DNA methylation leads to progression of Alzheimer’s disease. [126,132] Hyper methylation of APP in Alzheimer’s disease [176]. | Requires sophisticated equipment; complex data analysis. High cost; complexity in data analysis. | Improved MS sensitivity and resolution for low-abundance peptides. Advances in sequencing technologies for more efficient and cost-effective analysis. | [13,143] |
| Phosphorylation | Proximity Ligation Assay (PLA) | Detects phosphorylation in situ; used in signal transduction studies. | Phosphorylation of tau protein in Alzheimer’s disease [36,177]. | Limited to proteins with available specific antibodies. | More sensitive probes and automated image analysis. | [16] |
| Phosphoproteomics | Large-scale study of phosphorylated proteins; useful in disease research. | Phosphorylation of alpha-synuclein with the aid of kinases and phosphatases in PD [36]. | Complex sample preparation; data interpretation challenges. | Improved enrichment techniques for phosphopeptides. | [131] | |
| Sumoylation | SIMS (Sumoylation Identification by Mass Spectrometry) | Identifies sumoylation sites on proteins; useful in studying protein function and regulation. | Sumoylation of alpha-synuclein protein leads to progression of Parkinson’s disease [134,166]. | Requires advanced mass spectrometry techniques; complex sample preparation. | Enhanced sensitivity and accuracy in sumoylation site identification. | [134,166] |
| SuPrPA (SUMO Protease Protection Assay) | Studies SUMO conjugation/deconjugation dynamics. | Sumoylation of beta-secretases leads to increased amyloid-beta production leading to onset of Alzheimer’s disease [178]. | Limited by the availability and specificity of SUMO proteases. | Development of more specific and sensitive assays for SUMO dynamics. | [166] | |
| Ubiquitination | Ubiquitin Enrichment | Enriches ubiquitinated proteins; studies protein degradation and signaling pathways. | Ubiquitination of ligases result in neurodegenerative conditions [69,97]. | Potential for non-specific binding; requires careful control. | Development of more specific enrichment strategies. | [144] |
| TUBEs (Tandem Ubiquitin Binding Entities) | Enriches ubiquitinated proteins; studies ubiquitin-mediated processes. | PARK2 is a ubiquitin-signaling gene that is associated with PD [179]. | Potential for non-specific binding; requires careful control. | Development of more specific TUBEs for different ubiquitin chains. | [135] | |
| Nitration | Immunoprecipitation combined with Mass Spectrometry (IP-MS) | Detects and quantifies protein nitration; used in oxidative stress studies. | Tyrosine nitration is an earliest marker of Alzheimer’s disease. [139,141] | Specificity depends on antibody quality; complex sample preparation. | Improved antibody specificity and mass spectrometry techniques. | [141] |
| LC-MS/MS (Liquid Chromatography-Mass Spectrometry/Mass Spectrometry) | Quantitative analysis of protein nitration; studies nitrosative stress. | Nitration of alpha-synuclein leads to increased aggregation and PD pathogenesis [180]. | Complex sample preparation; requires high-resolution mass spectrometry. | Advances in LC-MS/MS for improved detection and quantification. | [138,139] | |
| Truncation | N-Terminal COFRADIC | Analyzes protein N-termini; identifies truncation events. | Truncation of Tau protein in Alzheimer’s disease. [143,144] | Technically challenging; requires specialized equipment. | Improved methods for N-terminal peptide enrichment and analysis. | [144] |
| C-Terminal Sequencing | Identifies protein C-terminal truncations; useful in studying proteolysis. | Truncation of alpha-synuclein and amyloid-beta leads to neurodegenerative conditions [181] | Limited availability of techniques for C-terminal analysis. | Development of more efficient C-terminal sequencing methods. | [143,181] | |
| O-GlcNAcylation | Chemoenzymatic Labeling | Detects O-GlcNAc modifications; studies cellular signaling and regulation. | Dysregulated O-GlcNAcylation of Alpha-synuclein leads to Parkinsonism. [82,90,94] | Requires specific enzymes and bioorthogonal chemistry. | Development of more efficient labeling and detection methods. | [149] |
| GlcNAz (Metabolic Labeling with Azide-Modified GlcNAc) | Incorporates azide-modified GlcNAc into proteins; studies O-GlcNAcylation dynamics. | O-GlcNAcylation of amyloid-beta have neuroprotective effects in Alzheimer’s disease [182]. | Requires cell permeable and metabolically incorporated azide-GlcNAc. | Improved azide-GlcNAc analogs for better incorporation and detection. | [81] | |
| Hydroxylation | Proximity Ligation Assay (PLA) | Detects hydroxylation in situ; used in hypoxia and collagen biosynthesis studies. | Proline Hydroxylation in the brain leads to activation of kinases leading to inflammation. [38,42] | Limited to proteins with available specific antibodies. | More sensitive probes and automated image analysis. | [155] |
| SILAC combined with Mass Spectrometry | Quantitative analysis of hydroxylation; studies protein stability and signaling. | Tyrosine hydroxylation leads to PD pathogenesis [183]. | Requires metabolic labeling; complex sample preparation. | Advances in SILAC and MS for improved quantification and sensitivity. | [154] | |
| Sulfenation | Biotin Switch Technique | Detects cysteine sulfenation; studies redox regulation and signaling. | Sulfenation of cysteine residues leads to onset of Parkinsonism. [156,157] | Potential for non-specific labeling; requires careful control. | Development of more specific reagents for sulfenic acid detection. | [156] |
| Resin-Assisted Capture (RAC) | Enriches cysteine-modified peptides; identifies sulfenation sites. | Sulfation of tau, Aβ, and α-synuclein aggregates aided by heparan sulfate proteoglycans (HSPGs) leads to neurodegenerative conditions [184]. | Requires specific capture resins; complex sample preparation. | Improved resin materials for more efficient capture and release. | [151,184] |
6. Conclusions
Funding
Conflicts of Interest
References
