SUPPRESSOR OF MAX2 LIKE 6, 7, and 8 Interact with DDB1 BINDING WD REPEAT DOMAIN HYPERSENSITIVE TO ABA DEFICIENT 1 to Regulate the Drought Tolerance and Target SUCROSE NONFERMENTING 1 RELATED PROTEIN KINASE 2.3 to Abscisic Acid Response in Arabidopsis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials and Growth Conditions
2.2. Seed Germination Assay
2.3. Drought Stress and Relative Water Content (RWC) Analysis
2.4. Yeast Two-Hybrid (Y2H) and Yeast One-Hybrid (Y1H) Assays
2.5. GST Pull-Down Assay
2.6. Bimolecular Fluorescence Complementation (BiFC) Assay
2.7. Cell-Free Degradation Assay
2.8. Constructs for Overexpression and Green Fluorescent Protein (GFP) Fluorescence Assay
2.9. Toluidine Blue Staining
2.10. Immunoblot Analysis
2.11. Quantitative Real-Time PCR (qRT-PCR) Assay
2.12. Transient Expression Assays and Dual-Luciferase Reporter Assay
2.13. Electrophoretic Mobility Shift Assay (EMSA)
2.14. Statistical Analysis
3. Results
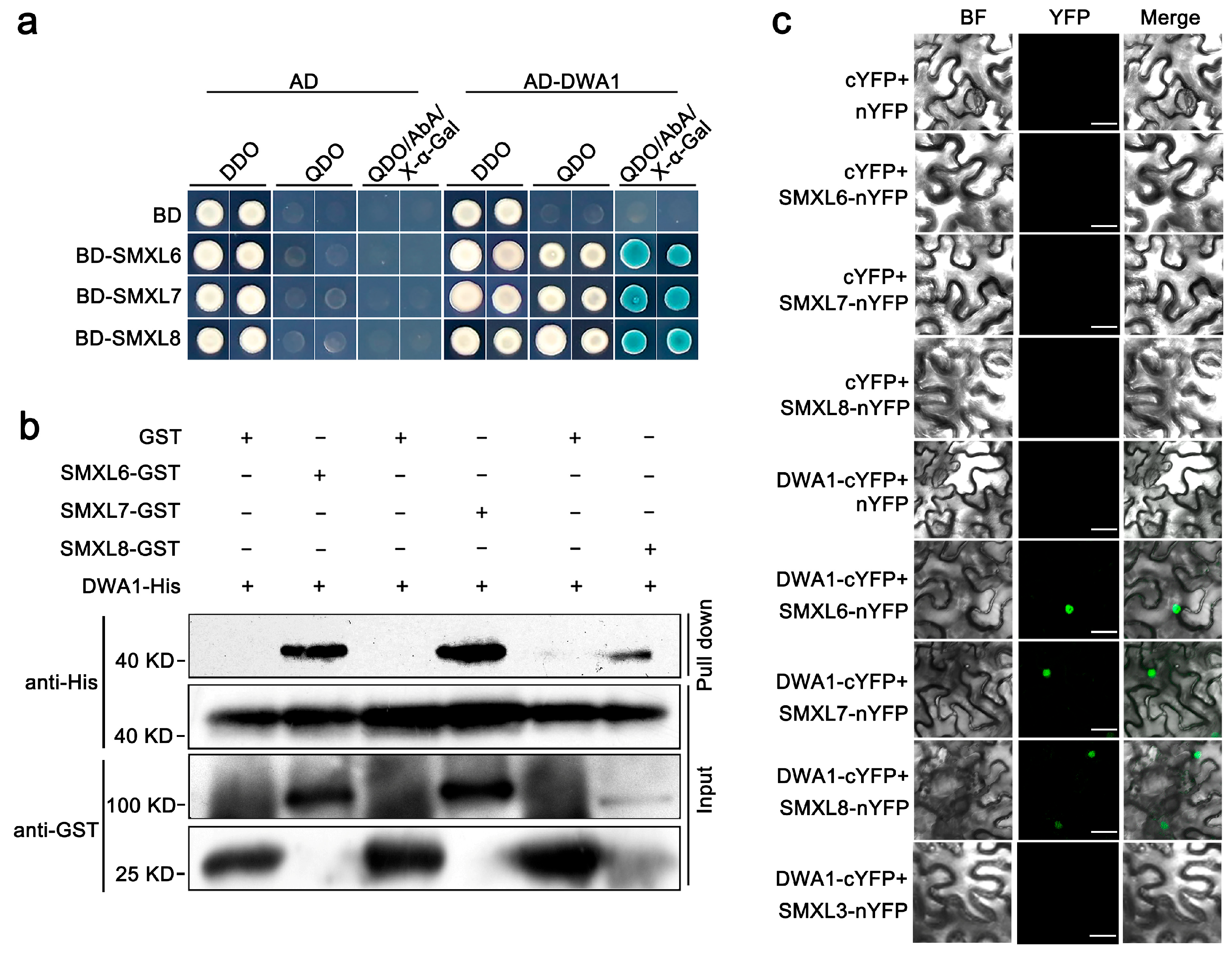
3.1. SMXL6,7,8 Proteins Physically Interact with the DWA1 Protein
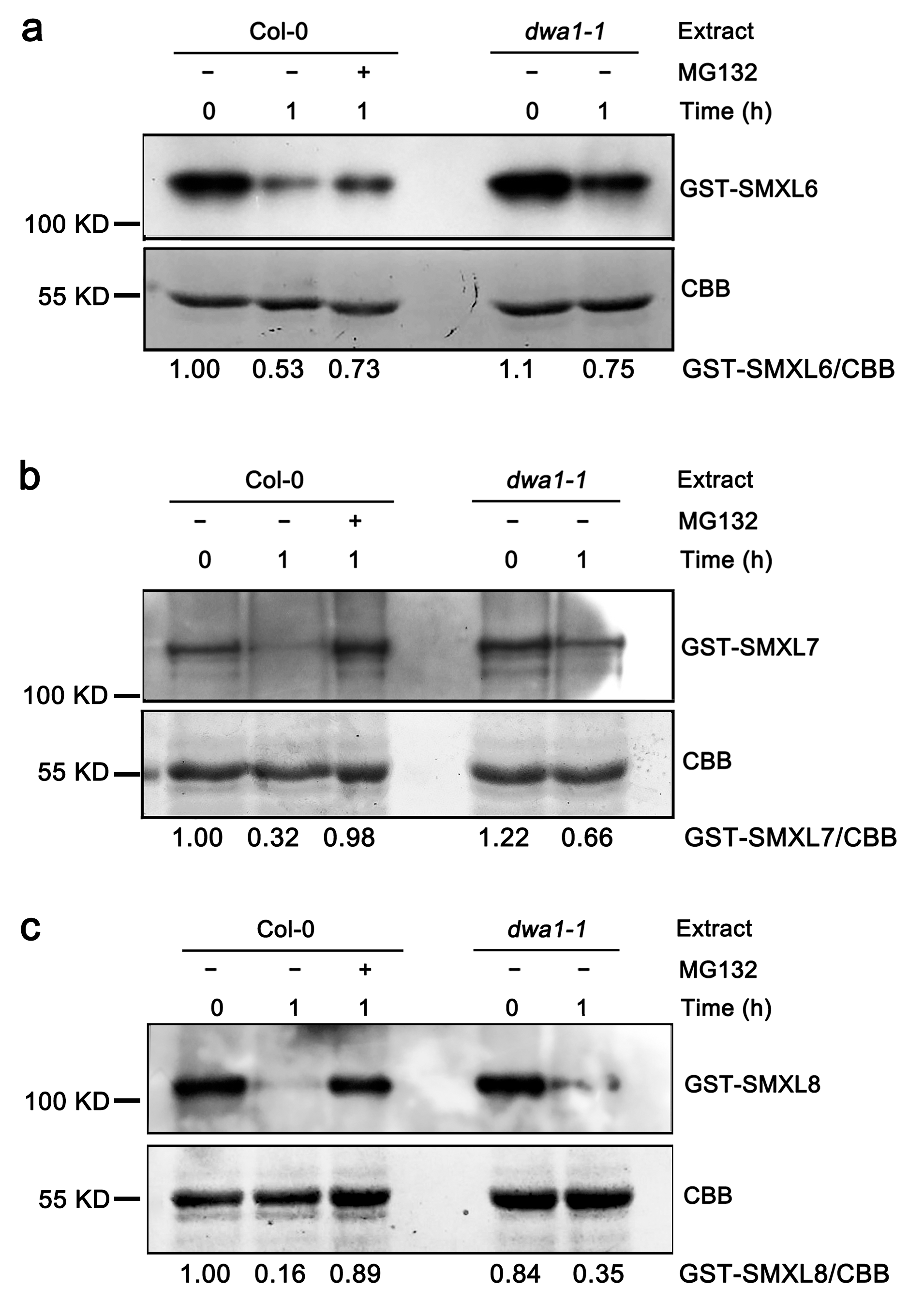
3.2. The Degradation of SMXL6,7,8 Proteins Partially Depend on DWA1
3.3. SMXL6, 7, and 8 Are Epistatic to DWA1 in Response to Drought Tolerance
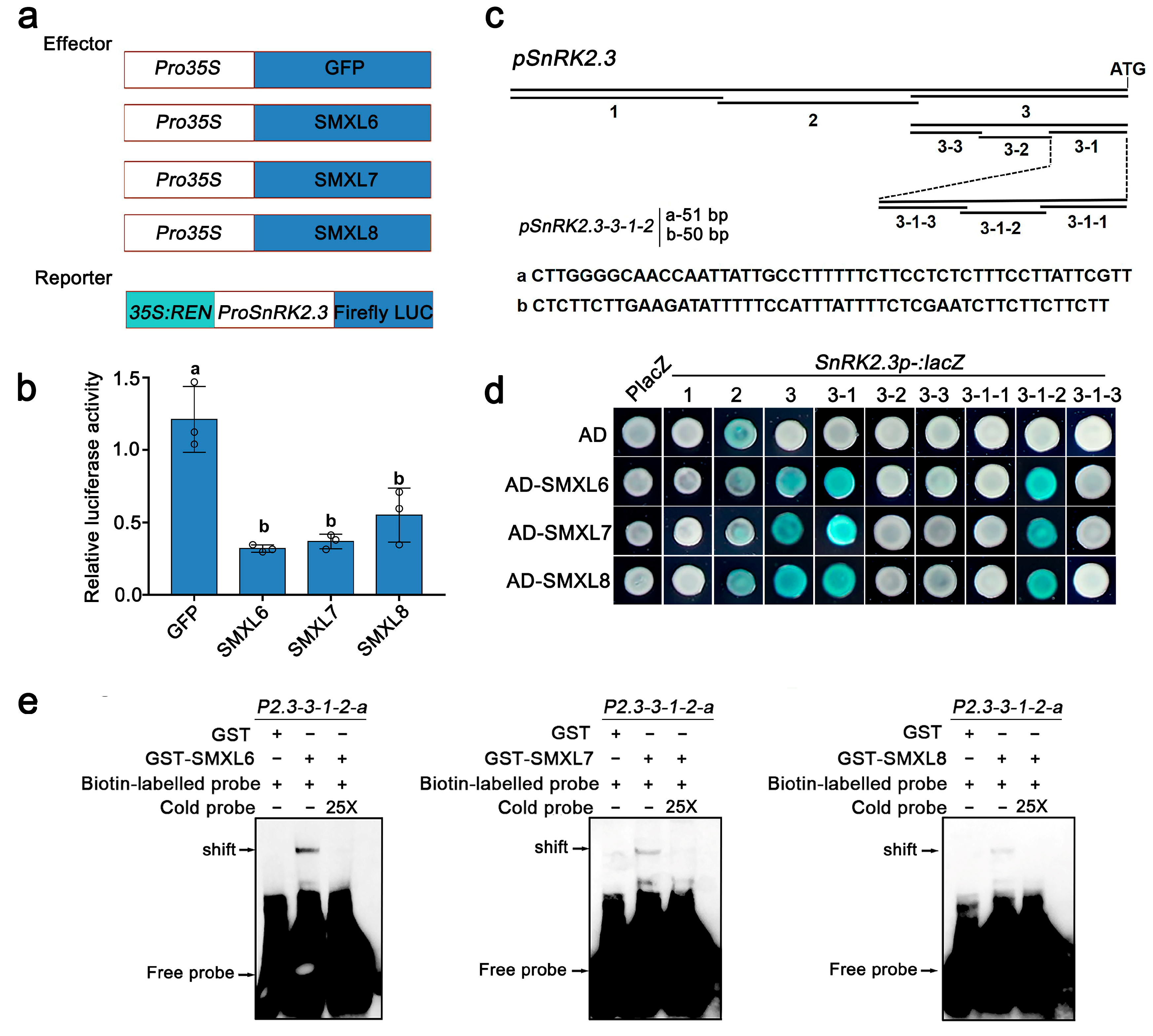
3.4. SMXL6,7,8 Directly Bind to the Promoter of SnRK2.3
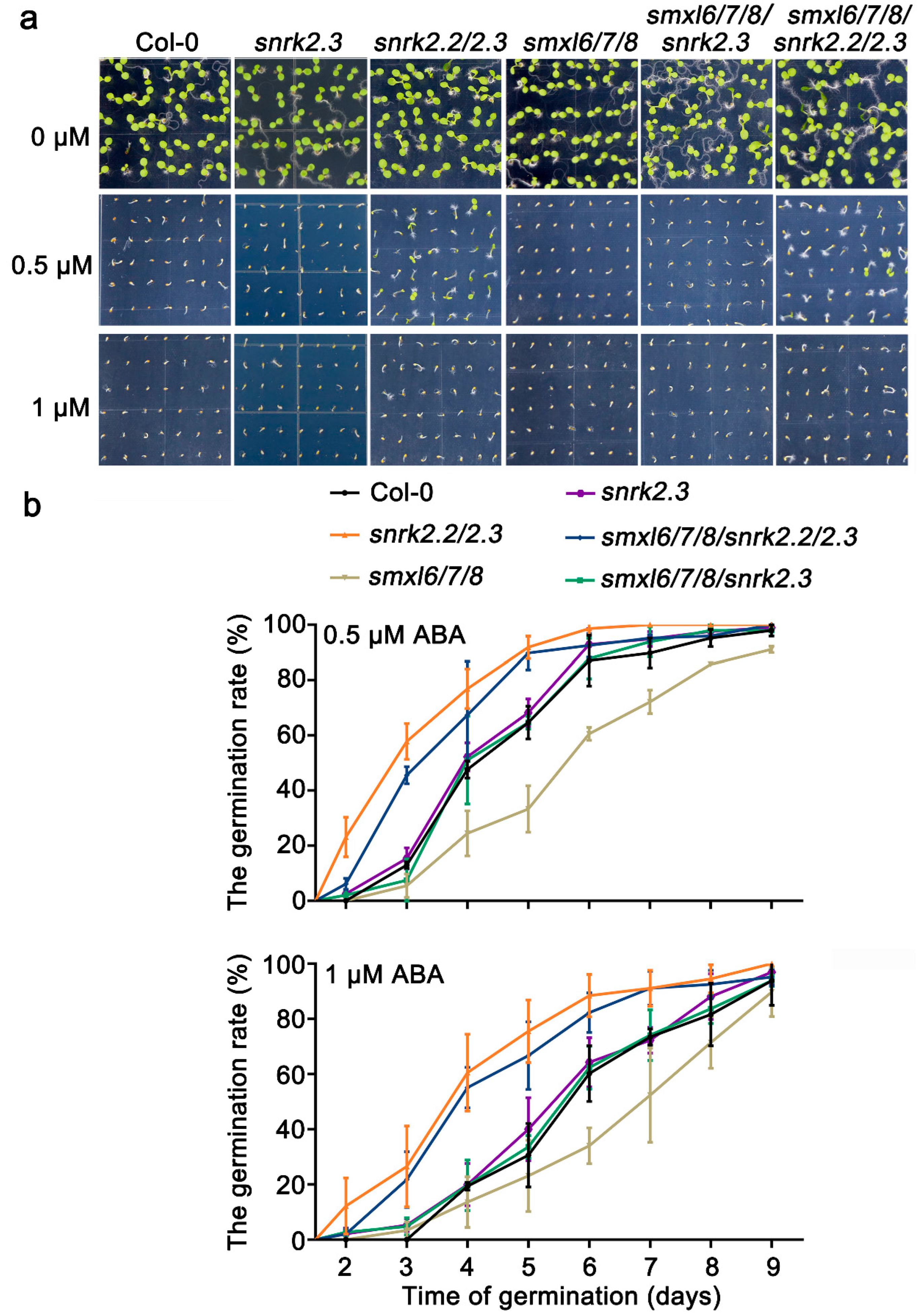
3.5. SnRK2.2/2.3 Act Downstream of SMXL6,7,8 in Response to ABA
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Khan, A.; Pan, X.D.; Najeeb, U.; Tan, D.K.Y.; Fahad, S.; Zahoor, R.; Luo, H.H. Coping with drought: Stress and adaptive mechanisms, and management through cultural and molecular alternatives in cotton as vital constituents for plant stress resilience and fitness. Biol. Res. 2018, 51, 47. [Google Scholar] [CrossRef] [PubMed]
- Liang, C.Z.; Wang, Y.Q.; Zhu, Y.N.; Tang, J.Y.; Hu, B.; Liu, L.C.; Ou, S.J.; Wu, H.K.; Sun, X.H.; Chu, J.F.; et al. OsNAP connects abscisic acid and leaf senescence by fine-tuning abscisic acid biosynthesis and directly targeting senescence-associated genes in rice. Proc. Natl. Acad. Sci. USA 2014, 111, 10013–10018. [Google Scholar] [CrossRef] [PubMed]
- Yamburenko, M.V.; Zubo, Y.O.; Vanková, R.; Kusnetsov, V.V.; Kulaeva, O.N.; Börner, T. Abscisic acid represses the transcription of chloroplast genes. J. Exp. Bot. 2013, 64, 4491–4502. [Google Scholar] [CrossRef]
