Comprehensive Proteomic Analysis of Common Bean (Phaseolus vulgaris L.) Seeds Reveal Shared and Unique Proteins Involved in Terminal Drought Stress Response in Tolerant and Sensitive Genotypes
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and Experiments
2.2. Sample Preparation and Digestion
2.3. Nano LC-MS/MS Analysis
2.4. Functional Enrichment Analysis
2.5. Quantitative PCR
3. Results
3.1. Proteomic Profiling of Genotypes in Response to Stress
3.2. Functional Analysis of Stress-Responsive Proteins
3.2.1. Gene Ontology Annotations
3.2.2. Biological Process (BP)
3.2.3. Cellular Components (CC)
3.2.4. Molecular Function (MF)
3.3. Elucidating Drought Stress-Altered Biological Processes
3.4. The Potential Biological Function of Identified Proteins
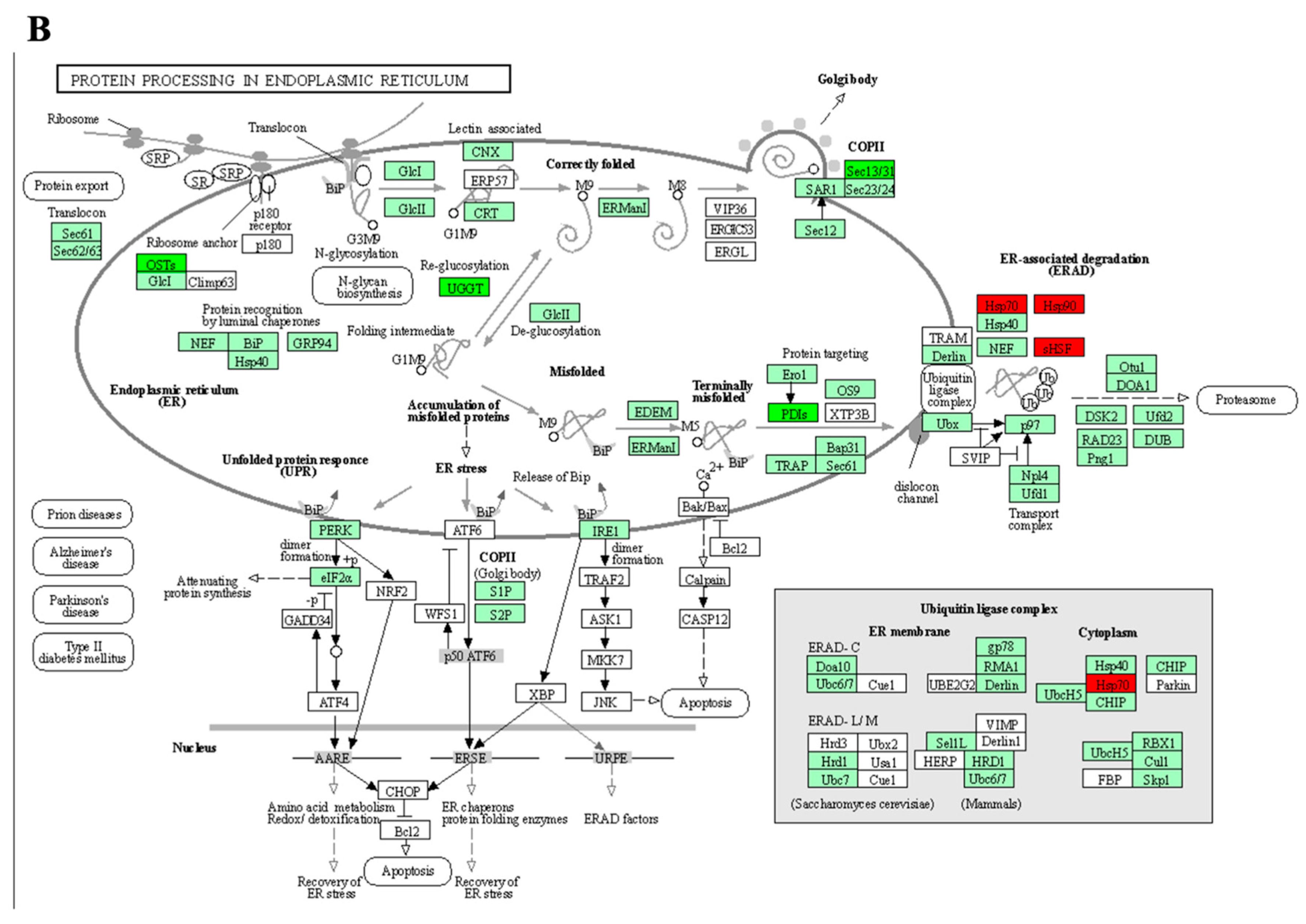
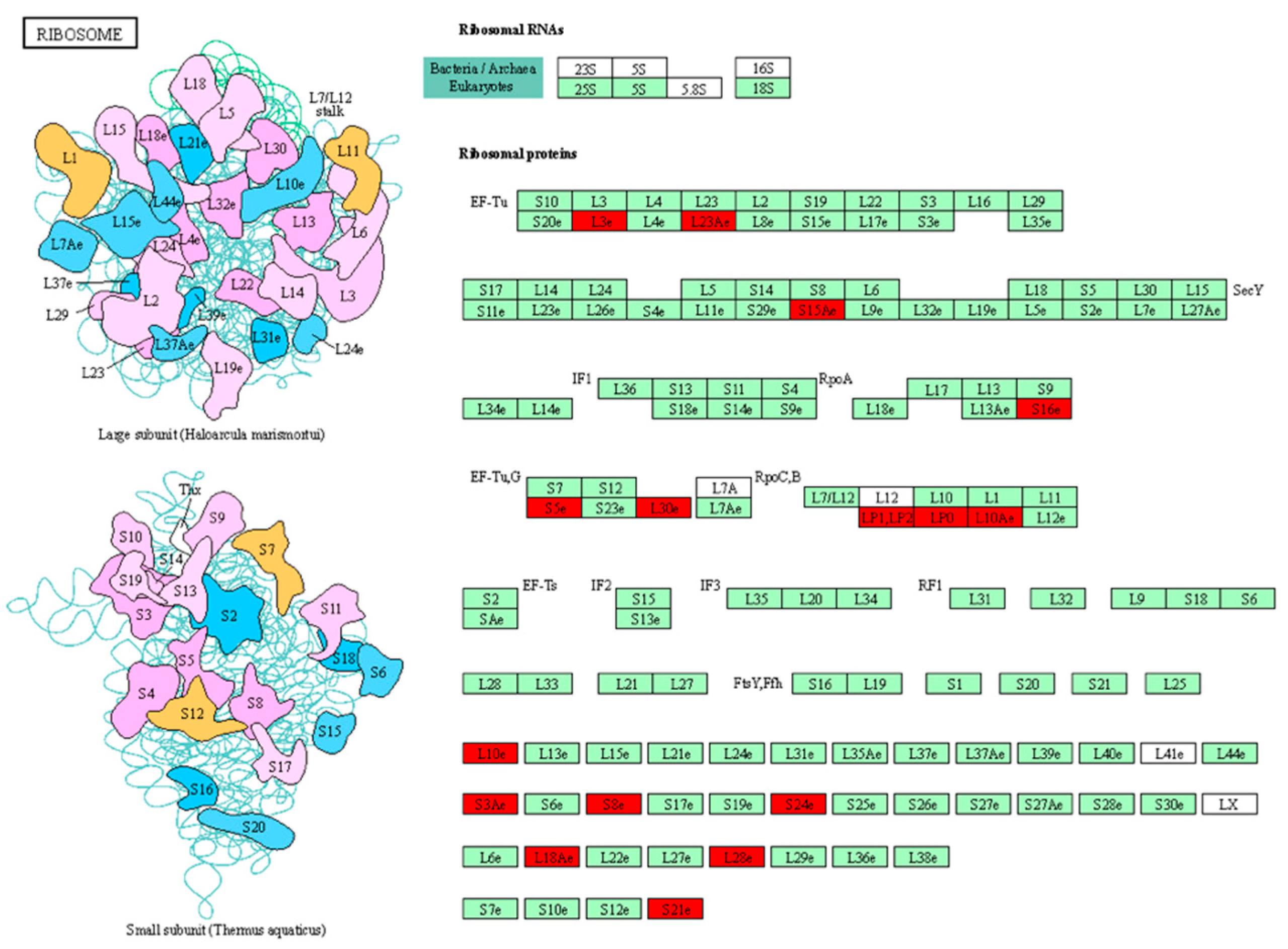
3.5. KEGG Pathways Enrichment
3.5.1. Tolerant Genotypes
3.5.2. Sensitive Genotypes
3.6. Corresponding Genes Expression in the Tolerant and Sensitive Genotypes
4. Discussion
4.1. Terminal Drought Stress and Abundance of Proteins
4.2. Heat Shock Proteins
4.3. Signal Transduction Mechanisms and Adaptive Responses during Stress
4.4. Protein Alterations in Plant Energy Balance for Drought Stress Resilience
4.5. Genotype Dependent Enrichment of Drought Stress Pathways
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Mamo, T.; Singh, A.; Singh, A.; Mahama, A.A. Chapter 8: Common Bean Breeding; Iowa State University Digital Press: Ames, IA, USA, 2023. [Google Scholar]
- Smith, M.R.; Veneklaas, E.; Polania, J.; Rao, I.M.; Beebe, S.E.; Merchant, A. Field drought conditions impact yield but not nutritional quality of the seed in common bean (Phaseolus vulgaris L.). PLoS ONE 2019, 14, e0217099. [Google Scholar] [CrossRef] [PubMed]
- Gebeyehu, S.; Wiese, H.; Schubert, S. Effects of drought stress on seed sink strength and leaf protein patterns of common bean genotypes. Afr. Crop Sci. J. 2011, 18, 75–88. [Google Scholar] [CrossRef][Green Version]
- Hummel, M.; Hallahan, B.F.; Brychkova, G.; Ramirez-Villegas, J.; Guwela, V.; Chataika, B.; Curley, E.; McKeown, P.C.; Morrison, L.; Talsma, E.F.; et al. Reduction in nutritional quality and growing area suitability of common bean under climate change induced drought stress in Africa. Sci. Rep. 2018, 8, 16187. [Google Scholar] [CrossRef] [PubMed]
- Hossain, Z.; Komatsu, S. Potentiality of soybean proteomics in untying the mechanism of flood and drought stress tolerance. Proteomes 2014, 2, 107–127. [Google Scholar] [CrossRef]