- Chen, L.; Kashina, A. Post-translational Modifications of the Protein Termini. Front. Cell Dev. Biol. 2021, 9, 719590. [Google Scholar] [CrossRef] [PubMed]
- Ramazi, S.; Zahiri, J. Post-translational modifications in proteins: Resources, tools and prediction methods. Database 2021, 2021, baab012. [Google Scholar] [CrossRef] [PubMed]
- Mann, M.; Jensen, O.N. Proteomic analysis of post-translational modifications. Nat. Biotechnol. 2003, 21, 255–261. [Google Scholar] [CrossRef]
- Waddell, J.; Banerjee, A.; Kristian, T. Acetylation in Mitochondria Dynamics and Neurodegeneration. Cells 2021, 10, 3031. [Google Scholar] [CrossRef] [PubMed]
- Shashi, V.; Magiera, M.M.; Klein, D.; Zaki, M.; Schoch, K.; Rudnik-Schöneborn, S.; Norman, A.; Lopes Abath Neto, O.; Dusl, M.; Yuan, X.; et al. Loss of tubulin deglutamylase CCP1 causes infantile-onset neurodegeneration. EMBO J. 2018, 37, e100540. [Google Scholar] [CrossRef]
- Bowie, J.U.; Post, C.B. The 29th Annual Symposium of the Protein Society, Barcelona, Spain, July 22–25. Protein Sci. 2015, 24 (Suppl. S1), 1–313. [Google Scholar]
- Duan, G.; Walther, D. The Roles of Post-translational modifications in the context of protein interaction networks. PLoS Comput. Biol. 2015, 11, e1004049. [Google Scholar] [CrossRef]
- Liu, Y.-P.; Zhang, T.-N.; Wen, R.; Liu, C.-F.; Yang, N. Role of Posttranslational Modifications of Proteins in Cardiovascular Disease. Oxidative Med. Cell. Longev. 2022, 2022, 3137329. [Google Scholar] [CrossRef]
- Zhong, Q.; Xiao, X.; Qiu, Y.; Xu, Z.; Chen, C.; Chong, B.; Zhao, X.; Hai, S.; Li, S.; An, Z.; et al. Protein posttranslational modifications in health and diseases: Functions, regulatory mechanisms, and therapeutic implications. Medcomm 2023, 4, e261. [Google Scholar] [CrossRef]
- Yang, Y.-H.; Wen, R.; Yang, N.; Zhang, T.-N.; Liu, C.-F. Roles of protein post-translational modifications in glucose and lipid metabolism: Mechanisms and perspectives. Mol. Med. 2023, 29, 93. [Google Scholar] [CrossRef]
- Gajjala, P.R.; Fliser, D.; Speer, T.; Jankowski, V.; Jankowski, J. Emerging role of post-translational modifications in chronic kidney disease and cardiovascular disease. Nephrol. Dial. Transplant. 2015, 30, 1814–1824. [Google Scholar] [CrossRef]
- Gupta, R.; Sahu, M.; Srivastava, D.; Tiwari, S.; Ambasta, R.K.; Kumar, P. Post-translational modifications: Regulators of neurodegenerative proteinopathies. Ageing Res. Rev. 2021, 68, 101336. [Google Scholar] [CrossRef]
- Huang, K.-Y.; Lee, T.-Y.; Kao, H.-J.; Ma, C.-T.; Lee, C.-C.; Lin, T.-H.; Chang, W.-C.; Huang, H.-D. dbPTM in 2019: Exploring disease association and cross-talk of post-translational modifications. Nucleic Acids Res. 2018, 47, D298–D308. [Google Scholar] [CrossRef] [PubMed]
- Walsh, G.; Jefferis, R. Post-translational modifications in the context of therapeutic proteins. Nat. Biotechnol. 2006, 24, 1241–1252. [Google Scholar] [CrossRef]
- Ong, S.-E.; Mann, M.A. practical recipe for stable isotope labeling by amino acids in cell culture (SILAC). Nat. Protoc. 2006, 1, 2650–2660. [Google Scholar] [CrossRef] [PubMed]
- Hunter, T. Signaling—And Beyond. Cell 2000, 100, 113–127. [Google Scholar] [CrossRef] [PubMed]
- Röhl, A.; Rohrberg, J.; Buchner, J. The chaperone Hsp90: Changing partners for demanding clients. Trends Biochem. Sci. 2013, 38, 253–262. [Google Scholar] [CrossRef]
- Sahasrabudhe, P.; Rohrberg, J.; Biebl, M.M.; Rutz, D.A.; Buchner, J. The Plasticity of the Hsp90 Co-chaperone System. Mol. Cell 2017, 67, 947–961.e5. [Google Scholar] [CrossRef]
- Cox, M.B.; Johnson, J.L. Evidence for Hsp90 Co-chaperones in Regulating Hsp90 Function and Promoting Client Protein Folding. In Chaperones: Methods and Protocols; Humana Press: New York, NY, USA, 2018; pp. 397–422. [Google Scholar]
- Rakhit, R.; Chakrabartty, A. Structure, folding, and misfolding of Cu, Zn superoxide dismutase in amyotrophic lateral sclerosis. Biochim. Biophys. Acta (BBA)-Mol. Basis Dis. 2006, 1762, 1025–1037. [Google Scholar] [CrossRef]
- Arai, T.; Hasegawa, M.; Akiyama, H.; Ikeda, K.; Nonaka, T.; Mori, H.; Mann, D.; Tsuchiya, K.; Yoshida, M.; Hashizume, Y.; et al. TDP-43 is a component of ubiquitin-positive tau-negative inclusions in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Biochem. Biophys. Res. Commun. 2006, 351, 602–611. [Google Scholar] [CrossRef]
- Withers, J.; Dong, X. Post-translational regulation of plant immunity. Curr. Opin. Plant Biol. 2017, 38, 124–132. [Google Scholar] [CrossRef] [PubMed]
- Malik, N.; Ferreira, B.I.; Hollstein, P.E.; Curtis, S.D.; Trefts, E.; Weiser Novak, S.; Yu, J.; Gilson, R.; Hellberg, K.; Fang, L.; et al. Induction of lysosomal and mitochondrial biogenesis by AMPK phosphorylation of FNIP1. Science 2023, 1979, 380. [Google Scholar] [CrossRef] [PubMed]
- Gavin, A.-C.; Bösche, M.; Krause, R.; Grandi, P.; Marzioch, M.; Bauer, A.; Schultz, J.; Rick, J.M.; Michon, A.-M.; Cruciat, C.-M.; et al. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 2002, 415, 141–147. [Google Scholar] [CrossRef] [PubMed]
- Shubchynskyy, V.; Boniecka, J.; Schweighofer, A.; Simulis, J.; Kvederaviciute, K.; Stumpe, M.; Mauch, F.; Balazadeh, S.; Mueller-Roeber, B.; Boutrot, F.; et al. Protein phosphatase AP2C1 negatively regulates basal resistance and defense responses to Pseudomonas syringae. J. Exp. Bot. Erw. 2017, 68, 1169–1183. [Google Scholar]
- Ji, F.; Zhou, M.; Zhu, H.; Jiang, Z.; Li, Q.; Ouyang, X.; Lv, Y.; Zhang, S.; Wu, T.; Li, L. Integrative Proteomic Analysis of Multiple Posttranslational Modifications in Inflammatory Response. Genom. Proteom. Bioinform. 2022, 20, 163–176. [Google Scholar] [CrossRef]
- Aillaud, C.; Bosc, C.; Peris, L.; Bosson, A.; Heemeryck, P.; Van Dijk, J.; Le Friec, J.; Boulan, B.; Vossier, F.; Sanman, L.E.; et al. Vaso-hibins/SVBP are tubulin carboxypeptidases (TCPs) that regulate neuron differentiation. Science 2017, 358, 1448–1453. [Google Scholar] [CrossRef] [PubMed]
- Brazil, D.P.; Hemmings, B.A. Ten years of protein kinase B signalling: A hard Akt to follow. Trends Biochem. Sci. 2001, 26, 657–664. [Google Scholar] [CrossRef]