- Park, S.-Y.; Fung, P.; Nishimura, N.; Jensen, D.R.; Fujii, H.; Zhao, Y.; Lumba, S.; Santiago, J.; Rodrigues, A.; Chow, T.-F.F.; et al. Abscisic Acid Inhibits Type 2C Protein Phosphatases via the PYR/PYL Family of START Proteins. Science 2009, 324, 1068–1071. [Google Scholar] [CrossRef]
- Vlad, F.; Rubio, S.; Rodrigues, A.; Sirichandra, C.; Belin, C.; Robert, N.; Leung, J.; Rodriguez, P.L.; Laurière, C.; Merlot, S. Protein Phosphatases 2C Regulate the Activation of the Snf1-Related Kinase OST1 by Abscisic Acid in Arabidopsis. Plant Cell 2009, 21, 3170–3184. [Google Scholar] [CrossRef]
- Hrabak, E.M.; Chan, C.W.M.; Gribskov, M.; Harper, J.F.; Choi, J.H.; Halford, N.; Kudla, J.; Luan, S.; Nimmo, H.G.; Sussman, M.R.; et al. The Arabidopsis CDPK-SnRK superfamily of protein kinases. Plant Physiol. 2003, 132, 666–680. [Google Scholar]
- Fujii, H.; Verslues, P.E.; Zhu, J.K. Identification of two protein kinases required for abscisic acid regulation of seed germination, root growth, and gene expression in Arabidopsis. Plant Cell 2007, 19, 485–494. [Google Scholar] [CrossRef]
- Nakashima, K.; Fujita, Y.; Kanamori, N.; Katagiri, T.; Umezawa, T.; Kidokoro, S.; Maruyama, K.; Yoshida, T.; Ishiyama, K.; Kobayashi, M.; et al. Three Arabidopsis SnRK2 Protein Kinases, SRK2D/SnRK2.2, SRK2E/SnRK2.6/OST1 and SRK2I/SnRK2.3, Involved in ABA Signaling are Essential for the Control of Seed Development and Dormancy. Plant Cell Physiol. 2009, 50, 1345–1363. [Google Scholar] [CrossRef] [PubMed]
- Cheng, C.H.; Wang, Z.J.; Ren, Z.Y.; Zhi, L.Y.; Yao, B.; Su, C.; Liu, L.; Li, X. SCFAtPP2-B11 modulates ABA signaling by facilitating SnRK2.3 degradation in Arabidopsis thaliana. PLoS Genet. 2017, 13, e1006947. [Google Scholar] [CrossRef]
- Fujita, Y.; Nakashima, K.; Yoshida, T.; Katagiri, T.; Kidokoro, S.; Kanamori, N.; Umezawa, T.; Fujita, M.; Maruyama, K.; Ishiyama, K.; et al. Three SnRK2 Protein Kinases are the Main Positive Regulators of Abscisic Acid Signaling in Response to Water Stress in Arabidopsis. Plant Cell Physiol. 2009, 50, 2123–2132. [Google Scholar] [CrossRef]
- Tian, S.J.; Mao, X.G.; Zhang, H.Y.; Chen, S.S.; Zhai, C.C.; Yang, S.M.; Jing, R.L. Cloning and characterization of TaSnRK2.3, a novel SnRK2 gene in common wheat. J. Exp. Bot. 2013, 64, 2063–2080. [Google Scholar] [CrossRef]