- Zadražnik, T.; Egge-Jacobsen, W.; Meglič, V.; Šuštar-Vozlič, J. Proteomic analysis of common bean stem under drought stress using in-gel stable isotope labeling. J. Plant Physiol. 2017, 209, 42–50. [Google Scholar] [CrossRef]
- López, C.M.; Pineda, M.; Alamillo, J.M. Transcriptomic Response to Water Deficit Reveals a Crucial Role of Phosphate Acquisition in a Drought-Tolerant Common Bean Landrace. Plants 2020, 9, 445. [Google Scholar] [CrossRef]
- Wu, J.; Wang, L.; Li, L.; Wang, S. De novo assembly of the common bean transcriptome using short reads for the discovery of drought-responsive genes. PLoS ONE 2014, 9, e109262. [Google Scholar] [CrossRef]
- Jorge, J.G.; Villalobos-López, M.A.; Chavarría-Alvarado, K.L.; Ríos-Meléndez, S.; López-Meyer, M.; Arroyo-Becerra, A. Genome-wide transcriptional changes triggered by water deficit on a drought-tolerant common bean cultivar. BMC Plant Biol. 2020, 20, 1–20. [Google Scholar] [CrossRef]
- Leitão, S.T.; Santos, C.; Araújo, S.d.S.; Rubiales, D.; Patto, M.C.V. Shared and tailored common bean transcriptomic responses to combined fusarium wilt and water deficit. Hortic. Res. 2021, 8, 149. [Google Scholar] [CrossRef]
- Liu, H.; Sultan, M.A.R.F.; Liu, X.L.; Zhang, J.; Yu, F.; Zhao, H.X. Physiological and comparative proteomic analysis reveals different drought responses in roots and leaves of drought-tolerant wild wheat (Triticum boeoticum). PLoS ONE 2015, 10, e0121852. [Google Scholar] [CrossRef]
- Ghatak, A.; Chaturvedi, P.; Weckwerth, W. Cereal Crop Proteomics: Systemic Analysis of Crop Drought Stress Responses Towards Marker-Assisted Selection Breeding. Front. Plant Sci. 2017, 8, 757. [Google Scholar] [CrossRef] [PubMed]
- Yahoueian, S.H.; Bihamta, M.R.; Babaei, H.R.; Bazargani, M.M. Proteomic analysis of drought stress response mechanism in soybean (Glycine max L.) leaves. Food Sci. Nutr. 2021, 9, 2010–2020. [Google Scholar] [CrossRef]
- Lima, E.; Silva, M.D.S.; de Abreu, C.; Mesquita, R.; Lobo, M.; Monteiro-Moreira, A.D.O.; Gomes-Filho, E.; Bertini, C.D.M. Research Article Differential proteomics in contrasting cowpea genotypes submitted to different water regimes. Evolution 2019, 18, 1–17. [Google Scholar] [CrossRef]
- Vessal, S.; Arefian, M.; Siddique, K.H.M. Proteomic responses to progressive dehydration stress in leaves of chickpea seedlings. BMC Genom. 2020, 21, 523. [Google Scholar] [CrossRef]