- Persad, S.; Attwell, S.; Gray, V.; Mawji, N.; Deng, J.T.; Leung, D.; Yan, J.; Sanghera, J.; Walsh, M.P.; Dedhar, S. Regulation of Protein Kinase B/Akt-Serine 473 Phosphorylation by Integrin-linked Kinase. J. Biol. Chem. 2001, 276, 27462–27469. [Google Scholar] [CrossRef]
- Patel, M.; Sachidanandan, M.; Adnan, M. Serine arginine protein kinase 1 (SRPK1): A moonlighting protein with theranostic ability in cancer prevention. Mol. Biol. Rep. 2019, 46, 1487–1497. [Google Scholar] [CrossRef]
- Kenwood, B.M.; Weaver, J.L.; Bajwa, A.; Poon, I.K.; Byrne, F.L.; Murrow, B.A.; Calderone, J.A.; Huang, L.; Divakaruni, A.S.; Tomsig, J.L.; et al. Identification of a novel mitochondrial uncoupler that does not depolarize the plasma membrane. Mol. Metab. 2014, 3, 114–123. [Google Scholar] [CrossRef]
- Liu, Y.-Z.; Chrivia, J.C.; Latchman, D.S. Nerve Growth Factor Up-regulates the Transcriptional Activity of CBP through Ac-tivation of the p42/p44MAPK Cascade. J. Biol. Chem. 1998, 273, 32400–32407. [Google Scholar] [CrossRef][Green Version]
- Alberini, C.M. Transcription Factors in Long-Term Memory and Synaptic Plasticity. Physiol. Rev. 2009, 89, 121–145. [Google Scholar] [CrossRef]
- Chrivia, J.C.; Kwok, R.P.S.; Lamb, N.; Hagiwara, M.; Montminy, M.R.; Goodman, R.H. Phosphorylated CREB binds specifically to the nuclear protein CBP. Nature 1993, 365, 855–859. [Google Scholar] [CrossRef]
- Illenberger, S.; Zheng-Fischhöfer, Q.; Preuss, U.; Stamer, K.; Baumann, K.; Trinczek, B.; Biernat, J.; Godemann, R.; Mandelkow, E.M.; Mandelkow, E. The endogenous and cell cycle-dependent phosphorylation of tau protein in living cells: Implications for Alzheimer’s disease. Mol. Biol. Cell 1998, 9, 1495–1512. [Google Scholar] [CrossRef] [PubMed]
- Noble, W.; Hanger, D.P.; Miller, C.C.J.; Lovestone, S. The Importance of Tau Phosphorylation for Neurodegenerative Diseases. Front. Neurol. 2013, 4, 83. [Google Scholar] [CrossRef]
- Kaelin, W.G.; Ratcliffe, P.J. Oxygen sensing by metazoans: The central role of the HIF hydroxylase pathway. Mol. Cell 2008, 30, 393–402. [Google Scholar] [CrossRef]
- Hutton, J.J.; Kaplan, A.; Udenfriend, S. Conversion of the amino acid sequence gly-pro-pro in protein to gly-pro-hyp by collagen proline hydroxylase. Arch. Biochem. Biophys. 1967, 121, 384–391. [Google Scholar] [CrossRef] [PubMed]
- Hausinger, R.P. Fe(II)/α-Ketoglutarate-Dependent Hydroxylases and Related Enzymes. Crit. Rev. Biochem. Mol. Biol. 2004, 39, 21–68. [Google Scholar] [CrossRef]
- Rappu, P.; Salo, A.M.; Myllyharju, J.; Heino, J. Role of prolyl hydroxylation in the molecular interactions of collagens. Essays Biochem. 2019, 63, 325–335. [Google Scholar] [PubMed]
- Salo, A.M.; Myllyharju, J. Prolyl and lysyl hydroxylases in collagen synthesis. Exp. Dermatol. 2021, 30, 38–49. [Google Scholar] [CrossRef]
- Kirchner, M.; Deng, H.; Xu, Y. Heterogeneity in proline hydroxylation of fibrillar collagens observed by mass spectrometry. PLoS ONE 2021, 16, e0250544. [Google Scholar] [CrossRef]
- Ferrante, F.; Giaimo, B.D.; Friedrich, T.; Sugino, T.; Mertens, D.; Kugler, S.; Gahr, B.M.; Just, S.; Pan, L.; Bartkuhn, M.; et al. Hydroxylation of the NOTCH1 intracellular domain regulates Notch signaling dynamics. Cell Death Dis. 2022, 13, 600. [Google Scholar] [CrossRef] [PubMed]
- Cai, Z.; Zhao, B.; Deng, Y.; Shangguan, S.; Zhou, F.; Zhou, W.; Li, X.; Li, Y.; Chen, G. Notch signaling in cerebrovascular diseases (Review). Mol. Med. Rep. 2016, 14, 2883–2898. [Google Scholar] [CrossRef] [PubMed]
- Jakovljevic, A.; Nikolic, N.; Paternò Holtzman, L.; Tournier, P.; Gaudin, A.; Cordaro, L.; Milinkovic, I. Involvement of the Notch signaling system in alveolar bone resorption. Jpn. Dent. Sci. Rev. 2023, 59, 38–47. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.Y.; Teyssier, C.; Strahl, B.D.; Stallcup, M.R. Role of Protein Methylation in Regulation of Transcription. Endocr. Rev. 2005, 26, 147–170. [Google Scholar] [CrossRef] [PubMed]
- Kouzarides, T. Chromatin Modifications and Their Function. Cell 2007, 128, 693–705. [Google Scholar] [CrossRef]
- Shi, Y.; Lan, F.; Matson, C.; Mulligan, P.; Whetstine, J.R.; Cole, P.A.; Casero, R.A.; Shi, Y. Histone Demethylation Mediated by the Nuclear Amine Oxidase Homolog LSD1. Cell 2004, 119, 941–953. [Google Scholar] [CrossRef]
- Lee, Y.-H.; Stallcup, M.R. Minireview: Protein Arginine Methylation of Nonhistone Proteins in Transcriptional Regulation. Mol. Endocrinol. 2009, 23, 425–433. [Google Scholar] [CrossRef]
- Kim, J.; Daniel, J.; Espejo, A.; Lake, A.; Krishna, M.; Xia, L.; Zhang, Y.; Bedford, M.T. Tudor, MBT and chromo domains gauge the degree of lysine methylation. Embo. Rep. 2006, 7, 397–403. [Google Scholar] [CrossRef]
- Scoumanne, A.; Zhang, J.; Chen, X. PRMT5 is required for cell-cycle progression and p53 tumor suppressor function. Nucleic Acids Res. 2009, 37, 4965–4976. [Google Scholar] [CrossRef]
- Carr, S.M.; Munro, S.; Sagum, C.A.; Fedorov, O.; Bedford, M.T.; La Thangue, N.B. Tudor-domain protein PHF20L1 reads lysine methylated retinoblastoma tumour suppressor protein. Cell Death Differ. 2017, 24, 2139–2149. [Google Scholar] [CrossRef]
- Huang, J.; Sengupta, R.; Espejo, A.B.; Lee, M.G.; Dorsey, J.A.; Richter, M.; Opravil, S.; Shiekhattar, R.; Bedford, M.T.; Jenuwein, T.; et al. p53 is regulated by the lysine demethylase LSD1. Nature 2007, 449, 105–108. [Google Scholar] [CrossRef]
- Balmik, A.A.; Chinnathambi, S. Methylation as a key regulator of Tau aggregation and neuronal health in Alzheimer’s disease. Cell Commun. Signal. 2021, 19, 51. [Google Scholar] [CrossRef]
- Sommerer, Y.; Dobricic, V.; Schilling, M.; Ohlei, O.; Sabet, S.S.; Wesse, T.; Fuß, J.; Franzenburg, S.; Franke, A.; Parkkinen, L.; et al. Entorhinal cortex epigenome-wide association study highlights four novel loci showing differential methylation in Alzheimer’s disease. Alzheimers Res. Ther. 2023, 15, 92. [Google Scholar] [CrossRef] [PubMed]
- Sacksteder, C.A.; Qian, W.-J.; Knyushko, T.V.; Wang, H.; Chin, M.H.; Lacan, G.; Melega, W.P.; Camp, D.G.; Smith, R.D.; Smith, D.J.; et al. Endogenously Nitrated Proteins in Mouse Brain: Links to Neurodegenerative Disease. Biochemistry 2006, 45, 8009–8022. [Google Scholar] [CrossRef] [PubMed]