- Lee, J.H.; Yoon, H.J.; Terzaghi, W.; Martinez, C.; Dai, M.Q.; Li, J.G.; Byun, M.O.; Deng, X.W. DWA1 and DWA2, two Arabidopsis DWD protein components of CUL4-based E3 ligases, act together as negative regulators in ABA signal transduction. Plant Cell 2010, 22, 1716–1732. [Google Scholar]
- Stanga, J.P.; Smith, S.M.; Briggs, W.R.; Nelson, D.C. SUPPRESSOR OF MORE AXILLARY GROWTH2 1 Controls Seed Germination and Seedling Development in Arabidopsis. Plant Physiol. 2013, 163, 318–330. [Google Scholar] [CrossRef]
- Yamada, Y.; Furusawa, S.; Nagasaka, S.; Shimomura, K.; Yamaguchi, S.; Umehara, M. Strigolactone signaling regulates rice leaf senescence in response to a phosphate deficiency. Planta 2014, 240, 399–408. [Google Scholar] [CrossRef] [PubMed]
- Ueda, H.; Kusaba, M. Strigolactone Regulates Leaf Senescence in Concert with Ethylene in Arabidopsis. Plant Physiol. 2015, 169, 138–147. [Google Scholar] [CrossRef] [PubMed]
- Brewer, P.B.; Koltai, H.; Beveridge, C.A. Diverse Roles of Strigolactones in Plant Development. Mol. Plant 2013, 6, 18–28. [Google Scholar] [CrossRef]
- Van Ha, C.; Leyva-González, M.A.; Osakabe, Y.; Tran, U.T.; Nishiyama, R.; Watanabe, Y.; Tanaka, M.; Seki, M.; Yamaguchi, S.; Van Dong, N.; et al. Positive regulatory role of strigolactone in plant responses to drought and salt stress. Proc. Natl. Acad. Sci. USA 2014, 111, 851–856. [Google Scholar] [CrossRef]
- Chi, C.; Xu, X.C.; Wang, M.Q.; Zhang, H.; Fang, P.P.; Zhou, J.; Xia, X.J.; Shi, K.; Zhou, Y.H.; Yu, J.Q. Strigolactones positively regulate abscisic acid-dependent heat and cold tolerance in tomato. Hortic. Res. 2021, 8, 237. [Google Scholar] [CrossRef]
- Umehara, M.; Hanada, A.; Yoshida, S.; Akiyama, K.; Arite, T.; Takeda-Kamiya, N.; Magome, H.; Kamiya, Y.; Shirasu, K.; Yoneyama, K.; et al. Inhibition of shoot branching by new terpenoid plant hormones. Nature 2008, 455, 195–200. [Google Scholar] [CrossRef]
- Jiang, L.; Liu, X.; Xiong, G.S.; Liu, H.H.; Chen, F.L.; Wang, L.; Meng, X.B.; Liu, G.F.; Yu, H.; Yuan, Y.D.; et al. DWARF 53 acts as a repressor of strigolactone signalling in rice. Nature 2013, 506, 7488. [Google Scholar] [CrossRef]
- Wang, L.; Wang, B.; Jiang, L.; Liu, X.; Li, X.L.; Lu, Z.F.; Meng, X.B.; Wang, Y.H.; Smith, S.M.; Li, J.Y. Strigolactone Signaling in Arabidopsis Regulates Shoot Development by Targeting D53-Like SMXL Repressor Proteins for Ubiquitination and Degradation. Plant Cell 2015, 27, 3128–3142. [Google Scholar] [CrossRef] [PubMed]