- Katam, R.; Sakata, K.; Suravajhala, P.; Pechan, T.; Kambiranda, D.M.; Naik, K.S.; Guo, B.; Basha, S.M. Comparative leaf proteomics of drought-tolerant and -susceptible peanut in response to water stress. J. Proteom. 2016, 143, 209–226. [Google Scholar] [CrossRef] [PubMed]
- Zargar, S.M.; Mahajan, R.; Nazir, M.; Nagar, P.; Kim, S.T.; Rai, V.; Masi, A.; Ahmad, S.M.; Shah, R.A.; Ganai, N.A.; et al. Common bean proteomics: Present status and future strategies. J. Proteom. 2017, 169, 239–248. [Google Scholar] [CrossRef]
- Ramalingam, A.; Kudapa, H.; Pazhamala, L.T.; Weckwerth, W.; Varshney, R.K. Proteomics and metabolomics: Two emerging areas for legume improvement. Front. Plant Sci. 2015, 6, 1116. [Google Scholar] [CrossRef]
- Zargar, S.M.; Nazir, M.; Rai, V.; Hajduch, M.; Agrawal, G.K.; Rakwal, R. Towards a common bean proteome atlas: Looking at the current state of research and the need for a comprehensive proteome. Front. Plant Sci. 2015, 6, 201. [Google Scholar] [CrossRef][Green Version]
- Papathanasiou, F.; Ninou, E.; Mylonas, I.; Baxevanos, D.; Papadopoulou, F.; Avdikos, I.; Sistanis, I.; Koskosidis, A.; Vlachostergios, D.N.; Stefanou, S.; et al. The Evaluation of Common Bean (Phaseolus vulgaris L.) Genotypes under Water Stress Based on Physiological and Agronomic Parameters. Plants 2022, 11, 2432. [Google Scholar] [CrossRef]
- Langat, C.; Ombori, O.; Leley, P.; Karanja, D.; Cheruiyot, R.; Gathaara, M.; Masila, B. Genetic variability of agronomic traits as potential indicators of drought tolerance in common beans (Phaseolus vulgaris L.). Int. J. Agron. 2019, 2019, 2360848. [Google Scholar] [CrossRef]
- Hageman, A.; Van Volkenburgh, E. Sink strength maintenance underlies drought tolerance in common bean. Plants 2021, 10, 489. [Google Scholar] [CrossRef]
- Thomas, A.; Beena, R.; Laksmi, G.; Kb, S.; Alex, S.; Mm, V. Changes in sucrose metabolic enzymes to water stress in contrasting rice genotypes. Plant Stress 2022, 5, 100088. [Google Scholar] [CrossRef]
- Sehgal, A.; Sita, K.; Siddique, K.H.M.; Kumar, R.; Bhogireddy, S.; Varshney, R.K.; HanumanthaRao, B.; Nair, R.M.; Prasad, P.V.V.; Nayyar, H. Drought or/and heat-stress effects on seed filling in food crops: Impacts on functional biochemistry, seed yields, and nutritional quality. Front. Plant Sci. 2018, 9, 1705. [Google Scholar] [CrossRef] [PubMed]