- Beckman, J.S.; Beckman, T.W.; Chen, J.; Marshall, P.A.; Freeman, B.A. Apparent hydroxyl radical production by peroxynitrite: Implications for endothelial injury from nitric oxide and superoxide. Proc. Natl. Acad. Sci.USA 1990, 87, 1620–1624. [Google Scholar] [CrossRef]
- Torreilles, F.; Salman-Tabcheh, S.; Guérin, M.-C.; Torreilles, J. Neurodegenerative disorders: The role of peroxynitrite. Brain Res. Rev. 1999, 30, 153–163. [Google Scholar] [CrossRef] [PubMed]
- Manzanza, N.D.O.; Sedlackova, L.; Kalaria, R.N. Alpha-Synuclein Post-translational Modifications: Implications for Patho-genesis of Lewy Body Disorders. Front. Aging Neurosci. 2021, 13, 690293. [Google Scholar] [CrossRef]
- Amagai, R.; Yoshioka, S.; Otomo, R.; Nagano, H.; Hashimoto, N.; Sakakibara, R.; Tanaka, T.; Okado-Matsumoto, A. Post-translational modification of lysine residues in erythrocyte α-synuclein. J. Biochem. 2023, 173, 177–184. [Google Scholar] [CrossRef]
- Takahashi, T.; Yamashita, H.; Nakamura, T.; Nagano, Y.; Nakamura, S. Tyrosine 125 of α-synuclein plays a critical role for dimerization following nitrative stress. Brain Res. 2002, 938, 73–80. [Google Scholar] [CrossRef]
- Gómez-Tortosa, E.; Gonzalo, I.; Newell, K.; Yébenes, J.; Vonsattel, J.; Hyman, B. Patterns of protein nitration in dementia with Lewy bodies and striatonigral degeneration. Acta Neuropathol. 2002, 103, 495–500. [Google Scholar] [CrossRef]
- Reynolds, M.R.; Berry, R.W.; Binder, L.I. Site-Specific Nitration and Oxidative Dityrosine Bridging of the τ Protein by Perox-ynitrite: Implications for Alzheimer’s Disease. Biochemistry 2005, 44, 1690–1700. [Google Scholar] [CrossRef] [PubMed]
- Horiguchi, T.; Uryu, K.; Giasson, B.I.; Ischiropoulos, H.; LightFoot, R.; Bellmann, C.; Richter-Landsberg, C.; Lee, V.M.-Y.; Trojanowski, J.Q. Nitration of Tau Protein Is Linked to Neurodegeneration in Tauopathies. Am. J. Pathol. 2003, 163, 1021–1031. [Google Scholar] [CrossRef] [PubMed]
- Haines, J.D.; Inglese, M.; Casaccia, P. Axonal Damage in Multiple Sclerosis. Mt. Sinai J. Med. A J. Transl. Pers. Med. 2011, 78, 231–243. [Google Scholar] [CrossRef]
- Kean, R.B.; Spitsin, S.V.; Mikheeva, T.; Scott, G.S.; Hooper, D.C. The Peroxynitrite Scavenger Uric Acid Prevents Inflammatory Cell Invasion into the Central Nervous System in Experimental Allergic Encephalomyelitis through Maintenance of Blood-Central Nervous System Barrier Integrity. J. Immunol. 2000, 165, 6511–6518. [Google Scholar] [CrossRef]
- Roos, G.; Messens, J. Protein sulfenic acid formation: From cellular damage to redox regulation. Free Radic. Biol. Med. 2011, 51, 314–326. [Google Scholar] [CrossRef] [PubMed]
- Reddie, K.G.; Carroll, K.S. Expanding the functional diversity of proteins through cysteine oxidation. Curr. Opin. Chem. Biol. 2008, 12, 746–754. [Google Scholar] [CrossRef]
- Gupta, V.; Carroll, K.S. Sulfenic acid chemistry, detection and cellular lifetime. Biochim. Biophys. Acta (BBA)-General. Subj. 2014, 1840, 847–875. [Google Scholar] [CrossRef]
- Meng, F.; Yao, D.; Shi, Y.; Kabakoff, J.; Wu, W.; Reicher, J.; Ma, Y.; Moosmann, B.; Masliah, E.; Lipton, S.A.; et al. Oxidation of the cysteine-rich regions of parkin perturbs its E3 ligase activity and contributes to protein aggregation. Mol. Neurodegener. 2011, 6, 34. [Google Scholar] [CrossRef]
- Nakamura, T.; Tu, S.; Akhtar, M.W.; Sunico, C.R.; Okamoto, S.I.; Lipton, S.A. Review Aberrant Protein S-Nitrosylation in Neuro-degenerative Diseases. Neuron 2013, 78, 596–614. [Google Scholar] [CrossRef]
- Karplus, P.A. A Primer on Peroxiredoxin Biochemistry. Free Radic. Biol. Med. 2015, 80, 183. [Google Scholar] [CrossRef] [PubMed]
- Karplus, P.A.; Poole, L.B. Peroxiredoxins as molecular triage agents, sacrificing themselves to enhance cell survival during a peroxide attack. Mol. Cell 2012, 45, 275–278. [Google Scholar] [CrossRef] [PubMed]
- Xiong, A.; Yang, Z.; Shen, Y.; Zhou, J.; Shen, Q. Transcription Factor STAT3 as a Novel Molecular Target for Cancer Prevention. Cancers 2014, 6, 926–957. [Google Scholar] [CrossRef] [PubMed]
- Sobotta, M.C.; Liou, W.; Stöcker, S.; Talwar, D.; Oehler, M.; Ruppert, T.; Scharf, A.N.D.; Dick, T.P. Peroxiredoxin-2 and STAT3 form a redox relay for H2O2 signaling. Nat. Chem. Biol. 2014, 11, 64–70. [Google Scholar] [CrossRef]
- Lee, S.R.; Yang, K.S.; Kwon, J.; Lee, C.; Jeong, W.; Rhee, S.G. Reversible inactivation of the tumor suppressor PTEN by H2O2. J. Biol. Chem. 2002, 277, 20336–20342. [Google Scholar] [CrossRef]
- Awata, H.; Huang, C.; Handlogten, M.E.; Miller, R.T. Interaction of the Calcium-sensing Receptor and Filamin, a Potential Scaffolding Protein. J. Biol. Chem. 2001, 276, 34871–34879. [Google Scholar] [CrossRef] [PubMed]
- Paulsen, C.E.; Truong, T.H.; Garcia, F.J.; Homann, A.; Gupta, V.; Leonard, S.E.; Carroll, K.S. Peroxide-dependent sulfenylation of the EGFR catalytic site enhances kinase activity. Nat. Chem. Biol. 2011, 8, 57–64. [Google Scholar] [CrossRef]
- Paulsen, C.E.; Carroll, K.S. Cysteine-Mediated Redox Signaling: Chemistry, Biology, and Tools for Discovery. Chem. Rev. 2013, 113, 4633–4679. [Google Scholar] [CrossRef]
- Scheibe, F.; Prüss, H.; Mengel, A.M.; Kohler, S.; Nümann, A.; Köhnlein, M.; Ruprecht, K.; Alexander, T.; Hiepe, F.; Meisel, A. Bortezomib for treatment of therapy-refractory anti-NMDA receptor encephalitis. Neurology 2017, 88, 366–370. [Google Scholar] [CrossRef]
- Hart, G.W.; Housley, M.P.; Slawson, C. Cycling of O-linked beta-N-acetylglucosamine on nucleocytoplasmic proteins. Nature 2007, 446, 1017–1022. [Google Scholar] [CrossRef]
- Ma, J.; Li, Y.; Hou, C.; Wu, C. O-GlcNAcAtlas: A database of experimentally identified O-GlcNAc sites and proteins. Glycobiology 2021, 31, 719–723. [Google Scholar] [CrossRef] [PubMed]