- Kohlen, W.; Charnikhova, T.; Lammers, M.; Pollina, T.; Tóth, P.; Haider, I.; Pozo, M.J.; Maagd, R.A.; Ruyter-Spira, C.; Bouwmeester, H.J.; et al. The tomato CAROTENOID CLEAVAGE DIOXYGENASE8(SlCCD8) regulates rhizosphere signaling, plant architecture and affects reproductive development through strigolactone biosynthesis. New Phytol. 2012, 196, 535–547. [Google Scholar] [CrossRef]
- Soundappan, I.; Bennett, T.; Morffy, N.; Liang, Y.Y.; Stang, J.P.; Abbas, A.; Leyser, O.; Nelson, D.C. SMAX1-LIKE/D53 Family Members Enable Distinct MAX2-Dependent Responses to Strigolactones and Karrikins in Arabidopsis. Plant Cell 2015, 27, 3143–3159. [Google Scholar] [CrossRef]
- Zhou, F.; Lin, Q.B.; Zhu, L.H.; Ren, Y.L.; Zhou, K.N.; Shabek, N.; Wu, F.Q.; Mao, H.B.; Dong, W.; Gan, L.; et al. D14–SCFD3-dependent degradation of D53 regulates strigolactone signalling. Nature 2013, 504, 406–410. [Google Scholar] [CrossRef] [PubMed]
- Li, W.Q.; Nguyen, K.H.; Chu, H.D.; Watanabe, Y.; Osakabe, Y.; Sato, M.; Toyooka, K.; Seo, M.; Tian, L.; Tian, C.J.; et al. Comparative functional analyses of DWARF14 and KARRIKIN INSENSITIVE 2 in drought adaptation of Arabidopsis thaliana. Plant J. 2020, 103, 111–127. [Google Scholar] [CrossRef]
- Bu, Q.Y.; Lv, T.X.; Shen, H.; Luong, P.; Wang, J.; Wang, Z.Y.; Huang, Z.G.; Xiao, L.T.; Engineer, C.; Kim, T.H.; et al. Regulation of Drought Tolerance by the F-Box Protein MAX2 in Arabidopsis. Plant Physiol. 2014, 164, 424–439. [Google Scholar] [CrossRef]
- An, J.P.; Li, R.; Qu, F.J.; You, C.X.; Wang, X.F.; Hao, Y.J. Apple F-Box Protein MdMAX2 Regulates Plant Photomorphogenesis and Stress Response. Front. Plant Sci. 2016, 7, 1685. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.Y.; Ren, Y.R.; Zheng, P.F.; Zhao, L.L.; You, C.X.; Wang, X.F.; Hao, Y.J. Cloning and functional identification of a strigolactone receptor gene MdD14 in apple. Plant Cell Tissue Organ Cult. (PCTOC) 2020, 140, 197–208. [Google Scholar] [CrossRef]
- Wang, Q.J.; Ni, J.; Shah, F.; Liu, W.B.; Wang, D.D.; Yao, Y.Y.; Hu, H.; Huang, S.W.; Hou, J.Y.; Fu, S.L.; et al. Overexpression of the Stress-Inducible SsMAX2 Promotes Drought and Salt Resistance via the Regulation of Redox Homeostasis in Arabidopsis. Int. J. Mol. Sci. 2019, 20, 837. [Google Scholar] [CrossRef]