- Sehgal, A.; Sita, K.; Bhandari, K.; Kumar, S.; Kumar, J.; Vara Prasad, P.V.; Siddique, K.H.M.; Nayyar, H. Influence of drought and heat stress, applied independently or in combination during seed development, on qualitative and quantitative aspects of seeds of lentil (Lens culinaris Medikus) genotypes, differing in drought sensitivity. Plant Cell Environ. 2019, 42, 198–211. [Google Scholar] [CrossRef] [PubMed]
- Subramani, M.; Urrea, C.A.; Kalavacharla, V. Comparative Analysis of Untargeted Metabolomics in Tolerant and Sensitive Genotypes of Common Bean (Phaseolus vulgaris L.) Seeds Exposed to Terminal Drought Stress. Metabolites 2022, 12, 944. [Google Scholar] [CrossRef] [PubMed]
- Subramani, M.; Urrea, C.A.; Habib, R.; Bhide, K.; Thimmapuram, J.; Kalavacharla, V. Comparative Transcriptome Analysis of Tolerant and Sensitive Genotypes of Common Bean (Phaseolus vulgaris L.) in Response to Terminal Drought Stress. Plants 2023, 12, 210. [Google Scholar] [CrossRef] [PubMed]
- Urrea, C.A.; Smith, J.R.; Porch, T.G. Release of drought-tolerant pinto SB-DT2 and small red SB-DT3 common bean germplasm from a shuttle breeding program between Nebraska and Puerto Rico. J. Plant Regist. 2022, 16, 400–409. [Google Scholar] [CrossRef]
- Triboi, A.-M.; Martre, P.; Triboï-Blondel, A.M. Environmentally-induced changes in protein composition in developing grains of wheat are related to changes in total protein content. J. Exp. Bot. 2003, 54, 1731–1742. [Google Scholar] [CrossRef]
- Ghanbari, A.A.; Shakiba, M.R.; Toorchi, M.; Choukan, R. Nitrogen changes in the leaves and accumulation of some minerals in the seeds of red, white and Chitti beans (Phaseolus vulgaris) under water deficit conditions. AJCS 2013, 7, 706–712. [Google Scholar]
- Wang, X.; Oh, M.W.; Komatsu, S. Characterization of S-adenosylmethionine synthetases in soybean under flooding and drought stresses. Biol. Plant. 2016, 60, 269–278. [Google Scholar] [CrossRef]
- Zhou, Y.; Li, H.; Chen, H.; Yang, X.; Yu, T.; Wang, Y.; Wang, Y.; Jiang, K.; Wang, Y.; Chen, Z.; et al. Proteomic Investigation of Molecular Mechanisms in Response to PEG-Induced Drought Stress in Soybean Roots. Plants 2022, 11, 1173. [Google Scholar] [CrossRef] [PubMed]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.-P.; Cueff, G.; Hegedus, D.D.; Rajjou, L.; Bentsink, L. A role for seed storage proteins in Arabidopsis seed longevity. J. Exp. Bot. 2015, 66, 6399–6413. [Google Scholar] [CrossRef] [PubMed]