- Wells, L.; Vosseller, K.; Cole, R.N.; Cronshaw, J.M.; Matunis, M.J.; Hart, G.W. Mapping Sites of O-GlcNAc Modification Using Affinity Tags for Serine and Threonine Post-translational Modifications. Mol. Cell. Proteom. 2002, 1, 791–804. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.H.; Kim, J.E.; Nam, H.W.; Ju, J.W.; Kim, H.S.; Kim, Y.S.; Cho, J.W. Modification of p53 with O-linked N-acetylglucosamine regulates p53 activity and stability. Nat. Cell Biol. 2006, 8, 1074–1083. [Google Scholar] [CrossRef]
- Hart, G.W. Dynamic o-linked glycosylation of nuclear and cytoskeletal proteins. Annu. Rev. Biochem. 1997, 66, 315–335. [Google Scholar] [CrossRef]
- Wells, L.; Gao, Y.; Mahoney, J.A.; Vosseller, K.; Rosen, A.; Hart, G.W. Dynamic O-Glycosylation of Nuclear and Cytosolic Proteins: Further characterization of the nucleocytoplasmic β-N-acetylglucosaminidase, O-GlcNAcase. J. Biol. Chem. 2002, 277, 1755–1761. [Google Scholar] [CrossRef]
- Bond, M.R.; Hanover, J.A. O-GlcNAc Cycling: A Link Between Metabolism and Chronic Disease. Annu. Rev. Nutr. 2013, 33, 205–229. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Ongusaha, P.P.; Miles, P.D.; Havstad, J.C.; Zhang, F.; So, W.V.; Kudlow, J.E.; Michell, R.H.; Olefsky, J.M.; Field, S.J.; et al. Phosphoinositide signalling links O-GlcNAc transferase to insulin resistance. Nature 2008, 451, 964–969. [Google Scholar] [CrossRef]
- Yang, X.; Qian, K. Protein O-GlcNAcylation: Emerging mechanisms and functions. Nat. Rev. Mol. Cell Biol. 2017, 18, 452. [Google Scholar] [CrossRef]
- Toleman, C.A.; Schumacher, M.A.; Yu, S.-H.; Zeng, W.; Cox, N.J.; Smith, T.J.; Soderblom, E.J.; Wands, A.M.; Kohler, J.J.; Boyce, M. Structural basis of O-GlcNAc recognition by mammalian 14-3-3 proteins. Proc. Natl. Acad. Sci. USA 2018, 115, 5956–5961. [Google Scholar] [CrossRef]
- Zhu, Y.; Liu, T.-W.; Madden, Z.; Yuzwa, S.A.; Murray, K.; Cecioni, S.; Zachara, N.; Vocadlo, D.J. Post-translational O-GlcNAcylation is essential for nuclear pore integrity and maintenance of the pore selectivity filter. J. Mol. Cell Biol. 2016, 8, 2–16. [Google Scholar] [CrossRef]
- Zachara, N.E.; O’Donnell, N.; Cheung, W.D.; Mercer, J.J.; Marth, J.D.; Hart, G.W. Dynamic O-GlcNAc Modification of Nucleo-cytoplasmic Proteins in Response to Stress A Survival Response Of Mammalian Cells. J. Biol. Chem. 2004, 279, 30133–30142. [Google Scholar] [CrossRef]
- Zou, L.; Yang, S.; Hu, S.; Chaudry, I.H.; Marchase, R.B.; Chatham, J.C. The protective effects of PUGNAc on cardiac function after trauma-hemorrhage are mediated via increased protein O-GlcNAc levels. Shock 2007, 27, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Lee, A.; Miller, D.; Henry, R.; Paruchuri, V.D.P.; O’Meally, R.N.; Boronina, T.; Cole, R.N.; Zachara, N.E. Combined Antibody/Lectin Enrichment Identifies Extensive Changes in the O-GlcNAc Sub-proteome upon Oxidative Stress. J. Proteome Res. 2016, 15, 4318–4336. [Google Scholar] [CrossRef]
- Zafar, S.; Shafiq, M.; Younas, N.; Schmitz, M.; Ferrer, I.; Zerr, I. Prion Protein Interactome: Identifying Novel Targets in Slowly and Rapidly Progressive Forms of Alzheimer’s Disease. J. Alzheimers Dis. 2017, 59, 265–275. [Google Scholar] [CrossRef]
- Pickart, C.M. Mechanisms Underlying Ubiquitination. Annu. Rev. Biochem. 2001, 70, 503–533. [Google Scholar] [CrossRef] [PubMed]
- Komander, D.; Rape, M. The ubiquitin code. Annu. Rev. Biochem. 2012, 81, 203–229. [Google Scholar] [CrossRef] [PubMed]
- Reyes-Turcu, F.E.; Horton, J.R.; Mullally, J.E.; Heroux, A.; Cheng, X.; Wilkinson, K.D. The Ubiquitin Binding Domain ZnF UBP Recognizes the C-Terminal Diglycine Motif of Unanchored Ubiquitin. Cell 2006, 124, 1197–1208. [Google Scholar] [CrossRef]
- Meluh, P.B.; Koshland, D. Evidence that the MIF2 gene of Saccharomyces cerevisiae encodes a centromere protein with homology to the mammalian centromere protein CENP-C. Mol. Biol. Cell 1995, 6, 793–807. [Google Scholar] [CrossRef]
- Ikeda, F.; Dikic, I. Atypical ubiquitin chains: New molecular signals. ‘Protein Modifications: Beyond the Usual Suspects’ Review Series. EMBO Rep. 2008, 9, 536–542. [Google Scholar] [CrossRef] [PubMed]
- Flotho, A.; Melchior, F. Sumoylation: A regulatory protein modification in health and disease. Annu. Rev. Biochem. 2013, 82, 357–385. [Google Scholar] [CrossRef]
- Pichler, A.; Fatouros, C.; Lee, H.; Eisenhardt, N. SUMO conjugation—A mechanistic view. Biomol. Concepts 2017, 8, 13–36. [Google Scholar] [CrossRef] [PubMed]
- Soares, E.S.; Prediger, R.D.; Brocardo, P.S.; Cimarosti, H.I. SUMO-modifying Huntington’s disease. IBRO Neurosci. Rep. 2022, 12, 203–209. [Google Scholar] [CrossRef] [PubMed]
- Hannoun, Z.; Fletcher, J.; Greenhough, S.; Medine, C.; Samuel, K.; Sharma, R.; Pryde, A.; Black, J.R.; Ross, J.A.; Wilmut, I.; et al. The comparison between conditioned media and serum-free media in human embryonic stem cell culture and differentiation. Cell Reprogram 2010, 12, 133–140. [Google Scholar] [CrossRef] [PubMed]
- Mukhopadhyay, D.; Dasso, M. Modification in reverse: The SUMO proteases. Trends Biochem. Sci. 2007, 32, 286–295. [Google Scholar] [CrossRef] [PubMed]
- Hay, R.T. Modifiying NEMO. Nat. Cell Biol. 2004, 6, 89–91. [Google Scholar] [CrossRef] [PubMed]
- Kuhn, P.H.; Koroniak, K.; Hogl, S.; Colombo, A.; Zeitschel, U.; Willem, M.; Volbracht, C.; Schepers, U.; Imhof, A.; Hoffmeister, A.; et al. Secretome protein enrichment identifies physiological BACE1 protease substrates in neurons. EMBO J. 2012, 31, 3157–3168. [Google Scholar] [CrossRef]
- Rogers, L.D.; Overall, C.M. Proteolytic Post-translational Modification of Proteins: Proteomic Tools and Methodology. Mol. Cell. Proteom. 2013, 12, 3532–3542. [Google Scholar] [CrossRef]
- Muzio, M.; Stockwell, B.R.; Stennicke, H.R.; Salvesen, G.S.; Dixit, V.M. An Induced Proximity Model for Caspase-8 Activation. J. Biol. Chem. 1998, 273, 2926–2930. [Google Scholar] [CrossRef] [PubMed]
- Stennicke, H.R.; Jürgensmeier, J.M.; Shin, H.; Deveraux, Q.; Wolf, B.B.; Yang, X.; Zhou, Q.; Ellerby, H.M.; Ellerby, L.M.; Bredesen, D.; et al. Pro-caspase-3 is a major physiologic target of Caspase-8. J. Biol. Chem. 1998, 273, 27084–27090. [Google Scholar] [CrossRef]
- Rohn, T.T.; Head, E.; Nesse, W.H.; Cotman, C.W.; Cribbs, D.H. Activation of Caspase-8 in the Alzheimer’s Disease Brain. Neurobiol. Dis. 2001, 8, 1006–1016. [Google Scholar] [CrossRef]