- Wang, L.; Wang, B.; Yu, H.; Guo, H.Y.; Lin, T.; Kou, L.Q.; Wang, A.Q.; Shao, N.; Ma, H.Y.; Xiong, G.S.; et al. Transcriptional regulation of strigolactone signalling in Arabidopsis. Nature 2020, 583, 277–281. [Google Scholar] [CrossRef]
- Yang, T.; Lian, Y.K.; Kang, J.H.; Bian, Z.Y.; Xuan, L.J.; Gao, Z.S.; Wang, X.Y.; Deng, J.M.; Wang, C.Y. The SUPPRESSOR of MAX2 1 (SMAX1)-Like SMXL6, SMXL7 and SMXL8 Act as Negative Regulators in Response to Drought Stress in Arabidopsis. Plant Cell Physiol. 2020, 61, 1477–1492. [Google Scholar] [CrossRef]
- Li, W.Q.; Nguyen, K.H.; Tran, C.D.; Watanabe, Y.; Tian, C.J.; Yin, X.J.; Li, K.; Yang, Y.; Guo, J.G.; Miao, Y.C.; et al. Negative roles of strigolactone-related SMXL6, 7 and 8 proteins in drought resistance in Arabidopsis. Biomolecules 2020, 10, 607. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.R.; Liu, Y.; Ma, M.D.; Zhou, Q.; Zhao, Y.P.; Zhao, B.B.; Wang, B.B.; Wei, H.B.; Wang, H.Y. Arabidopsis FHY3 and FAR1 integrate light and strigolactone signaling to regulate branching. Nat. Commun. 2020, 11, 1955. [Google Scholar] [CrossRef]
- Liu, J.; Cheng, X.L.; Liu, P.; Sun, J.Q. miR156-targeted SBP-Box transcriptionfactors interact with DWARF53 to regulate TEOSINTE BRANCHED1 and BARREN STALK1 expression in Bread Wheat. Plant Physiol. 2017, 174, 1931–1948. [Google Scholar] [CrossRef] [PubMed]
- Song, X.G.; Lu, Z.F.; Yu, H.; Shao, G.N.; Xiong, J.S.; Meng, X.B.; Jing, Y.H.; Liu, G.F.; Xiong, G.S.; Duan, J.B.; et al. IPA1 functions as a downstream transcription factor repressed by D53 in strigolactone signaling in rice. Cell Res. 2017, 27, 1128–1141. [Google Scholar] [CrossRef]
- Tang, J.Y.; Chu, C.C. Strigolactone Signaling: Repressor Proteins Are Transcription Factors. Trends Plant Sci. 2020, 25, 960–963. [Google Scholar] [CrossRef]
- Stirnberg, P.; van de Sande, K.; Leyser, H.M.O. MAX1 and MAX2 control shoot lateral branching in Arabidopsis. Development 2002, 129, 1131–1141. [Google Scholar] [CrossRef] [PubMed]