- Jan, N.; Rather, A.M.-U.; John, R.; Chaturvedi, P.; Ghatak, A.; Weckwerth, W.; Zargar, S.M.; Mir, R.A.; Khan, M.A.; Mir, R.R. Proteomics for abiotic stresses in legumes: Present status and future directions. Crit. Rev. Biotechnol. 2022, 43, 171–190. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; James, A.T.; Yang, A.; Jones, A.; Mendoza-Porras, O.; Bétrix, C.-A.; Ma, H.; Colgrave, M.L. A comparative proteomic study of drought-tolerant and drought-sensitive soybean seedlings under drought stress. Crop. Pasture Sci. 2016, 67, 528. [Google Scholar] [CrossRef]
- Ghaffari, M.; Toorchi, M.; Valizadeh, M.; Komatsu, S. Differential response of root proteome to drought stress in drought sensitive and tolerant sunflower inbred lines. Funct. Plant Biol. 2013, 40, 609–617. [Google Scholar] [CrossRef] [PubMed]
- Nongpiur, R.; Soni, P.; Karan, R.; Singla-Pareek, S.L.; Pareek, A. Histidine kinases in plants. Plant Signal. Behav. 2012, 7, 1230–1237. [Google Scholar] [CrossRef]
- Li, C.; Chen, S.; Wang, Y. Physiological and proteomic changes of Castanopsis fissa in response to drought stress. Sci. Rep. 2023, 13, 12567. [Google Scholar] [CrossRef]
- Barral, P.; Suárez, C.; Batanero, E.; Alfonso, C.; Alché, J.D.D.; Rodríguez-García, M.I.; Villalba, M.; Rivas, G.; Rodríguez, R. An olive pollen protein with allergenic activity, Ole e 10, defines a novel family of carbohydrate-binding modules and is potentially implicated in pollen germination. Biochem. J. 2005, 390 Pt 1, 77–84. [Google Scholar] [CrossRef]
- Simpson, C.; Thomas, C.; Findlay, K.; Bayer, E.; Maule, A.J. An arabidopsis GPI-anchor plasmodesmal neck protein with callose binding activity and potential to regulate cell-to-cell trafficking. Plant Cell 2009, 21, 581–594. [Google Scholar] [CrossRef]
- Prinsi, B.; Negri, A.S.; Failla, O.; Scienza, A.; Espen, L. Root proteomic and metabolic analyses reveal specific responses to drought stress in differently tolerant grapevine rootstocks. BMC Plant Biol. 2018, 18, 126. [Google Scholar] [CrossRef] [PubMed]
- Priya, M.; Dhanker, O.P.; Siddique, K.H.M.; HanumanthaRao, B.; Nair, R.M.; Pandey, S.; Singh, S.; Varshney, R.K.; Prasad, P.V.V.; Nayyar, H. Drought and heat stress-related proteins: An update about their functional relevance in imparting stress tolerance in agricultural crops. Theor. Appl. Genet. 2019, 132, 1607–1638. [Google Scholar] [CrossRef] [PubMed]
- Kregel, K.C. Invited Review: Heat shock proteins: Modifying factors in physiological stress responses and acquired thermotolerance. J. Appl. Physiol. 2002, 92, 2177–2186. [Google Scholar] [CrossRef]