- Mikhailova, A.G.; Nekrasov, A.N.; Zinchenko, A.A.; Rakitina, T.V.; Korzhenevsky, D.A.; Lipkin, A.V.; Razguljaeva, O.A.; Ovchinnikova, M.V.; Gorlenko, V.A.; Rumsh, L.D. Truncated Variants of Serratia proteamaculans Oligopeptidase B Having Different Activities. Biochemistry 2015, 80, 1331–1343. [Google Scholar] [CrossRef] [PubMed]
- Fortini, M.E. Notch signaling: The core pathway and its posttranslational regulation. Dev. Cell 2009, 16, 633–647. [Google Scholar] [CrossRef] [PubMed]
- Conover, C.A.; Oxvig, C. PAPP-A: A promising therapeutic target for healthy longevity. Aging Cell 2017, 16, 205–209. [Google Scholar] [CrossRef] [PubMed]
- Oxvig, C.; Conover, C.A. The Stanniocalcin-PAPP-A-IGFBP-IGF Axis. J. Clin. Endocrinol. Metab. 2023, 108, 1624–1633. [Google Scholar] [CrossRef] [PubMed]
- Voutsadakis, I.A. Mutations of p53 associated with pancreatic cancer and therapeutic implications. Ann. Hepatobiliary Pancreat Surg. 2021, 25, 315–327. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.J.; Xu, Y.F.; Cook, C.; Gendron, T.F.; Roettges, P.; Link, C.D.; Lin, W.L.; Tong, J.; Castanedes-Casey, M.; Ash, P.; et al. Aberrant cleavage of TDP-43 enhances aggregation and cellular toxicity. Proc. Natl. Acad. Sci. USA 2009, 106, 7607–7612. [Google Scholar] [CrossRef]
- Johnson, B.A.; Murray, E.D.; Clarke, S.; Glass, D.B.; Aswad, D.W. Protein carboxyl methyltransferase facilitates conversion of atypical L-isoaspartyl peptides to normal L-aspartyl peptides. J. Biol. Chem. 1987, 262, 5622–5629. [Google Scholar] [CrossRef]
- Chatterjee, T.; Das, G.; Chatterjee, B.K.; Dhar, J.; Ghosh, S.; Chakrabarti, P. The role of isoaspartate in fibrillation and its prevention by Protein-L-isoaspartyl methyltransferase. Biochim. Biophys. Acta (BBA)-General. Subj. 2020, 1864, 129500. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Lyutvinskiy, Y.; Herukka, S.K.; Soininen, H.; Rutishauser, D.; Zubarev, R.A. Prognostic polypeptide blood plasma biomarkers of Alzheimer’s disease progression. J. Alzheimers Dis. 2014, 40, 659–666. [Google Scholar] [CrossRef]
- Yang, H.; Wittnam, J.L.; Zubarev, R.A.; Bayer, T.A. Shotgun brain proteomics reveals early molecular signature in presymp-tomatic mouse model of Alzheimer’s disease. J. Alzheimers Dis. 2013, 37, 297–308. [Google Scholar] [CrossRef]
- Yang, H.; Lyutvinskiy, Y.; Soininen, H.; Zubarev, R.A. Alzheimer’s disease and mild cognitive impairment are associated with elevated levels of isoaspartyl residues in blood plasma proteins. J. Alzheimers Dis. 2011, 27, 113–118. [Google Scholar] [CrossRef] [PubMed]
- Fukuda, H.; Shimizu, T.; Nakajima, M.; Mori, H.; Shirasawa, T. Synthesis, aggregation, and neurotoxicity of the Alzheimer’s Aβ1-42 amyloid peptide and its isoaspartyl isomers. Bioorg Med. Chem. Lett. 1999, 9, 953–956. [Google Scholar] [CrossRef] [PubMed]
- Johnson, B.A.; Shirokawa, J.M.; Geddes, J.W.; Choi, B.H.; Kim, R.C.; Aswad, D.W. Protein L-isoaspartyl methyltransferase in postmortem brains of aged humans. Neurobiol. Aging 1991, 12, 19–24. [Google Scholar] [CrossRef]
- Wang, J.; Zhang, Y.R.; Shen, X.N.; Han, J.; Cui, M.; Tan, L.; Dong, Q.; Zubarev, R.A.; Yu, J.T. Deamidation-related blood biomarkers show promise for early diagnostics of neurodegeneration. Biomark. Res. 2022, 10, 91. [Google Scholar] [CrossRef] [PubMed]
- Patterson, K.; Molloy, L.; Qu, W.; Clark, S. DNA Methylation: Bisulphite Modification and Analysis. J. Vis. Exp. 2011, 56, e3170. [Google Scholar] [CrossRef]
- Aebersold, R.; Mann, M. Mass-spectrometric exploration of proteome structure and function. Nature 2016, 537, 347–355. [Google Scholar] [CrossRef] [PubMed]
- Loo, J.A. Electrospray ionization mass spectrometry: A technology for studying noncovalent macromolecular complexes. Int. J. Mass. Spectrom. 2000, 200, 175–186. [Google Scholar] [CrossRef]
- Gillette, M.A.; Carr, S.A. Quantitative analysis of peptides and proteins in biomedicine by targeted mass spectrometry. Nat. Methods 2013, 10, 28–34. [Google Scholar] [CrossRef]
- Hendriks, I.A.; Lyon, D.; Young, C.; Jensen, L.J.; Vertegaal, A.C.O.; Nielsen, M.L. Site-specific mapping of the human SUMO proteome reveals co-modification with phosphorylation. Nat. Struct. Mol. Biol. 2017, 24, 325–336. [Google Scholar] [CrossRef]
- Humphrey, S.J.; Yang, G.; Yang, P.; Fazakerley, D.J.; Stöckli, J.; Yang, J.Y.; James, D.E. Dynamic adipocyte phosphoproteome reveals that akt directly regulates mTORC2. Cell Metab. 2013, 17, 1009–1020. [Google Scholar] [CrossRef] [PubMed]
- Olova, N.; Krueger, F.; Andrews, S.; Oxley, D.; Berrens, R.V.; Branco, M.R.; Reik, W. Correction to: Comparison of whole-genome bisulfite sequencing library preparation strategies identifies sources of biases affecting DNA methylation data. Genome Biol. 2018, 19, 33. [Google Scholar] [CrossRef] [PubMed]
- Hendriks, I.A.; D’souza, R.C.; Yang, B.; Verlaan-De Vries, M.; Mann, M.; Vertegaal, A.C. Uncovering global SUMOylation signaling networks in a site-specific manner. Nat. Struct. Mol. Biol. 2014, 21, 927–936. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Sun, M.; Hu, Y.; Shan, Y.; Liang, Z.; Zhang, L.; Zhang, Y. Antibody-free enrichment method for proteome-wide analysis of endogenous SUMOylation sites. Anal. Chim. Acta 2021, 1154, 338324. [Google Scholar] [CrossRef] [PubMed]
- Hjerpe, R.; Aillet, F.; Lopitz-Otsoa, F.; Lang, V.; England, P.; Rodriguez, M.S. Efficient protection and isolation of ubiquitylated proteins using tandem ubiquitin-binding entities. Embo. Rep. 2009, 10, 1250–1258. [Google Scholar] [CrossRef] [PubMed]
- Kim, W.; Bennett, E.J.; Huttlin, E.L.; Guo, A.; Li, J.; Possemato, A.; Sowa, M.E.; Rad, R.; Rush, J.; Comb, M.J. Systematic and Quantitative As-sessment of the Ubiquitin-Modified Proteome. Mol. Cell. 2011, 44, 325–340. [Google Scholar] [CrossRef]
- Damgaard, R.B. The ubiquitin system: From cell signalling to disease biology and new therapeutic opportunities. Cell Death Differ. 2021, 28, 423–426. [Google Scholar] [CrossRef] [PubMed]