- Qi, L.J.; Liu, S.; Li, C.; Fu, J.Y.; Jing, Y.J.; Cheng, J.K.; Li, H.; Zhang, D.; Wang, X.J.; Dong, X.J.; et al. PHYTOCHROME-INTERACTING FACTORS Interact with the ABA Receptors PYL8 and PYL9 to Orchestrate ABA Signaling in Darkness. Mol. Plant 2020, 13, 414–430. [Google Scholar] [CrossRef] [PubMed]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, T.; Tanaka, H.; Machida, C.; Watanabe, M.; Machida, Y. A new method for rapid visualization of defects in leaf cuticle reveals five intrinsic patterns of surface defects in Arabidopsis. Plant J. 2004, 37, 139–146. [Google Scholar] [CrossRef]
- Xu, D.Q.; Li, J.G.; Gangappa, S.N.; Hettiarachchi, C.; Lin, F.; Andersson, M.X.; Jiang, Y.; Deng, X.W.; Holm, M. Convergence of Light and ABA Signaling on the ABI5 Promoter. PLoS Genet. 2014, 10, e1004197. [Google Scholar] [CrossRef]
- Wang, Z.Y.; Xiong, L.M.; Li, W.B.; Zhu, J.K.; Zhu, J.H. The Plant Cuticle Is Required for Osmotic Stress Regulation of Abscisic Acid Biosynthesis and Osmotic Stress Tolerance in Arabidopsis. Plant Cell 2011, 23, 1971–1984. [Google Scholar] [CrossRef]
- Fujii, H.; Zhu, J.K. Arabidopsis mutant deficient in 3 abscisic acid-activated protein kinases reveals critical roles in growth, reproduction, and stress. Proc. Natl. Acad. Sci. USA 2009, 106, 8380–8385. [Google Scholar] [CrossRef]
- Lin, Z.; Li, Y.; Wang, Y.B.; Liu, X.L.; Ma, L.; Zhang, Z.J.; Mu, C.; Zhang, Y.; Peng, L.; Xie, S.J.; et al. Initiation and amplification of SnRK2 activation in abscisic acid signaling. Nat. Commun. 2021, 12, 2456. [Google Scholar] [CrossRef]
- Soma, F.; Takahashi, F.; Suzuki, T.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Plant Raf-like kinases regulate the mRNA population upstream of ABA-unresponsive SnRK2 kinases under drought stress. Nat. Commun. 2020, 11, 137. [Google Scholar] [CrossRef]
- Biedermann, S.; Hellmann, H. WD40 and CUL4-based E3 ligases: Lubricating all aspects of life. Trends Plant Sci. 2011, 16, 38–46. [Google Scholar] [CrossRef]
- Seo, K.I.; Lee, J.H.; Nezames, C.D.; Zhong, S.W.; Song, E.; Byun, M.O.; Deng, X.W. ABD1 is an Arabidopsis DCAF substrate receptor for CUL4-DDB1-Based E3 ligases that acts as a negative regulator of abscisic acid signaling. Plant Cell 2014, 26, 695–711. [Google Scholar] [CrossRef]
- Irigoyen, M.L.; Iniesto, E.; Rodriguez, L.; Puga, M.I.; Yanagawa, Y.; Pick, E.; Strickland, E.; Paz-Ares, J.; Wei, N.; De Jaeger, G.; et al. Correction to: Targeted Degradation of Abscisic Acid Receptors Is Mediated by the Ubiquitin Ligase Substrate Adaptor DDA1 in Arabidopsis. Plant Cell 2014, 26, 712–728. [Google Scholar] [CrossRef]