- Yano, M.; Nakamuta, S.; Wu, X.; Okumura, Y.; Kido, H.; Trinklein, N.D.; Murray, J.I.; Hartman, S.J.; Botstein, D.; Myers, R.M.; et al. A Novel Function of 14-3-3 Protein: 14-3-3ζ Is a Heat-Shock–related Molecular Chaperone That Dissolves Thermal-aggregated Proteins. Mol. Biol. Cell 2006, 17, 4769–4779. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Liang, C.; Yang, S.; Song, J.; Li, X.; Dai, X.; Wang, F.; Juntawong, N.; Tan, F.; Zhang, X.; et al. iTRAQ-based quantitative proteomic analysis of heat stress-induced mechanisms in pepper seedlings. PeerJ 2021, 9, e11509. [Google Scholar] [CrossRef] [PubMed]
- Xiong, L.; Schumaker, K.S.; Zhu, J.-K. Cell signaling during cold, drought, and salt stress. Plant Cell 2002, 14, S165–S183. [Google Scholar] [CrossRef]
- Lata, C.; Muthamilarasan, M.; Prasad, M. Drought stress responses and signal transduction in plants. In Elucidation of Abiotic Stress Signaling in Plants: Functional Genomics Perspectives; Springer: New York, NY, USA, 2015; Volume 2, pp. 195–225. [Google Scholar] [CrossRef]
- Wang, L.; Jin, X.; Li, Q.; Wang, X.; Li, Z.; Wu, X. Comparative proteomics reveals that phosphorylation of β carbonic anhydrase 1 might be important for adaptation to drought stress in Brassica napus. Sci. Rep. 2016, 6, 39024. [Google Scholar] [CrossRef] [PubMed]
- Ashoub, A.; Beckhaus, T.; Berberich, T.; Karas, M.; Brüggemann, W. Comparative analysis of barley leaf proteome as affected by drought stress. Planta 2013, 237, 771–781. [Google Scholar] [CrossRef]
- Ibrahim, M.I.; Ramadan, A.M.; Amer, M.; Khan, T.K.; Mohamed, N.G.; Said, O.A. Deciphering the enigma of RNA editing in the ATP1_alpha subunit of ATP synthase in Triticum aestivum. Saudi J. Biol. Sci. 2023, 30, 103703. [Google Scholar] [CrossRef]
- Babayev, H.; Mehvaliyeva, U.; Aliyeva, M.; Feyziyev, Y.; Guliyev, N. The study of NAD-malic enzyme in Amaranthus cruentus L. under drought. Plant Physiol. Biochem. 2014, 81, 84–89. [Google Scholar] [CrossRef]
- Ebskamp, M.J.M.; van der Meer, I.M.; Spronk, B.A.; Weisbeek, P.J.; Smeekens, S.C. Accumulation of Fructose Polymers in Transgenic Tobacco. Bio. Technol. 1994, 12, 272–275. [Google Scholar] [CrossRef] [PubMed]
- Gundaraniya, S.A.; Ambalam, P.S.; Tomar, R.S. Metabolomic Profiling of Drought-Tolerant and Susceptible Peanut (Arachis hypogaea L.) Genotypes in Response to Drought Stress. ACS Omega 2020, 5, 31209–31219. [Google Scholar] [CrossRef] [PubMed]