- Tyther, R.; McDonagh, B.; Sheehan, D. Proteomics in investigation of protein nitration in kidney disease: Technical challenges and perspectives from the spontaneously hypertensive rat. Mass. Spectrom. Rev. 2011, 30, 121–141. [Google Scholar] [CrossRef]
- Radi, R. Protein tyrosine nitration: Biochemical mechanisms and structural basis of functional effects. Acc. Chem. Res. 2013, 46, 550–559. [Google Scholar] [CrossRef]
- Kansanen, E.; Bonacci, G.; Schopfer, F.J.; Kuosmanen, S.M.; Tong, K.I.; Leinonen, H.; Woodcock, S.R.; Yamamoto, M.; Carlberg, C.; Ylä-Herttuala, S.; et al. Electrophilic nitro-fatty acids activate NRF2 by a KEAP1 cysteine 151-independent mechanism. J. Biol. Chem. 2011, 286, 14019–14027. [Google Scholar] [CrossRef]
- Feeney, M.B.; Schöneich, C. Proteomic approaches to analyze protein tyrosine nitration. Antioxid. Redox Signal. 2013, 19, 1247–1256. [Google Scholar] [CrossRef]
- Medeiros, R.; Sousa, B.; Rossi, S.; Afonso, C.; Bonino, L.; Pitt, A.; López, E.; Spickett, C.; Borthagaray, G. Identification and relative quantification of 3-nitrotyrosine residues in fibrinogen nitrated in vitro and fibrinogen from ischemic stroke patient plasma using LC-MS/M.S. Free. Radic. Biol. Med. 2021, 165, 334–347. [Google Scholar] [CrossRef]
- Ong, S. Mass spectrometric-based approaches in quantitative proteomics. Methods 2003, 29, 124–130. [Google Scholar] [CrossRef] [PubMed]
- Stes, E.; Laga, M.; Walton, A.; Samyn, N.; Timmerman, E.; De Smet, I.; Goormachtig, S.; Gevaert, K. A COFRADIC Protocol To Study Protein Ubiquitination. J. Proteome Res. 2014, 13, 3107–3113. [Google Scholar] [CrossRef]
- Ma, B.; Khan, K.S.; Xu, T.; Amada, J.X.; Guo, Z.; Yan, Y.; Cheng, A.S.L.; Ng, B.W.L. Targeted Protein O-GlcNAcylation Using Bifunctional Small Molecules. bioRxiv 2023. [Google Scholar] [CrossRef]
- Wu, Z.L.; Tatge, T.J.; Grill, A.E.; Zou, Y. Detecting and Imaging O-GlcNAc Sites Using Glycosyltransferases: A Systematic Approach to Study O-GlcNAc. Cell Chem. Biol. 2018, 25, 1428–1435.e3. [Google Scholar] [CrossRef]
- Gorelik, A.; van Aalten, D.M.F. Tools for functional dissection of site-specific O-GlcNAcylation. RSC Chem. Biol. 2020, 1, 98–109. [Google Scholar] [CrossRef] [PubMed]
- Yoo, T.Y.; Mitchison, T.J. O-GlcNAc modification of nuclear pore complexes accelerates bidirectional transport. J. Cell Biol. 2021, 220, e202010141. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Qi, J.; Pei, J.; Zhang, M.; Shang, Y.; Li, Z.; Wang, Y.; Guo, J.; Sun, K.; Fan, J.; et al. O-GlcNAc modification mediates aquaporin 3 to coordinate endometrial cell glycolysis and affects embryo implantation. J. Adv. Res. 2022, 37, 119–131. [Google Scholar] [CrossRef] [PubMed]
- Söderberg, O.; Gullberg, M.; Jarvius, M.; Ridderstråle, K.; Leuchowius, K.-J.; Jarvius, J.; Wester, K.; Hydbring, P.; Bahram, F.; Larsson, L.-G.; et al. Direct observation of individual endogenous protein complexes in situ by proximity ligation. Nat. Methods 2006, 3, 995–1000. [Google Scholar] [CrossRef]
- Darmanis, S.; Nong, R.Y.; Vänelid, J.; Siegbahn, A.; Ericsson, O.; Fredriksson, S.; Bäcklin, C.; Gut, M.; Heath, S.; Gut, I.G.; et al. ProteinSeq: High-performance proteomic analyses by proximity ligation and next generation sequencing. PLoS ONE 2011, 6, e25583. [Google Scholar] [CrossRef]
- Cubeñas-Potts, C.; Srikumar, T.; Lee, C.; Osula, O.; Subramonian, D.; Zhang, X.-D.; Cotter, R.J.; Raught, B.; Matunis, M.J. Identification of SUMO-2/3-modified proteins associated with mitotic chromosomes. Proteomics 2015, 15, 763–772. [Google Scholar] [CrossRef] [PubMed]
- Edelmann, M.J.; Shack, L.A.; Naske, C.D.; Walters, K.B.; Nanduri, B. SILAC-Based Quantitative Proteomic Analysis of Human Lung Cell Response to Copper Oxide Nanoparticles. PLoS ONE 2014, 9, e114390. [Google Scholar] [CrossRef]
- Chen, X.; Wei, S.; Ji, Y.; Guo, X.; Yang, F. Quantitative proteomics using SILAC: Principles, applications, and developments. Proteomics 2015, 15, 3175–3192. [Google Scholar] [CrossRef] [PubMed]
- Fredriksson, S.; Gullberg, M.; Jarvius, J.; Olsson, C.; Pietras, K.; Gústafsdóttir, S.M.; Östman, A.; Landegren, U. Protein detection using proximity-dependent DNA ligation assays. Nat. Biotechnol. 2002, 20, 473–477. [Google Scholar] [CrossRef]
- Akter, S.; Fu, L.; Jung, Y.; Conte, M.L.; Lawson, J.R.; Lowther, W.T.; Sun, R.; Liu, K.; Yang, J.; Carroll, K.S. Chemical proteomics reveals new targets of cysteine sulfinic acid reductase. Nat. Chem. Biol. 2018, 14, 995–1004. [Google Scholar] [CrossRef] [PubMed]
- Poole, L.B. The basics of thiols and cysteines in redox biology and chemistry. Free. Radic. Biol. Med. 2015, 80, 148–157. [Google Scholar] [CrossRef]
- Brandi, J.; Noberini, R.; Bonaldi, T.; Cecconi, D. Advances in enrichment methods for mass spectrometry-based proteomics analysis of post-translational modifications. J. Chromatogr. A 2022, 1678, 463352. [Google Scholar] [CrossRef] [PubMed]
- Ren, R.J.; Dammer, E.B.; Wang, G.; Seyfried, N.T.; Levey, A.I. Proteomics of protein post-translational modifications implicated in neurodegeneration. Transl. Neurodegener. 2014, 3, 23. [Google Scholar] [CrossRef]
- Humphrey, S.J.; James, D.E.; Mann, M. Protein phosphorylation: A major switch mechanism for metabolic regulation. Trends Endocrinol. Metab. 2015, 26, 676–687. [Google Scholar] [CrossRef]
- Rani, N.; Sahu, M.; Ambasta, R.K.; Kumar, P. Triaging between post-translational modification of cell cycle regulators and their therapeutics in neurodegenerative diseases. Ageing Res. Rev. 2023, 94, 102174. [Google Scholar] [CrossRef]
- Kurtishi, A.; Rosen, B.; Patil, K.S.; Alves, G.W.; Møller, S.G. Cellular proteostasis in neurodegeneration. Mol. Neurobiol. 2019, 56, 3676–3689. [Google Scholar] [CrossRef] [PubMed]
- Jean Beltran, P.M.; Federspiel, J.D.; Sheng, X.; Cristea, I.M. Proteomics and integrative omic approaches for understanding host–pathogen interactions and infectious diseases. Mol. Syst. Biol. 2017, 13, 922. [Google Scholar] [CrossRef]