- Li, D.K.; Zhang, L.; Li, X.Y.; Kong, X.G.; Wang, X.Y.; Li, Y.; Liu, Z.B.; Wang, J.M.; Li, X.F.; Yang, Y. AtRAE1 is involved in degradation of ABA receptor RCAR1 and negatively regulates ABA signalling in Arabidopsis. Plant Cell Environ. 2018, 41, 231–244. [Google Scholar] [CrossRef]
- Chen, Q.; Liu, R.J.; Wu, Y.R.; Wei, S.W.; Wang, Q.; Zheng, Y.N.; Xia, R.; Shang, X.L.; Yu, F.F.; Yang, X.Y.; et al. ERAD-related E2 and E3 enzymes modulate the drought response by regulating the stability of PIP2 aquaporins. Plant Cell 2021, 33, 2883–2898. [Google Scholar] [CrossRef]
- Zhao, J.F.; Zhao, L.L.; Zhang, M.; Zafar, S.A.; Fang, J.J.; Li, M.; Zhang, W.H.; Li, X.Y. Arabidopsis E3 Ubiquitin Ligases PUB22 and PUB23 Negatively Regulate Drought Tolerance by Targeting ABA Receptor PYL9 for Degradation. Int. J. Mol. Sci. 2017, 18, 1841. [Google Scholar] [CrossRef]
- You, J.; Zong, W.; Li, X.K.; Ning, J.; Hu, H.H.; Li, X.H.; Xiao, J.H.; Xiong, L.Z. The SNAC1-targeted gene OsSRO1c modulates stomatal closure and oxidative stress tolerance by regulating hydrogen peroxide in rice. J. Exp. Bot. 2013, 64, 569–583. [Google Scholar] [CrossRef] [PubMed]
- Tan, W.R.; Zhang, D.W.; Zhou, H.P.; Zheng, T.; Yin, Y.H.; Lin, H.H. Transcription factor HAT1 is a substrate of SnRK2.3 kinase and negatively regulates ABA synthesis and signaling in Arabidopsis responding to drought. PLoS Genet. 2018, 14, e1007336. [Google Scholar] [CrossRef]
- Zhu, Y.F.; Huang, P.C.; Guo, P.C.; Chong, L.; Yu, G.B.; Sun, X.L.; Hu, T.; Li, Y.; Hsu, C.C.; Tang, K.; et al. CDK8 is associated with RAP2.6 and SnRK2.6 and positively modulates abscisic acid signaling and drought response in Arabidopsis. New Phytol 2020, 228, 1573–1590. [Google Scholar] [CrossRef]
- Zhao, Y.; Chan, Z.L.; Gao, J.H.; Xing, L.; Cao, M.J.; Yu, C.M.; Hu, Y.L.; You, J.; Shi, H.T.; Zhu, Y.F.; et al. ABA receptor PYL9 promotes drought resistance and leaf senescence. Proc. Natl. Acad. Sci. USA 2016, 113, 1949–1954. [Google Scholar] [CrossRef] [PubMed]
- Fujita, Y.; Yoshida, T.; Yamaguchi-Shinozaki, K. Pivotal role of the AREB/ABF-SnRK2 pathway in ABRE-mediated transcription in response to osmotic stress in plants. Physiol Plant. 2013, 147, 15–27. [Google Scholar] [CrossRef] [PubMed]