- Guo, R.; Shi, L.; Jiao, Y.; Li, M.; Zhong, X.; Gu, F.; Liu, Q.; Xia, X.; Li, H. Metabolic responses to drought stress in the tissues of drought-tolerant and drought-sensitive wheat genotype seedlings. AoB Plants 2018, 10, ply016. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.; Wang, Y.; Sun, H. Targeted and untargeted metabolomics reveals deep analysis of drought stress responses in needles and roots of Pinus taeda seedlings. Front. Plant Sci. 2023, 13, 1031466. [Google Scholar] [CrossRef]
- He, X.; Wang, C.; Wang, H.; Li, L.; Wang, C. The Function of MAPK Cascades in Response to Various Stresses in Horticultural Plants. Front. Plant Sci. 2020, 11, 952. [Google Scholar] [CrossRef]
- Lei, L.; Shi, J.; Chen, J.; Zhang, M.; Sun, S.; Xie, S.; Li, X.; Zeng, B.; Peng, L.; Hauck, A.; et al. Ribosome profiling reveals dynamic translational landscape in maize seedlings under drought stress. Plant J. 2015, 84, 1206–1218. [Google Scholar] [CrossRef]
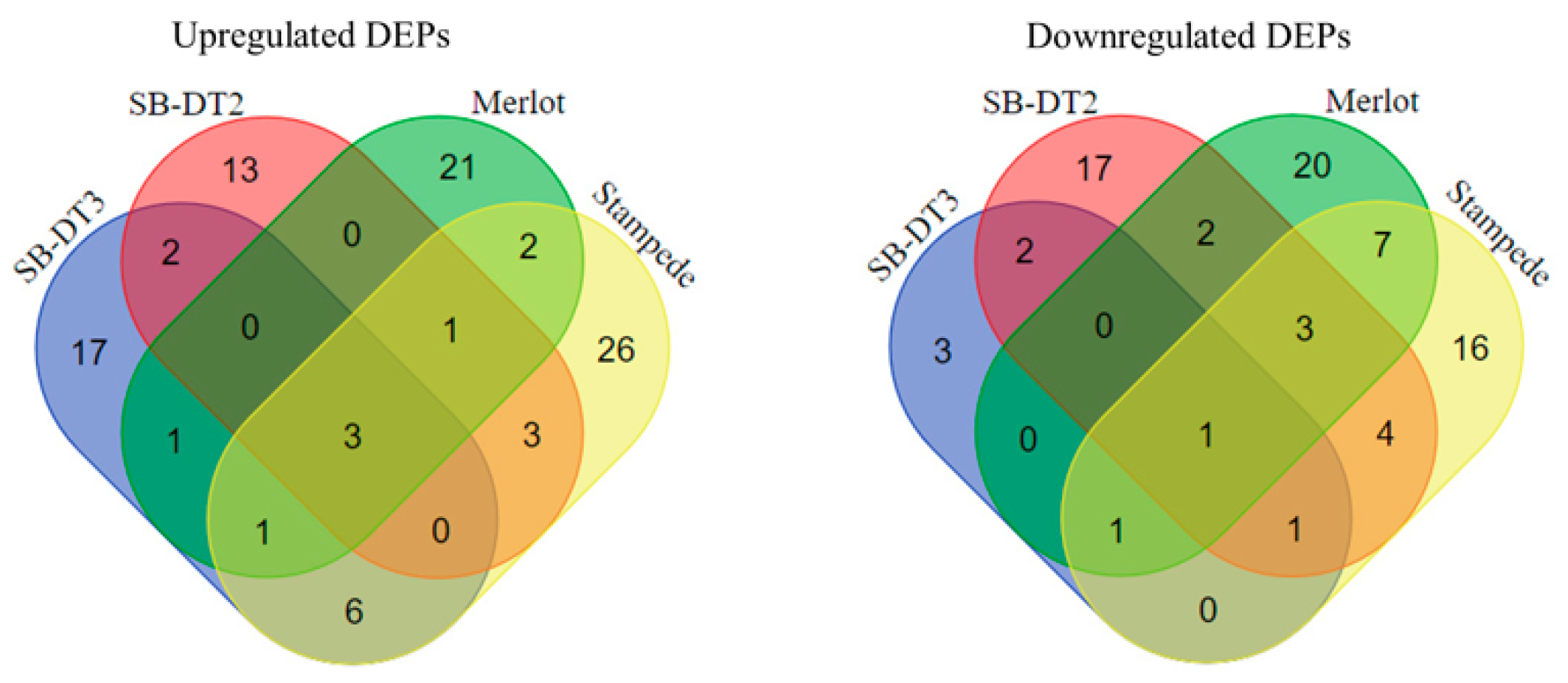
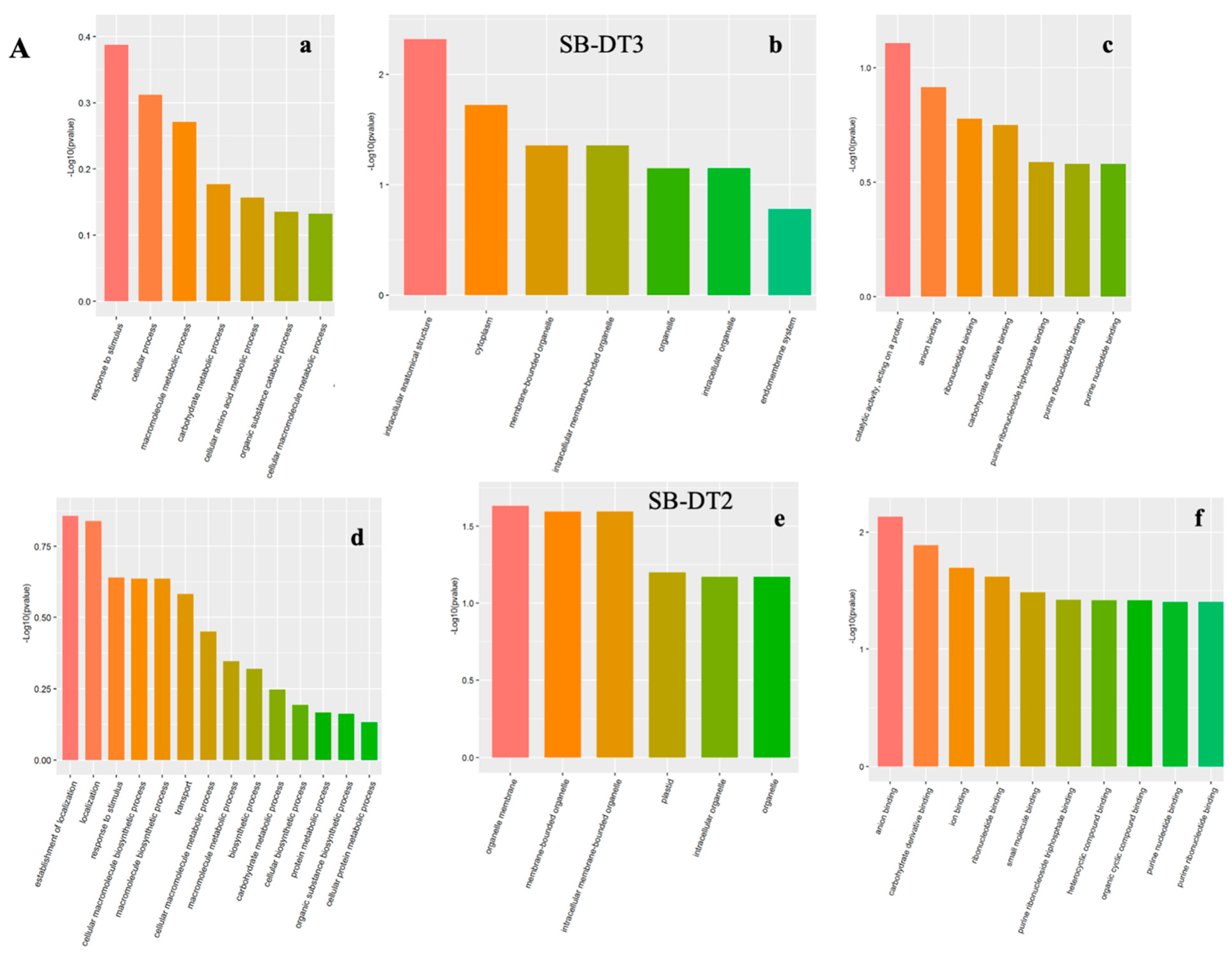
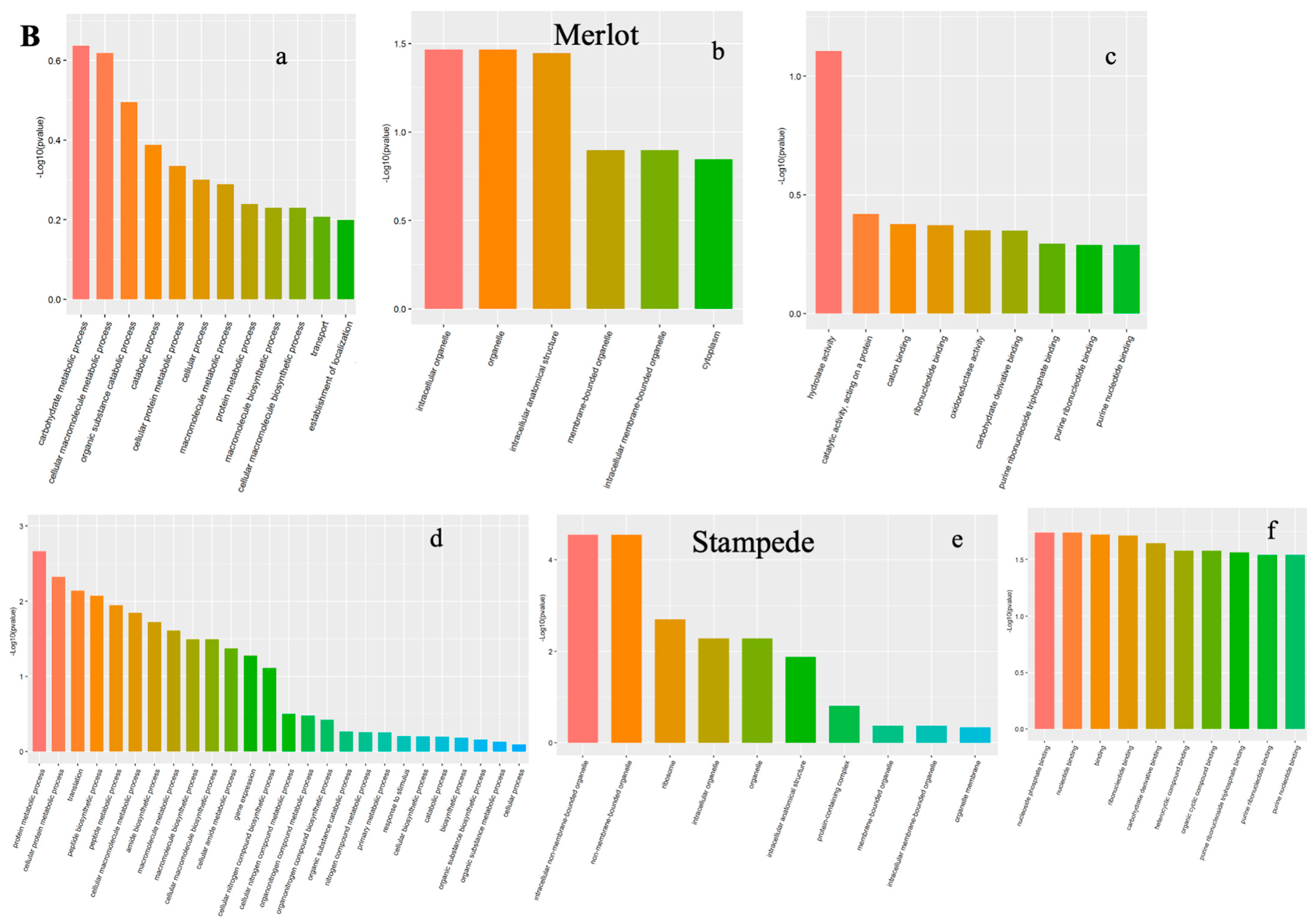

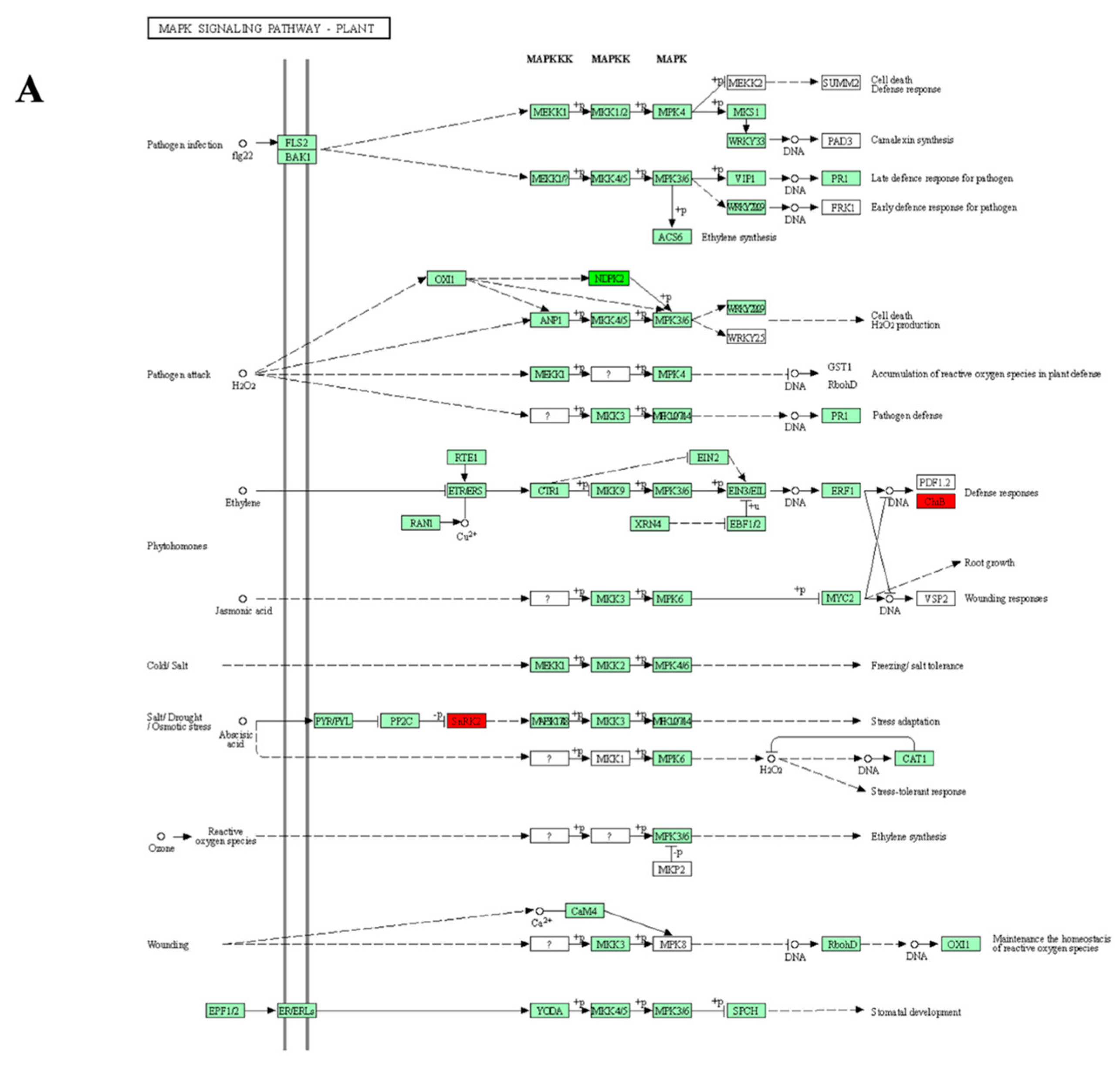
| Proteins | Protein ID | SB-DT3 (FC) | SB-DT2 (FC) |
|---|---|---|---|
| Glycosyltransferase (EC 2.4.1.-) | V5N8Q0 | 0.66549411 | 0.504210124 |
| Protein disulfide-isomerase (EC 5.3.4.1) | V7AXU4 | 0.389970224 | 0.265331957 |
| MPN domain-containing protein | V7B979 | 0 | 0.246595377 |
| HATPase_c domain-containing protein | V7C4I2 | 2.695036446 | 2.224064578 |
| X8 domain-containing protein | V7C5Q7 | 4.00817599 | 0 |
| ATP-dependent 6-phosphofructokinase (ATP-PFK) (Phosphofructokinase) (EC 2.7.1.11) (Phosphohexokinase) | V7CAY4 | 0.642129979 | 0.771054996 |
| WD_REPEATS_REGION domain-containing protein | V7CUC0 | 1.415665913 | 1.60895747 |
| Proteins | Protein ID | Merlot (FC) | Stampede (FC) |
| Alpha amylase inhibitor-1 | A0T2V3 | 1.499790976 | 1.398607793 |
| Thioredoxin-dependent peroxiredoxin (EC 1.11.1.24) | Q9FE12 | 0.512757684 | 0.662087976 |
| 1,4-alpha-glucan branching enzyme (EC 2.4.1.18) | Q9XIS5 | 0.38915774 | 0.731928462 |
| 60S acidic ribosomal protein Po | V7C7B8 | 2.252100651 | 3.055876326 |
| Proteasome subunit beta | T2DN03 | 0.660628218 | 0.684574202 |
| Proteasome subunit alpha type | V7AHS3 | 0.691908505 | 0.71020376 |
| NADH dehydrogenase (ubiquinone) flavoprotein 1, mitochondrial (EC 7.1.1.2) | V7BMQ3 | 0.62391639 | 0.597474222 |