- Beck, H.C.; Nielsen, E.C.; Matthiesen, R.; Jensen, L.H.; Sehested, M.; Finn, P.; Grauslund, M.; Hansen, A.M.; Jensen, O.N. Quantitative proteomic analysis of post-translational modifications of human histones. Mol. Cell Proteom. 2006, 5, 1314–1325. [Google Scholar] [CrossRef] [PubMed]
- Dekker, F.J.; van den Bosch, T.; Martin, N.I. Small molecule inhibitors of histone acetyltransferases and deacetylases are po-tential drugs for inflammatory diseases. Drug Discov. Today 2014, 19, 654–660. [Google Scholar] [CrossRef] [PubMed]
- Gupta, R.; Kumar, P. Computational Analysis Indicates That PARP1 Acts as a Histone Deacetylases Interactor Sharing Common Lysine Residues for Acetylation, Ubiquitination, and SUMOylation in Alzheimer’s and Parkinson’s Disease. ACS Omega 2021, 6, 5739–5753. [Google Scholar] [CrossRef]
- Zhang, S.; Meng, Y.; Zhou, L.; Qiu, L.; Wang, H.; Su, D.; Zhang, B.; Chan, K.; Han, J. Targeting epigenetic regulators for inflammation: Mechanisms and intervention therapy. MedComm 2022, 3, e173. [Google Scholar] [CrossRef]
- Xu, L.; Feng, J.; Tang, H.; Dong, Y.; Shu, M.; Chen, X. Chidamide epigenetically represses autophagy and exerts co-operative antimyeloma activity with bortezomib. Cell Death Dis. 2020, 11, 297. [Google Scholar] [CrossRef]
- Rastgoo, N.; Pourabdollah, M.; Abdi, J.; Reece, D.; Chang, H. Dysregulation of EZH2/miR-138 axis contributes to drug resistance in multiple myeloma by downregulating RBPMS. Leukemia 2018, 32, 2471–2482. [Google Scholar] [CrossRef]
- Nara, M.; Teshima, K.; Watanabe, A.; Ito, M.; Iwamoto, K.; Kitabayashi, A.; Kume, M.; Hatano, Y.; Takahashi, N.; Iida, S.; et al. Bortezomib Reduces the Tumorigenicity of Multiple Myeloma via Downregulation of Upregulated Targets in Clon-ogenic Side Population Cells. PLoS ONE 2013, 8, e56954. [Google Scholar] [CrossRef]
- Zhou, L.; Zhang, Q.; Zhang, P.; Sun, L.; Peng, C.; Yuan, Z.; Cheng, J. c-Abl-mediated Drp1 phosphorylation promotes oxidative stress-induced mitochondrial fragmentation and neuronal cell death. Cell Death Dis. 2017, 8, e3117. [Google Scholar] [CrossRef]
- Lan, W.; Santofimia-Castaño, P.; Swayden, M.; Xia, Y.; Zhou, Z.; Audebert, S.; Camoin, L.; Huang, C.; Peng, L.; Jiménez-Alesanco, A.; et al. ZZW-115–dependent inhibition of NUPR1 nuclear translocation sensitizes cancer cells to genotoxic agents. J. Clin. Investig. 2020, 5, e138117. [Google Scholar] [CrossRef]
- Santofimia-Castaño, P.; Lan, W.; Bintz, J.; Gayet, O.; Carrier, A.; Lomberk, G.; Neira, J.L.; González, A.; Urrutia, R.; Soubeyran, P.; et al. Inactivation of NUPR1 promotes cell death by coupling ER-stress responses with necrosis. Sci. Rep. 2018, 8, 16999. [Google Scholar] [CrossRef] [PubMed]
- Santofimia-Castaño, P.; Xia, Y.; Lan, W.; Zhou, Z.; Huang, C.; Peng, L.; Soubeyran, P.; Velázquez-Campoy, A.; Abián, O.; Rizzuti, B.; et al. Ligand-based design identifies a potent NUPR1 inhibitor exerting anticancer activity via necroptosis. J. Clin. Investig. 2019, 129, 2500–2513. [Google Scholar] [CrossRef] [PubMed]
- Tenreiro, S.; Eckermann, K.; Outeiro, T.F. Protein phosphorylation in neurodegeneration: Friend or foe? Front. Mol. Neurosci. 2014, 7, 42. [Google Scholar] [CrossRef] [PubMed]
- Rasmi, Y.; Shokati, A.; Hassan, A.; Aziz, S.G.-G.; Bastani, S.; Jalali, L.; Moradi, F.; Alipour, S. The role of DNA methylation in progression of neurological disorders and neurodegenerative diseases as well as the prospect of using DNA methylation inhibitors as therapeutic agents for such disorders. IBRO Neurosci. Rep. 2023, 14, 28–37. [Google Scholar] [CrossRef]
- Kawahata, I.; Finkelstein, D.I.; Fukunaga, K. Pathogenic Impact of α-Synuclein Phosphorylation and Its Kinases in α-Synucleinopathies. Int. J. Mol. Sci. 2022, 23, 6216. [Google Scholar] [CrossRef] [PubMed]
- Mandel, N.; Agarwal, N. Role of SUMOylation in Neurodegenerative Diseases. Cells 2022, 11, 3395. [Google Scholar] [CrossRef]
- Schmidt, M.F.; Gan, Z.Y.; Komander, D.; Dewson, G. Ubiquitin signalling in neurodegeneration: Mechanisms and therapeutic opportunities. Cell Death Differ. 2021, 28, 570–590. [Google Scholar] [CrossRef]
- Nakamura, T.; Oh, C.-K.; Zhang, X.; Lipton, S.A. Protein S-nitrosylation and oxidation contribute to protein misfolding in neurodegeneration. Free. Radic. Biol. Med. 2021, 172, 562–577. [Google Scholar] [CrossRef]
- Tsoi, P.S.; Quan, M.D.; Ferreon, J.C.; Ferreon, A.C.M. Aggregation of Disordered Proteins Associated with Neurodegeneration. Int. J. Mol. Sci. 2023, 24, 3380. [Google Scholar] [CrossRef]
- Lee, B.E.; Suh, P.-G.; Kim, J.-I. O-GlcNAcylation in health and neurodegenerative diseases. Exp. Mol. Med. 2021, 53, 1674–1682. [Google Scholar] [CrossRef] [PubMed]
- Tabrez, S.; Jabir, N.R.; Shakil, S.; Greig, N.H.; Alam, Q.; Abuzenadah, A.M.; Damanhouri, G.A.; Kamal, M.A. A Synopsis on the Role of Tyrosine Hydroxylase in Parkinson’s Disease. CNS Neurol. Disord.-Drug Targets 2012, 11, 395–409. [Google Scholar] [CrossRef] [PubMed]
- Yamada, M.; Hamaguchi, T. The sulfation code for propagation of neurodegeneration. J. Biol. Chem. 2018, 293, 10841–10842. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zafar, S.; Fatima, S.I.; Schmitz, M.; Zerr, I. Current Technologies Unraveling the Significance of Post-Translational Modifications (PTMs) as Crucial Players in Neurodegeneration. Biomolecules 2024, 14, 118. https://doi.org/10.3390/biom14010118
Zafar S, Fatima SI, Schmitz M, Zerr I. Current Technologies Unraveling the Significance of Post-Translational Modifications (PTMs) as Crucial Players in Neurodegeneration. Biomolecules. 2024; 14(1):118. https://doi.org/10.3390/biom14010118
Chicago/Turabian StyleZafar, Saima, Shehzadi Irum Fatima, Matthias Schmitz, and Inga Zerr. 2024. "Current Technologies Unraveling the Significance of Post-Translational Modifications (PTMs) as Crucial Players in Neurodegeneration" Biomolecules 14, no. 1: 118. https://doi.org/10.3390/biom14010118
APA StyleZafar, S., Fatima, S. I., Schmitz, M., & Zerr, I. (2024). Current Technologies Unraveling the Significance of Post-Translational Modifications (PTMs) as Crucial Players in Neurodegeneration. Biomolecules, 14(1), 118. https://doi.org/10.3390/biom14010118