- Cai, Z.Y.; Liu, J.J.; Wang, H.J.; Yang, C.J.; Chen, Y.X.; Li, Y.C.; Pan, S.J.; Dong, R.; Tang, G.L.; Barajas-Lopez, J.D.; et al. GSK3-like kinases positively modulate abscisic acid signaling through phosphorylating subgroup III SnRK2s in Arabidopsis. Proc. Natl. Acad. Sci. USA 2014, 111, 9651–9656. [Google Scholar] [CrossRef]
- Chang, Y.N.; Wang, Z.; Ren, Z.; Wang, C.H.; Wang, P.; Zhu, J.K.; Li, X.; Duan, C.G. NUCLEAR PORE ANCHOR and EARLY IN SHORT DAYS 4 negatively regulate abscisic acid signaling by inhibiting Snf1-related protein kinase2 activity and stability in Arabidopsis. J. Integr. Plant Biol. 2022, 64, 2060–2074. [Google Scholar] [CrossRef] [PubMed]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lian, Y.; Lian, C.; Wang, L.; Li, Z.; Yuan, G.; Xuan, L.; Gao, H.; Wu, H.; Yang, T.; Wang, C. SUPPRESSOR OF MAX2 LIKE 6, 7, and 8 Interact with DDB1 BINDING WD REPEAT DOMAIN HYPERSENSITIVE TO ABA DEFICIENT 1 to Regulate the Drought Tolerance and Target SUCROSE NONFERMENTING 1 RELATED PROTEIN KINASE 2.3 to Abscisic Acid Response in Arabidopsis. Biomolecules 2023, 13, 1406. https://doi.org/10.3390/biom13091406
Lian Y, Lian C, Wang L, Li Z, Yuan G, Xuan L, Gao H, Wu H, Yang T, Wang C. SUPPRESSOR OF MAX2 LIKE 6, 7, and 8 Interact with DDB1 BINDING WD REPEAT DOMAIN HYPERSENSITIVE TO ABA DEFICIENT 1 to Regulate the Drought Tolerance and Target SUCROSE NONFERMENTING 1 RELATED PROTEIN KINASE 2.3 to Abscisic Acid Response in Arabidopsis. Biomolecules. 2023; 13(9):1406. https://doi.org/10.3390/biom13091406
Chicago/Turabian StyleLian, Yuke, Chengfei Lian, Lei Wang, Zhimin Li, Guoqiang Yuan, Lijuan Xuan, Huanhuan Gao, Haijun Wu, Tao Yang, and Chongying Wang. 2023. "SUPPRESSOR OF MAX2 LIKE 6, 7, and 8 Interact with DDB1 BINDING WD REPEAT DOMAIN HYPERSENSITIVE TO ABA DEFICIENT 1 to Regulate the Drought Tolerance and Target SUCROSE NONFERMENTING 1 RELATED PROTEIN KINASE 2.3 to Abscisic Acid Response in Arabidopsis" Biomolecules 13, no. 9: 1406. https://doi.org/10.3390/biom13091406
APA StyleLian, Y., Lian, C., Wang, L., Li, Z., Yuan, G., Xuan, L., Gao, H., Wu, H., Yang, T., & Wang, C. (2023). SUPPRESSOR OF MAX2 LIKE 6, 7, and 8 Interact with DDB1 BINDING WD REPEAT DOMAIN HYPERSENSITIVE TO ABA DEFICIENT 1 to Regulate the Drought Tolerance and Target SUCROSE NONFERMENTING 1 RELATED PROTEIN KINASE 2.3 to Abscisic Acid Response in Arabidopsis. Biomolecules, 13(9), 1406. https://doi.org/10.3390/biom13091406