| Alpha-galactosidase (EC 3.2.1.22) (Melibiase) | V7B2C4 | 0.757427659 | 0.755447862 |
| Epimerase domain-containing protein | V7BF01 | 0.685465562 | 0.648430837 |
| Mitochondrial Rho GTPase (EC 3.6.5.-) | V7BIE7 | 1.397530527 | 1.285438844 |
| CYTOSOL_AP domain-containing protein | V7AYA6 | 0.699524201 | 0.726445174 |
| Genotypes | Pathways (p < 0.05) |
|---|---|
| SB-DT3 | Fructose and mannose metabolism Butanoate metabolism Taurine and hypotaurine metabolism |
| SB-DT2 | Oxidative phosphorylation MAPK signaling pathway—plant Tyrosine metabolism Phenylalanine metabolism Protein processing in endoplasmic reticulum Isoflavonoid biosynthesis |
| Merlot | Plant–pathogen interaction Proteasome Biotin metabolism |
| Stampede | Ribosome Proteasome Galactose metabolism Plant–pathogen interaction |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Subramani, M.; Urrea, C.A.; Tamatamu, S.R.; Sripathi, V.R.; Williams, K.; Chintapenta, L.K.; Todd, A.; Ozbay, G. Comprehensive Proteomic Analysis of Common Bean (Phaseolus vulgaris L.) Seeds Reveal Shared and Unique Proteins Involved in Terminal Drought Stress Response in Tolerant and Sensitive Genotypes. Biomolecules 2024, 14, 109. https://doi.org/10.3390/biom14010109
Subramani M, Urrea CA, Tamatamu SR, Sripathi VR, Williams K, Chintapenta LK, Todd A, Ozbay G. Comprehensive Proteomic Analysis of Common Bean (Phaseolus vulgaris L.) Seeds Reveal Shared and Unique Proteins Involved in Terminal Drought Stress Response in Tolerant and Sensitive Genotypes. Biomolecules. 2024; 14(1):109. https://doi.org/10.3390/biom14010109
Chicago/Turabian StyleSubramani, Mayavan, Carlos A. Urrea, Sowjanya R. Tamatamu, Venkateswara R. Sripathi, Krystal Williams, Lathadevi K. Chintapenta, Antonette Todd, and Gulnihal Ozbay. 2024. "Comprehensive Proteomic Analysis of Common Bean (Phaseolus vulgaris L.) Seeds Reveal Shared and Unique Proteins Involved in Terminal Drought Stress Response in Tolerant and Sensitive Genotypes" Biomolecules 14, no. 1: 109. https://doi.org/10.3390/biom14010109
APA StyleSubramani, M., Urrea, C. A., Tamatamu, S. R., Sripathi, V. R., Williams, K., Chintapenta, L. K., Todd, A., & Ozbay, G. (2024). Comprehensive Proteomic Analysis of Common Bean (Phaseolus vulgaris L.) Seeds Reveal Shared and Unique Proteins Involved in Terminal Drought Stress Response in Tolerant and Sensitive Genotypes. Biomolecules, 14(1), 109. https://doi.org/10.3390/biom14010109







